Abstract
Several chromosomal regions have been linked to bipolar disorder (BD). However, the search for specific genes has been hampered by inconsistent findings, partly due to genetic and phenotypic heterogeneity. We focused on lithium-responsive bipolar patients, a subgroup thought to be more homogeneous and conducted a multistage study including an initial linkage study followed up by fine mapping and gene expression. Our sample consisted of 36 families (275 genotyped individuals, 132 affected) recruited through probands who were responders to long-term lithium treatment. We conducted a genome-wide scan with 811 microsatellite markers followed by fine mapping. Gene expression studies of candidate regions were conducted on six post-mortem prefrontal brain regions of 20 individuals (8 BD and 12 controls). We identified regions 3p25, 3p14 and 14q11 as showing the highest genome-wide linkage signal (LOD 2.53, 2.04 and 3.19, respectively). Fine mapping provided further support for 3p25, while only modest support was found in the other two regions. We identified a group of synaptic, mitochondrial and apoptotic genes with altered expression patterns in BD. Analysis of an independent microarray dataset supported the implication of synapse-related and mitochondrial genes in BD. In conclusion, using two complementary strategies, we found evidence of linkage to lithium-responsive BD on 3p25, 3p14 and 14q11 as well as significantly dysregulated genes on these regions suggesting altered synaptic and mitochondrial function in BD. Further studies are warranted to demonstrate the functional role of these genes in BD.
Keywords: Bipolar disorder, gene expression, linkage, lithium responsive, synapse-related genes
Introduction
Bipolar disorder (BD) is a highly familial psychiatric illness, with heritability estimates ranging from 60% to 85% (Smoller & Finn, 2003). However, as in other genetically complex psychiatric disorders, the search for susceptibility genes has been a rather gradual and challenging task. Over the years, a number of genomic regions have been linked to BD, yet failure to consistently replicate those findings has characterized most individual studies. Furthermore, recent meta-analyses of genome scans have yielded conflicting and inconclusive results (Badner & Gershon, 2002; McQueen et al. 2005; Segurado et al. 2003). The inconsistency in study findings may be driven, in part, by genetic and phenotypic heterogeneity. Hence, investigating specific BD clinical subtypes with distinct clinical characteristics and increased familial transmission may facilitate gene-mapping efforts, by focusing on, possibly, more homogeneous subgroups with stronger genetic loading. Some of these familial BD features include suicidal behaviour, psychosis, comorbid anxiety, substance abuse/dependence, earlier age of onset, rapid cycling and response to lithium prophylaxis (Grof et al. 2002; Potash et al. 2007; Saunders et al. 2008; Schulze et al. 2006).
Lithium is a mood stabilizer that has been extensively studied and used for several decades as first-choice in the prophylactic treatment of BD; and to date, it continues to be widely used in mood disorders (Baldessarini et al. 2002; Baldessarini & Tondo, 2000). However, its efficacy tends to vary between BD patients. A series of phenotypic studies of responders to lithium have shown that : (1) the response to lithium appears to be longitudinally stable (Berghofer et al. 2008); (2) lithium responders suffer from a more typical recurrent illness with full remission between episodes and low rates of comorbid conditions (Alda 2004); and (3) first-degree relatives with BD respond to lithium as well (Grof et al. 2002). Taken together, these studies suggest that response to lithium prophylaxis may be considered a BD clinical subtype with less genetic heterogeneity and stronger genetic effects.
In this study, we continued our efforts investigating molecular factors associated with BD by combining linkage and gene expression analyses. In the first stage of this study, we conducted a second genome-wide scan analysis using over 800 microsatellite markers in 36 lithium-responsive BD families. This analysis was followed by fine mapping of identified candidate regions and investigation of altered patterns of brain expression of all genes mapping to linked regions using an independent sample of BD subjects and controls.
Methods
Subjects
Families included in this study were ascertained through probands with BD who were followed prospectively and responded unequivocally to prophylactic lithium treatment. A total of 36 probands, 19 males and 17 females, were recruited from specialized clinics at McMaster University, Hamilton; University of Ottawa; and Dalhousie University, Halifax. They all met criteria for bipolar I (n=25) or bipolar II (n=11) disorder according to both Research Diagnostic Criteria (RDC; Spitzer et al. 1978) and DSM-IV (APA, 1994). None of the probands had any other Axis I or Axis II disorders. Their mean age (±S.D.) was 46.0±13.6 yr, and their mean age at onset was 24.5±7.9 yr. Their clinical course had been characterized by a high number of manic and depressive episodes before lithium (8.2±10.1) and by a full stability on lithium monotherapy for 14.4±6.8 yr on average. The criteria used to define excellent response to lithium were: (a) diagnosis of primary episodic BD; (b) high recurrence risk prior to lithium treatment; and (c) no recurrences during the 3-yr minimum observation time on lithium monotherapy with an average plasma concentration over 0.6 mequiv./l (Turecki et al. 2001).
Families were included in this study if they met the following criteria : (1) at least four members (including the proband) were willing to participate in an interview and provide a blood sample; (2) at least two of the four members were affected; and (3) participants were aged ≥16 yr at the time of recruitment. Overall, 36 multiplex families were included, accounting for a total of 275 individuals. Of these, 132 were considered affected.
Best-estimate diagnoses were made by a panel of experienced psychiatrists who blindly reviewed the data from the Schedule for Affective Disorders and Schizophrenia – Lifetime version (SADS-L; Endicott & Spitzer, 1978) interviews and available medical records. Diagnoses were based on RDC (Spitzer et al. 1978). All families were Caucasian of European descent.
For the purpose of linkage analysis, the affected phenotype included individuals with the following diagnoses: bipolar I and bipolar II disorders, schizoaffective disorder bipolar type, and recurrent major depression with the additional criterion of functional incapacitation during at least one episode. These criteria are based on our previous family (Grof et al. 1994) and linkage (Turecki et al. 2001) study. Other psychiatric conditions were rare in these families, with rates comparable to those in the general population.
Approval for the different stages of this study was obtained from local institutional review boards, and for post-mortem studies, informed consent was obtained from next of kin.
Genotyping
Genome-wide scan
Genomic DNA was extracted from peripheral blood samples following standard procedures (Sambrook et al. 1989). A genome-wide scan was performed using a total of 811 fluorescent-labelled polymerase chain reaction (PCR) markers from the ABI PRISM Linkage Mapping Set v. 2.5-HD5 (Applied Biosystems, USA). PCR products were analysed by capillary electrophoresis using the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). These set of markers were selected from the Généthon map, had an average heterozygosity of 0.77 and provided a 5-cM average resolution.
Individual genotypes were assigned using GeneScan analysis software (v. 3.7) and Genotyper software (v. 3.7) (Applied Biosystems). Mendelian inconsistencies were assessed using PedCheck software (O’Connell & Weeks, 1998). Inconsistent genotypes were re-examined and those with ambiguous genotypes were deleted prior to analysis.
Fine mapping
Fine mapping of candidate regions was performed by genotyping additional fluorescent-labelled micro-satellite markers from either the Marshfield or deCODE genetic maps. The average resolution for the markers used was of 1.8 cM. Fifteen markers were genotyped in both chromosome 3 regions, eight of these between markers D3S1263 (36.10 cM) and D3S1266 (52.60 cM), and seven between markers D3S1300 (80.32 cM) and D3S1566 (97.75 cM). On chromosome 14, seven markers were genotyped between markers D14S1023 (8.28 cM) and D14S1040 (31.75 cM). Markers were analysed as outlined above. The average marker heterozygosity was 0.72; and the average inter-marker resolution for both regions on chromosome 3 was 1.5 and 1.7, the latter in the more centromeric region; whereas, on chromosome 14 it was 2.2 cM. Individual genotypes were assigned using GeneMapper software (v. 3.7) (Applied Biosystems).
Stastistical analysis
Linkage was tested by two-point LOD score analysis, with genetic models based on our previous analyses of mode of inheritance in this population (Alda et al. 1997). We used the MLINK program from the FASTLINK computer package (Cottingham et al. 1993). A total of five genetic models with sex-specific penetrance values were used to maximize the evidence of linkage: (a) Dominant 1, allele frequency (q)=0.006, male penetrance (fM)=0.4, female penetrance (fF)=0.7 ; (b) Dominant 2, q=0.012, fM=0.2, fF=0.35; (c) Recessive 1, q=0.110, fM=0.35, fF=0.65; (d) Recessive 2, q=0.160, fM=0.18, fF=0.33; (e) Intermediate, q=0.012, fM=0.4 and fF=0.7 for homozygotes, and fM=0.2 and fF=0.35 for heterozygotes. The two dominant and two recessive models differed from each other in the degree of sex-specific penetrance, with one model having a lower penetrance and the other a higher penetrance. For all models, the rates of phenocopies for males and females were 0.005 and 0.009, respectively.
Empirical p values were calculated for loci with LOD scores >1.0 by computer simulations. First we generated 10000 replicates of the sample under the hypothesis of no linkage using the SIMULATE program (Terwilliger et al. 1993). We then analysed the simulated data using the MSIM program from the SLINK package (Weeks et al. 1990). In addition, we analysed the data under the assumption of heterogeneity using the ANALYZE package (ftp://ftp.ebi.ac.uk/pub/software/linkage_and_mapping/linkage_cpmc_olumbia/analyze/).
Brain tissue donors
Brain tissue samples were obtained from the Quebec Suicide Brain Bank (www.douglasrecherche.qc.ca/suicide). Dorsolateral [Brodmann area (BA) 8/9 and BA 46] and ventromedial (BA 10, BA 44, BA 45, BA 47) prefrontal cortices were sampled at 4 °C and snapfrozen in liquid nitrogen before storage at −80 °C following standard procedures (Bird & Vonsattel, 1993). These brain regions were selected given their hypothesized involvement in mood disorders (Drevets et al. 2008; Savitz & Drevets, 2009). Diagnostic characterization of all subjects was made by psychological autopsies using structured diagnostic instruments based on DSM-IV criteria, as described previously (Dumais et al. 2005). The sample investigated in this study consisted of a case group (BD, n=8) that included seven subjects with BD and one subject with schizoaffective disorder, bipolar type; as well as a control group (n=12) with no history of suicidal behaviour, nor major mood and/or psychotic disorders. Demographic and clinical characteristics, including post-mortem intervals (PMI) and pH measures of the sample can be found in Supplementary Table S1 (available online). All deaths in the BD group were due to suicide, except for one death secondary to a workplace accident. In the control group, all subjects died suddenly from cardiac-related problems and automobile accidents with no extended agonal period prior to death.
Microarray analysis
RNA sample integrity was measured using a high-resolution electrophoresis system (Agilent 2100 Bioanalyzer, Agilent Technologies, USA). RNA samples included in the study had an A260/280 ratio >1.9 and a 28S/18S peak height ratio >1.6. Samples were processed using the Human Genome U133 Plus 2.0 Array (http://www.affymetrix.com). GeneChip data analysis was performed using Genesis 2.0 software (Gene Logic Inc., USA), Microarray Analysis Suite version 5.0 (MAS 5.0) and Data Mining Tool 2.0. All transcripts represented in the GeneChip data were globally normalized and scaled to a signal intensity of 100. The RNA and microarray quality control parameters used to filter arrays prior to analysis included raw Q values (noise), the number of ‘present’ calls for genes across arrays and β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 5′/3′ signal ratios.
Data analysis
A list of genes mapping to candidate regions was obtained using the public database Ensembl (http://www.ensembl.org) and the probe sets annotated to these genes were obtained using NetAffx Analysis Center (www.affymetrix.com).
Gene expression analyses of only the genes on this list were performed using Partek Genomics Suite software (http://www.partek.com/). Candidate regions were defined by flanking markers with LOD scores≥1. The margins were further extended by 5Mb on each side to reduce the chance of excluding genes potentially relevant to BD, particularly given the fact that we used maximized LOD scores. For each of the brain regions investigated, a probe set had to be ‘present’ or detectable in at least 75% of the subjects in at least one of the two groups to reduce the chances of false-positive results and to exclude non-specific probe sets. ‘Present’ genes were determined using the MAS 5.0 normalization algorithm, and expression values used in the analyses were obtained using the robust multi-array (RMA) normalization algorithm. All gene expression values were floored to 1 and then log2-transformed prior to statistical analysis.
Significant differences between the two groups were determined by ANCOVA analysis on Partek with PMI and pH as covariates. Genes were considered significantly differentially expressed if the following criteria were met: a p value ≤0.05 and an absolute fold-change value ≥1.3. Of these genes, those with consistent patterns of expression in at least two brain regions, or in the same brain region, but demonstrated by at least two probe sets of the same gene, were selected for validation by real-time PCR.
Reverse transcription and real-time PCR
Total RNA was extracted from frozen brain tissue using the RNeasy Lipid Tissue Mini kit (Qiagen Inc., Canada). RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies) and only those samples with an RNA integrity number (RIN) >5 were included. These samples were independent from those used for the microarray analysis, and were taken from adjacent tissue dissections. Synthesis of cDNA was performed using oligo(dT) priming (Invitrogen, The Netherlands). PCR reactions were run in quadruplicate using an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). We used 10-μl volume reactions, with 5 ng cDNA, 5 μl Power SYBR Green Master Mix (Applied Biosystems), and 4 pmol primers (IDT). Relative expression values were calculated using the comparative threshold cycle (Ct) method in SDS software version 2.2.1 (Applied Biosystems) with GAPDH as an endogenous control. Baseline and threshold fluorescence levels were automatically computed by the SDS software. Samples were excluded from the analysis if the standard deviations of the Ct value were>0.3, reflecting excessive variability among the replicates. Aone-tailed Student’s t test was used to compare relative expression values between groups. Pearson’s correlation coefficient was used to determine the correlation between the microarray and real-time PCR comparative fold changes. Statistical analyses were performed using SPSS v. 12.0 (SPSS Inc., USA).
Replication sample analysis
A dataset from the Stanley Medical Research Institute (SMRI) (www.stanleyresearch.org/dnn/) was used to follow-up our gene expression findings. This dataset had been processed using the Human Genome U133 Plus 2.0 Array (http://www.affymetrix.com) in the prefrontal cortical region BA 46. We followed the same procedure as in our sample to analyse this dataset. After applying quality control parameters, the final dataset used consisted of 23 BD individuals and 24 unaffected controls. No significant differences were found in age (p=0.634), PMI (p=0.530) or pH (p=0.110). Demographic and clinical characteristics of this dataset can be found in Supplementary Table S2 (online). Information about collection, diagnostic assessment, processing and storage of these samples can be found in the SMRI Array Collection site (www.stanleyresearch.org/dnn/BrainResearchLaboratorybrBrainCollection/ArrayCollection/tabid/89/Default.aspx).
Results
Genome-wide scan
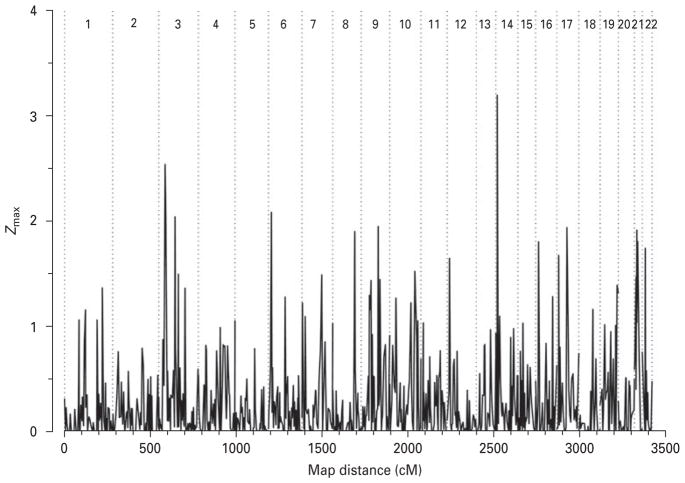
Maximized two-point LOD score analysis revealed 35 loci on 13 chromosomes with LOD scores >1 under one of the five models used (Table 1). Of these, marker D14S990 on chromosome 14q11.2 provided the highest LOD score (3.19) under the recessive model with lower penetrance. The empirical p value associated with this result was 0.0001. The next two largest LOD scores obtained in our study were on chromosome 3. Marker D3S2338 located on 3p25.1 provided a maximum LOD score of 2.53 (empirical p=0.0003) under the intermediate model; whereas marker D3S3697 on 3p14.1 showed a maximum LOD score of 2.04 (empirical p=0.0012) under the dominant model with lower penetrance. Other markers with suggestive evidence of linkage (Lander & Kruglyak, 1995) were observed on chromosomes 17q12 and 21q21.1, with markers D17S927 (LOD 1.94, empirical p=0.0013) and D21S1922 (LOD 1.91, empirical p=0.0021), respectively. The test of linkage under the assumption of heterogeneity gave similar Z scores with an additional finding on chromosome 6p (marker D5S1953, Z=2.08 under the recessive model with lower penetrance) (Fig. 1)
Table 1.
Maximized two-point LOD scores over five genetic models used
| Chromosome | Markera | Position (cM)b | Modelc | LOD score | Theta | p value |
|---|---|---|---|---|---|---|
| 1 | D1S435 | 128.90 | REC2 | 1.15 | 0.05 | 0.0115 |
| 1 | D1S218 | 196.50 | DOM1 | 1.05 | 0.20 | 0.0141 |
| 1 | D1S2692 | 227.30 | REC2 | 1.13 | 0.05 | 0.0103 |
| 3 | D3S2338 | 36.30 | INTER | 2.53 | 0.05 | 0.0003 |
| 3 | D3S3659 | 40.70 | DOM1 | 1.11 | 0.20 | 0.0138 |
| 3 | D3S3697 | 94.50 | DOM2 | 2.04 | 0.00 | 0.0012 |
| 3 | D3S3634 | 114.00 | DOM2 | 1.50 | 0.10 | 0.0037 |
| 3 | D3S3637 | 152.90 | DOM2 | 1.36 | 0.05 | 0.0069 |
| 4 | D4S2930 | 212.20 | DOM2 | 1.10 | 0.15 | 0.0134 |
| 7 | D7S2459 | 120.70 | DOM1 | 1.49 | 0.20 | 0.0064 |
| 9 | D9S169 | 48.20 | REC2 | 1.11 | 0.05 | 0.0109 |
| 9 | D9S161 | 50.30 | REC2 | 1.26 | 0.10 | 0.0080 |
| 9 | D9S1853 | 53.00 | REC2 | 1.16 | 0.05 | 0.0125 |
| 9 | D9S1781 | 99.10 | REC1 | 1.02 | 0.10 | 0.0202 |
| 9 | D9S271 | 109.80 | REC2 | 1.16 | 0.00 | 0.0128 |
| 9 | D9S1682 | 132.90 | DOM2 | 1.08 | 0.15 | 0.0155 |
| 10 | D10S185 | 123.30 | DOM2 | 1.31 | 0.15 | 0.0077 |
| 12 | D12S99 | 13.90 | REC1 | 1.24 | 0.15 | 0.0089 |
| 14 | D14S990 | 8.20 | REC2 | 3.19 | 0.00 | 0.0001 |
| 14 | D14S275 | 21.90 | REC1 | 1.06 | 0.20 | 0.0176 |
| 16 | D16S404 | 16.70 | INTER | 1.80 | 0.00 | 0.0020 |
| 16 | D16S516 | 98.30 | DOM1 | 1.28 | 0.15 | 0.0072 |
| 17 | D17S927 | 58.80 | REC1 | 1.94 | 0.00 | 0.0013 |
| 17 | D17S1868 | 65.10 | DOM1 | 1.03 | 0.15 | 0.0133 |
| 19 | D19S884 | 26.00 | REC1 | 1.02 | 0.15 | 0.0170 |
| 19 | D19S210 | 104.90 | REC1 | 1.04 | 0.15 | 0.0182 |
| 21 | D21S1256 | 8.60 | REC1 | 1.47 | 0.00 | 0.0036 |
| 21 | D21S1922 | 18.00 | REC1 | 1.91 | 0.00 | 0.0021 |
| 21 | D21S1884 | 18.70 | REC1 | 1.02 | 0.15 | 0.0152 |
| 21 | D21S1914 | 23.00 | REC1 | 1.76 | 0.10 | 0.0022 |
| X | DXS1060 | 10.10 | REC1 | 1.12 | 0.15 | 0.0145 |
| X | DXS1061 | 41.70 | DOM1 | 1.24 | 0.15 | 0.0081 |
| X | DXS991 | 86.90 | INTER | 1.37 | 0.05 | 0.0057 |
| X | DXS8009 | 148.40 | DOM1 | 1.11 | 0.15 | 0.0063 |
| X | DXS8069 | 190.40 | INTER | 1.18 | 0.20 | 0.0086 |
Markers with LOD scores >1.
Positions are based on the Généthon map and were taken from NCBI.
REC1, Autosomal recessive with higher penetrance; REC2, autosomal recessive with lower penetrance; DOM1, autosomal dominant with higher penetrance; DOM2, autosomal dominant with lower penetrance; INTER, intermediate.
Fig. 1.
Genome-wide scan heterogeneity LOD scores.
Fine mapping
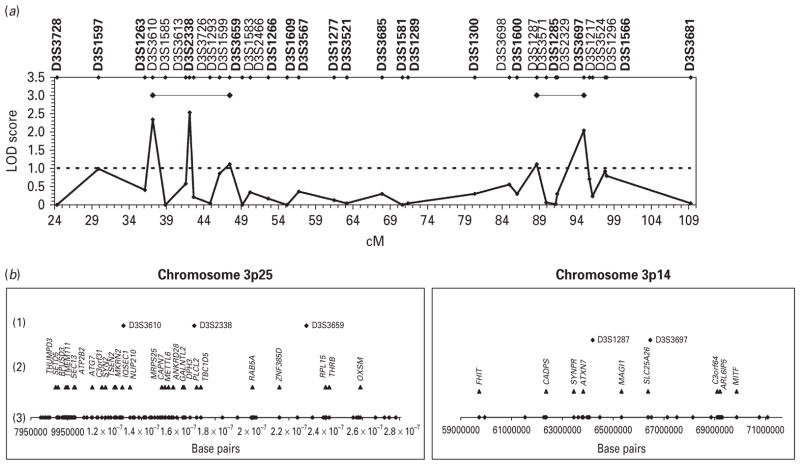
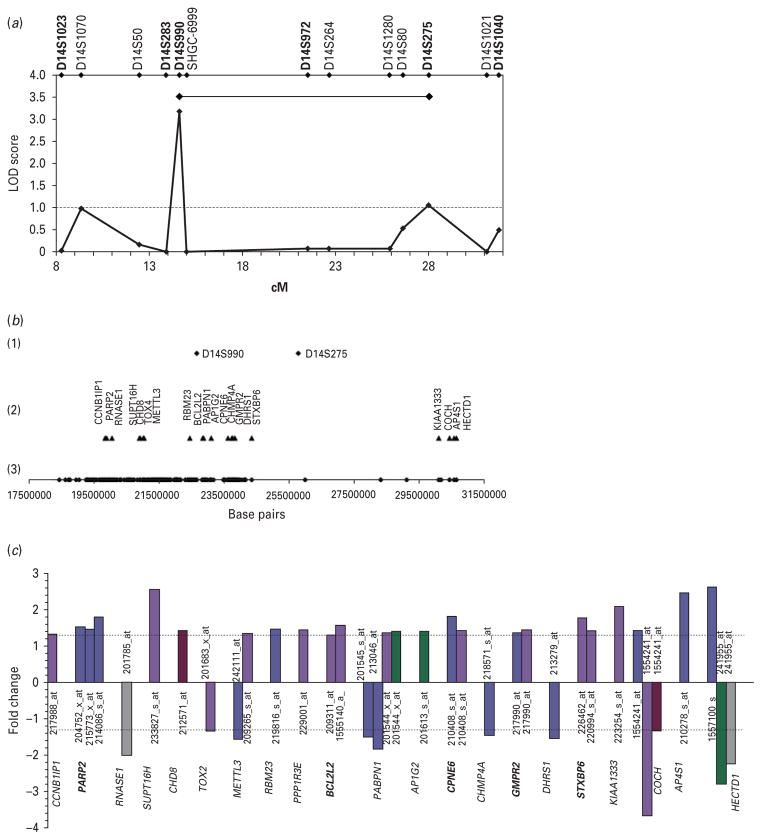
To better define candidate regions for further study, we genotyped additional microsatellite markers surrounding the loci on 14q11.2, 3p25.1, and 3p14.1, as these showed the highest LOD scores in our genome-wide scan. Fine mapping of the more telomeric region in chromosome 3 (3p25.3-p24.1) provided the strongest evidence of linkage. The highest LOD score (2.35) was obtained with marker D3S3610 (Fig. 2a) under the intermediate model. By combining the information provided by both genome-wide and fine-mapping markers, we identified a region delineated by markers D3S3610 and D3S3659, both with LOD scores >1 (Fig. 2a). Modest evidence of linkage in the second, more centromeric region of chromosome 3 (3p14.2-p14.1) was obtained with marker D3S1287 (1.11) (Fig. 2a). A refined region between markers D3S1287 and D3S3697, both with LOD scores >1, was established (Fig. 2a). While fine mapping of the region on chromosome 14 (14q11.2-q12) provided the least evidence for linkage (Fig. 4a), it is worth noting that no negative LOD scores were seen in this region and therefore the 14q region was still used for subsequent analyses.
Fig. 2.
(a) Maximized two-point LOD scores for genome-wide (in bold) and fine-mapping markers in chromosome 3. Candidate regions are indicated by black horizontal lines. (b) Microsatellite markers and genes on candidate regions 3p25 and 3p14. (1) Microsatellite markers with LOD scores >1; (2) differentially expressed genes after controlling for pH and PMI effects (p≤0.05) ; (3) physical location of the 114 (3p25) and 31 (3p14) genes included in brain expression analyses.
Fig. 4.
(a) Maximized two-point LOD scores for genome-wide (in bold) and fine-mapping markers in chromosome 14. Candidate region is indicated by a black horizontal line. (b) Microsatellite markers and genes on candidate region 14q11. (1) Microsatellite markers with LOD scores >1; (2) differentially expressed genes after controlling for pH and PMI effects (p≤0.05) ; (3) physical location of the 286 genes included in brain expression analyses. (c) Gene probe sets differentially expressed in various brain regions. Brodmann areas (BA) are indicated as follows : BA 8/9, light blue ; BA10, dark blue ; BA 44, violet ; BA 45, green; BA 46, red; BA 47, grey. Dotted lines indicate absolute fold change of 1.3. Genes selected for validation are in bold.
Gene expression and real-time PCR validation
In order to further characterize the three candidate regions showing evidence of linkage to lithium-responsive BD, we investigated the differential expression of genes mapping to these regions using brain samples from six prefrontal brain regions of BD cases and controls. The more telomeric candidate region (3p25) spanned ~20 Mb and included 114 annotated genes, whereas the more centromeric region (3p14) covered ~12 Mb and comprised 31 annotated genes (Fig. 2b). The region on chromosome 14 (14q11) spanned ~13Mb and contained 286 genes (Fig. 4b).
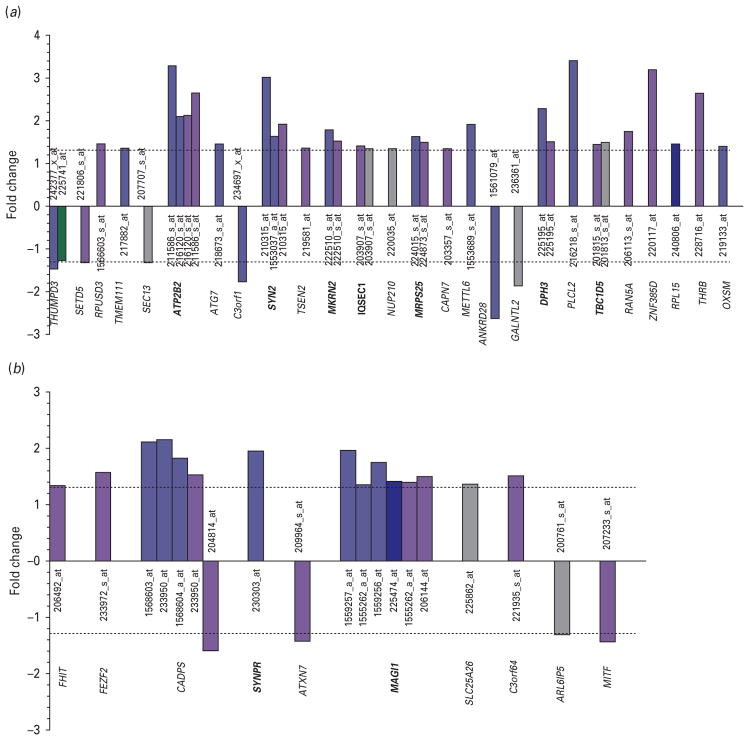
Our results showed 26 genes significantly and differentially expressed in candidate region 3p25 (Figs 2b, 3a), 10 genes in region 3p14 (Figs 2b, 3b), and 21 genes in region 14q11 (Fig. 4b, c) (p ≤0.05 and absolute fold change ≥1.3). Of these genes, 14 showed either consistent patterns of expression in at least two brain regions or within the same brain region but demonstrated by at least two of its probe sets, and therefore were selected for validation (Table 2). Of note, one of the genes selected for validation, synaptoporin (SYNPR) did not meet the two-probe set criterion. However, as it was represented in our array exclusively by one single probe set and given its role in synaptic function, we chose to include it in our followup studies.
Fig. 3.
Gene probe sets differentially expressed in various brain regions. (a) Chromosome 3p25; (b) chromosome 3p14. Brodmann areas (BA) are indicated as follows: BA 8/9, light blue; BA10, dark blue ; BA 44, violet ; BA 45, green; BA47, grey. Dotted lines indicate absolute fold change of 1.3. Genes selected for validation are in bold.
Table 2.
Validation of selected differentially expressed genes by real-time PCR
| Gene | BA | Probe set | Microarray
|
Real-time PCR
|
Correlation
|
|||
|---|---|---|---|---|---|---|---|---|
| FC | p | FC | pa | r | p | |||
| ATP2B2 | 8/9 | 211586_s_at | 3.28 | 0.009 | 1.58 | 0.049 | 0.212 | 0.532 |
| 44 | 211586_s_at | 2.64 | 0.001 | 1.11 | 0.241 | 0.405 | 0.120 | |
| 44 | 216120_s_at | 2.12 | 0.007 | 0.516 | 0.041** | |||
| SYN2b | 8/9 | 210315_at | 3.02 | 0.015 | 1.23 | 0.253 | 0.440 | 0.176 |
| 44 | 210315_at | 1.91 | 0.013 | 1.33 | 0.051 | 0.689 | 0.003** | |
| SYN2a | 8/9 | 1553037_a_at | 1.62 | 0.035 | −1.05 | 0.419 | 0.083 | 0.808 |
| 44 | 1553037_a_at | 1.24 | 0.092 | 1.09 | 0.406 | 0.103 | 0.695 | |
| MKRN2 | 8/9 | 222510_s_at | 1.78 | 0.010 | 1.45 | 0.087 | 0.474 | 0.141 |
| 44 | 222510_s_at | 1.52 | <0.001 | 1.32 | 0.139 | 0.538 | 0.032** | |
| IQSEC1 | 44 | 203907_s_at | 1.40 | <0.001 | −1.21 | 0.179 | −0.183 | 0.498 |
| 47 | 203907_s_at | 1.34 | 0.021 | −1.29 | 0.053 | −0.478 | 0.084 | |
| MRPS25 | 8/9 | 224015_s_at | 1.63 | 0.009 | 1.23 | 0.184 | 0.092 | 0.788 |
| 44 | 224873_s_at | 1.49 | 0.003 | 1.47 | 0.014 | 0.464 | 0.070 | |
| DPH3 | 8/9 | 225195_at | 2.28 | 0.034 | 1.38 | 0.257 | −0.118 | 0.73 |
| 44 | 225195_at | 1.51 | 0.017 | 1.65 | 0.010 | 0.602 | 0.011** | |
| TBC1D5 | 44 | 201815_s_at | 1.43 | 0.001 | 1.43 | 0.016 | 0.564 | 0.018** |
| 47 | 201813_s_at | 1.49 | 0.030 | 1.43 | 0.013 | 0.447 | 0.109 | |
| SYNPR | 8/9 | 230303_at | 1.94 | 0.013 | 1.17 | 0.204 | 0.42 | 0.199 |
| MAGI1 | 8/9 | 1559257_a_at | 1.96 | 0.018 | 1.17 | 0.246 | 0.08 | 0.815 |
| 1555262_a_at | 1.35 | 0.030 | 0.035 | 0.918 | ||||
| 1559256_at | 1.75 | 0.042 | 0.295 | 0.378 | ||||
| 44 | 1555262_a_at | 1.40 | 0.006 | 1.38 | 0.074 | 0.631 | 0.007** | |
| 206144_at | 1.49 | 0.021 | 0.419 | 0.094 | ||||
| PARP2 | 8/9 | 204752_x_at | 1.53 | 0.005 | −1.15 | 0.163 | 0.075 | 0.826 |
| 215773_x_at | 1.48 | 0.019 | −0.113 | 0.741 | ||||
| 214086_s_at | 1.80 | 0.037 | 0.015 | 0.966 | ||||
| BCL2L2 | 44 | 1555140_a_at | 1.58 | 0.011 | 1.27 | 0.124 | 0.537 | 0.026** |
| 209311_at | 1.32 | 0.001 | 0.309 | 0.227 | ||||
| CPNE6 | 8/9 | 210408_s_at | 1.83 | 0.003 | 1.17 | 0.288 | 0.333 | 0.317 |
| 44 | 210408_s_at | 1.42 | 0.036 | 1.50 | 0.014 | 0.823 | <0.001** | |
| GMPR2 | 8/9 | 217990_at | 1.38 | 0.011 | 1.15 | 0.317 | 0.143 | 0.675 |
| 44 | 217990_at | 1.45 | <0.001 | 1.23 | 0.046 | 0.341 | 0.196 | |
| STXBP6 | 44 | 226462_at | 1.78 | 0.003 | 1.66 | 0.001 | 0.821 | <0.001** |
| 220994_s_at | 1.43 | 0.039 | 0.845 | <0.001** | ||||
BA, Brodmann area; FC, fold change; r, Pearson’s r correlation between microarray and real-time PCR data.
One-tailed.
Statistically significant correlations.
The real-time PCR data demonstrated similar patterns of expression as those seen by microarray analysis (Table 2). We found statistically significant correlations for nine of the 14 genes investigated (Table 2). The strongest correlations were seen for : SYN2b (Synapsin IIb) on 3p25; MAGI1 (membrane-associated guanylate kinase, WW and PDZ domain containing 1) on 3p14, STXBP6 (syntaxin binding protein 6) and CPNE6 (neuronal copine VI) both on 14q11. Other genes with significant correlations included ATP2B2 (ATPase, Ca2+ transporting, plasma membrane 2) on 3p25; BCL2L2 (BCL2-like 2) on 14q11; MKRN2 (makorin, ring finger protein, 2), DPH3 (DPH3, KTI11 homolog), and TBC1D5 (TBC1 domain family, member 5) on region 3p25. All nine genes were found up-regulated in BD, particularly in BA 8/9 and BA 44. Of note, six of these nine genes were also nominally significant by real-time-PCR: ATP2B2, SYN2, DPH3, TBC1D5, CPNE6, STXBP6. Whereas MRPS25 (mitochondrial ribosomal protein S25) showed only a marginally significant correlation, this gene was found significantly up-regulated in BD by microarray anlaysis and real-time PCR (Table 2).
In the case of SYN2, two sets of primers were designed to target both of its transcripts (SYN2a and SYN2b). Whereas SYN2b was validated, SYN2a failed to validate. Similarly, SYNPR (synaptoporin) also failed to validate.
We followed up our results by analysing a dataset of BD and control subjects publicly available through the SMRI Brain Collection, which was comparable to ours (Supplementary Table S2, online). Several of the genes identified in our sample were also differentially expressed in the SMRI dataset. Whereas one of them reached statistical significance (MRPS25), four other genes (SYN2, MAGI1, CPNE6, IQSEC1), three of which code for proteins involved in synaptic processes, were marginally significant (Supplementary Table S3, online). Interestingly, their patterns of expression were consistent in the two samples. IQSEC1 was significantly up-regulated in both microarray samples but gave conflicting results by real-time PCR in our sample.
Discussion
In this study, we conducted a linkage analysis of lithium-responsive BD families and identified candidate regions on chromosomes 3 and 14. We then investigated brain expression patterns of genes mapping to these regions in post-mortem brain tissue from BD patients, and identified a group of synaptic, apoptotic, mitochondrial and neurogenesis genes. Analysis of an independent dataset from the SMRI provided further support for synaptic and mitochondrial genes in BD.
The first part of our study revealed several chromosomal regions of potential interest in BD (Table 1). Of these, the most promising signals were on 3p25, 3p14 and 14q11; therefore, we focused our efforts on these three regions. However, other regions to consider in future studies are 6p, 17q, and 21q. Fine mapping of regions on chromosomes 3 and 14 provided further support for 3p25, while only modest additional support was found for 3p14 and 14q11. The latter might be explained by lower marker density in these two regions compared to region 3p25, particularly on 14q11 where the lack of markers in publicly available genetic maps precluded higher marker coverage. Alternatively, the signals on regions 3p14 and 14q11 may represent false-positive findings. However, support for these regions has been provided by studies performed on similar homogeneous groups such as early-onset BD (Etain et al. 2006) and an eastern Cuban population (Marcheco-Teruel et al. 2006). Of the three candidate regions, 3p25 is probably the one with most support for linkage in BD. This result partially overlaps with findings from our previous genome scan (Turecki et al. 2001). Furthermore, loci adjacent to this region including 3p23, 3p22 and 3p21 have been implicated in BD by other genome-scan and genome-wide association studies (GWAS) (Fallin et al. 2004; Perlis et al. 2009; Scott et al. 2009; Wellcome Trust Case Control Consortium, 2007). A recently published GWAS of lithium response in BD pointed to five chromosomal regions, including 3p22, as showing suggestive evidence of association with lithium response in two separate cohorts (Perlis et al. 2009).
Few groups have conducted genetic studies of BD in families of lithium responders. Morissette et al. (1999) studied two large families recruited through BD probands from the Saguenay-Lac-St-Jean population. One of their criteria included lithium response. Their most prominent findings pointed to chromosomal region 12q23-q24, which was later confirmed by the same group after recruiting more families and genotyping more markers (Shink et al. 2005). However, a direct comparison of our findings with theirs is difficult given that in our study lithium response was assessed prospectively, while it is not clear how it was defined in their studies.
Investigating the brain expression patterns of genes mapping to candidate regions, we identified nine genes showing significant correlations between microarray and real-time PCR analyses with the majority of these genes being also nominally significant in realtime PCR in BD. Interestingly, these genes may be broadly divided in either having a role in synaptic function (SYN2, STXBP6, ATP2B2, CPNE6 and MAGI1), apoptotic regulation (BCL2L2) or neurogenesis (MKRN2). The other two genes are DPH3 and TBC1D5. Furthermore, all synapse-related genes, with the exception of ATP2B2, showed the strongest correlations between microarray and real-time PCR expression levels. Of note, MRPS25 was found differentially expressed by both microarray and real-time PCR. Analysis of the SMRI dataset provided further support for dysregulation of synaptic genes, particularly SYN2, as well as for the mitochondrial MRPS25 gene in the prefrontal cortex of BD.
Most of the synapse-related genes found in this study encode proteins located in the presynaptic terminal. SYN2 (3p25) is the one that has been most extensively investigated, particularly in schizophrenia where decreased expression of this gene has been seen (Dyck et al. 2009; Vawter et al. 2002). SYN2 encodes a neuronal phosphoprotein associated with the cytoplasmic surface of the synaptic vesicle membrane (Greengard et al. 1993). It has been involved in synaptogenesis, maintenance of synapses and the modulation of neurotransmitter release (Ferreira et al. 1995; Jovanovic et al. 2000). Therefore, dysregulation of this gene may have a profound effect on disorders such as BD. A previous study by our group also found increased expression of SYN2 in suicide victims with major depression in BA 45 (Klempan et al. 2009). However, these results are in contrast with those by Vawter et al (2002) where decreased expression of SYN2 was seen in the hippocampus of patients with BD and schizophrenia.
MAGI1 (3p14), located in the cytoplasmic membrane at the cell–cell junction appears to be an important component of neuronal junctions (Laura et al. 2002) and have a role in signal transduction in the brain (Shiratsuchi et al. 1998). CPNE6 (14q11), which has been localized to the cell body and dendrites of mouse brains, appears to have a role in post-synaptic events as a calcium sensor and in synaptic plasticity as well (Nakayama et al. 1998). Therefore, dysregulation of these genes may result in impaired synaptic function.
Other relevant synaptic genes found in our study but not in the SMRI dataset are mentioned herein. STXBP6 (14q11) is a gene involved in the formation of the SNARE complex, which plays an important role in the transport of vesicles to the cell membrane for exocytosis and therefore altered expression of this gene may impair the synaptic vesicle cycle (Scales et al. 2002). Calcium influx to the synaptic vesicle is important for the release of neurotransmitter to the synaptic cleft ; therefore dysregulation of ATP2B2 (3p25), which plays a critical role in intracellular calcium homeostasis by maintaining a low intracellular calcium concentration (DeMarco et al. 2002) may impair neurotransmission. In addition, this gene has also been involved in synaptic plasticity (Empson et al. 2007).
Further evidence supporting altered synaptic function in BD comes from a study performed by Ryan et al. (2006). Although, their findings showed decreased expression of synaptic genes in BA 11 (none of which was studied here), their results point to alterations in the synapse as possible mechanism in BD.
MRPS25 encodes for one of more than 70 mitochondrial ribosomal proteins (Kenmochi et al. 2001). Findings from our sample and the SMRI dataset add further support for mitochondrial dysfunction as a possible pathophysiological mechanism in BD, which has been previously proposed by Kato & Kato (2000). To our knowledge, this gene has not been identified in previous studies of BD.
Impaired apoptotic regulation might also be implicated in BD. Altered expression of BCL2L2 (14q11) was observed in our study. This gene contributes to reduced cell apoptosis under cytotoxic conditions (Gibson et al. 1996). Additionally, studies in mice have indicated an anti-apoptotic role in regulating the survival of NGF- and BDNF-dependent neurons (Middleton et al. 2001). Dysregulation of MKRN2 (14q11) may result in impaired neurogenesis as it has been suggested that it inhibits neurogenesis on its human ortholog in Xenopus laevis (Yang et al. 2008). However, neither of these genes was found to be significant in the SMRI dataset.
The strengths of this study include combining two complementary genetic strategies to find susceptibility genes for BD. The systematic and prospective evaluation of lithium response in recruited families may have resulted in decreased clinical and possibly genetic heterogeneity. In addition, the investigation of altered patterns of expression in most prefrontal cortices of BD subjects allowed for the identification of region-specific abnormalities. Taken together, this two-stage approach allowed us to first identify regions harbouring genes of interest to BD in a well characterized sample; and second, to screen and prioritize genes on those candidate regions based on their patterns of expression using a post-mortem sample. However, there are several limitations to this study. Although both of the cohorts had a diagnosis of BD, they were not recruited following the same criterion of lithium response. The design used for linkage analysis did not allow us to determine if the chromosomal regions identified are related to lithium response or to BD. Moreover, the fine-mapping resolution achieved in this study was limited by the number of available markers in public databases, which may have resulted in overlooking other regions of interest.
Given that the sample used in gene expression analyses was comprised mostly of males and was of French-Canadian origin, our results may not be generalized to other populations. In addition, as most of the BD individuals died by suicide, it is difficult to say whether or not the altered expression patterns are related to BD, suicide or both. Similarly, as some of our subjects had used mood stabilizers including lithium, and other substances, the altered levels of expression observed might have been the effect of psychotropic medication or substance use. Of note, microarray analyses were not corrected for multiple testing ; however, using both fold changes and p values to determine gene expression differences, as well as controlling for pH and PMI effects may have added stringency to our analyses. Furthermore, we also selected genes for validation based on consistency of expression patterns across brain regions or within the same brain region as demonstrated by various probe sets of the same gene.
Last, our study may have suffered from decreased power, given the relatively small sample used in both the family and the expression studies. Nevertheless, our results are consistent with results from a few other genetic studies of BD and are supported, partly, by analysis of an independent microarray dataset.
In summary, using linkage and gene expression analyses we found evidence of linkage to lithium responsive BD in regions 3p25, 3p14 and 14q11, as well as a group of differentially expressed genes mapping to these regions, suggesting altered synaptic and mitochondrial function in BD. Impaired apoptosis and neurogenesis regulation might be also implicated in BD. Further studies are warranted to verify the validity and significance of our results in other populations as well as to determine the function of identified genes and the effect of mood stabilizers (i.e. lithium) on their expression.
Supplementary Material
Acknowledgments
We thank our patients and their family members for participating in the study. Ms. Julie Garnham, RN, and Ms. Claire Slaney, RN, helped coordinate part of the study and performed some of the diagnostic assessment along with Drs Patrizia Cavazzoni, Anne Duffy, Claire O’Donovan, Martina Ruzickova, Marina Sokolenko, and Petr Zvolsky Jr. Drs. Naan Cheng and Tomas Hajek performed parts of the statistical analyses. Dr Lindsay Blair carried out part of the genotyping for the linkage study. Dr Tim Klempan helped in part of the gene expression studies. We also thank the SMRI for providing the dataset for replication analysis. The study was funded by CIHR grant 64410.
Footnotes
Note
Supplementary material accompanies this paper on the Journal’s website (http://journals.cambridge.org/pnp).
Statement of Interest
None.
References
- Alda M. The phenotypic spectra of bipolar disorder. European Neuropsychopharmacology. 2004;14 (Suppl 2):S94–S99. doi: 10.1016/j.euroneuro.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Alda M, Grof E, Cavazzoni P, Duffy A, et al. Autosomal recessive inheritance of affective disorders in families of responders to lithium prophylaxis? Journal of Affective Disorders. 1997;44:153–157. doi: 10.1016/s0165-0327(97)00042-6. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Molecular Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- Baldessarini RJ, Tondo L. Does lithium treatment still work? evidence of stable responses over three decades. Archives of General Psychiatry. 2000;57:187–190. doi: 10.1001/archpsyc.57.2.187. [DOI] [PubMed] [Google Scholar]
- Baldessarini RJ, Tondo L, Hennen J, Viguera AC. Is lithium still worth using? an update of selected recent research. Harvard Review of Psychiatry. 2002;10:59–75. [PubMed] [Google Scholar]
- Berghofer A, Alda M, Adli M, Baethge C, et al. Long-term effectiveness of lithium in bipolar disorder : a multicenter investigation of patients with typical and atypical features. Journal of Clinical Psychiatry. 2008;69:1860–1868. doi: 10.4088/jcp.v69n1203. [DOI] [PubMed] [Google Scholar]
- Bird ED, Vonsattel JP. The development of a brain bank. Journal of Neural Transmission (Suppl) 1993;39:17–23. [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. American Journal of Human Genetics. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- DeMarco SJ, Chicka MC, Strehler EE. Plasma membrane Ca2+ ATPase isoform 2b interacts preferentially with Na+/H+ exchanger regulatory factor 2 in apical plasma membranes. Journal of Biological Chemistry. 2002;277:10506–10511. doi: 10.1074/jbc.M111616200. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders : implications for neurocircuitry models of depression. Brain Structure and Function. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumais A, Lesage AD, Alda M, Rouleau G, et al. Risk factors for suicide completion in major depression: a case-control study of impulsive and aggressive behaviors in men. American Journal of Psychiatry. 2005;162:2116–2124. doi: 10.1176/appi.ajp.162.11.2116. [DOI] [PubMed] [Google Scholar]
- Dyck BA, Skoblenick KJ, Castellano JM, Ki K, et al. Behavioral abnormalities in synapsin II knockout mice implicate a causal factor in schizophrenia. Synapse. 2009;63:662–672. doi: 10.1002/syn.20643. [DOI] [PubMed] [Google Scholar]
- Empson RM, Garside ML, Knopfel T. Plasma membrane Ca2+ ATPase 2 contributes to short-term synapse plasticity at the parallel fiber to Purkinje neuron synapse. Journal of Neuroscience. 2007;27:3753–3758. doi: 10.1523/JNEUROSCI.0069-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A diagnostic interview : the schedule for affective disorders and schizophrenia. Archives of General Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- Etain B, Mathieu F, Rietschel M, Maier W, et al. Genome-wide scan for genes involved in bipolar affective disorder in 70 European families ascertained through a bipolar type I early-onset proband: supportive evidence for linkage at 3p14. Molecular Psychiatry. 2006;11:685–694. doi: 10.1038/sj.mp.4001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, et al. Genomewide linkage scan for bipolar-disorder susceptibility loci among Ashkenazi Jewish families. American Journal of Human Genetics. 2004;75:204–219. doi: 10.1086/422474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A, Han HQ, Greengard P, Kosik KS. Suppression of synapsin II inhibits the formation and maintenance of synapses in hippocampal culture. Proceedings of the National Academy of Sciences USA. 1995;92:9225–9229. doi: 10.1073/pnas.92.20.9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson L, Holmgreen SP, Huang DC, Bernard O, et al. bcl-w, a novel member of the bcl-2 family, promotes cell survival. Oncogene. 1996;13:665–675. [PubMed] [Google Scholar]
- Greengard P, Valtorta F, Czernik AJ, Benfenati F. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science. 1993;259:780–785. doi: 10.1126/science.8430330. [DOI] [PubMed] [Google Scholar]
- Grof P, Alda M, Grof E, Zvolsky P, et al. Lithium response and genetics of affective disorders. Journal of Affective Disorders. 1994;32:85–95. doi: 10.1016/0165-0327(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Grof P, Duffy A, Cavazzoni P, Grof E, et al. Is response to prophylactic lithium a familial trait? Journal of Clinical Psychiatry. 2002;63:942–947. doi: 10.4088/jcp.v63n1013. [DOI] [PubMed] [Google Scholar]
- Jovanovic JN, Czernik AJ, Fienberg AA, Greengard P, et al. Synapsins as mediators of BDNF-enhanced neurotransmitter release. Nature Neuroscience. 2000;3:323–329. doi: 10.1038/73888. [DOI] [PubMed] [Google Scholar]
- Kato T, Kato N. Mitochondrial dysfunction in bipolar disorder. Bipolar Disorders. 2000;2:180–190. doi: 10.1034/j.1399-5618.2000.020305.x. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Suzuki T, Uechi T, Magoori M, et al. The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders. Genomics. 2001;77:65–70. doi: 10.1006/geno.2001.6622. [DOI] [PubMed] [Google Scholar]
- Klempan TA, Sequeira A, Canetti L, Lalovic A, et al. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Molecular Psychiatry. 2009;14:175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits : guidelines for interpreting and reporting linkage results. Nature Genetics. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Laura RP, Ross S, Koeppen H, Lasky LA. MAGI-1: a widely expressed, alternatively spliced tight junction protein. Experimental Cell Research. 2002;275:155–170. doi: 10.1006/excr.2002.5475. [DOI] [PubMed] [Google Scholar]
- Marcheco-Teruel B, Flint TJ, Wikman FP, Torralbas M, et al. A genome-wide linkage search for bipolar disorder susceptibility loci in a large and complex pedigree from the eastern part of Cuba. American Journal of Medical Genetics, B: Neuropsychiatric Genetics. 2006;141B:833–843. doi: 10.1002/ajmg.b.30314. [DOI] [PubMed] [Google Scholar]
- McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. American Journal of Human Genetics. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton G, Wyatt S, Ninkina N, Davies AM. Reciprocal developmental changes in the roles of Bcl-w and Bcl-x(L) in regulating sensory neuron survival. Development. 2001;128:447–457. doi: 10.1242/dev.128.3.447. [DOI] [PubMed] [Google Scholar]
- Morissette J, Villeneuve A, Bordeleau L, Rochette D, et al. Genome-wide search for linkage of bipolar affective disorders in a very large pedigree derived from a homogeneous population in Quebec points to a locus of major effect on chromosome 12q23-q24. American Journal of Medical Genetics. 1999;88:567–587. doi: 10.1002/(sici)1096-8628(19991015)88:5<567::aid-ajmg24>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Yaoi T, Yasui M, Kuwajima G. N-copine: a novel two C2-domain-containing protein with neuronal activity-regulated expression. FEBS Letters. 1998;428:80–84. doi: 10.1016/s0014-5793(98)00497-9. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Smoller JW, Ferreira MA, McQuillin A, et al. A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. American Journal of Psychiatry. 2009;166:718–725. doi: 10.1176/appi.ajp.2009.08111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potash JB, Toolan J, Steele J, Miller EB, et al. The bipolar disorder phenome database: a resource for genetic studies. American Journal of Psychiatry. 2007;164:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, et al. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Molecular Psychiatry. 2006;11:965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning. A Laboratory Manual 2. New York: Cold Spring Harbor Press; 1989. [Google Scholar]
- Saunders EH, Scott LJ, McInnis MG, Burmeister M. Familiality and diagnostic patterns of subphenotypes in the National Institutes of Mental Health bipolar sample. American Journal of Medical Genetics, B: Neuropsychiatric Genetics. 2008;147B:18–26. doi: 10.1002/ajmg.b.30558. [DOI] [PubMed] [Google Scholar]
- Savitz J, Drevets WC. Bipolar and major depressive disorder : neuroimaging the developmental-degenerative divide. Neuroscience and Biobehavioral Reviews. 2009;33:699–771. doi: 10.1016/j.neubiorev.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales SJ, Hesser BA, Masuda ES, Scheller RH. Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. Journal of Biological Chemistry. 2002;277:28271–28279. doi: 10.1074/jbc.M204929200. [DOI] [PubMed] [Google Scholar]
- Schulze TG, Hedeker D, Zandi P, Rietschel M, et al. What is familial about familial bipolar disorder? Resemblance among relatives across a broad spectrum of phenotypic characteristics. Archives of General Psychiatry. 2006;63:1368–1376. doi: 10.1001/archpsyc.63.12.1368. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proceedings of the National Academy of Sciences USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part III : bipolar disorder. American Journal of Human Genetics. 2003;73:49–62. doi: 10.1086/376548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shink E, Morissette J, Sherrington R, Barden N. A genome-wide scan points to a susceptibility locus for bipolar disorder on chromosome 12. Molecular Psychiatry. 2005;10:545–552. doi: 10.1038/sj.mp.4001601. [DOI] [PubMed] [Google Scholar]
- Shiratsuchi T, Futamura M, Oda K, Nishimori H, et al. Cloning and characterization of BAI-associated protein 1 : a PDZ domain-containing protein that interacts with BAI1. Biochemical and Biophysical Research Communications. 1998;247:597–604. doi: 10.1006/bbrc.1998.8603. [DOI] [PubMed] [Google Scholar]
- Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. American Journal of Medical Genetics, C: Seminars in Medical Genetics. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Endicott J, Robins E. Research diagnostic criteria : rationale and reliability. Archives of General Psychiatry. 1978;35:773–782. doi: 10.1001/archpsyc.1978.01770300115013. [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Speer M, Ott J. Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genetic Epidemiology. 1993;10:217–224. doi: 10.1002/gepi.1370100402. [DOI] [PubMed] [Google Scholar]
- Turecki G, Grof P, Grof E, D’Souza V, et al. Mapping susceptibility genes for bipolar disorder : a pharmacogenetic approach based on excellent response to lithium. Molecular Psychiatry. 2001;6:570–578. doi: 10.1038/sj.mp.4000888. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Thatcher L, Usen N, Hyde TM, et al. Reduction of synapsin in the hippocampus of patients with bipolar disorder and schizophrenia. Molecular Psychiatry. 2002;7:571–578. doi: 10.1038/sj.mp.4001158. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM. SLINK: a general simulation program for linkage analysis. American Journal of Human Genetics. 1990;47:A204. [Google Scholar]
- Wellcome Trust Case Control Consortium . Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PH, Cheung WK, Peng Y, He ML, et al. Makorin-2 is a neurogenesis inhibitor downstream of phosphatidylinositol 3-kinase/Akt (PI3K/Akt) signal. Journal of Biological Chemistry. 2008;283:8486–8495. doi: 10.1074/jbc.M704768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.