Abstract
Chromatin remodeling during DNA double-strand break (DSB) repair is required to facilitate access to and repair of DSBs. This remodeling requires increased acetylation of histones and a shift in nucleosome organization to create open, relaxed chromatin domains. However, the underlying mechanism driving changes in nucleosome structure at DSBs is poorly defined. Here, we demonstrate that histone H2A.Z is exchanged onto nucleosomes at DSBs by the p400 remodeling ATPase. H2A.Z exchange at DSBs shifts the chromatin to an open conformation, and is required for acetylation and ubiquitination of histones and for loading of the brca1 complex. H2A.Z exchange also restricts single-stranded DNA production by nucleases and is required for loading of the Ku70/80 DSB repair protein. H2A.Z exchange therefore promotes specific patterns of histone modification and reorganization of the chromatin architecture, leading to the assembly of a chromatin template which is an efficient substrate for the DSB repair machinery.
INTRODUCTION
DNA double-strand break (DSB) repair requires the loading of DNA repair proteins onto the damaged chromatin. DSB repair is initiated by the ATM-dependent phosphorylation of histone H2AX (γH2AX), creating domains of γH2AX which extend for 100s of kilobases along the chromatin (Bonner et al., 2008). The mdc1 protein then binds to γH2AX (Stucki et al., 2005) and concentrates other repair proteins, including ATM, the MRN complex and the RNF8 and RNF168 ubiquitin ligases, onto the chromatin (Sun et al., 2010). Further processing of the damaged chromatin then occurs through histone ubiquitination by RNF8/RNF168 (Doil et al., 2009; Huen et al., 2007; Mailand et al., 2007), the loading of repair factors such as CtIP and brca1 (Kim et al., 2007; Sartori et al., 2007) and processing of the DNA ends.
DSB repair also requires remodeling of the local chromatin structure to enable the DNA repair machinery to access sites of DNA damage (Lukas et al., 2011; Xu and Price, 2011). The NuA4 chromatin remodeling complex plays a key role in this process (Downs et al., 2004; Lukas et al., 2011). 2 subunits of NuA4 - the Tip60 acetyltransferase (Sun et al., 2009) and the p400 motor ATPase (Xu et al., 2010) – are important for remodeling chromatin structure at DSBs. Tip60 acetylates histones H2A and H4 (Downs et al., 2004; Kusch et al., 2004; Murr et al., 2006), creating hyperacetylated chromatin domains which extend away from the DSB (Xu et al., 2010). p400, like Tip60, is recruited to DSBs as part of the NuA4 complex (Chan et al., 2005; Gevry et al., 2007; Xu et al., 2010). Loss of p400’s ATPase activity leads to defects in chromatin remodeling at DSBs and an increase in genomic instability (Xu et al., 2010). The positioning of NuA4 at DSBs therefore leads to increased histone acetylation by the Tip60 sub-unit (Downs et al., 2004; Kusch et al., 2004; Murr et al., 2006; Sun et al., 2009), which, in combination with the ATPase activity of p400 (Xu et al., 2010), creates open, relaxed chromatin domains at the DSB. Subsequent studies indicate that these open chromatin domains are required for ubiquitination of the chromatin and the loading of the brca1 protein at DSBs (Murr et al., 2006; Xu et al., 2010). The ATPase activity of the p400 sub-unit of NuA4 is therefore critical for DSB repair.
Although the formation of open chromatin domains at DSBs requires both histone acetylation by Tip60 and the ATPase activity of p400 (Xu et al., 2010), how p400 employs its ATPase activity to alter nucleosome stability at DSBs is not known. p400 belongs to the Ino80 family of remodeling ATPases, which are implicated in mediating exchange of the histone variant H2A.Z onto the chromatin (Clapier and Cairns, 2009; Conaway and Conaway, 2008). Histone H2A.Z has only 60% homology to H2A but plays a key role in gene expression, chromosome segregation and gene silencing (Marques et al., 2010; Zlatanova and Thakar, 2008). H2A.Z can be acetylated by several acetyltransferases, including Tip60 (Babiarz et al., 2006; Keogh et al., 2006; Millar et al., 2006; Zlatanova and Thakar, 2008), although H2A.Z acetylation occurs mainly after exchange onto the chromatin (Babiarz et al., 2006; Keogh et al., 2006; Millar et al., 2006). Studies in yeast indicate that loss of H2A.Z leads to increased sensitivity to DNA damaging agents and increased genomic instability (Morillo-Huesca et al., 2010; Papamichos-Chronakis et al., 2011). In drosophila, H2Av (which combines the function of H2AX and H2A.Z), is acetylated by Tip60 after DNA damage, a process which stimulates removal of H2Av by the p400/domino ATPase (Kusch et al., 2004). Further, H2A.Z plays a central role in regulating repair of persistent DSBs in yeast (Kalocsay et al., 2009), indicating a key role for H2A.Z in the DNA damage response (DDR).
Here, we demonstrate that p400 utilizes its ATPase activity to exchange H2A.Z onto nucleosomes at DSBs. Exchange of H2A.Z alters nucleosome function and creates open, relaxed chromatin domains which extend away from the DSB. Further, H2A.Z exchange is essential for the subsequent acetylation and ubiquitination of the damaged chromatin template. Finally, the altered chromatin conformation resulting from H2A.Z exchange plays a pivotal role in regulating CtIP function and implies a key role for H2A.Z in regulating repair through the HR and NHEJ repair pathways.
RESULTS
H2A.Z is exchanged onto the chromatin at DSBs
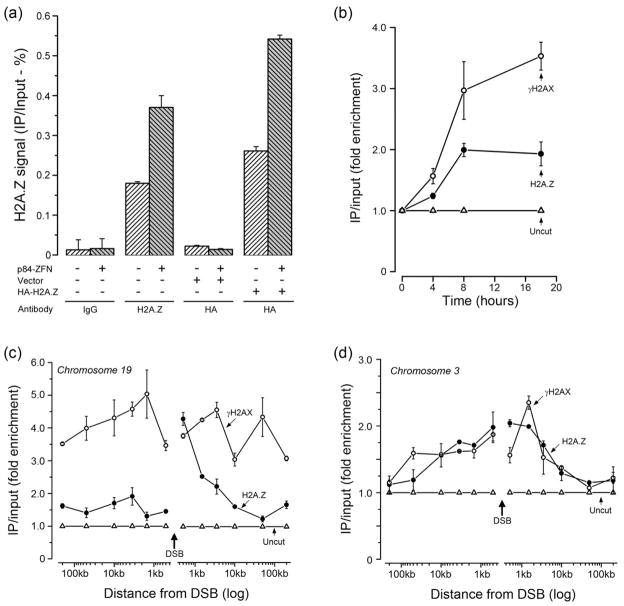
H2A.Z exchange was monitored using the p84-ZFN Zinc Finger Nuclease (ZFN) (Urnov et al., 2005), which creates a unique DSB on chromosome 19 (supplementary figure 1a) and which activates the DDR (Brunet et al., 2009; Xu et al., 2010). ChIP using H2A.Z antibody (but not IgG) demonstrated specific accumulation of H2A.Z at the p84-ZFN DSB (figure 1a). In addition, ChIP using HA antibody to an exogenously expressed HA-H2A.Z protein (supplementary figure 1b), demonstrated that the HA-H2A.Z protein was also exchanged at the DSB (figure 1a). The accumulation of H2A.Z was rapid (within 4 hrs), and occurred with a similar time course to the appearance of γH2AX (figure 1b). Phosphorylation of H2AX and exchange of H2A.Z were reversible, with both returning to near basal levels by 96hr (supplementary figure 1d), at which point >90% of the DSBs are repaired (supplementary figure 1a). Further, although both γH2AX and H2A.Z were increased at the p84-ZFN DSB (figure 1b), the level of histones H2AX and H3 were unaltered by DSB induction (supplementary figure 1f), implying that H2A.Z is not exchanged for H2AX. Finally, overall H2A.Z protein levels in the cell were not increased in response to DNA damage (supplementary figure 1c). Figure 1 therefore demonstrates that H2A.Z is rapidly and reversibly exchanged onto the chromatin at DSBs created by the p84-ZFN.
Figure 1. H2A.Z is exchanged onto nucleosomes at DSBs.
(a) 293T cells or 293T cells expressing vector or HA-H2A.Z were transfected with p84-ZFN. Cells were processed for ChIP 18hr later using either IgG, ChIP grade H2A.Z antibody or HA-antibody (for HA-H2A.Z). Real Time quantitative PCR (RT-qPCR) utilized primers located 1.5kb to the right of the DSB. ChIP signals were calculated as percent IP DNA/Input DNA. Results ± SD (n = 3). (b) 293T cells were transfected with p84-ZFN (●,○) or vector (Δ). Cells were processed for ChIP using antibodies to γH2AX (○) or H2A.Z (●), followed by RT-qPCR with primers located 1.5kb to the right of the DSB. ChIP signals were calculated as IP DNA/Input DNA and expressed as fold enrichment relative to the uncut (minus p84-ZFN; Δ) sample. Results ± SD (n = 3). (c) 293T cells were transiently transfected with vector (Δ) or the p84-ZFN (○,●). 18hr post-transfection, cells were processed for ChIP using antibodies to γH2AX (○) or H2A.Z (●), followed by RT-qPCR using the indicated primer pairs. The relative ChIP signal from the uncut, vector transfected cells is shown (Δ). Results ± SD (n = 3). (d) 293T cells were transfected with vector (Δ) or K230-ZFN (○,●), which creates a unique DSB at position 46.16Mb on chromosome 3. 18hr post-transfection, cells were processed for ChIP using antibodies to γH2AX (○) or H2A.Z (●), followed by RT-qPCR using the indicated primer pairs. The ChIP signal from the uncut, vector transfected cells is shown (Δ). Results ± SD (n = 3). See figures S1 and S2.
To determine if H2A.Z exchange was localized to the DSB, or spreads along the chromatin similar to γH2AX, the distribution of γH2AX and H2A.Z around the DSB was monitored. ChIP analysis demonstrates that γH2AX domains extended for at least 200kb on either side of the p84-ZFN DSB (figure 1c). In contrast, H2A.Z exchange had a highly asymmetrical distribution, with high levels of H2A.Z incorporated 0.5kb–3.5kb to the right of the DSB, with lower (<2 fold) but significant levels of H2A.Z exchange to the left of the break (figure 1c). A potential explanation for this distribution is that nucleosomes within transcribed genes are frequently enriched for H2A.Z (Jin et al., 2009; Zhang et al., 2005). p84-ZFN creates a DSB in intron 1 of the actively transcribed PPP1R12C gene (Urnov et al., 2005), which is located within a region of high gene density on chromosome 19 (supplementary figure 2a). Analysis of the levels of endogenous H2A.Z at the p84-ZFN target site prior to DSB production demonstrates high levels of pre-existing H2A.Z immediately to the left of the DSB, with much lower levels to the right (supplementary figure 2c). The nonuniform distribution of H2A.Z across the PPP1R12C gene therefore indicates that regions with high levels of H2A.Z exchange after DSB production (i.e. to the right of the DSB; figure 1c) have inherently lower levels of endogenous H2A.Z (supplementary figure 2c). To further examine how the level of endogenous H2A.Z impacts the subsequent damage induced exchange, a second ZFN, K230-ZFN, was used (Lee et al., 2010). K230-ZFN targets an intergenic region of chromosome 3 which contains few genes (supplementary figure 2b). This region of chromosome 3 contains a low, uniform distribution of H2A.Z across the K230-ZFN target site, which is distinct from that seen near the p84-ZFN (supplementary figure 2d). Further, DSBs created by K230-ZFN (supplementary figure 3a) induced both H2A.Z exchange and phosphorylation of H2AX (figure 1d). Importantly, unlike the p84-ZFN, H2A.Z exchange at the K230-ZFN DSB created symmetrical domains of H2A.Z which spread evenly from the DSB, and which overlapped with the γH2AX domains. This is consistent with the low levels of H2A.Z present on the chromatin at the K230-ZFN break site (supplementary figure 2d). The spreading of H2A.Z and γH2AX at the K230-ZFN site was restricted to 10–50kb on either side of the K230-ZFN break, compared to >200kb (for γH2AX) with the p84-ZFN (figure 1c and 1d). The distance that H2A.Z (and γH2AX) domains propagate from individual DSBs is therefore dependent on the chromosomal location and potentially differs between genes and intergenic regions. This is consistent with previous reports that spreading of γH2AX is heterogeneous (Iacovoni et al., 2010). Further, the presence of H2A.Z on nucleosomes prior to DSB production has a significant impact on the relative increase in H2A.Z density along the chromatin at DSBs. H2A.Z is therefore exchanged onto the chromatin at DSBs at 2 different chromosomal locations and occupies chromatin domains which overlap with, but are not contiguous with, γH2AX domains.
p400 exchanges H2A.Z at DSBs
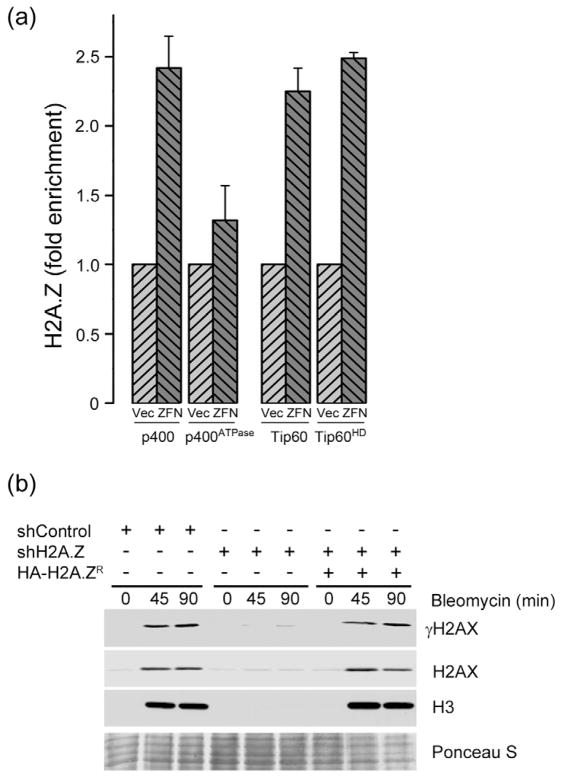
The formation of open chromatin domains at DSBs required both acetylation of histone H4 by the Tip60 acetyltransferase and the ATPase activity of p400 (Xu et al., 2010). Well characterized cells (Xu et al., 2010) with inactivating mutations in p400’s ATPase domain (p400ATPase) or Tip60’s acetyltransferase domain (Tip60HD; supplementary figures 3b and 3c) were used determine if Tip60 or p400 were required for H2A.Z exchange. Inactivating Tip60’s acetyltransferase activity or p400’s ATPase activity did not alter the ability of p84-ZFN to induce DSBs (supplementary figure 3a). However, inactivation of p400’s ATPase activity significantly reduced the exchange of H2A.Z at DSBs (figure 2a), demonstrating that p400’s ATPase activity is required for H2A.Z exchange. Surprisingly, inactivation of Tip60’s acetyltransferase activity did not inhibit H2A.Z exchange by p400 (figure 2a). Tip60 can directly acetylate H2A.Z (Babiarz et al., 2006; Keogh et al., 2006; Millar et al., 2006; Zlatanova and Thakar, 2008) and acetylation of the drosophila H2A.Z homolog H2Av by Tip60 is required for removal of H2Av from damage sites (Kusch et al., 2004). However, studies in yeast have shown that, in the absence of DNA damage, Tip60 acetylates H2A.Z after H2A.Z has been exchanged onto chromatin (Keogh et al., 2006; Millar et al., 2006). Our results are therefore consistent with previous yeast work, and indicate that H2A.Z exchange at DSBs in mammalian cells does not require the acetyltransferase activity of Tip60 (figure 2a). However, this does not rule out the possibility that acetylation of H2A.Z or histone H4 by other acetyltransferases is important for H2A.Z exchange by p400.
Figure 2. H2A.Z exchange mediates decreased nucleosome stability at DSBs.
(a) 293T cells expressing p400 or catalytically inactive p400ATPase, Tip60 or catalytically inactive Tip60HD were transfected with vector (Vec) or p84-ZFN (ZFN). 18hr post-transfection, cells were processed for ChIP using H2A.Z antibody, followed by RT-qPCR using primer pairs located 1.5kb to the right of the DSB. Results ± SD (n = 3). (b) 293T cells expressing a non-specific shRNA (shControl), shRNA targeting H2A.Z (shH2A.Z) or shH2A.Z expressing cells rescued by expression of an shRNA resistant HA-H2A.Z (HA-H2A.ZR) were incubated with bleomycin (5μM) for the indicated times (min). Cell pellets were extracted in 1.0M NaCl and released histones detected by western blot analysis for γH2AX, H2AX and H3. Ponceau S staining was used to demonstrate equal loading. See figure S3.
DSBs create open, relaxed chromatin domains in which the histone-histone and histone-DNA interactions are reduced, a process which requires Tip60 and p400 (Kusch et al., 2004; Murr et al., 2006; Xu et al., 2010). Consequently, when cells are exposed to DNA damage and the chromatin is extracted in 1.0M NaCl, histones can be specifically removed from the chromatin adjacent to the DSB (Xu et al., 2010). Bleomycin was used to create DSBs and nuclei were extracted in 1.0M NaCl (figure 2b). DNA damage increased the NaCl solubility of histones γH2AX, H2AX and H3 within damaged chromatin (figure 2b), a process which we confirmed requires the ATPase activity of the p400 protein (supplementary figure 3d (Xu et al., 2010)). Importantly, when H2A.Z was depleted with shRNA (supplementary figure 1b), the DNA-damage dependent increase in NaCl solubility of histones was eliminated (figure 2b). Further, re-expression of an shRNA resistant HA-H2A.Z protein (supplementary figure 1b) rescued the loss of the endogenous H2A.Z (figure 2b), confirming the specificity of the shRNA vector. We interpret this to mean that exchange of H2A.Z by p400 decreases the stability of the histone-histone and histone-DNA interactions within nucleosomes at the DSB, creating open, relaxed chromatin domains.
H2A.Z exchange promotes acetylation and ubiquitination of the chromatin
Because H2A.Z is important for gene transcription (Svotelis et al., 2009), we established if H2A.Z depletion altered the expression or activity of key DDR proteins. However, activation of ATM’s kinase activity (supplementary figure 4a), phosphorylation of H2AX (supplementary figures 4b and 4c), and the recruitment of mdc1, CtIP and 53BP1 to DSBs (supplementary figures 4d and 6c–e) were independent of H2A.Z. H2A.Z depletion does not impact the expression or activation of key components of the DDR.
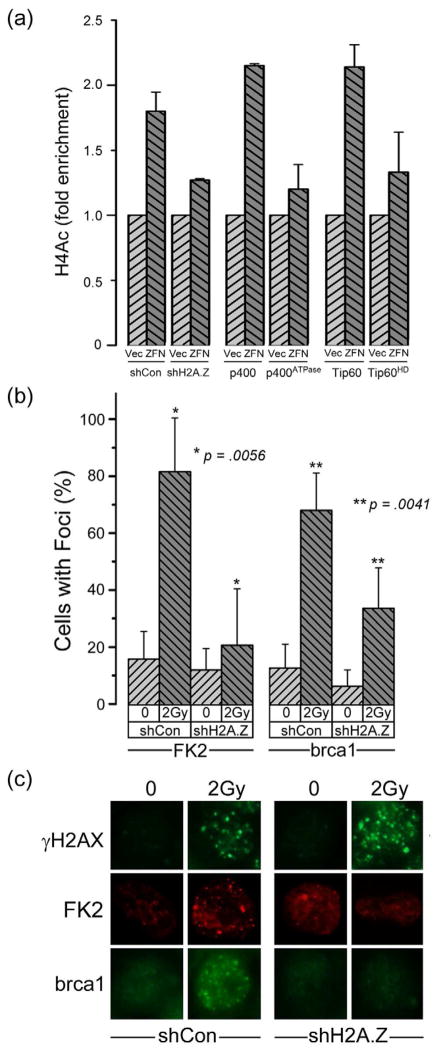
The exchange of H2A.Z at DSBs may alter nucleosome structure in a way which facilitates further processing and modification of the damaged chromatin template. To test this, we determined if 2 chromatin modifications required for DSB repair, the acetylation of histone H4 by Tip60 (Murr et al., 2006; Xu et al., 2010), and ubiquitination of the chromatin by RNF8 (Kolas et al., 2007; Mailand et al., 2007), required H2A.Z. Using p84-ZFN and ChIP analysis, figure 3a demonstrates that acetylation of histone H4 at DSBs required Tip60’s acetyltransferase activity, as expected. Importantly, depletion of H2A.Z or inactivation of p400’s ATPase activity significantly impaired histone H4 acetylation by Tip60 (figure 3a). Although previous studies have shown that histone acetylation at DSBs can shift chromatin to a more open conformation (Downs et al., 2004; Murr et al., 2006; Xu et al., 2010), we have now shown that H2A.Z exchange by p400 is required for, and precedes, the acetylation of histone H4 by Tip60.
Figure 3. H2A.Z exchange is required for acetylation and ubiquitination of the chromatin.
(a) 293T cells expressing a non-specific shRNA (shCon), shRNA to H2A.Z (shH2A.Z), or expressing p400, p400ATPase, Tip60 or Tip60HD were transfected with vector (Vec) or p84-ZFN (ZFN). 18hr post-transfection, cells were processed for ChIP using antibody to acetylated histone H4 (H4Ac), followed by RT-qPCR using primers located 1.5kb to the right of the DSB. Results ± SD (n = 3). (b) 293T cells expressing a non-specific shRNA (shCon) or shRNA targeting H2A.Z (shH2A.Z) were irradiated (2Gy). Ubiquitin (detected with FK2 antibody) and brca1 foci were detected by immunofluorescent staining. Cells with >5 foci were counted as positive (results ± SD, n = ~100). P-values calculated using a t-test. (c) Selected images from (b). See figure S4.
Next, we examined if H2A.Z-mediated changes in chromatin structure were required for chromatin ubiquitination (Kolas et al., 2007; Mailand et al., 2007). Depletion of H2A.Z blocked the formation of ubiquitin foci at DSBs (figure 3b and 3c). Further, loading of brca1, which is dependent on prior ubiquitination of the chromatin by RNF8, was also reduced in H2A.Z depleted cells (figure 3b and 3c). The p400-mediated exchange of H2A.Z onto nucleosomes at DSBs is therefore required for acetylation and ubiquitination of the chromatin and for loading of the brca1 complex at DSBs.
H2A.Z is required for loading the Ku70/80 complex
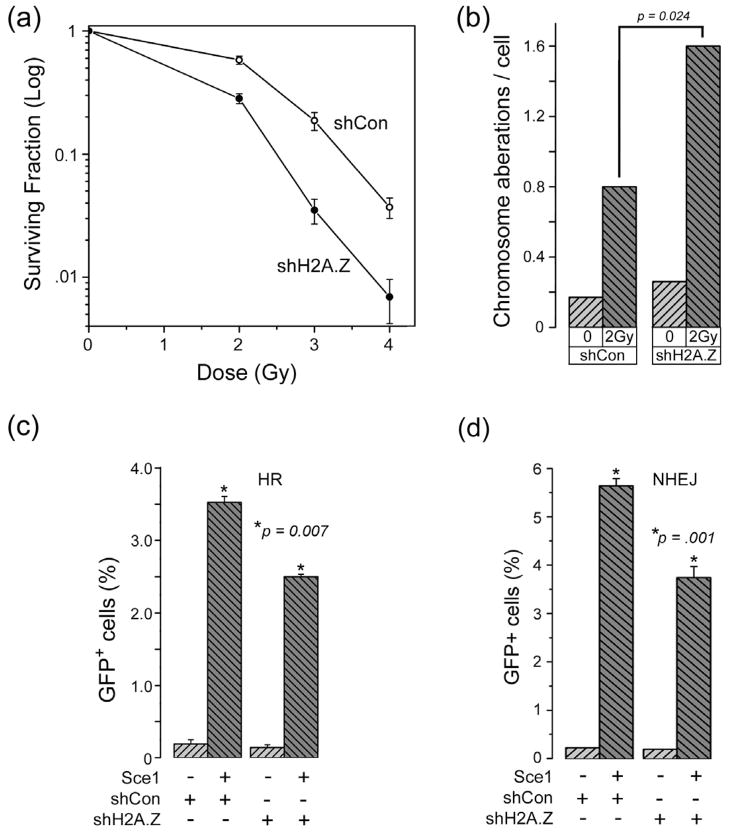
Next, we determined how H2AZ exchange impacted the mechanism of DSB repair. Cells in which H2A.Z expression was reduced by shRNA (supplementary figure 1b) exhibited increased sensitivity to ionizing radiation (IR) (figure 4a), a process which could be complemented by re-expression of an shRNA resistant HA-H2A.Z construct (supplementary figure 5a). Further, cells lacking H2A.Z accumulated more chromosomal aberrations after IR exposure (figure 4b), consistent with a defect in DNA repair. In addition, and similar to previous studies in cells lacking p400 expression (Xu et al., 2010), H2A.Z-deficient cells exhibited an attenuated G2 checkpoint (supplementary figure 5b) following irradiation. Loss of H2A.Z therefore leads to increased genomic instability and increased sensitivity to IR.
Figure 4. Histone H2A.Z is an essential component of the DNA damage response.
(a) 293T cells expressing a non-specific shRNA (○) or shRNA targeting H2A.Z (●) were irradiated and clonogenic cell survival measured. Results ± SD (n = 4). (b) 293T cells expressing a non-targeting shRNA (shCon) or shRNA targeting H2A.Z (shH2A.Z) were irradiated. 25hr later, cells were treated with colcemid, and the number of chromosome aberrations per cell counted. p was calculated using at t-test. (c) Cells with the stably integrated GFP-HR reporter system expressing a non-specific shRNA (shCon) or shRNA targeting H2A.Z (shH2A.Z) were transfected with the I-Sce1 nuclease to introduce DSBs, and GFP positive cells monitored by FACS. Results ± SD (n = 3). p-values calculated using a t-test. (d) Cells stably expressing an NHEJ-GFP reporter system were transfected with a non-specific vector (shCon) or shRNA targeting H2A.Z (shH2A.Z). Cells were transfected with the I-Sce1 nuclease and GFP positive cells measured. Results expressed ± SD (n = 3). p-values calculated using a t-test. See figure S5.
Because brca1 is important for the homologous recombination (HR) repair pathway, we examined HR activity using a well characterized GFP-HR reporter system (Pierce and Jasin, 2005). H2A.Z depletion did not alter cell cycle kinetics (supplementary figure 6b). However, H2A.Z depleted cells exhibited reduced levels of HR (figure 4c), consistent with the reduction in brca1 loading at DSBs (figure 3b). Cells in which HR is inhibited can compensate by increasing the activity of the non-homologous end-joining (NHEJ) pathway. Surprisingly, NHEJ activity was also decreased in H2A.Z depleted cells (figure 4d). Further, cells in which the H2A.Z exchange protein p400 was inactivated also exhibited reduced levels of NHEJ (supplementary figure 5c) and HR (supplementary figure 5d). Loss of H2A.Z exchange therefore leads to defects in both HR and NHEJ mediated repair.
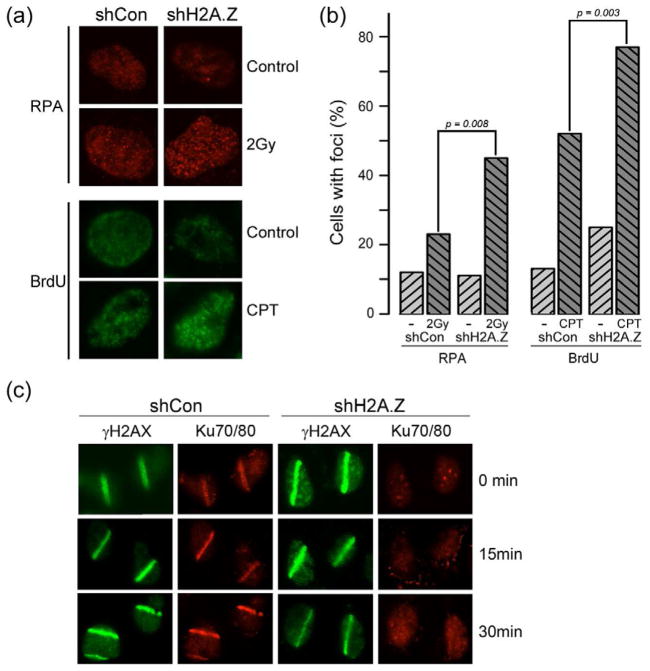
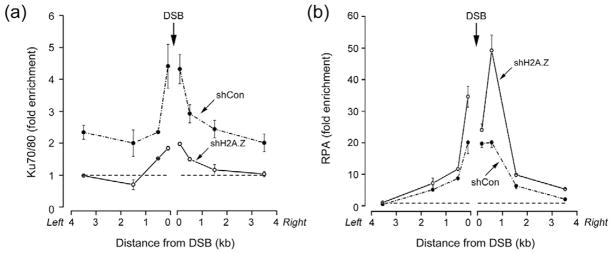
To further examine the repair defect in H2A.Z-deficient cells, we examined the production of single-strand DNA (ssDNA) after DNA damage. Loss of H2A.Z led to an increase in the accumulation of the ssDNA binding protein RPA32 at DSBs (figure 5b). Further, using BrdU labeling to specifically detect ssDNA demonstrated increased amounts of ssDNA in H2A.Z depleted cells after DNA damage (figure 5a and 5b). The Ku70/80 complex binds preferentially to blunt DNA ends (Dynan and Yoo, 1998; Foster et al., 2011) and promotes DSB repair through NHEJ (Huertas, 2010; Kass and Jasin, 2010; Paull, 2010). Therefore, we examined if loading of the Ku70/80 complex at DSBs was impacted by the increase in ssDNA detected in H2A.Z-deficient cells. Ku70/80 colocalizes with γH2AX at laser induced sites of DNA damage (figure 5c). Importantly, Ku70/80 was not detectably recruited to sites of laser damage in H2A.Z deficient cells (figure 5c), even though H2A.Z depleted cells express normal levels of Ku70 (supplementary figure 6a). To determine if the loss of Ku70/80 binding and increased RPA detected at DSBs by immunofluorescent techniques (figure 5) reflected a loss of Ku70/80 directly at the DSB, the recruitment of Ku70/80 and RPA32 to the p84-ZFN DSB was monitored. Both Ku70/80 (figure 6a) and RPA (figure 6b) were localized to the DSB, with peak binding for both RPA and Ku70/80 located within approximately 500bp of the DSB. No RPA was detected further than 1.5kb from the DSB (figure 6b). However, significant loading of the Ku70/80 complex was also detected on the chromatin domains flanking the DSB (figure 6a). This is similar to other DSB repair proteins, which accumulate both directly at the DSB and on the adjacent chromatin domains (Lukas et al., 2011). Why Ku70 associates with the flanking chromatin domains is currently unclear, but could be important for maintaining DNA-PKcs activity or for concentrating Ku70/80 at the site of damage.
Figure 5. H2A.Z is required to retain the Ku70/80 complex at DSBs.
(a) Upper panel: U2OS cells expressing shCon or shH2AZ were irradiated (2Gy/60mins), fixed and RPA32 detected by immunofluorescent staining. Lower panel: U2OS cells expressing shCon or shH2AZ were prelabeled with BrdU then treated with camptothecin (CPT: 1μM) for 60 mins. Cells were fixed under non-denaturing conditions and BrdU in ssDNA detected with antiBrdU antibody. (b) Quantitation of (a). Cells with > 5 foci were counted as positive, with an average of 100 cells counted per experiment. p-values calculated using a t-test. (c) Laser striping was used to create DNA damage in U2OS cells expressing shCon or shH2AZ. Cells were pre-extracted (see methods) immediately after completion of laser striping (0 minutes) or allowed to recover for 15 or 30 minutes. γH2AX and Ku70/80 were detected by immunofluorescent staining. See figure S6 and S7.
Figure 6. H2A.Z regulates loading of Ku70/80 and RPA at DSBs.
293T cells expressing shCon or shH2A.Z were transfected with p84-ZFN. 18hr post-transfection, cells were processed for ChIP using antibodies to (a) Ku70/80 or (b) RPA32. RT-qPCR was carried out using primer pairs indicated. Broken line indicates ChIP signal from control (uncut) cells. Results ± SD (n = 3).
Depletion of H2A.Z had opposing effects on RPA and Ku70/80 loading at the DSB. In the absence of H2A.Z, Ku70/80 was lost from the flanking chromatin domains and there was a significant reduction of Ku70/80 loading directly at the DSB (figure 6a). Conversely, loss of H2A.Z led to an increase in RPA32 binding at the break (figure 6b). Figures 5 and 6 therefore demonstrate that, in the absence of H2A.Z exchange, there is a loss of Ku70/80 binding at the DSB and a corresponding increase in both RPA binding and ssDNA. H2A.Z-mediated alterations in nucleosome structure therefore promote Ku70/80 retention at DSBs and restrict the production of ssDNA at DSBs.
Loss of CtIP rescues Ku70/80 binding in H2A.Z depleted cells
Several DSB repair proteins have been shown to either restrict (e.g. 53BP1) or promote (e.g. CtIP, brca1) ssDNA production (Bunting et al., 2010; Sartori et al., 2007; Yun and Hiom, 2009). Of these, CtIP is particularly important for creating the ssDNA intermediates which are required for HR mediated repair (Huertas, 2010; Paull, 2010; Sartori et al., 2007). However, both CtIP and 53BP1 were correctly recruited to DSBs in the absence of H2A.Z (supplementary figures 6c, 6d and 6e), indicating that H2A.Z does not affect the loading of 53BP1 or CtIP. Brca1 recruitment is reduced in H2A.Z depleted cells (figure 3b and 3c), and brca1-CtIP interaction can regulate CtIP activity and ssDNA production (Bunting et al., 2010; Yun and Hiom, 2009). However, depletion of brca1 (supplementary figure 7a) did not alter recruitment of Ku70/80 to sites of DNA damage (supplementary figure 7b). Finally, loss of 53BP1 did not impact Ku70/80 loading at sites of damage (supplementary figure 7c and 7d). Therefore, loss of H2A.Z does not impact the loading of either 53BP1 or CtIP at DSBs, indicating that the reduction in Ku70/80 binding in H2A.Z deficient cells is independent of 53BP1 and brca1 status.
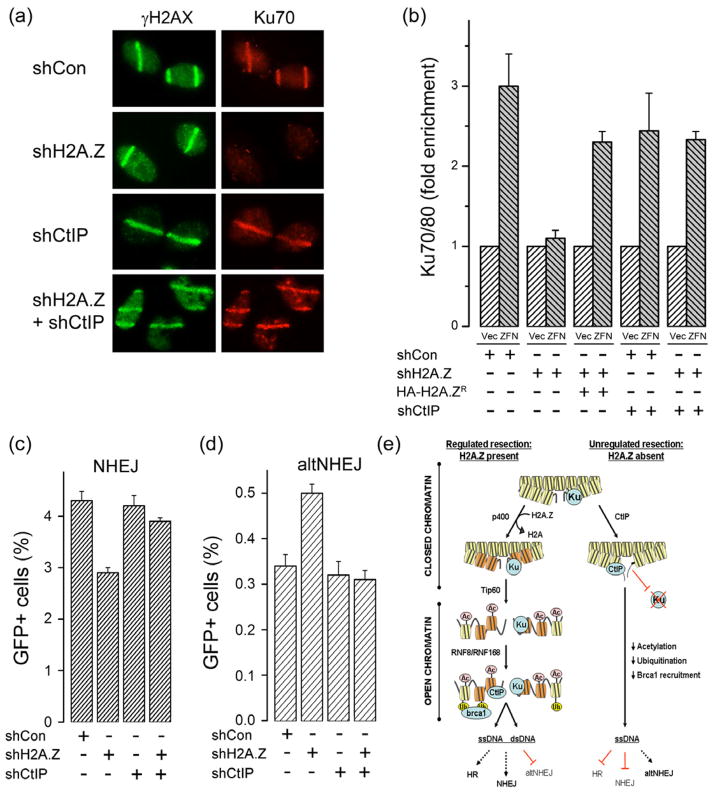
Next, we examined if blocking ssDNA production by CtIP restored recruitment of Ku70/80 in the absence of H2A.Z. Depletion of H2A.Z (supplementary figure 7e), but not CtIP, abolished Ku70/80 binding (figure 7a), indicating that CtIP is not required for Ku70/80 binding at DSBs. Importantly, depletion of CtIP in a H2A.Z depleted background rescued Ku70/80 binding (figure 7a). Similar results were obtained using p84-ZFN generated DSBs. Ku70/80 binding was significantly reduced in H2A.Z deficient cells, but could be rescued by expression of a shRNA resistant H2A.Z protein (figure 7b). Importantly, depletion of CtIP in a H2A.Z depleted background rescued Ku70/80 binding at the DSB (figure 7b). Because Ku70/80 binding was restored by silencing of CtIP in H2A.Z depleted cells, this argues against a direct H2A.Z-Ku70/80 interaction or an indirect role for H2A.Z in loading of Ku70/80 at DSBs. Rather, this indicates a model in which exchange of H2A.Z alters the nucleosome conformation and thereby restrains or limits the ability of CtIP to resect the DNA. Without H2A.Z, CtIP carries out unregulated resection, increasing ssDNA and eliminating the blunt ended, double strand DNA required for binding of Ku70/80 to DSBs. This model implies that loss of H2A.Z should decrease the activity of the Ku-dependent NHEJ pathway. In fact, H2A.Z depleted cells have reduced NHEJ activity (figure 7c). Importantly, co-depletion of CtIP and H2A.Z restores normal NHEJ (figure 7c), consistent with the restoration of Ku70 binding upon loss of CtIP (figure 7a and 7b). Further, cells which lack Ku70 utilize the alternate NHEJ (alt-NHEJ) for repair. The alt-NHEJ pathway utilizes regions of microhomology on ssDNA to rejoin the DNA ends, a process which requires CtIP (to create ssDNA) but not Ku70/80 (Bennardo et al., 2008; Langerak et al., 2011; Zhang and Jasin, 2011). Strikingly, H2A.Z depleted cells exhibited an increase in activity of the alt-NHEJ pathway (figure 7d), consistent with a loss of Ku70 binding at DSBs in H2A.Z deficient cells. Importantly, the increased alt-NHEJ activity in H2A.Z depleted cells was suppressed when CtIP was depleted (figure 7d). This is consistent with the observation that depletion of CtIP in a H2A.Z null background restores both Ku70 binding and NHEJ activity (figure 7a and 7c). H2A.Z exchange therefore plays a critical role in creating open chromatin structures which direct the correct modification of the chromatin, and functions to restrict unscheduled end-processing of the damaged template by CtIP.
Figure 7. Loss of CtIP restores Ku70/80 binding and NHEJ in H2A.Z depleted cells.
(a) Laser striping was used to create DNA damage in U2OS cells expressing a non-specific shRNA (shCon) or shRNA targeting either H2A.Z, CtIP or both. Cells were stained for γH2AX and Ku70 15 minutes after laser striping. (b) 293T cells were transfected with non-specific shRNA (shCon), or shRNA targeting either H2A.Z or CtIP. 293T cells expressing an shRNA resistant HA-H2A.Z protein (HA-H2A.ZR) were also used. Cells were subsequently transfected with vector (Vec) or p84-ZFN (ZFN) and ChIP analysis carried out using antibody to Ku70/80 and primers located 500bp to the right of the DSB. Results ± SD (n = 3). (c) Cells stably expressing the NHEJ-GFP reporter system and either a non-specific shRNA (shCon) or shRNA targeting H2A.Z, CtIP or both were transfected with the I-Sce1 endonuclease (+). The percent of GFP positive cells is shown. Results expressed ± SD (n = 3). (d) Cells with the integrated altNHEJ-GFP reporter system were transfected with a non-specific shRNA (shCon) or shRNA targeting H2A.Z, CtIP or both. Following transfection of I-Sce1 endonuclease (+), the percent of GFP positive cells was measured. Results expressed ± SD (n = 3). (e) Model for function of H2A.Z in DSB repair. Legend: Yellow = H2A nucleosomes; Orange = H2A.Z nucleosomes. Ku = Ku70/80; Ac = acetylation; Ub = ubiquitination. See figure S7.
DISCUSSION
We have identified the histone variant H2A.Z as a new component of the DNA damage response network. Unlike other DDR proteins, which are recruited to the chromatin, H2A.Z is actively exchanged onto nucleosomes at DSBs by the p400 motor ATPase. However, because H2A.Z can also regulate gene transcription (Gevry et al., 2009), loss of H2A.Z could indirectly affect DSB repair through alterations in gene expression. Several lines of evidence indicate that this is unlikely. First, the levels and activation of most DSB repair proteins are unaltered by loss of H2A.Z expression (supplementary figures 4 and 6). Second, cells expressing a p400ATPase mutant retain H2A.Z in the chromatin, but lack H2A.Z exchange at DSBs (figure 2). Importantly, this p400ATPase mutant phenocopies the effect of H2A.Z depletion on HR and NHEJ (supplementary figure 5). Third, the loss of Ku70 binding and the reduction in NHEJ seen in the absence of H2A.Z are complemented by co-depletion of CtIP (figure 7a–d). That is, cells lacking both CtIP and H2A.Z carry out normal NHEJ and Ku70 loading despite the lack of significant chromatin associated H2A.Z (figure 7). Fourth, H2A.Z is directly exchanged at the DSB. Taken together, these results indicate that H2AZ functions directly at the DSB, rather than mediating its effects on DNA repair through an indirect mechanism involving altered gene expression profiles.
H2A.Z has ~60% homology to H2A (and H2AX), and diverges significantly from H2A in the domain which interacts with adjacent H3–H4 dimers (Zlatanova and Thakar, 2008). Previous work has shown that nucleosomes containing H2A.Z are less stable than those containing H2A (Jin and Felsenfeld, 2007; Jin et al., 2009; Zhang et al., 2005; Zlatanova and Thakar, 2008), which is consistent with the observation here that H2A.Z exchange promotes the formation of open, relaxed chromatin in response to DNA damage. This suggests a model in which exchange of H2A.Z by p400 at DSBs alters histone-histone interactions both within and between adjacent nucleosomes, creating open, relaxed chromatin domains. This shift in nucleosome organization may then expose the n-terminal of histone H4 for acetylation by Tip60, and reveal cryptic ubiquitination sites on the chromatin for ubiquitination by RNF8 (Doil et al., 2009; Huen et al., 2007). This combination of H2A.Z exchange and DNA damage induced histone modification then promotes transition to an open, relaxed chromatin conformation at the site of damage. Eventually, this leads to the formation of H2A.Z domains which extend away from the DSB, and which largely overlap with the previously described domains of γH2AX (Bonner et al., 2008; Rogakou et al., 1999). DNA damage induced foci therefore contain both high levels of DSB repair proteins and an open, relaxed chromatin structure, both of which are essential for correct processing of the chromatin template for DSB repair.
The exchange of H2A.Z onto nucleosomes functioned to restrain or limit end resection mediated by CtIP. 53BP1 is a negative regulator of end resection (Bunting et al., 2010), and is recruited to DSBs through a complex mechanism involving both RNF8-dependent ubiquitination (Doil et al., 2009; Mailand et al., 2007) and methylation of histone H4 (Pei et al., 2011). Here, H2A.Z depletion did not alter loading of 53BP1 at DSBs, despite the decrease in chromatin ubiquitination and the loss of brca1 loading. This may indicate that H4 methylation or other mechanisms for 53BP1 recruitment can compensate for the loss of ubiquitination in H2A.Z defective cells. In either case, H2A.Z does not appear to directly regulate loading of 53BP1 at DSBs. In addition, CtIP recruitment was not affected by loss of H2A.Z, suggesting that H2A.Z-mediated changes in nucleosome architecture impact CtIP’s activity rather than its localization. This could occur through H2A.Z-directed changes in histone modifications or histone binding proteins at the DSB, or reflect topological constraints on CtIP activity due to the altered nucleosome structure. In fact, studies on transcription (Zlatanova and Thakar, 2008) indicate that the nucleosome free region at the transcriptional start site (TSS) of many genes is flanked by H2A.Z-nucleosomes (Jin et al., 2009; Zhang et al., 2005). These well positioned H2AZ nucleosomes fix the position of nucleosomes upstream and downstream of the TSS and maintain the nucleosome free DNA sequence for e.g. transcription factor binding. Because nucleosomes are ejected from the DNA at the DSBs (Berkovich et al., 2007; Tsukuda et al., 2005), DSBs also represent a nucleosome free region. Our results indicate that H2A.Z nucleosomes are positioned either side of the nucleosome free region at the DSB, creating a structure analogous to that seen at the TSS (Jin et al., 2009; Zhang et al., 2005; Zlatanova and Thakar, 2008). The H2A.Z nucleosomes at the DSB may therefore function to limit spreading of the nucleosome free region at DSBs and fix the position of the nucleosomes up and downstream of the break. Further, the presence of H2A.Z nucleosomes at the boundary of the nucleosome free region may limit the ability of CtIP to resect the DNA. H2A.Z on the chromatin at DSBs may therefore serve as a boundary factor which defines the limits of the nucleosome free regions, restricts the extent of DNA resection by CtIP and functions to reorganize the remaining nucleosomes adjacent to the DSB. This H2A.Z-dependent reorganization of the nucleosomes then allows the cell to further process the chromatin (e.g. through acetylation and ubiquitination) and recruit appropriate repair proteins to the DSB. Once the cell has fully processed the DSB, it can then either proceed with CtIP-mediated end resection (which may potentially require either modification or removal of H2A.Z) or commit to Ku70-mediated NHEJ repair.
We propose the following model (figure 7e). Previous work indicates that p400 and Tip60 are recruited to DSBs as part of the NuA4 remodeling complex (Downs et al., 2004; Murr et al., 2006; Xu et al., 2010) through interaction with the mdc1 protein (Xu and Price, 2011; Xu et al., 2010). The p400 motor ATPase subunit of NuA4 then catalyzes the rapid exchange of H2A.Z (figure 7e). The exchange of H2A.Z, combined with the acetylation of the H4, shifts the chromatin into an open, relaxed conformation which facilitates further modification of the chromatin through ubiquitination and loading of brca1 complexes. When H2A.Z exchange is blocked, chromatin remains compact and is neither acetylated nor ubiquitinated and brca1 is not loaded at the DSB (figure 7e). Further, without H2A.Z to protect the damaged DNA ends and to reorganize the chromatin, CtIP carries out unregulated end processing. This leads to increased ssDNA production, loss of Ku70/80 binding and eventually to defects in both HR and NHEJ mediated repair. Although the increase in ssDNA could potentially feed into the HR pathway, it is likely that the loss of brca1 loading and the altered chromatin architecture (which could impact e.g. homology searching in the sister chromatid), prevent the efficient processing of the ssDNA into the HR pathway.
These results therefore provide new insight into the molecular mechanisms involved in remodeling chromatin structure at DSBs, and reveal that a series of molecular events, involving sequential H2A.Z exchange, histone acetylation and chromatin ubiquitination, function together to create an open, acetylated chromatin domain at sites of DNA damage. The exchange of H2A.Z by the NuA4 complex therefore promotes reorganization of the chromatin architecture to create a chromatin template which is an efficient substrate for the DSB repair machinery.
EXPERIMENTAL PROCEEDURES
Cell culture
293T, U2OS and HeLa cells expressing Tip60wt, Tip60HD, p400 and p400ATPase, clonogenic cell survival assays and analysis of metaphase spreads are described in (Sun et al., 2005; Xu et al., 2010; Yang et al., 2001). shRNA sequences, antibodies, nucleosome stability assay and western blot analysis are described in the supplementary methods.
HR, NHEJ and alt-NHEJ reporter systems
U2OS cells expressing an HR reporter (Pierce and Jasin, 2005), HEK293 cells expressing the alt-NHEJ reporter (Bennardo et al., 2008) or HeLa cells expressing a NHEJ reporter (Bennardo et al., 2008) were infected individually with the pSuper control plasmid, shRNA to H2A.Z or CtIP or sequentially infected by shRNA to H2A.Z and CtIP. 5 days post-infection, cells were transfected with the I-SceI plasmid using lipofectamine 2000 (Invitrogen, CA) and GFP positive cells counted 48hr later using a BD FACscan cell analyzer with data analysis using the Flow Jo software package.
DSB production and ChIP assays
293T cells were transfected with either p84-ZFN or K230-ZFN and allowed to recover for 6–18hr as indicated. Cells were fixed in 1% methanol-free formaldehyde for 10 minutes to cross link proteins, then lyzed in ChIP buffer (Cell Signaling Technology, MA), sonicated (Fisher Scientific Sonic 250) and cleared by centrifugation. For ChIP (Xu et al., 2010) purified chromatin was immunoprecipitated with ChIP grade antibodies, the DNA isolated and associated DNA amplified by RT-qPCR as described in supplementary methods.
Laser microirradiation and immunofluorescence
For laser microirradiation, cells were incubated with Hoechst dye 33258 (10μg/ml) for 5 min at 37°C (Rogakou et al., 1999). Laser microirradiation utilized an inverted confocal microscope (Leica TCS SP5; Leica Microsystems Inc, USA) equipped with a 37°C heating chamber and a 30mW 405nm diode laser focused through a 40x-Plan Apochromat/1.25 NA oil objective. Laser output was set to 40–60% of maximum power to generate DNA damage restricted to the laser path without noticeable cytotoxicity For immunofluorescent analysis, cells were fixed in PBS/paraformaldehyde (4%), blocked with fetal bovine serum (2%) and permeabilized in bovine serum albumen (0.2%) containing saponin (0.2%) for 15min. For RPA and Ku70/80, cells were pre-extracted in buffer D (10mM PIPES pH 7.0; 100mM NaCl; 300mM sucrose; 3mM MgCl2; 0.5% Triton-X100) prior to fixing. For BrdU staining (Raderschall et al., 1999), cells were incubated in BrdU (10μM; 16hr) followed by camptothecin (1μM) for 1 hour. Cells were then washed, incubated in buffer D (10 minutes) and fixed as described above. Slides were mounted with Fluoromount-G (Southern Biotech, AL) and images collected with a Zeiss AxioImager Z1 microscope equipped with an Axiocam MRc Rev.3 Color Digital Camera and Plan APO 63X/1.4 oil M27 lens (magnification 63X, aperture 1.4). Acquisition software and image processing utilized the Zeiss AxioVision software package (Zeiss Imaging, NY).
Supplementary Material
Highlights.
H2A.Z is exchanged onto nucleosomes at DSBs by the p400 remodeling ATPase.
H2A.Z exchange creates open, relaxed chromatin domains at DSBs.
H2A.Z exchange promotes histone acetylation and chromatin ubiquitination.
H2A.Z restricts end resection and promotes the correct processing of damaged chromatin.
Acknowledgments
We thank J. Stark for altNHEJ cells, F. Urnov and J-S Kim for ZFNs, R. Baer for CtIP antibody and J. Gaudreau for shRNA vectors to H2A.Z. We thank Haico Van Attikum for helpful discussions. This work was supported by grants from NCI (CA64585 and CA93602) and the DOD Breast Cancer Program to BDP.
Footnotes
Author contributions. BDP and YX conceived the research, wrote the paper and carried out data analysis. YX carried out the majority of the experiments, participated in data analysis and planned experiments. MA and YH carried out laser striping experiments and contributed to data analysis. CX and OG carried out some of the ChIP experiments and contributed to protocol development. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes & development. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS genetics. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci U S A. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53 --> p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2008 doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of Chromatin-Modifying Activities to Phosphorylated Histone H2A at DNA Damage Sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic acids research. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SS, Balestrini A, Petrini JH. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Molecular and cellular biology. 2011;31:4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevry N, Svotelis A, Larochelle M, Gaudreau L. Nucleosome mapping. Methods Mol Biol. 2009;543:281–291. doi: 10.1007/978-1-60327-015-1_19. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nature structural & molecular biology. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. Embo J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS genetics. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Marques M, Laflamme L, Gervais AL, Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010;5:267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes & development. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M, Clemente-Ruiz M, Andujar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PloS one. 2010;5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA repair. 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Jasin M. Measuring recombination proficiency in mouse embryonic stem cells. Methods in molecular biology. 2005;291:373–384. doi: 10.1385/1-59259-840-4:373. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–936. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svotelis A, Gevry N, Gaudreau L. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem Cell Biol. 2009;87:179–188. doi: 10.1139/O08-138. [DOI] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–267. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kuang Y, Montes De Oca R, Hays T, Moreau L, Lu N, Seed B, D’Andrea AD. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001;98:3435–3440. doi: 10.1182/blood.v98.12.3435. [DOI] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nature structural & molecular biology. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.