Abstract
Purpose
HER2-amplified breast cancer is sometimes clinically insensitive to HER2 targeted treatment with trastuzumab. Laboratory models of resistance have causally implicated changes in HER2 expression and activation of the PI3K-AKT pathway. We conducted a prospective tissue acquisition study to determine if there is evidence for these lesions in metastatic tumors that have progressed on trastuzumab-containing therapy.
Experimental Design
From 2/2007 to 11/2011, 63 patients with HER2-amplified breast cancer with recurrence of disease after adjuvant trastuzumab therapy or WHO- defined progression of metastatic disease on a trastuzumab-containing regimen were prospectively enrolled and underwent tumor biopsy. Specimens were analyzed for activating mutations in PIK3CA and HER2 by Sequenom® and analyzed for HER2 and PTEN status by immunohistochemistry (IHC).
Results
In 53/60 cases (88%, 3 cases not evaluable for HER2), HER2 overexpression persisted in the metastatic tumor following trastuzumab exposure. Among the 7 cases lacking HER2 overexpression, repeat analysis of the pre-treatment tumor failed to confirm HER2 overexpression in 5 cases. Among cases evaluable for PTEN (56) or PI3K mutation (45), absent or significantly diminished PTEN expression was noted in 33 (59%) and activating mutations in PIK3CA in 13 (29%). The combined rate of PTEN loss and PIK3CA mutation in the trastuzumab-refractory tumors was 71% compared to 44% (p=0.007) in an unexposed cohort of 73 HER2-amplified tumors.
Conclusions
In this series of prospectively collected trastuzumab-refractory human breast cancers, loss of HER2 overexpression was rare while activation of the PI3K-AKT pathway through loss of PTEN or PIK3CA mutation was frequently observed.
Introduction
Breast cancer is a heterogeneous disease which may be classified by molecular profiles into subtypes with unique behaviors and responsiveness to targeted therapies (1, 2). HER2 is amplified in ~20% of invasive breast cancer and is associated with a more aggressive biology, increased risk for progression of the disease and decreased overall survival (3, 4). HER2 is a member of the ErbB family of receptor tyrosine kinases and predominantly exerts its oncogenic functions by stimulating the PI3K/AKT/mTOR pathway.
Direct pharmacologic targeting of HER2 was first realized with trastuzumab (Herceptin), a humanized recombinant, monoclonal antibody that binds to its extracellular domain (5). Upon binding, trastuzumab downregulates the ligand-independent HER2 dimerization and growth factor signaling cascades downstream of HER2 including the PI3K/AKT/mTOR pathway (6–8). Trastuzumab has been demonstrated to mediate several antitumor mechanisms including induction of an immune response to tumor through antibody-dependent cellular cytotoxicity (ADCC), blockade of cleavage of the HER2 receptor, as well as downregulating ligand independent HER2 dimers. Both preclinical and clinical evidence demonstrate that the antitumor effects trastuzumab exerts are confined to tumors in which HER2 is amplified or overexpressed.
Despite the considerable efficacy of trastuzumab, many patients with metastatic HER2-amplified breast cancer either do not respond or have a limited duration of benefit. Almost all eventually have progressive disease even after responding to trastuzumab plus chemotherapy regimens (9–11). The molecular basis for resistance, either de novo or acquired, has been difficult to elucidate in part because of the difficulty in obtaining tumors samples after tumor progression and in part because trastuzumab has multiple modes of pharmacologic action (12). A number of laboratory models of resistance to trastuzumab have been reported. Among these, hyperactivation of the PI3K/AKT/mTOR pathway has been described through several different molecular lesions. In particular, Nagata et al. demonstrated that loss of the PTEN tumor suppressor could reduce the antitumor effect of trastuzumab in cell culture and animal models of HER2 amplified breast cancer and was associated with lack of responsiveness to trastuzumab in combination with paclitaxel in a retrospective analysis of primary tumor samples from patients who received this combination for metastatic disease (13, 14). Extending this work, Berns et al. demonstrated that mutational activation of PIK3CA also diminishes the antitumor effects of trastuzumab in models of HER2 amplified breast cancer (15). Together, activation of the PI3K pathway in response to either PIK3CA mutation or loss of PTEN expression was shown to be associated with reduced benefit from trastuzumab in retrospective studies.
Alteration of HER2 expression has also been implicated as a mechanism of resistance to anti-HER2 therapy in laboratory models. This has been suggested to occur through outright loss of HER2 overexpression or through induction of a cleaved form of HER2 known as p95-HER2 that lacks the extracellular domain to which trastuzumab binds (16, 17). In the case of the former, complete loss of HER2 expression has not been explained mechanistically and cannot be easily distinguished in vivo from false positive HER2 tests. With respect to overexpression of p95-HER2, appearance of this species has been associated with resistance in small retrospective studies but has been difficult to confirm in larger cohorts of patients due to the lack of a reliable method for identifying p95-HER2 from FFPE samples. In addition to these, it has been suggested though not proven that certain activating mutations in HER2 might reduce tumor responsiveness to trastuzumab.
Identification of the mechanisms of resistance to trastuzumab therapy has important implications for the rational selection of subsequent targeted therapies. If hyperactivation of PI3K/AKT/mTOR signaling causes resistance, inhibitors of this pathway would be beneficial. In the case of loss of outright HER2 overexpression, a move away from HER2 targeted therapy and towards conventional chemotherapy would be appropriate. To establish whether there is corroborating evidence for these mechanisms of resistance in patients with actual resistant cancer, we prospectively acquired tissue samples (research biopsies) from metastatic sites in patients with progression of disease on trastuzumab to assess expression of HER2 and PTEN and to determine whether PIK3CA was mutated.
Patients and Methods
Patient identification and tumor collection
Patients with breast cancers in which HER2 was overexpressed or amplified (score 3+ by IHC or amplified by FISH) and either recurrence of disease after receiving adjuvant trastuzumab therapy or WHO-defined progression of metastatic disease while receiving a trastuzumab-containing regimen were prospectively enrolled on an IRB approved tissue collection protocol from February 2007 through November 2011. All patients underwent biopsy of at least a single local or distant site to document progressive disease. Mutational analysis was performed on fresh frozen specimens. Formalin fixed paraffin embedded (FFPE) blocks of both the pre-treatment primary tumor and the progressive disease (local or distant) were obtained for assessment of HER2 status and PTEN analysis by immunohistochemistry (IHC). The presence of tumor, in both frozen samples and FFPE tissue sections, was confirmed by the study pathologist.
An unselected cohort of 73 primary HER2+ tumors from patients who had surgery between 2000 and 2006 were identified from the Breast Service database and subject to the same analyses as a comparison for HER2+ tumors not exposed to trastuzumab. The mean and median age of the patients in this cohort at the time of surgery was 50 yrs and 49 yrs (range 28–79 yrs) respectively. They all had surgery that was followed by standard adjuvant therapy but none received trastuzumab. HER2 was amplified in all 73 lesions whereas ER was positive in 41/73 (56%) lesions and PR in 30/73 (41%) lesions (Table 2).
Table 2.
Receptor status of primary versus post-trastuzumab tumors
| Treated cohort of HER2+ lesions | Untreated cohort of HER2+ lesions | ||
|---|---|---|---|
| Primary or pre-therapy | Secondary or post-therapy | ||
| HER2 | |||
| pos | 63/63 (100%) | 53/60 (88%) | 73/73 (100%) |
| neg | - | 7/60 (12%) | - |
| na | - | 3/63 | - |
| ER | |||
| pos | 31/63 (49%) | 32/59 (54%) | 41/73 (56%) |
| neg | 32/63 (51%) | 27/59 (46%) | 32/73 (44%) |
| na | - | 4/63 (6%) | - |
| PR | |||
| pos | 15/63 (24%) | 14/58 (24%) | 30/73 (41%) |
| neg | 48/63 (76%) | 44/58 (76%) | 43/73 (59%) |
| na | - | 5/63 (8%) | - |
Immunohistochemistry
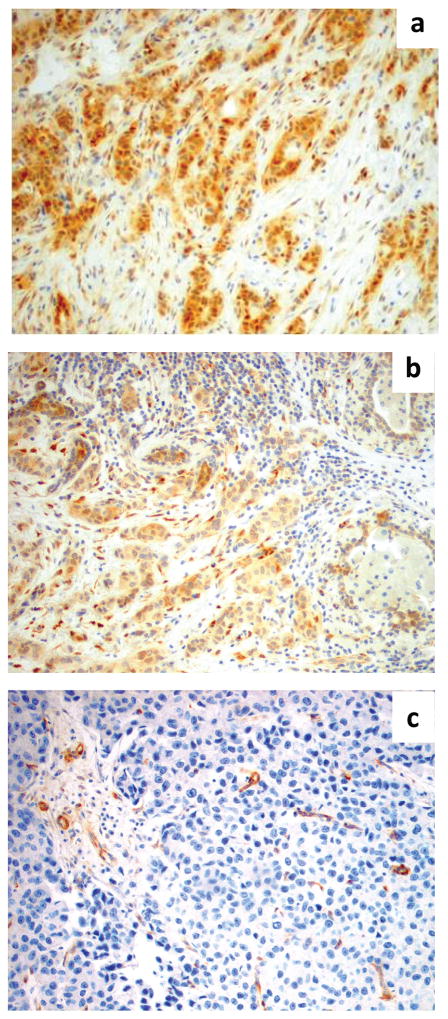
ER, PR and HER2 expression were evaluated by IHC according to standard clinical protocols. PTEN expression was assessed by IHC as previously described (18). Briefly, after incubation for 20 minutes at 98°C in Target Retrieval Solution pH 9 (EDTA, pH 9, Dako Cytomation), the sections were incubated for 30 minutes at room temperature with the monoclonal mouse anti-human PTEN antibody (dilution 1:100, clone 6H2.1, Dako) then with peroxidase-labeled polymer conjugated to goat anti-mouse immunoglobulins (EnVision/HRP, Dako). The chromogenic reaction was carried out with 3,3′-diaminobenzidine chromogen solution. All staining was reviewed and scored by 2 independent observers. For ER and PR, any nuclear positivity was scored as positive. HER2 was scored per ASCO/CAP guidelines (19). For PTEN immunohistochemistry, MDA-468 and MCF7 cell lines were used for negative and positive controls. Scoring was performed as previously described (18). The surrounding normal epithelium served as an internal control and tumor immunoreactivity was scored accordingly: score 0 = no immunoreaction; score 1 = reduced intensity of immunoreaction compared to normal epithelium; score 2 = intensity equal to normal epithelium (Figure 1).
Figure 1.
PTEN scoring of tumors by IHC. Depicted are photomicrographs of PTEN scoring: a) score 2 = intensity of the immunoreaction in the tumor equal to the adjacent normal epithelium (100X); b) score 1 = intensity of the immunoreaction reduced in the tumor as compared to the adjacent normal epithelium (100X); c) score 0 = PTEN immunoreaction is absent in the tumor and present in the adjacent normal epithelium (100X).
HER2 amplification assessed by IHC was confirmed by FISH analysis. Tumor specimens were classified amplified if the ratio of ERBB2 copy number signals to chromosome 17 centromere signal was >2 upon enumeration of 20 interphase nuclei.
Mutational analysis
Extraction of genomic DNA was performed using QuickGeneTM DNA tissue kit (Fujifilm). A minimum of 5mg of fresh frozen tumor was suspended in 360 μl of tissue lysis buffer (MDT) plus 40 μl of proteinase K and incubated for 48 hours at 55°C. Lysis buffer LDT (360μl) was then added and samples were incubated at 70°C for 30 minutes. Samples were then washed with ethanol 100% and wash buffer. Elution was in 50 μl of EDTA buffer. Quantification of DNA by Nanodrop® Fluorospectrometer showed an average yield of 20 μg (Picogreen ave yield 250ng).
Genotyping was performed by Sequenom® MassARRAY® system. The iPLEX® Gold genotyping assay is a single-base primer extension assay. After multiplexed PCR amplification of ~100 bp fragments with primers bracketing the mutation of interest, extension primers designed immediately adjacent to the mutation site prompt extension by one nucleotide depending on the template sequence. Purified primer extension reactions are loaded on a matrix pad of a SpectroCHIP (Sequenom) for analysis. The difference in mass between extended products as measured by laser desorption/ionization time-of-flight mass spectrometer (SpectroREADER; Sequenom) is used to make a genotype call comparing wild-type and mutant alleles. The multiplexed assays and primers were designed for analysis of PIK3CA hotspot mutations in exon 9 and 20 and for analysis of HER2 mutations (D767M, L755S, R896C) using Assay Design 3.1.
Statistical analysis
The association between mutation status and biological features was assessed using Fisher Exact test and Chi-Square test for categorical data analysis and Student t- test for mean values comparison. Exact 95% confidence intervals were calculated for each proportion. P-value of less than 0.05 was considered statistically significant. Statistical analyses were performed using SAS software, version 9.1 (SAS Institute, Cary, NC).
Results
Samples from metastatic lesions of 63 patients who were clinically resistant to trastuzumab therapy were analyzed (Supplementary Figure 1) and 60 had sufficient material for additional analysis. The mean and median ages of patients at primary diagnosis were 47 yrs and 45 yrs (range 24–68 yrs) respectively. Of these patients, 21 (35%) had received trastuzumab in combination with standard chemotherapy in the adjuvant management of primary breast cancer and 42 (65%) had received one or more lines of chemotherapy in combination with trastuzumab as palliative therapy for metastatic disease (Table 1). The histology of the primary breast cancer was invasive ductal carcinoma in 58 (92%) of the cases. As an entry criterion for the study, HER2 was found to be overexpressed in the primary tumor by at least one positive test (score 3+ by IHC in 16 samples or amplified by FISH >2.0 copies in 47 samples). Among this cohort of resistant HER2+ tumors, estrogen receptor (ER) was scored positive in 31 (49%) of the samples (Table 2). As primary tumors were not available for the majority of these cases, a comparison cohort of HER2+ primary tumors from patients who underwent surgery between 2000 and 2006 at MSKCC was utilized. None of these patients received preoperative systemic therapy or trastuzumab in the adjuvant setting. The tumors were all HER2+ by IHC and similar to the metastatic cohort with respect to ER status (Table 2).
Table 1.
Characteristics of HER2+ tumors recurring on or after trastuzumab therapy
| n = 63 | |
|---|---|
| Therapeutic regimen | |
| Only adjuvant trastuzumab | 21 |
| Several lines of chemotherapy | 42 |
| Metastatic tissue site | |
| Ipsilateral breast/axilla/chest wall | 39 (62%) |
| Bone | 1 (2%) |
| Liver | 13 (21%) |
| Lung | 4 (6%) |
| Brain | 6 (9%) |
Changes in HER2 expression status
We evaluated HER2 expression status in the 60 metastatic tumors with documented resistance to trastuzumab therapy. In all of these cases, the primary tumor was classified as HER2+. HER2 overexpression/amplification was maintained after trastuzumab exposure in 53 of 60 (88%) evaluable cases with trastuzumab-refractory lesions (Table 2). In 7/60 (12%), HER2 status apparently changed from amplified to non-amplified. Among these 7 cases, the original FISH scores were positive for 5 cases but all 5 were close to the standard cut-off of 2.0 (Supplementary Table 1). In 3 of the 7 cases, sufficient material was available for repeat HER2 testing on the pre-trastuzumab sample and this analysis did not confirm evidence for overexpression. Samples were not available for repeat testing on the other 4 cases (Supplementary Table 1).
PIK3CA mutations
Among the 63 patients with demonstrated clinical resistance to trastuzumab, analysis for PIK3CA mutations was successful in 45 cases. 13/45 (29%) were found to harbor activating PIK3CA mutations in their metastatic lesions (Table 3). The spectrum of mutations in PIK3CA included 6 cases of exon 9 (helical domain) mutants and 4 cases of exon 20 (kinase domain) mutants. In 2 samples a mutation was identified in the C2 domain (C420R) and in 1 sample a mutation was identified in the ABD domain (R88Q). By comparison, in the cohort of 73 primary HER2+ tumors not exposed to trastuzumab, 13 (18%) were observed to have an activating PIK3CA mutation. This approximates the rate seen in other HER2+ cohorts such as in the TCGA analysis where 23% of the HER2+ tumors had activating PIK3CA mutations (30% with any PIK3CA mutation) (20, 21, 22).
Table 3.
PI3K alterations in trastuzumab refractory lesions versus untreated HER2+ cohort
| PIK3CA mutation or PTEN absent/reduced expression | P value | ||
|---|---|---|---|
| Trastuzumab refractory cohort | Primary untreated HER2+ cohort | ||
| PIK3CA | 13/45 (29%, 95%CI 16–44%) | 13/73 (18%, 95%CI 10–29%) | .176 |
| PTEN | 33/56 (59%, 95% CI 46–71%) | 25/73 (34%, 95% CI 24–46%) | .007 |
| PIK3CA or PTEN | 29/41 (71%, 95% CI 54–83%) | 32/73 (44%, 95% CI 32–56%) | .007 |
HER2 mutation
As an exploratory analysis, known activating mutations in HER2 that have been previously associated with resistance to HER2 targeted therapies were also screened in metastatic lesions with sufficient material. Of the 44 analyzed cases, only 1 case demonstrated a mutation in HER2 (L755S). The role of this mutation in mediating resistance to trastuzumab is uncertain and this case was notable for concurrent diminished expression of PTEN.
PTEN loss
Fifty six of the metastatic tumors were evaluable for PTEN by IHC (Table 3). Absent or significantly reduced PTEN was observed in 33/56 (59%) refractory metastatic lesions and was more common among these cases when compared to the cohort of HER2-amplified primary breast cancers not exposed to trastuzumab (59% vs 34%, p=0.007). For cases where both PIK3CA and PTEN were evaluable, 29/41 (71%) showed lesions in either PIK3CA or PTEN or both. This was also significantly more common than the untreated cohort of HER2 amplified breast cancers (71% vs 44%, p=0.007).
Pre-treatment primary breast tumor and post- treatment metastatic FFPE sections were available for 26 cases. Reduction or loss of PTEN expression between the primary and metastatic site was noted in 9/26 (35%) cases raising the possibility of acquired loss of PTEN with exposure to trastuzumab based therapy (Figure 2, Supplementary Table 2, and Supplementary Figure 2). Considerably fewer primary tumor samples were available for PIK3CA mutation analysis although two cases were identified with a mutation present in the trastuzumab exposed tumor and not the primary tumor (Supplementary Table 3). Several cases of both PTEN reduction and PIK3CA mutation were identified (Supplementary Table 4) further suggesting a heightened dependence on this pathway in cases of trastuzumab resistance. ER and PR status were not different with respect to PIK3CA/PTEN status (Supplementary Table 5).
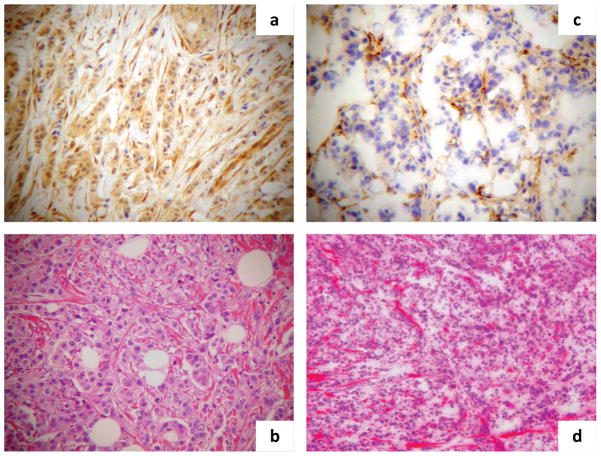
Figure 2.
PTEN IHC changes between pre-trastuzumab and post-trastuzumab samples. Depicted are photomicrographs of PTEN change from normal to loss of expression: a) PTEN expression by IHC in the primary breast invasive carcinoma with nuclear and cytoplasmic staining equal to the adjacent normal stroma (100X); b) HE staining of the primary lesion (100X); c) PTEN complete loss of expression in the secondary invasive carcinoma with absence of any staining in the lesion as compared to the adjacent normal stroma (100X); d) HE staining of the secondary lesion (100X).
Discussion
Therapeutic targeting of HER2 results in dramatic clinical efficacy almost exclusively in patients in which the protein is overexpressed as a result of gene amplification. Although a number of different drugs have been devised to target HER2, the antibody trastuzumab has had the biggest impact on patient survival to date. Unfortunately, targeting HER2 with trastuzumab does not result in the cure of patients with detectable metastatic breast cancer and a significant number of patients receiving trastuzumab in the adjuvant setting eventually develop metastatic disease. The molecular basis for resistance or incomplete activity has been difficult to ascertain because of the absence of a firm understanding of the mechanism of trastuzumab action and the lack of tumor tissue from patients who develop resistance. Two of the major mechanistic hypotheses behind trastuzumab refractory disease have been mutational activation of the PI3K/AKT pathway and changes in the HER2 molecule itself. In particular, mutational activation in PIK3CA, loss of PTEN, increased expression of p95-HER2, and loss of expression of HER2 have been proposed to contribute to resistance to trastuzumab. In this study of specimens from patients with tumors progressing on trastuzumab therapy, we found evidence for changes that activate PI3K/AKT signaling, but little evidence for loss of HER2 expression.
In the context of immunologic and antibody-based cancer therapy, tumor downregulation of the targeted epitope is a well-established mechanism for escape from the drug itself. Loss of HER2 expression has been proposed as the cause of trastuzumab resistance in one preclinical model, although how this would confer drug resistance while promoting tumor growth is unclear. In an accompanying neoadjuvant study comparing pre-trastuzumab core needle biopsies with the post-chemotherapy plus trastuzumab surgical specimens, tumors in eight of twenty five patients with residual disease had no evidence of HER2 amplification (16). In our study, we find that loss of HER2 overexpression in resistant tumors is a very rare event.
Several possible explanations could account for the greater frequency of HER2 loss in their study. First, cancer cells remaining after neoadjuvant therapy are unlikely to reflect the biology of progressive metastatic tumors. Residual tumors may reflect an inadequate duration of therapy for complete eradication and cannot be assumed to be trastuzumab refractory as is the case with progressive metastatic disease. Second, consideration needs to be given to the accuracy of the HER2 test itself. Even with centralized testing, there remains a false positive rate for HER2 testing and this is likely to contribute to at least a portion of the discordant findings in the neoadjuvant setting. In support of this point, a recently reported retrospective comparison of primary and metastatic tumors described how variability in testing could account for a significant proportion of the discordance observed between primary and metastatic samples (23).
In the setting of disease progression, as opposed to disease responsiveness and potential cure, our data suggests that the oncogenic driver is not lost. This is consistent with other examples of resistance to targeted therapy studied including ER, AR, BCR- ABL, mutant EGFR, mutant BRAF, and mutant ALK. For most of these, the tumor becomes resistant to the targeted drug but is still dependent on the target. This is likely to be the case in HER2-amplified breast cancers with trastuzumab resistance, given that targeting of HER2 by other means is effective in a large fraction of these patients. Many such agents, including pertuzumab, lapatinib, neratinib, and 17-AAG have been shown to be active despite prior refractoriness to trastuzumab.
A large body of evidence suggests that HER2 overexpression drives the growth of breast cancer predominantly by activating the PI3K/AKT signaling pathway via HER2- HER3 heterodimerization (7, 24). As trastuzumab can partially downregulate this dimer it can attenuate PI3K/AKT driven oncogenic signaling. If this effect is necessary for the antitumor effects of the antibody, mutations of downstream components of the pathway that cause PI3K/AKT signaling to be HER2 independent would be predicted to cause trastuzumab resistance. In our study, we found a preponderance of tumors (71%) with either a mutation in PI3K and/or loss of expression of PTEN (7 cases showed both PIK3CA mutation and PTEN loss/reduction of expression; supplementary Table 4). This significantly exceeds the frequency expected with HER2+ breast tumors that lack exposure to trastuzumab as in our untreated cohort (p=0.007) and in other studies. These data support preclinical studies that show that trastuzumab resistant models are sensitive to PI3K and AKT inhibitors (7, 25).
Although mutations in PI3K appear to be less frequent in cohorts of HER2 amplified breast cancer not exposed to trastuzumab, the underlying rate of 18–25% reported in several studies supports that activation of the pathway may play a role in de novo resistance to the drug. This analysis of refractory HER2+ tumors demonstrates that the majority of HER2+ tumors that progress on trastuzumab containing regimens are likely to have additional lesions hyperactivating the PI3K/AKT pathway. Such lesions may have been present prior to trastuzumab exposure or “acquired” as the tumor became resistant. Our study only analyzed one portion of the primary tumor and could have missed areas of genotypic heterogeneity where the PI3K pathway lesion was present. However, a recent analysis suggests that such intratumoral heterogeneity is not common for PIK3CA mutations in breast cancer (26). Regardless of the specific cause, in a few cases in our series, comparison of the pretreatment and post-trastuzumab specimen suggests a mutation was selected for with exposure to the drug.
This would suggest that prior studies that relied entirely on pretreatment primary tumor samples might have underestimated the true prevalence of PI3K/AKT alterations contributing to trastuzumab resistance. Importantly, these data support current clinical efforts designed to co-target the PI3K/AKT pathway along with HER2 itself to achieve more durable and potent antitumor effects. As targeting the PI3K/AKT pathway alone is expected to induce feedback signaling through the HER2/HER3 dimer, combinations of PI3K/AKT inhibitors with HER2 inhibitors may be significantly more effective than either alone in the resistant, metastatic setting (27–29).
A major challenge in translational medicine is to understand if biological principles derived from laboratory models are clinically relevant in human disease. In solid tumor malignancies, the burden of this validation has often been placed on primary tumor samples. While such samples retain a large repository of information, they may not capture the dynamic nature of metastatic tumors. Our view is that the progressive tumor contains vital information on the current biologic state of the tumor and is invaluable to understanding the disease despite the hurdles posed by interventional procedures needed to obtain these specimens. In this study, we utilized voluntary biopsies to highlight which mechanisms of resistance are most relevant for trastuzumab refractory HER2 amplified breast cancer. The insights we obtained should help to shape subsequent therapeutic approaches. In this case, continued targeting of HER2 along with co-targeting of the PI3K/AKT pathway appears to be rational. Beyond our findings for HER2 driven breast cancer, the study also underscores the feasibility of study designs that incorporate metastatic solid tumor biopsies as no related adverse events occurred for the duration of this study. We believe that this approach should be pursued more frequently with the intent towards, ultimately enabling greater personalization of treatment.
Supplementary Material
Acknowledgments
We gratefully acknowledge individual support from Julie Laub, Walsh Benefits, and The Cary Grossman Research Fellowship in Breast Cancer to support this research. SC is supported by a Mentored Clinician Scientist Training Award (NCI-K08 CA134833), LN and NR are supported by a Breast Program grant (NCI-P01 CA094060).
Footnotes
Conflict of Interest: None of the authors have an actual, potential, or perceived conflicts of interest to disclose.
Disclosures of Potential Conflicts of Interest
None of the authors have an actual, potential, or perceived conflicts of interest to disclose.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 5.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4285–9. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuello M, Ettenberg SA, Clark AS, Keane MM, Posner RH, Nau MM, et al. Down-regulation of the erbB-2 receptor by trastuzumab (herceptin) enhances tumor necrosis factor-related apoptosis- inducing ligand-mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB-2. Cancer research. 2001;61:4892–900. [PubMed] [Google Scholar]
- 7.Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand- independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer cell. 2009;15:429–40. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer research. 2002;62:4132–41. [PubMed] [Google Scholar]
- 9.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–48. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 10.Pegram M. Can we circumvent resistance to ErbB2-targeted agents by targeting novel pathways? Clin Breast Cancer. 2008;8 (Suppl 3):S121–30. doi: 10.3816/cbc.2008.s.008. [DOI] [PubMed] [Google Scholar]
- 11.Pohlmann PR, Mayer IA, Mernaugh R. Resistance to Trastuzumab in Breast Cancer. Clin Cancer Res. 2009;15:7479–91. doi: 10.1158/1078-0432.CCR-09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev Anticancer Ther. 2011;11:263–75. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer cell. 2004;6:117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 14.Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177:164756. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 16.Mittendorf EA, Wu Y, Scaltriti M, Meric-Bernstam F, Hunt KK, Dawood S, et al. Loss of HER2 amplification following trastuzumab-based neoadjuvant systemic therapy and survival outcomes. Clin Cancer Res. 2009;15:7381–8. doi: 10.1158/1078-0432.CCR-09-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. Journal of the National Cancer Institute. 2007;99:628–38. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 18.Sakr RA, Barbashina V, Morrogh M, Chandarlapaty S, Andrade VP, Arroyo CD, et al. Protocol for PTEN expression by immunohistochemistry in formalin-fixed paraffin-embedded human breast carcinoma. Appl Immunohistochem Mol Morphol. 2010;18:371–4. doi: 10.1097/PAI.0b013e3181d50bd5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 20.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer research. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 21.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer research. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature. 2012 doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niikura N, Liu J, Hayashi N, Mittendorf EA, Gong Y, Palla SL, et al. Loss of Human Epidermal Growth Factor Receptor 2 (HER2) Expression in Metastatic Sites of HER2-Overexpressing Primary Breast Tumors. J Clin Oncol. 2012;30:593–9. doi: 10.1200/JCO.2010.33.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8933–8. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS ONE. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalinsky K, Heguy A, Bhanot UK, Patil S, Moynahan ME. PIK3CA mutations rarely demonstrate genotypic intratumoral heterogeneity and are selected for in breast cancer progression. Breast Cancer Res Treat. 2011;129:635–43. doi: 10.1007/s10549-011-1601-4. [DOI] [PubMed] [Google Scholar]
- 27.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011 doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5021–6. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.