Abstract
Gpr171 is an orphan G protein-coupled receptor putatively related to the P2Y family of purinergic receptors (P2YRs) for extracellular nucleotides, a group of mediators previously shown to regulate hematopoietic progenitor cells (HPCs). No information is currently available on the ligand responsible for Gpr171 activation and its biological role remains unknown. We reconstructed Gpr171 phylogenesis in mice and confirmed that Gpr171 is evolutionally related to members of a P2Y gene-cluster localized on mouse chromosome 3. As a first step towards unveiling a role for Gpr171, we investigated its expression profile in murine hematopoietic cells. As opposed to other P2YRs, we found that Gpr171 expression is downregulated in monocytes and granulocytes, suggesting a negative role in myeloid lineage-specification. To test Gpr171 functional role, we next enforced Gpr171 expression in a myeloblastic cell line (32D cells) and in primary Sca-1+ hematopoietic progenitors (HPCs), and observed a decreased expression of myeloid markers upon induction of Gpr171, as well as an increased generation of colonies in vitro. Conversely, Gpr171 silencing induced opposite results, diminishing the expression of myeloid markers and the clonogenic potential of 32D cells. In vivo, mice transplanted with HPCs over-expressing Gpr171 displayed a significant reduction in the percentage of Mac-1+Gr-1− cells. As a preliminary step in the investigation of Gpr171 role in murine hematopoiesis, our findings indicate that the orphan receptor Gpr171 negatively regulates myeloid differentiation. Together with phylogenic analyses, our data suggest that Gpr171 may have followed a separate evolutionary pathway as compared to other P2YRs belonging to the same gene-cluster.
Keywords: Gpr171, GPRs, P2 receptors, myeloid differentiation
Introduction
Lifelong hematopoiesis is built on the fine balance between the capability of hematopoietic stem cells to undergo either self-renewal or multi-lineage differentiation [1]. Despite intense investigation, the molecular mechanisms underlying the multi-step process that leads from committed progenitors to terminally differentiated cells are still poorly understood. Over the past years, a few transcription factors have been identified as master regulators of lineage specification [2–4], such as Gata2 [5, 6], indispensable for megakaryocyte/erythrocyte development. Beside the critical role played by these factors in driving differentiation, transcriptional programs are also modulated by environmental factors through the activity of membrane receptors, such as G protein-coupled receptors (GPRs). GPRs, also known as seven-spanning membrane receptors, represent one of the largest receptor families in nature [7]. They are involved in the molecular decoding of extracellular signals generated by a wide variety of chemically diverse compounds, incuding small molecules, as well as peptides and proteins. Because of their role in communication between cells and the extracellular environment, seven-spanning membrane receptors have been the focus of intense investigation, aimed to identify their functional role and potential involvement in diseases [8]. GPR ligands comprise growth factors, cytokines, and chemokines, thus making GPRs one of the most investigated pharmaceutical targets [9, 10].
First identified as a putative member of the GPR family, Gpr171 (initially named H963) still remains an orphan receptor, and its ligand, signaling pathway and fuction are yet to be identified [11]. Based on sequence homology, Gpr171 has been suggested to belong to a particular subfamily of GPRs, named P2Y receptors, which are activated by extracellular nucleotides [12]. Beside the widely acknowledged role of triphosphate-nucleotides in cell metabolism, these molecules are also ubiquitous regulators of a great variety of biologic functions, comprising cell proliferation, differentiation, as well as migration and cell death [13, 14]. For instance, extracellular nucleotides and their cognate receptors have being implicated in the regulation of hematopoietic stem/progenitor cells, as well as in the function of terminally differentiated elements in the peripheral blood (PB) [15–17]. Usually compartmentalized inside the cytoplasm, upon mechanical, chemical or infective stress nucleotides can be released into the extracellular microenvironment [18], where they interact with purinergic membrane receptors [19–21]. Purinergic receptors comprise two subfamilies, classified as membrane channel-receptors (P2X) and 7-spanning receptors (P2Y receptors or P2YRs) [22]. As members of the GPR family, P2YRs transduction of intracellular signals proceeds through coupling to G proteins [19]. Gq/11 represents the G protein mainly activated by P2YRs, although in the case of P2Y12, P2Y13, and P2Y14 Gi/o proteins have been shown to be recruited [23].
The data recently provided by the sequencing of mammalian genomes permitted the identification of new members of this family. The present study represents a preliminary attempt to investigate the role of Gpr171 in hematopoietic progenitor cells (HPCs). Our findings indicate a peculiar evolutionary history for Gpr171 as compared to other members of the P2Y family and suggest that Gpr171 may act as a negative regulator of myeloid differentiation, both in vitro and in vivo.
Materials and Methods
Sequence analyses
Sequence alignments and phylogenetic analyses were performed using the program CLC MainWorkbench 5.5. Phylogenetic trees were created using the Neighbor-Joining (NJ) method and 1,000 bootstraps replications were conducted to evaluate the reliability of the trees. The putative transmembrane domains were predicted using the program TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) [24]. The following accession numbers were entered for alignment analyses of amino acid sequences: Gpr87_Mouse (NP_115775), Gpr99_Mouse (NP_001001490), Gpr171_Mouse (NP_775574), P2ry1_Mouse (NP_032798), P2ry2_Mouse (NP_032799), P2ry4_Mouse (NP_065646), P2ry6_Mouse (NP_898991), P2ry12_Mouse (NP_081847), P2ry13_Mouse (NP_083084), P2ry14_Mouse (NP_573463), Sucnr1_Mouse (NP_115776).
Cell line and mice
The hematopoietic progenitor cell line 32D was obtained from ATCC and maintained in RPMI 1640 medium, supplemented with 10%FBS and rIL-3 (5ng/mL). For studies in vivo, wild type (WT) C57BL/6, CD45.1 mice (6 to 12 weeks of age) were utilized for this study. Animals were housed and bred in specific pathogen-free conditions at Baylor College of Medicine.
Construction of retroviral vectors
The plasmids containing the genes described in the present study were purchased from ATCC and subsequently subcloned into the retroviral expression vector MSCV-IRES-eGFP. Retroviral particles were packaged by co-transfection of 293t cells with MSCV-GOI-IRES-eGFP and pCL-Eco plasmids as previously described [25]. 48 hours after transfection, culture supernatants were collected and their viral titer was determined by infecting NIH3T3 cells. As a control, the mock construct MSCV-IRES-eGFP was used. In 32D cells, Gpr171 expression was induced by direct nucleofection according to manufacturer’s instructions (Amaxa Cell Line Nucleofector Kit C, Lonza).
Retroviral transduction of bone marrow progenitor cells and transplantation
The enforced expression of Gpr171 was assayed by retroviral transduction of WT HPCs. WT donor mice (C57BL/6, CD45.2, 6–12 weeks of age) were i.p. injected with 5-Fluorouracil at 150 mg/Kg. Six days later, BM cells were harvested from tibias, femurs, and hips and enriched by immuno-magnetic separation (AutoMACS, Miltenyi Biotech, Auburn, CA) for Sca-1+ HPCs. Sca-1+ cells were seeded in 24-well plates (500,000 cells/well) in StemPro medium (GIBCO, Carlsbad, CA), supplemented with nutrient supplement 40X (GIBCO), L-Glutamine, Penicillin/Streptomycin, Polybrene (4 μg/mL), rmTPO (R&D, Minneapolis, MN), and rmSCF (R&D) and spin-infected (at 1,100 rpm for 2 hours, at room temperature) in the presence of retroviral vectors (MSCV-IRES-eGFP as a control, or MSCV-Gpr171-IRES-eGFP). After transduction, cells were harvested and transplanted into lethally irradiated C57BL/6, CD45.1 recipient mice (50,000 – 75,000 cells/mouse). PB engraftment and lineage distribution of CD45.2 GFP+ cells were analyzed at 5, 12, 22 and 28 weeks after transplantation.
Gpr171 silencing
A total of 106 32D cells were washed twice in PBS and resuspended in nucleofector solution (Amaxa Cell Line Nucleofector Kit C, Lonza) and mixed with siRNAs specific for Gpr171 silencing (SASI_Mm02_00345246 and SASI_Mm02_00345246_AS, Sigma-Aldrich). Cells were treated with Nucleofector program E-32, according to manufactuter’s instructions. Nucleofected cells were immediately removed from the cuvette by adding pre-warmed RPMI + 10% FBS medium and incubated at 37°C, 5% CO2. After 48 hours, cells were used for real-time PCR analysis, CFU-C in vitro assays and phenotypic analysis.
Real-time PCR analysis
Total RNA was isolated from treated cells using RNAeasy Micro Plus Kit (Qiagen) First strand synthesis was performed with Improm2 (Promega) and real-time PCR was performed with Taqman probe sets (Applied Biosystems) on a ABI Prism 7700 Sequence Detector for 40 cycles. A mouse internal control (Gusb) was included in every reaction for normalization and expression was measured for each assay relative to the Gusb internal standard (ΔCt). The fold change was calculated from the formula 2ΔΔCt. The error bars represent the corresponding SEM.
In vitro Colony-Forming-Unit (CFU-C) assay
For CFU-C assays, primary Sca-1+ cells were cultured at 37°C, 5%CO2 for 48 hours. GFP+Sca-1+ cells were then sorted (on a MoFlo flow cytometer) into 96-well plates (1 cell/well), containing M3434 MethoCult medium (Stem Cell Technologies, Vancouver, Canada), and incubated at 37°C, 5%CO2. After 12–14 days of culture, hematopoietic colonies were counted.
Flow-cytometry
32D cells or primary Sca-1+ cells were transduced with retroviral vectors as described, maintained in liquid culture for at least 48hrs and then stained with the following antibodies for phenotypic analysis: anti-Mac1 PE-Cy7 or APC, anti-CD16/32 PE, anti-Gr-1 APC or PE, biotinilated anti-F4/80 (followed by anti-biotin Pacific Blue, PB), and anti-CD14 PE. PB from transplanted mice was collected from the retro-orbital sinus at 5, 12, 22 and 28 weeks after transplantation. As previously described [26], the samples were pre-treated with Red Blood Cell – lysis buffer (TrisCl pH 7.8 and Ammonium Chloride, 1:9) and then stained for 20 minutes on ice with monoclonal antibodies, according to the following cocktail: anti-CD45.2 APC-conjugated, anti-CD4 PB, anti-CD8 PB, anti-B220 PB, anti-B220 PE-Cy7, anti-Mac1 PE-Cy7, and anti-Gr1 PE. All samples were resuspended in Hanks’ buffer containing Propidium Iodide (PI, 1 μg/mL), and analyzed on a LSRII cytofluorimeter (BD bioscience, Franklin Lakes, NJ).
Statistics
Results of experimental data are reported as mean ± SEM. Statistical significance was determined by two-tailed Student’s t test. P values are indicated as *p<0.05 and **p<0.01.
Results
Gpr171 is evolutionally related to P2YRs and its expression is suppressed upon myeloid differentiation
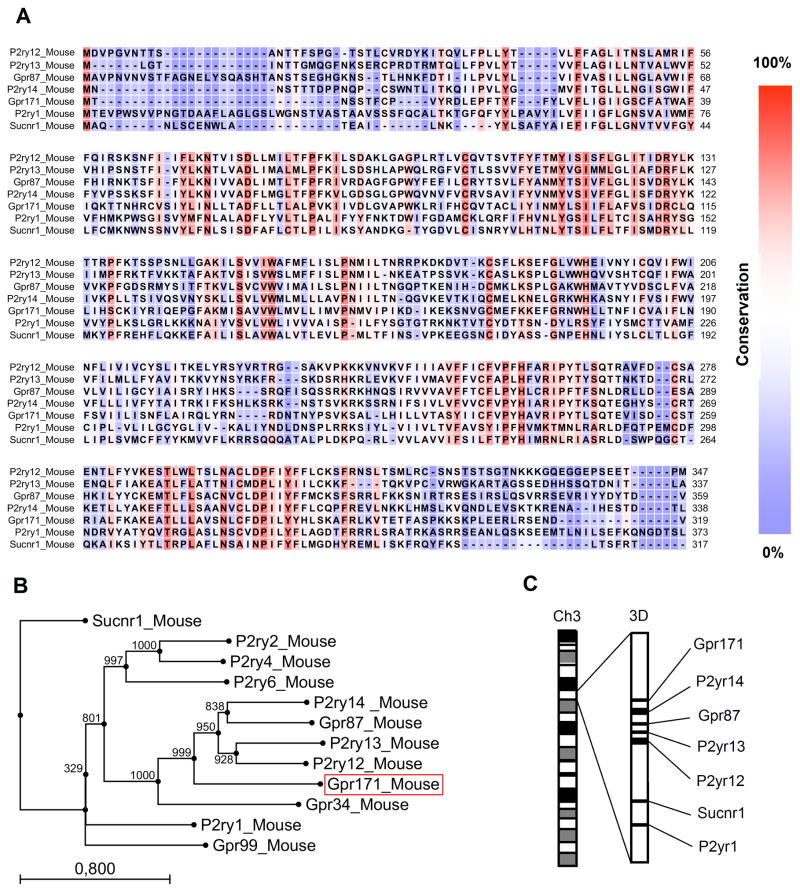
Based on sequence homology, it has been suggested that human GPR171 is evolutionally and structurally related to the GPR subfamily of purinergic P2YRs for extracelluar nucleotides [27]. Similarly to the human gene, when we aligned the amino acid sequences of murine Gpr171 and other murine P2YRs, we found a high degree of similarity, especially in correspondence of the sequences encoding for transmembrane domains (TM), particularly TM3, TM6, and TM7 (Fig. 1A). By reconstructing the phylogenetic tree of P2YRs encoded by the murine genome, we also found that the murine Gpr171 is evolutionally related to P2Y genes, particularly P2ry12, P2yr13, P2yr14, and Gpr87 (Fig. 1B). Interestingly, these five genes co-localize on chromosome 3, as depicted in Fig. 1C. Since co-localization of genes in the same chromosomal region is a strong indication of local gene amplification, the clustering of Gpr171, P2yr14, Gpr87, P2ry13, and P2ry12 is highly suggestive of events of gene duplication, followed by divergent evolution. Reconstructing the evolution of orphan receptors, such as Gpr171, could also help identify its cognate ligand, since receptors evolved from a common ancestor might share, at least partly, ligand-specificity.
FIG. 1.
Phylogenetic analysis of Gpr171. (A) Alignment of amino acid sequences of murine Gpr171 with other murine P2Y genes localized on chromosome 3. The seven transmembrane domains (indicated as TM) are overlined. As emphasized by the color scheme, the sequences display the highest degree of conservation in their TM3, TM6, and TM7 domains. (B) Phylogenetic tree illustrating how the murine orphan receptor Gpr171 is related to other members of the P2YR family. Alignment of amino acid sequences and tree reconstruction were performed using CLC Main Workbench 5. Trees were created using the Neighbor-Joining (NJ) method and 1,000 bootstraps replications were conducted. The length of each branch is proportional to evolutionary divergence. (C) Schematic representation of the reciprocal localization of Gpr171, Gpr87, P2ry12, P2ry13, P2y14, Sucnr1, and P2ry1 on chromosome 3. Co-localization of genes in the same chromosomal region strongly indicates that they evolved by gene duplication from the same ancestor gene.
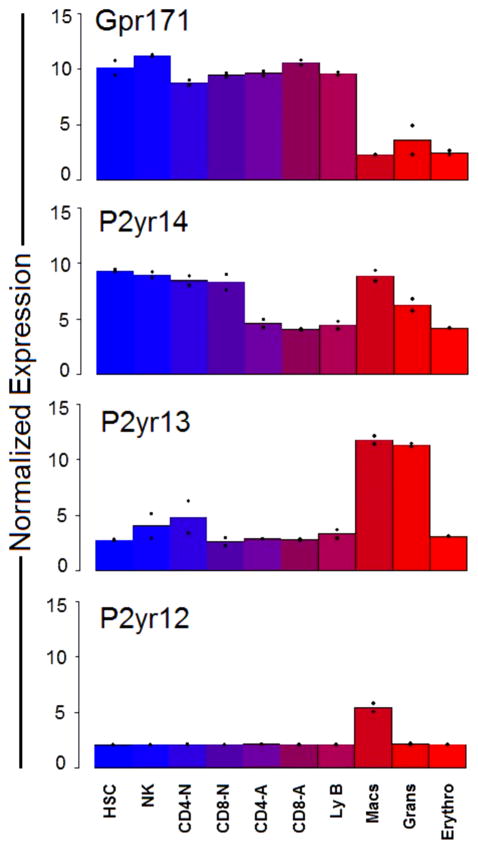
In an attempt to identify a functional role for Gpr171, we first consulted a database previously generated in our laboratory, comprising the genetic expression profile of HSCs, as well as their differentiated progeny [25]. By bio-informatic analysis, genes that are selectively expressed in the HSC compartment were discriminated from genes uniquely involved in the specification of the differentiated lineages (including granulocytes, macrophages, erythrocytes, NK cells, B and T lymphocytes). Using this approach specific lineage “fingerprints” were defined, comprising a combination of genes uniquely expressed in a particular hematopoietic population [25]. From the comparison of the expression profile of HSCs and their differentiated progeny, Gpr171 emerged as a gene consistently expressed in the lymphoid lineage (activated and naïve T Lymphocytes, B Lymphocytes, and NK cells), as wells as in the HSC compartment (identified as Side Population c-Kit+ Lineage− Sca-1+ cells, SPKLS cells [25, 26]). In contrast, Gpr171 expression appeared to be drastically suppressed in cells that underwent myeloid differentiation (monocytes, granulocytes, and erythocytes; Fig. 2, top panel). Interestingly, when we analyzed the expression profile of genes highly related to Gpr171, such as P2ry12, P2ry13, and P2ry14, we found a strikingly different pattern. As opposed to Gpr171, all these genes appear to be consistently up-regulated in cells belonging to the myeloid lineage and down-regulated in activated T cells, as well as B cells (Fig. 2). These findings suggest that an early bifurcation in the phylogenetic tree might have occurred, originating, on one side, Gpr171 and, on the other, P2ry12, P2ry13, P2ry14, and Gpr87 (Fig. 1B). The divergent evolution resulting from this bifurcation could have contributed to specializing Gpr171 in functions different from those described for the genes originating from the sibling branch.
FIG. 2.
Expression levels of Gpr171, P2ry14, P2ry13, and P2y12 in murine hematopoietic cells. The normalized (log2) expression intensity of Gpr171 expression (top panel) in myeloid cells (macrophages, granulocytes, and erythrocytes) is significantly decreased compared to the expression of other members of the same P2ry cluster on chromosome 3 (P2ry14, P2ry13, and P2y12).
Gpr171 is silenced in differentiating 32D cells and its enforced expression down-regulates myeloid differentiation
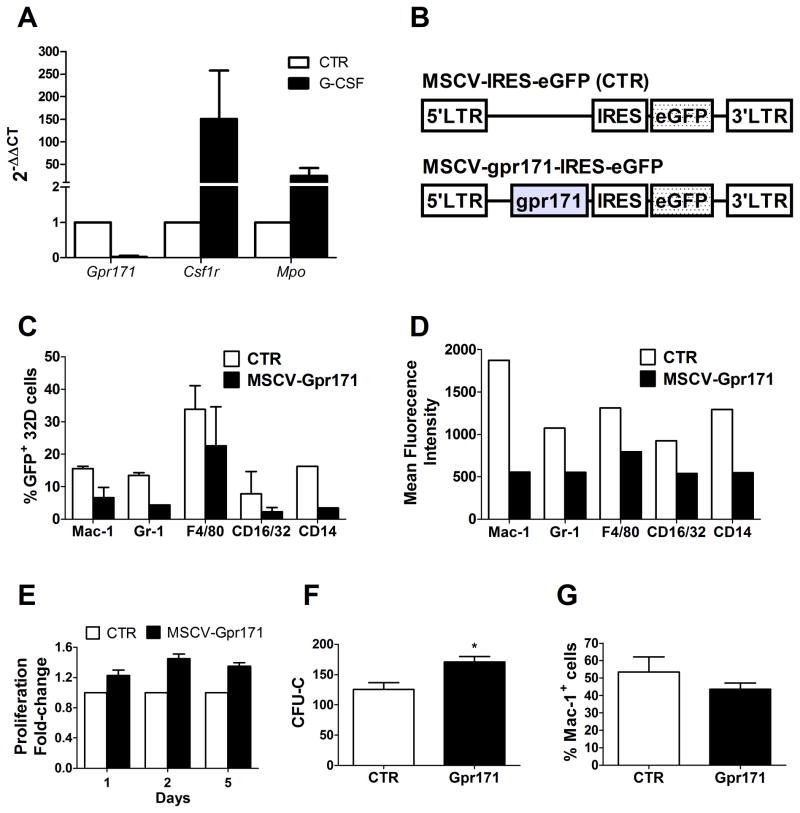
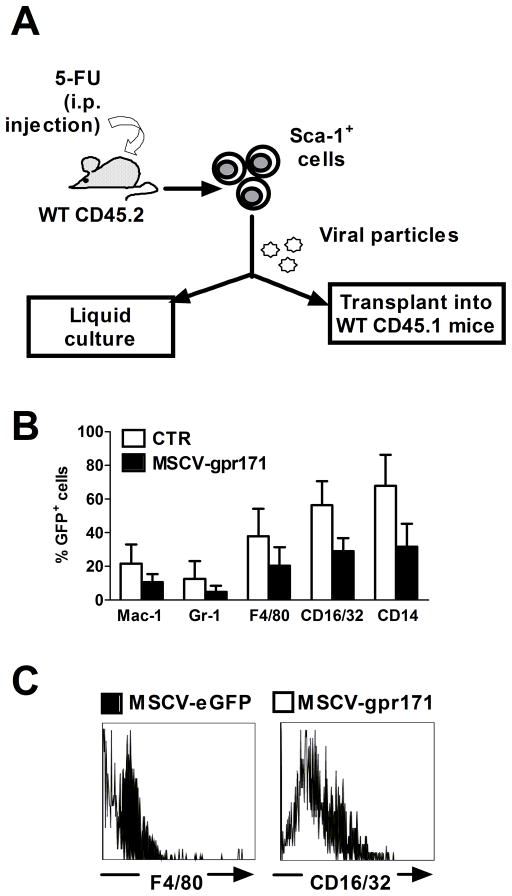
Data mining is an exceptional source of information about unknown, orphan receptors. Nonetheless, the ultimate validation of data provided by bio-informatic analysis is based on functional studies. Gene expression profiling in the hematopoietic system suggested a negative correlation between Gpr171 and myeloid differentiation (Fig. 2). In order to test this hypothesis, we first studied how Gpr171 expression was modulated during differentiation in an immortalized monoblast-like cell line (32Dcl.3 cells, henceforth referred to as 32D cells), widely used as a model for hematopoietic progenitor cells (HPCs). 32D cells proliferate as undifferentiated blasts when maintained in culture medium containing IL-3. On the contrary, IL-3 withdrawal and G-CSF treatment have been demonstrated to promote granulocytic differentiation in these cells [28]. To assess the relationship between Gpr171 and myeloid differentiation, 32D cells were cultured in the presence of G-CSF and in the absence of IL-3. As expected, differentiating 32D cells presented after three days an increase of in surface myeloid markers (such as Mac-1 and Gr-1, data not shown). In addition, G-CSF–treated cells presented a significant increase in the relative expression of two genes involved in myeloid differentiation, namely the colony stimulating factor 1-receptor Csf1r and the myeloperoxidase Mpo, whereas Gpr171 expression was almost completely silenced (Fig. 3A). In order to confirm the negative relationship between myeloid differentiation and Gpr171, we enforced Gpr171 expression in 32D cells using a vector derived from Mouse Stem Cell Virus (MSCV) (as shown in Fig. 3B). Phenotypic analyses of markers typically associated with myeloid differentiation showed that both the expression and the mean fluorescence intensity (MFI) of Mac-1, Gr-1, F4/80, CD16/32, and CD14 appeared to be markedly decreased upon over-expression of Gpr171 in 32D cells (Fig. 3C–D). In addition, we observed that the enforced expression of Gpr171 in 32D cells increases cell proliferation in liquid culture (Fig. 3E), as well as colony formation (Fig. 3F).
FIG. 3.
Gpr171 expression in 32D cells. (A) Relative expression of Gpr171, Csf1r, and Mpo in 32D cells after 3 days of culture under differentiating conditions (in the presence of G-CSF). (B) Schematic representation of the MSCV retroviral expression vector utilized to transduce hematopoietic cells. The mock construct (top) expresses the reporter gene GFP, while the Gpr171 construct (bottom) allows the coordinated expression of both Gpr171 and GFP. (C) Phenotypic characterization of transduced 32D cells: percentage of 32D cells expressing myeloid markers are decreased after induction of Gpr171 expression. (D) Mean Fluorescence Intensity (MFI) of myeloid markers in 32D cells over-expressing Gpr171. (E) Proliferation is increased in Gpr171-overexpressing 32D cells, as emerged over a 5-days time-course. Data are expressed as proliferation fold-change between MSCV-Gpr171 transduced cells and control cells. (F) Colony formation (expressed as CFU-Cs) is increased in 32D overexpressing Gpr171. (G) Modulation of Mac-1 expression under conditions driving myeloid differentiation in 32D cells. Transduced 32D cells were cultured for 8 days in the absence of IL-3. Results are reported as mean ± SEM of at least triplicate experiments. *p< .05, **p<.01.
In order to assess the capability of Gpr171 over-expression to counteract myeloid differentiation, we enforced Gpr171 expression and cultured transfected 32D cells in the absence of IL-3: a condition known to hinder self-renewal and promote cell differentiation. After 8 days, the percentage of Mac-1 positive cells in the Gpr171 over-expressing cell population was 20% less than the control, suggesting that even under conditions that allow differentiation of 32D cells the enforced expression of Gpr171 decelerates myeloid commitment (Fig. 3G). Taken together, these initial experiments suggest that Gpr171 may play a role in counteracting myeloid commitment by inhibiting cell growth and down-modulating the expression of myeloid markers in a cell line model of myeloblastic precursors.
Gpr171 silencing promotes myeloid differentiation in 32D cells
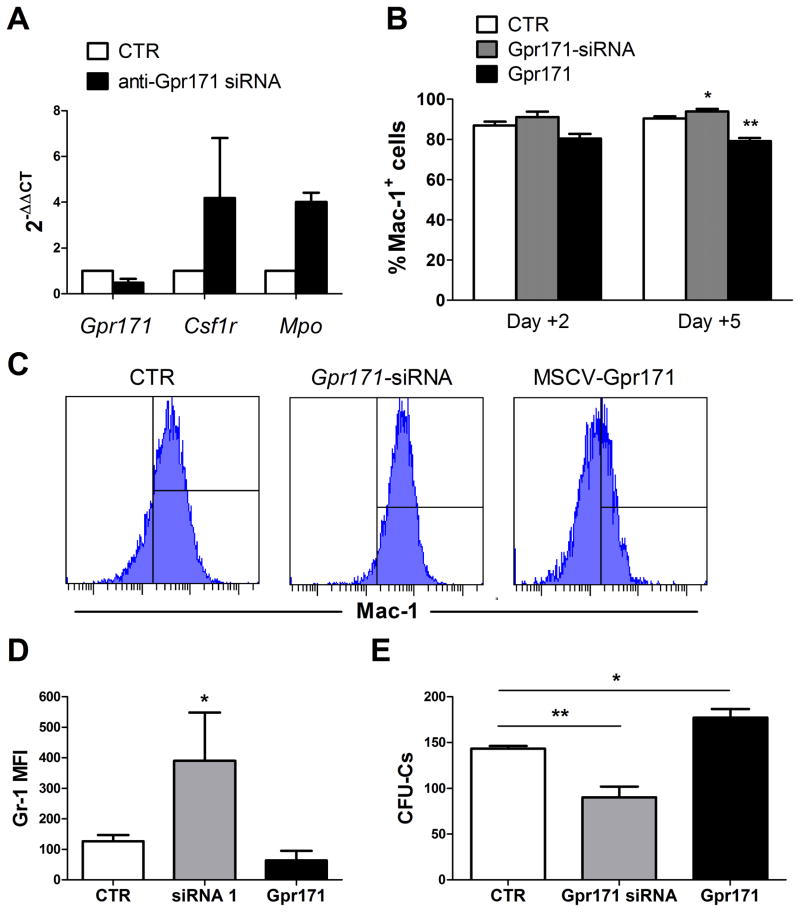
To further test the role of Gpr171 in myelopoesis, we transduced 32D cells with a mix of siRNAs specifically designed to silence Gpr171 expression. After 48 hours, we found a 52% reduction in Gpr171 expression in transduced 32D cells. Concomitantly, treated cells presented an increase in the relative expression of Csf1r and Mpo, two genes linked to myeloid differentiation (Fig. 4A). In addition, the membrane expression of both Mac-1 was increased in 32D cells transduced with anti-Gpr171 siRNAs, as assessed by flow cytometry (Fig. 4B–C). Similarly, the mean fluorescence intensity for Gr-1 was also increased in 32D cells transduced with anti-Gpr171 siRNAs (Fig. 4D). In contrast, Gpr171 over-expression induced the opposite effect, down-regulating Mac-1 expression in transduced cells Fig. 4B–C).
FIG. 4.
(A) Relative gene expression of Gpr171 and myeloid differentiation genes (Csfr1 and Mpo) in 32D cells transduced with anti-Gpr171 siRNAs. Cells were analyzed 48hrs post-transduction and mRNA levels were assessed by Real-Time PCR. (B) Percentages of Mac-1+ 32D cells after transduction with MSCV-Gpr171 or anti-Gpr171 siRNAs. Cells were analyzed by flow-cytomtry 2 and 5 days post-transduction. (C) Representative histograms of Mac-1 expression in 32D cells 96 hours after transduction with MSCV-Gpr171 or anti-Gpr171 siRNAs. (D) Mean Fluorescence Intensity (MFI) of myeloid marker Gr-1 in 32D cells transduced with MSCV-Gpr171 or anti-Gpr171 siRNAs. (E) 32D colony formation (espressed as CFU-Cs) is decreased after transduction with anti-Gpr171 siRNAs.
Functionally, the up-regulation of differentiation markers was paralleled by a decreased proliferative capacity of Gpr171-silenced 32D cells (38% reduction compared to the control), as displayed by in vitro clonogenic assays (Fig. 4E). Conversely, when Gpr171 over-expression was induced in 32D cells, a 20% increase in CFU-C numbers was recorded (Fig. 4E). Taken together, these findings contribute to define the role of Gpr171 in hematopoiesis and suggest that the expression of Gpr171 negatively regulates myeloid differentiation.
Gpr171 enforced expression in primary mouse HPCs inhibits the expression of myeloid markers both in vitro and in vivo
In order to test the effects of Gpr171 over-expression in primary murine cells, we retrovirally transduced Sca-1 – enriched HPCs, according to the experimental model depicted in Fig. 5A. Similarly to what we observed in 32D cells, Gpr171 over-expression in Sca-1+ progenitors suppressed the membrane expression of myeloid markers, such as Mac-1, Gr-1, F4/80, CD16/32, and CD14 (Fig. 5B–C).
FIG. 5.
Gpr171 over-expression in Sca-1+ cells in vitro. (A) Schematic representation of the experimental model utilized to study Gpr171 role in murine hematopoiesis. Sca-1+ HSCs/HPCs were collected from WT CD45.2 donor mice previously injected with 5-FU and transduced with the retroviral MSCV-Gpr171 vector or with the empty control vector. Transduced cells were then utilized for both in vitro and in vivo studies. (B) Percentages of GFP+, primary murine hematopoietic cells expressing markers associated with myeloid differentiation are decreased after induction of Gpr171 expression. (C) Representative flow plots showing decreased expression of two markers associated with terminal monocyte differentiation (F4/80 and CD16/32), following Gpr171 over-expression. Results are reported as mean ± SEM of at least triplicate experiments.
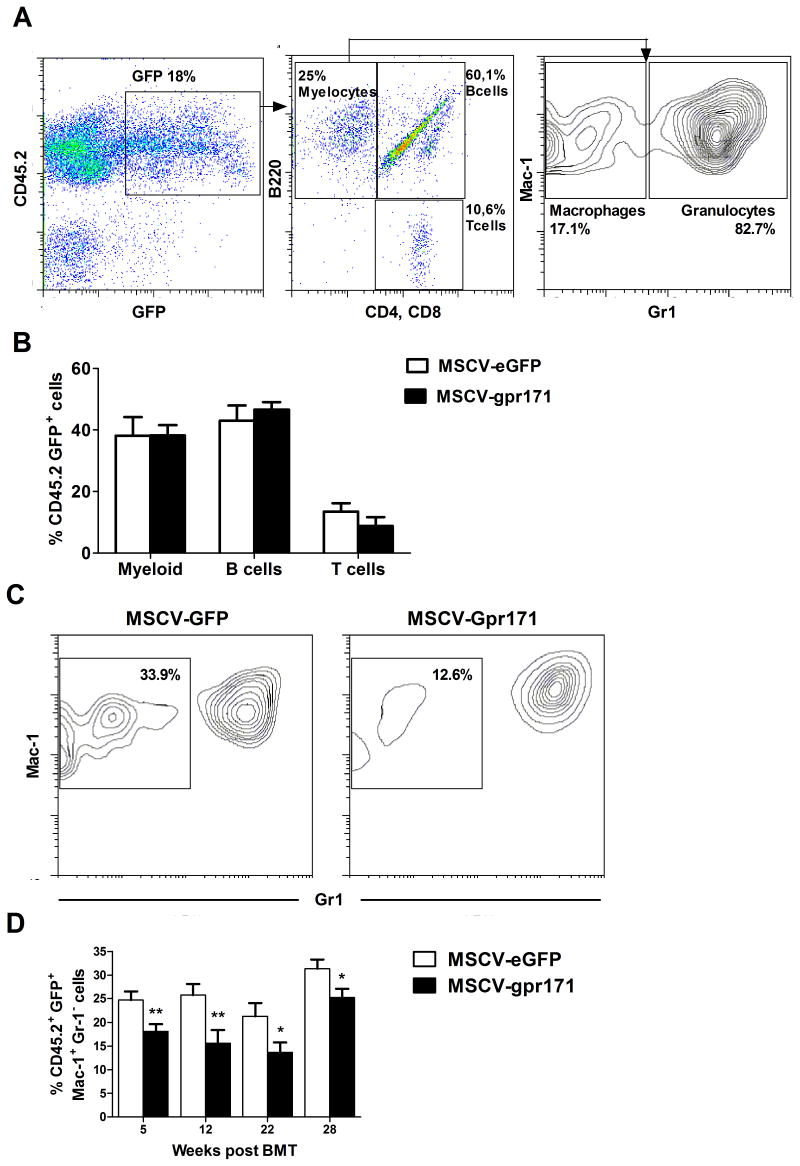
In order to investigate how powerfully Gpr171 can inhibit myeloid differentiation, we retrovirally transduced Sca-1+ HPCs and transplanted them into lethally irradiated mice. PB reconstitution was assessed by flow cytometry at different time points (5, 12, 22, and 28 weeks after transplant), according to the model depicted in Fig. 6A (left panel). Transduction efficiency, as inferred from the percentage of donor-derived CD45.2 GFP+ cells, was about the same at 5 weeks after transplantation in both control and experimental mice (21.18% vs 17.19% for MSCV-Gpr171 and MSCV control, respectively, data not shown).
FIG. 6.
Gpr171 over-expression in vivo. (A) Schematic representation of the PB analysis performed periodically on transplanted animals. First, transduced (GFP+), donor-derived (CD45.2+) blood cells were gated (top), then cells were divided in Mac-1+ myeloid cells, B220+ B lymphocytes, and CD4+CD8+ T lymphocytes (middle). Finally, myeloid cells were separated in monocytes and granulocytes, according to Gr-1 expression (bottom). (B) Lineage distribution of Mac-1+ Myeloid cells, B220+ B Lymphocytes, and CD4+CD8+ T Lymphocytes in the PB of transplanted mice, as assessed by flow cytometry at 12 weeks post-transplant. (C) Representative density plots displaying the relative percentages of PB GFP+Mac-1+Gr-1− monocytes and PB GFP+Mac-1+Gr-1+ granulocytes, as assessed by flow cytometry at 5 weeks after transplantation (control group on the left and Gpr171 group on the right). (D) Time-course analysis of the percentage of PB GFP+Mac-1+Gr-1− monocytes, as assessed by flow cytometry at 5, 12, 22, and 28 weeks after transplantation. Results are reported as mean ± SEM of at least triplicate experiments. *p< .05, **p<.01.
Next, we analyzed how Gpr171 enforced expression affects the contribution to the different hematopoietic lineages in vivo (Fig. 6A). At all the time points considered, we observed no significant skewing in the percentages of myeloid cells (Mac-1+ cells), B lymphocytes (B220+ cells), and T lymphocytes (CD4+CD8+ cells) in the subset of donor-derived, GFP+ cells (Fig. 6B). However, when we dissected the myeloid compartment on the basis of Mac-1 and Gr-1 expression, we found that the monocytic population (Mac-1+Gr-1−) was consistently reduced upon Gpr171 over-expression (Fig. 6C–D). At 5 weeks post-transplantation, we observed a 1.4-fold decrease in the percentage of GFP+Mac-1+Gr-1− cells in the Gpr171-group, compared to the control (Fig. 6D). This divergence became even more evident at 12 weeks after transplantation (when GFP+Mac-1+Gr-1− cells displayed a 1.7-fold decrease upon Gpr171 over-expression) and was maintained up to 28 weeks post transplantation (Fig. 5D). This findings confirm that Gpr171 plays a role in lineage specification and it is capable of inhibiting monocyte differentiation in vivo, as well as in vitro.
Discussion
Gpr171 is a novel, putative GPR member, structurally and evolutionally related to P2YRs. Since Gpr171 was first described, no progress has been made in deciphering its biological role and its physiological ligand [11]. In this report, we investigated Gpr171 function in the murine hematopoietic system and found that its expression negatively correlates with myeloid differentiation, both in vitro and in vivo. In addition, despite the similarity that Gpr171 shares with P2YRs, our data contribute to highlight a negative role for Gpr171 in myelopoiesis, as opposed to other members of the family highly expressed in granulocytes and monocytes.
Recently, Simon and Barnard found in humans a striking similarity between GPR171 (previously known as H963) and some members of the family of human P2YRs for extracellular nucleotides [27]. By mapping the exact chromosomal localization of all P2YRs so far identified, the authors found five orphan GPCRs (among which was GPR171), that are localized in the same region of chromosome 3, where the human genes for GPR87, P2Y12, P2Y13, and P2Y14 cluster together. Furthermore, the GPR171 sequence shares specific motifs in the transmembrane domains TM6 and TM7, found to be highly conserved among P2YRs. Based on the evidence provided by evolutionary and structural similarity, the orphan receptor Gpr171 likely represents a new member of the P2Y receptor-family, involved in the transduction of signals generated by extracellular nucleotides. Despite the phylogenetic relationship between Gpr171 and other members of the P2Y family, our results in the murine system suggest that Gpr171 might have followed a divergent evolutionary pathway, causing a substantial deviation in terms of biological function. We showed that P2ry12, P2ry13, and P2ry14, although part of the same P2Y cluster on chromosome 3, display the opposite expression pattern of Gpr171. Indeed, even minor mutations in the amino acid sequence of a receptor can give rise to significant evolutionary detours, leading to a different agonist specificity or signaling pathway. Hence, unveiling the physiological agonist for Gpr171, as well as developing specific antagonists, will help achieve a deeper understanding of the pharmacologic behavior of this, so far, orphan receptor and of its relationship to the other members of the family.
In this study, we investigated the expression profile of Gpr171 in hematopoietic cells (including the compartment of SPKLS HSCs) and found that Gpr171 is significantly suppressed in cells undergoing myeloid differentiation. In contrast, other P2YRs belonging to the same gene-cluster (e.g. P2y12, P2y13, and P2y14) present the opposite pattern of expression, with the highest levels recorded in differentiated myeloid cells (Fig. 2). Such a difference might be the result of an early bifurcation in the phylogenetic tree leading to the divergent evolution of Gpr171 and other genes of the same cluster on Chromosome 3. At the functional level, the enforced expression of Gpr171 in both 32D cells and primary Sca-1+ HPCs decreased the membrane expression of monocyte-specific antigens, such as Mac-1, CD14, F4/80, and CD16/32. In line with these findings, Gpr171 silencing induced opposite results, diminishing the expression of myeloid markers and the clonogenic potential of 32D cells. In vivo, when Sca-1+ HPCs transduced with MSCV-Gpr171 retrovirus were transplanted into lethally irradiated mice, a specific decrease in the percentage of Mac-1+Gr-1− monocytes was consistently observed in the myeloid fraction of Gpr171 over-expressing mice over a period of 28 weeks post-transplantation.
Taken together, these findings support the hypothesis that the orphan receptor Gpr171 negatively regulates myeloid differentiation and, together with phylogenic analyses, suggest that Gpr171 may have followed a separate evolutionary pathway as compared to other P2Y receptors of the same chromosomal cluster.
In the future, a deeper understanding of the role of Gpr171 in murine hematopoiesis is expected to arise from additional in vivo studies. The definitive determination of the role of Gpr171 in the hematopoietic system will involve the characterization of Gpr171 knockout mice, as well as the repression of Gpr171 expression in hematopoietic reconstitution assays. In addition, future investigations of the biological role of Gpr171 will greatly benefit from the identification of its physiological ligand. Based on the structural similarity that Gpr171 shares with P2Y receptors, we expect that its ligand may belong to the family of extracellular nucleotides. This group of molecules has recently emerged to play a role in immunity, inflammation, as well as in the regulation of the hematopoietic system both in animal models and humans [15–17, 29]. Over the past years, the differentiation of myeloid, lymphoid, mekacaryocitic, and erythroid progenitors has been shown to be influenced by extracellular nucleotides through the activation of purinergic receptors [30]. Interestingly, depending on the specific hematopoietic lineage, different P2 receptors appear to play a role in differentiation and the identification of novel orphan receptors will contribute to a deeper understanding of hematopoietic differentiation. The results emerged from our preliminary investigation show that Gpr171, as a putative novel member of the P2Y family, may be involved in the differentiation and proliferation of myeloid cells, suggesting a novel mechanism by which extracellular nucleotides can tune immunological responses [31].
Acknowledgments
L.R. was supported by a grant from the Italian Association for Leukemias and Lymphomas, section of Bologna (BolognaAIL) and the work was supported by grants DK58192, EB005173, and HL081007 from the NIH and 0740020N from the AHA.
Footnotes
Author Disclosure Statement
No competing financial interests exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huang X, Cho S, Spangrude GJ. Hematopoietic stem cells: generation and self-renewal. Cell Death Differ. 2007;14:1851–1859. doi: 10.1038/sj.cdd.4402225. [DOI] [PubMed] [Google Scholar]
- 2.Loose M, Patient R. Global genetic regulatory networks controlling hematopoietic cell fates. Curr Opin Hematol. 2006;13:229–236. doi: 10.1097/01.moh.0000231419.15654.7f. [DOI] [PubMed] [Google Scholar]
- 3.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 4.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 5.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 6.Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson MD, Siminovitch KA, Cole DE. G protein-coupled receptor pharmacogenetics. Methods Mol Biol. 2008;448:139–185. doi: 10.1007/978-1-59745-205-2_8. [DOI] [PubMed] [Google Scholar]
- 8.Kostenis E. G proteins in drug screening: from analysis of receptor-G protein specificity to manipulation of GPCR-mediated signalling pathways. Curr Pharm Des. 2006;12:1703–1715. doi: 10.2174/138161206776873734. [DOI] [PubMed] [Google Scholar]
- 9.Maekawa T, Ishii T. Chemokine/receptor dynamics in the regulation of hematopoiesis. Intern Med. 2000;39:90–100. doi: 10.2169/internalmedicine.39.90. [DOI] [PubMed] [Google Scholar]
- 10.Oh DY, Kim K, Kwon HB, Seong JY. Cellular and molecular biology of orphan G protein-coupled receptors. Int Rev Cytol. 2006;252:163–218. doi: 10.1016/S0074-7696(06)52003-0. [DOI] [PubMed] [Google Scholar]
- 11.Wittenberger T, Schaller HC, Hellebrand S. An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors. J Mol Biol. 2001;307:799–813. doi: 10.1006/jmbi.2001.4520. [DOI] [PubMed] [Google Scholar]
- 12.Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, et al. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacological reviews. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci. 2007;64:1471–83. doi: 10.1007/s00018-007-6497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnstock G, Williams M. P2 purinergic receptors: modulation of cell function and therapeutic potential. J Pharmacol Exp Ther. 2000;295:862–869. [PubMed] [Google Scholar]
- 15.Lemoli RM, Ferrari D, Fogli M, Rossi L, Pizzirani C, Forchap S, et al. Extracellular nucleotides are potent stimulators of human hematopoietic stem cells in vitro and in vivo. Blood. 2004;104:1662–1670. doi: 10.1182/blood-2004-03-0834. [DOI] [PubMed] [Google Scholar]
- 16.Rossi L, Manfredini R, Bertolini F, Ferrari D, Fogli M, Zini R, et al. The extracellular nucleotide UTP is a potent inducer of hematopoietic stem cell migration. Blood. 2007;109:533–542. doi: 10.1182/blood-2006-01-035634. [DOI] [PubMed] [Google Scholar]
- 17.Di Virgilio F, Chiozzi P, Ferrari D, Falzoni S, Sanz JM, Morelli A, et al. Nucleotide receptors: an emerging family of regulatory molecules in blood cells. Blood. 2001;97:587–600. doi: 10.1182/blood.v97.3.587. [DOI] [PubMed] [Google Scholar]
- 18.Lazarowski ER. Vesicular and conductive mechanisms of nucleotide release. Purinergic Signal. 2012;8:359–373. doi: 10.1007/s11302-012-9304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubyak GR, el-Moatassim C. Signal transduction via P2-purinergic receptors for extracellular ATP and other nucleotides. Am J Physiol. 1993;265:C577–606. doi: 10.1152/ajpcell.1993.265.3.C577. [DOI] [PubMed] [Google Scholar]
- 20.von Kugelgen I. Pharmacological profiles of cloned mammalian P2Y-receptor subtypes. Pharmacol Ther. 2006;110:415–432. doi: 10.1016/j.pharmthera.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 21.von Kugelgen I, Wetter A. Molecular pharmacology of P2Y-receptors. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:310–23. doi: 10.1007/s002100000310. [DOI] [PubMed] [Google Scholar]
- 22.Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci. 2007;64:1471–83. doi: 10.1007/s00018-007-6497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbracchio MP, Burnstock G. Purinergic signalling: pathophysiological roles. Jpn J Pharmacol. 1998;78:113–145. doi: 10.1254/jjp.78.113. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993:374. [Google Scholar]
- 25.Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry A. 2009;75:14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon J, Barnard EA. The P2Y nucleotide receptors in the human genome. Acta Biol Hung. 2003;54:191–201. doi: 10.1556/ABiol.54.2003.2.8. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder T, Just U. Notch signalling via RBP-J promotes myeloid differentiation. The EMBO journal. 2000;19:2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi L, Salvestrini V, Ferrari D, Di Virgilio F, Lemoli RM. The sixth sense: hematopoietic stem cells detect danger through purinergic signaling. Blood. doi: 10.1182/blood-2012-04-422378. In press. [DOI] [PubMed] [Google Scholar]
- 30.Sak K, Boeynaems JM, Everaus H. Involvement of P2Y receptors in the differentiation of haematopoietic cells. J Leukoc Biol. 2003;73:442–447. doi: 10.1189/jlb.1102561. [DOI] [PubMed] [Google Scholar]
- 31.Bours MJ, Dagnelie PC, Giuliani AL, Wesselius A, Di Virgilio F. P2 receptors and extracellular ATP: a novel homeostatic pathway in inflammation. Front Biosci. 2011;3:1443–1456. doi: 10.2741/235. [DOI] [PubMed] [Google Scholar]