Abstract
LRRFIP1 binds cytoplasmic double-stranded DNA and RNA and interacts with FLI, the mammalian homolog of Drosophila flightless I, through a highly conserved 87-amino acid domain. Upon binding nucleic acid ligands, LRRFIP1 recruits and activates β-catenin, leading to the IRF3-dependent production of type I interferon. However, the molecular mechanism of LRRFIP1 signaling is not well understood. Here we show that the FLI-interacting domain of LRRFIP1 forms a classic parallel, homodimeric coiled coil with ten heptad repeats and 22 helical turns. The coiled coil domain is also a dimer in solution. However, a longer LRRFIP1 construct spanning the coiled coil and DNA binding domains assembles into higher order oligomers in solution. The structure of LRRFIP1-CC constitutes a valuable tool for probing the mechanism of LRRFIP1 signaling and for structural studies of larger LRRFIP1 constructs.
Keywords: X-ray crystallography, innate immune receptor, nucleic acid binding protein, coiled-coil homodimer, DNA-binding protein, leucine zipper, heptad repeat
Introduction
In vertebrates, innate immune receptors detect broadly conserved microbial structures throughout the cell. The rapidly ensuing inflammatory response is the first line of defense against infection and controls the activation of the adaptive immune response. Microbial nucleic acids are one of the principal classes of ligands for innate immune receptors. Toll-like receptors (TLRs) 3, 7, 8 and 9 recognize microbial nucleic acids in endolysosomal compartments (Barton et al., 2006; Kawai and Akira, 2010). In the cytoplasm, RIG-I recognizes the 5′-triphosphate-capped blunt ends of plus-stranded RNA viruses, while MDA5 recognizes long cytosolic double-stranded RNA (dsRNA) delivered or generated during viral infection (Hornung et al., 2006; Kato et al., 2006; Pichlmair et al., 2006). Cytosolic double-stranded DNA (dsDNA) is recognized by AIM2, DAI (ZBP1) and RNA polymerase III (Ablasser et al., 2009; Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). However, these receptors do not fully account for the production of type I interferon in response to cytosolic GC-rich dsDNA (Yang et al., 2010). LRRFIP1 has been proposed as the missing receptor for cytosolic dsDNA. LRRFIP1 is a cytoplasmic protein that binds dsRNA, GC-rich Z-form dsDNA and AT-rich B-form dsDNA (Suriano et al., 2005; Wilson et al., 1998). LRRFIP1 also interacts with FLI, the mammalian homolog of Drosophila flightless I (Fli-I), a gelsolin-family actin binding protein (Fong and de Couet, 1999; Liu and Yin, 1998; Wilson et al., 1998). Upon binding viral dsRNA or bacterial dsDNA, LRRFIP1 recruits and activates β-catenin (Lee and Stallcup, 2006), which leads to IRF3-dependent production of type I interferon (Yang et al., 2010).
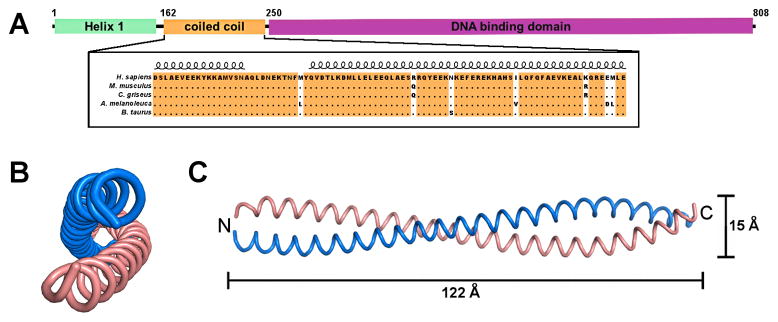
LRRFIP1 exists as a number of different isoforms, which may be differentially expressed and regulated (Fong and de Couet, 1999; Suriano et al., 2005). LRRFIP1 isoform 1 contains an N-terminal domain of unknown function, a conserved 87-amino acid domain predicted to be a coiled coil (Fong and de Couet, 1999; Liu and Yin, 1998) and a nucleic acid binding domain (Fig. 1A) (Wilson et al., 1998). The coiled coil domain, which is found in all LRRFIP genes, is highly conserved across mammalian species and is required for interaction with the leucine-rich repeat (LRR) domain of FLI (Fong and de Couet, 1999; Liu and Yin, 1998).
Fig. 1.
Crystal structure of LRRFIP1-CC. (A) Domain structure of LRRFIP1. LRRFIP1 contains three domains, an N-terminal helical region of unknown function, a central coiled coil (CC) domain that interacts with FLI, and a C-terminal DNA binding or nucleic acid recognition domain (DBD). (B) End-on and (C) side views of the LRRFIP1-CC homodimer. Chains A and B are shown in pink and blue, respectively. LRRFIP1-CC is a classic parallel, homodimeric coiled coil spanning 122 Å in length.
In the absence of structural information for LRRFIP1, the molecular mechanism of signal generation by LRRFIP1 upon binding dsRNA or dsDNA remains unknown. We report here the crystal structure of the coiled coil domain of LRRFIP1, LRRFIP1-CC. The protein forms a classic parallel, homodimeric coiled coil with ten heptad repeats and 22 helical turns. LRRFIP1-CC is also a dimer in solution. The LRRFIP1-CC structure constitutes a valuable tool for structural studies of larger LRRFIP1 constructs.
Materials and Methods
Cloning of the coiled-coil domain of LRRFIP1
A gene encoding the coiled-coil domain (residues 162–249, LRRFIP1-CC) of human LRRFIP1 isoform 3 (Open Biosystems clone ID 40027218) was cloned into the pET21a vector (Novagen) in frame with the vector’s C-terminal six-histidine purification tag using the Nde I and Hind III restriction sites. Genes encoding the DNA-binding domain of LRRFIP1 (residues 250–808, LRRFIP1-DBD) and the coiled-coil and DNA-binding domains of LRRFIP1 (residues 162–808, LRRFIP1-CC-DBD) were each cloned into the pET21a vector (Novagen) in frame with the vector’s C-terminal six-histidine purification tag using the Nde I and Sac I restriction sites.
Expression and purification of LRRFIP1 constructs
The LRRFIP1-CC expression vector was transformed into Escherichia coli Rosetta cells (Novagen) and cultured in Luria Broth (LB) supplemented with 0.1 g/l ampicillin. Cells were induced during log-phase growth with 0.4 mM IPTG for 4 h at 37 C. Cells were lysed at 4 C in lysis buffer, 50 mM NaH2PO4 pH 8.0, 0.3 M NaCl, 5% glycerol, with protease inhibitors (Roche). After centrifugation for 30 min at 40 krpm the clarified cell lysate was loaded onto a HisTrap FF nickel-affinity column (GE Healthcare). LRRFIP1-CC eluted from the column at 0.25 M imidazole pH 8.0. LRRFIP1-CC was further purified on a Superdex 200 (10/300) size-exclusion column (GE Healthcare) in 20 mM HEPES pH 7.5, 0.15 M NaCl. LRRFIP1-CC yield was 40–80 mg per liter of cell culture.
LRRFIP1-DBD and LRRFIP1-CC-DBD were purified by nickel-affinity and size-exclusion chromatography as described above, except that DNase I was included during cell lysis and that the proteins were purified by ion exchange chromatography on a MonoQ column in 20 mM MES pH 6.5 and elution from 0 to 1 M NaCl to remove bound genomic DNA prior to the final size-exclusion step. Additionally, while LRRFIP1-DBD could be purified on a Superdex 200 size-exclusion column, LRRFIP1-CC-DBD was purified on a Superose 6 size-exclusion size-exclusion column (GE Healthcare). The protein yield for LRRFIP1-CC and LRRFIP1-CC-DBD yield was 20–30 mg per liter of cell culture.
LRRFIP1-CC chemical crosslinking assay
LRRFIP1-CC (0.5 g/l in 10 mM HEPES pH 7.5, 0.15 M NaCl) was incubated with various concentrations of ethylene glycol bis(succinimidylsuccinate) (EGS) from 10 μM to 1 mM for 30 min at 25 C. The sample was analyzed by SDS- PAGE and by Western Blotting using an anti-tetrahistidine antibody (QIAgen).
Circular Dichroism (CD) Spectroscopy
LRRFIP1-CC was analyzed at 1.5 g/l in 10 mM NaH2PO4. Each measurement was the average of three scans in steps of 0.2 nm at 25°C. The differential CD spectrum of the complex was obtained by subtracting the buffer spectrum before conversion to molar ellipticity values. CD measurements were performed on a Jasco J-810 spectropolarimeter (Welltech Enterprises).
Multi-angle Light Scattering
Purified LRRFIP1-CC (8.8 g/l), LRRFIP1-DBD (4 g/l), or LRRFIP1-CC-DBD (2 g/l) was injected onto a Superdex 200 (10/300) column equilibrated in 10 mM HEPES pH 7.5, 0.15 mM NaCl and coupled to a DAWN EOS spectrometer and OPTILAB DSP interferometric refractometer (Wyatt Technology). Each protein was detected as it eluted from the column with a UV detector at 280 nm, a light scattering detector at 690 nm, and a refractive index detector. The molar mass of the protein sample was determined from the Debye plot of light scattering intensity versus scattering angle. Data processing was performed with ASTRA software (Wyatt Technology).
Crystallization of LRRFIP1-CC
Crystals of LRRFIP1-CC were grown by hanging drop vapor diffusion at 16 C. An 8.8 g/l solution of LRRFIP1-CC in 20 mM HEPES pH 7.5, 0.15 M NaCl was mixed with an equal volume of the reservoir solution, 15% PEG 8000, 0.1 M MES pH 6.5, 0.2 M sodium acetate. Crystals were flash frozen in liquid nitrogen in reservoir solution supplemented with 20% glycerol. Crystallographic data were collected at 100 K. Crystals diffracted to 2.89 Å resolution and belonged to space group C2, with four molecules per asymmetric unit. See Table 1 for crystallographic data collection statistics.
Table 1.
Crystallographic data collection and refinement statistics.
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 152.4, 71.4, 68.0 |
| β (°) | 110.1 |
| Resolution (Å)a | 50.0-2.89 (2.94 - 2.89) |
| Rmergea (%) | 10.4 (58.8) |
| I/σIa | 11.9 (1.8) |
| Completeness (%)a | 99.7 (95.5) |
| Redundancya | 3.5 (3.3) |
| Refinement and model quality | |
| Resolution range (Å) | 50.0 - 2.89 |
| No. reflections (working set) | 14690 |
| No. reflections (test set) | 775 |
| Rwork, Rfree | 23.5, 24.8 |
| No. atoms | |
| Protein | 2626 |
| Water | 23 |
| Average B-factors (residual after TLS refinementb) | 72.7 |
| Protein (Å2) | 73.7 |
| Water (Å2) | 64.5 |
| RMSc deviations from ideal values | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.9 |
| Ramachandran plot | |
| Favored regions (%) | 100.0 |
| Outliers (%) | 0.0 |
Highest resolution shell (2.94 – 2.89 Å) is shown in parentheses
See PDB entry for TLS refinement parameters.
RMS, root mean square
Rfree, Rwork with 5% of Fobs sequestered before refinement
Structure determination of LRRFIP1-CC
The structure of LRRFIP1-CC was determined by molecular replacement with Phaser (McCoy et al., 2007) using chains A and B of spindle pole body component SPC42 (Protein Data Bank code 2Q6Q) as the search model. Notably, SPC42 shares only 12% sequence identity with LRRFIP1-CC. Two SPC42 coiled-coil heterodimers were placed into the LRRFIP1-CC asymmetric unit. After refinement of the resulting solution with REFMAC5 (Murshudov et al., 1997), Rfree was 41%. Initial difference maps showed additional density near the N-terminus, and a composite omit map revealed some side chain features of LRRFIP1-CC. However, the register of the LRRFIP1-CC amino acid sequence remained ambiguous. To determine the register of the coiled coil, the amino acid sequence was aligned to a heptad repeat sequence to identify the putative hydrophobic leucine zipper characteristic of coiled coil proteins with hydrophobic residues occupying positions a and d, and acidic and basic residues at positions e and g, respectively, to form electrostatic interactions (Table 2). Crystal packing constrained the relative locations of these residue pairs such that only two models could be generated using COOT (Emsley and Cowtan, 2004) with the sequence of LRRFIP1-CC threaded into the molecular replacement solution in different registers. The atomic coordinates and temperature factors of the two models were refined with REFMAC5, initially with non-crystallographic symmetry restraints applied to all four subunits. Only one of the two models showed improvement in Rwork and Rfree during refinement. The better model was further improved with cycles of positional refinement followed by model building using B-factor sharpening (set to −15 Å2) in COOT to emphasize high-resolution data and improve electron density. α-helical restraints were maintained throughout model building using the “Regularization and Refinement Parameters” dialog in COOT. Rigid-body motions of the four subunits were modeled with REFMAC5 in terms of TLS tensors for translation, libration, and screw-rotation (Winn et al., 2001). Solvent molecules were added with the automated solvent building module of ARP/wARP (Langer et al., 2008), over five ARP/REFMAC refinement cycles with the geometry weighted at 0.01. See Table 1 for complete data collection and refinement statistics.
Table 2.
Alignment of the heptad repeats within the LRRFIP1-CC amino acid sequence (162-MDSLAEVEEK YKKAMVSNAQ LDNEKTNFMY QVDTLKDMLL ELEEQLAESR RQYEEKNKEF EREKHAHSIL QFQFAEVKEA LKQREEMLEK-249).
| a | b | c | d | e | f | g |
|---|---|---|---|---|---|---|
| D162 | S163 | |||||
| L164 | A165 | E166 | V167 | E168 | E169 | K107 |
| Y171 | K172 | K173 | A174 | M175 | V176 | S177 |
| N178 | A179 | Q180 | L181 | D182 | N183 | E184 |
| K185 | T186 | N187 | F188 | M189 | Y190 | Q191 |
| V192 | D193 | T194 | L195 | K196 | D197 | M198 |
| L199 | L200 | E201 | L202 | E203 | E204 | Q205 |
| L206 | A207 | E208 | S209 | R210 | R211 | Q212 |
| Y213 | E214 | E215 | K216 | N217 | K218 | E219 |
| F220 | E221 | R222 | E223 | K224 | H225 | A226 |
| H227 | S228 | I229 | L230 | Q231 | F232 | Q233 |
| F234 | A235 | E236 | V237 | K238 | E239 | A240 |
| L241 | K242 | Q243 | R244 | E245 | E246 | M247 |
| L248 | E249 |
Results and Discussion
Overall structure of LRRFIP1-CC
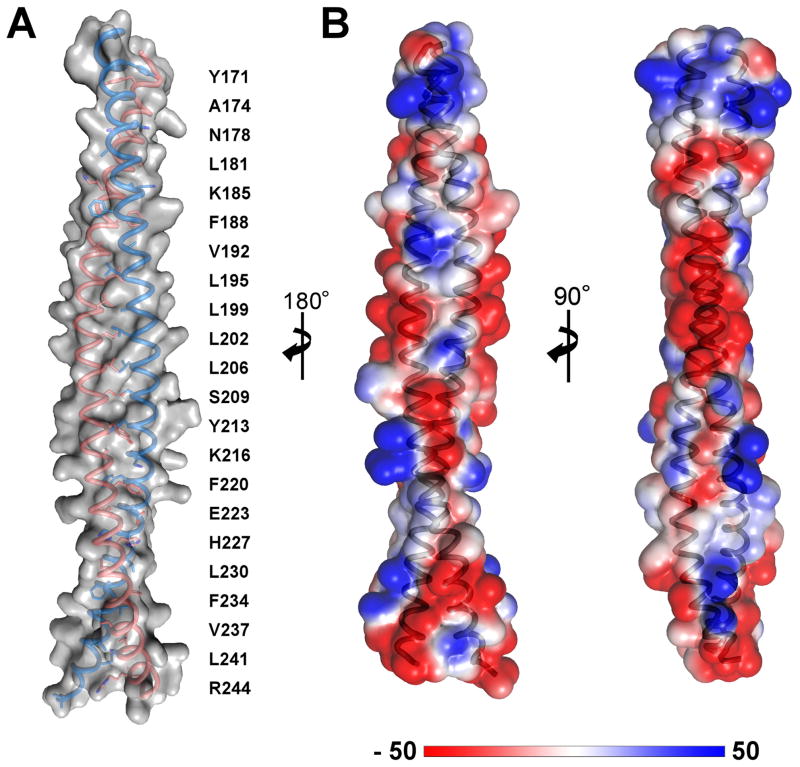
The structure of LRRFIP1-CC at 2.89 Å resolution reveals a classic parallel, homodimeric coiled coil with a total length of 122 Å (Fig. 1B, C). The crystals belonged to space group C2, with four molecules (two coiled coil dimers) in the asymmetric unit. The atomic models of chains A and B span residues 168–248 and 167–249, respectively. Chains C and D span residues 170–243 due to some disorder at the N- and C-termini. The dimer formed by chains A and B contains ten complete heptad repeats and 22 helical turns and a left-handed superhelical twist (Fig. 2A; Table 2). The buried surface area in the A–B dimer is approximately 4600 Å2. No stutters or packing defects were observed along the coil, resulting in a conformation with presumably only limited flexibility. Indeed, the root mean square deviation (rmsd) of Cα positions between the four chains in the asymmetric unit is between 0.33 and 0.87 Å, consistent with a rigid structure for LRRFIP1-CC.
Fig. 2.
Organization and physicochemical properties of the LRRFIP1-CC dimer. (A) The dimer contains ten complete heptad repeats with 22 helical turns. The residues along the hydrophobic leucine zipper (positions a and d of the coiled coil- see Table 2) are indicated. Chains A and B are shown. (B) Electrostatic surface potential of the LRRFIP1-CC dimer. The surface has mostly negative electrostatic surface potential, particularly in the central portion of the dimer. Electrostatic surface potential calculations were performed with the PyMOL plugin APBS (Baker et al., 2001) assuming physiological ionic strength. The units in the color bar are kB T ec−1, where T = 310 K, kB is the Boltzmann constant, and ec is the charge of an electron.
We note that the molecular surface of LRRFIP1-CC contains a patch with negative electrostatic potential. The negatively charged surface spans approximately 225 Å2 per monomer and is due to clustering of several acidic aspartate and glutamate residues (Asp193, Asp197, Glu201, Glu203, Glu204, Glu208) in the central region of each monomer (Fig. 2B). In the absence of any structural information it is unknown whether FLI contains a complementary positive surface.
LRRFIP1-CC eluted from a size-exclusion column with the elution volume of a 50 kDa globular protein, suggesting that LRRFIP1-CC may be an oligomer in solution (Fig. 3A). To determine the oligomeric state of LRRFIP1-CC, we performed chemical crosslinking with EGS and multi-angle laser light scattering (MALS). Crosslinking resulted in a single clear shift in the electrophoretic mobility of LRRFIP1-CC. The amount of light scattering from the protein corresponded to a 25 kDa molecular mass, consistent with a dimeric state in solution (Fig. 3B–C). The circular dichroism spectrum of LRRFIP1-CC contained two local minima, at 208 nm and 222 nm, consistent with an entirely α-helical secondary structure (Fig. 3D). Together, the biophysical data for LRRFIP1-CC in solution show as expected that the coiled coil homodimer observed in the crystal is stable in solution.
Fig. 3.
Biophysical parameters of LRRFIP1 in solution. (A) LRRFIP1-CC elutes from a Superdex 200 (10/300) size-exclusion column as a monodisperse peak with an elution volume corresponding to a 50-kDa globular protein. (B) Chemical crosslinking of LRRFIP1-CC results in a shift from monomer to dimer with increasing concentration of EGS crosslinker. (C) Multi-angle light scattering (MALS) confirms that LRRFIP1-CC is a 25-kDa homodimer in solution. (D) The circular dichroism spectrum of LRRFIP1-CC contains two local minima, at 208 and 222 nm, consistent with mostly α-helical secondary structure. (E) EMSA of increasing concentrations of LRRFIP1-DBD mixed with 1 μM TAR38 DNA. The amount of DNA bound increases with increasing concentrations of LRRFIP1-DBD based on staining with SYBR green (top). An additional band in the well suggests that higher concentrations of LRRFIP1-DBD may induce oligomerization or aggregation in the presence of DNA. Coomassie staining (bottom) shows a shift in mobility of LRRFIP1-DBD in the presence of DNA. (F) DLS measurements of LRRFIP1-DBD in the presence of DNA. Addition of TAR38 DNA increases the hydrodynamic radius of LRRFIP1-DBD/DNA complex, indicating that DNA binding does not increase foldedness or compaction of the protein.
Biophysical properties of the LRRFIP1 DNA binding domain in the presence and absence of DNA
Dimerization of LRRFIP1 through the coiled coil domain may play an important role in determining the oligomeric state of full-length LRRFIP1 during signaling. As a first step towards testing this hypothesis, we used MALS to determine the oligomeric state of the C-terminal DNA-binding domain (DBD) alone (LRRFIP1-DBD) and in the context of the coiled coil domain (LRRFIP1-CC-DBD). LRRFIP1-DBD was monomeric, despite eluting from the size-exclusion column as a 250-kDa globular protein (Fig. S1A–B), suggesting that the DBD is elongated, flexible or disordered. SAXS analysis confirmed that LRRFIP1-DBD is mostly unfolded or has a very elongated shape (Fig. S2A), and circular dichroism (CD) measurements showed that LRRFIP1-DBD has very little secondary structure (Fig. S2B). LRRFIP1-CC-DBD was also unfolded and/or elongated (Fig. S2A) but was unexpectedly mostly oligomeric, or a mixture of oligomeric species, with an average molecular mass of 220 kDa based on MALS. A fraction of the protein formed large soluble aggregates, which had a molecular weight of approximately 1 MDa and eluted in the void volume of the size-exclusion column (Fig. S1C–D).
To determine whether LRRFIP1-DBD, despite being unfolded, can bind DNA we performed an electrophoretic mobility gel shift assay (EMSA) under non-denaturing conditions. The electrophoretic mobility of a 38-nucleotide DNA hairpin (TAR38) was shifted in the presence of increasing concentrations of LRRFIP1-DBD, consistent with a Kd value of approximately 1 μM (Fig. 3E). CD measurements showed that DNA binding did not result in additional secondary structure content in LRRFIP1-DBD (Fig. S2B). Moreover, dynamic light scattering data show that the hydrodynamic radius of LRRFIP1-DBD/DNA complexes increased as TAR38 was titrated in, reaching up to 11.2 nm compared to 2.4 nm for TAR38 DNA alone and 6.5 nm for unliganded LRRFIP1-DBD (Fig. 3F). Even with an extended polymer model for LRRFIP1-DBD, multiple copies of the protein must be present in the DNA-bound complex to account for the 11.2-nm hydrodynamic radius. Thus, the DLS data suggest that LRRFIP1 binds DNA in an extended conformation lacking secondary structure, and that LRRFIP1-DBD forms higher order oligomers upon binding TAR38 DNA. Together these data suggest that the coiled coil domain induces assembly of LRRFIP1 into higher order oligomers in the context of the DNA binding domain, which binds DNA in an extended conformation.
Crystal contacts between LRRFIP1-CC dimers
The two LRRFIP1-CC dimers in the crystallographic asymmetric unit pack against each other, forming an extensive packing interface with a buried surface area of 2,260 Å2 (Fig. S3A). The energy stored at this interface was calculated with PISA (Krissinel and Henrick, 2007) at 13.7 kcal/mol. The interface is stabilized by a hydrogen-bonding network involving Glu203 and Arg211, which each contribute four hydrogen bonds across dimer pairs (Fig. S3B–C). In chains B and C, Glu203 forms hydrogen bonds with the amide moiety and carbonyl backbone of Gln191, as well as the hydroxyl group of Thr194 in chains D and A, respectively. Arg211 is stabilized by the amide side chains of Asn183 and Asn187, and by the hydroxyl group of Thr187. This network of hydrogen bonds surrounds a narrow, slightly hydrophobic pocket that includes Tyr190 and Ala207. Notably, this pocket is only sufficiently large to accommodate Ala, Ser, or Cys side chains, confirming that our atomic model was built with the correct heptad register. If the register were off by 3, 4, or 7 residues in either direction, the pocket would have to accommodate Glu, Glu or Leu (upstream), or Arg, Arg, or Glu (downstream), respectively. None of these residues types would fit into the pocket without steric clashes.
Structural similarity to other coiled coil proteins
The Dali server (Holm et al., 2008) was used to search for proteins with structural similarity to LRRFIP1-CC. Interestingly, the structures with the ten highest similarity scores, with a Z-score of 6.8–7.0, included two nucleic acid binding proteins, Saccharomyces cerevisiae Sir4 (silent information regulator 4) and Arabidopsis Mom1 helicase. Sir4 and Mom1 are large proteins, with 1,358 and 2,001 amino acids respectively. Like LRRFIP1, their coiled coil domains impose dimerization but are not responsible for nucleic acid binding. Despite being twenty amino acids shorter than LRRFIP1-CC, the C-terminal coiled coil domain of Sir4 can accommodate binding to multiple partners including Sir2, Sir3 and histones (Chang et al., 2003). Notably, the dimerization of Sir4 has been proposed to facilitate formation of chromatin by polyvalent interactions of the Sir proteins with DNA. Moreover, the region immediately N-terminal to the Sir4 coiled coil domain appears disordered and flexible, is readily proteolysed, and is thought to play a role in regulating the interactions of Sir4 with its partners (Chang et al., 2003). Based on these parallels between Sir4 and LRRFIP1, we speculate that the dimerization imposed by the LRRFIP1 coiled coil may contribute to signaling amplification by promoting divalent or multivalent interactions with DNA ligands.
Mom1 is, like Sir4, a silencing gene whose dimerization is important for transcriptional suppression, but in contrast to Sir4 or LRRFIP1, the coiled coil of Mom1 is antiparallel (Nishimura et al., 2012). Other structures similar to LRRFIP1-CC include three actin-binding proteins (Drosophila Shroom, scallop myosin heavy chain and chicken α-tropomyosin), and two SNARE membrane fusion proteins (rat VAMP-2 and mouse VAMP-4). The SD2 domain from Shroom (PDB entry 3THF) forms a three-segment antiparallel coiled coil. The “body” segment containing the central 85 residues superposes on LRRFIP1-CC with an rmsd of 2.24 Å for the Cα atoms. Myosin heavy chain (PDB entry 3BAT) and tropomyosin (PDB entry 1IC2) contain parallel coiled coils that superpose on LRRFIP1-CC with Cα-atom rmsd values of 1.85 and 2.18 Å, respectively. There are no clear functional parallels between LRRFIP1 and Shroom, myosins or SNAREs.
Conclusions
The strict sequence conservation of the coiled coil domain from human LRRFIP1 across all eukaryotes suggests that LRRFIP1-CC plays a key role in the function of LRRFIP1 as a cytosolic nucleic acid sensor. It is an emerging theme that innate immune sensors assemble into dimeric or multimeric signaling platforms to recruit and activate their downstream signaling partners (reviewed in (Berke and Modis, 2012; Botos et al., 2011; Ferrao et al., 2012). The dimeric state of LRRFIP1-CC in solution and the multimeric state of the LRRFIP1-CC-DBD construct suggest that full length LRRFIP1 also forms dimers or larger oligomers. Moreover, the acidic patches on the surface of the coiled coil homodimer could mediate the interaction of LRRFIP1-CC with FLI. Biophysical characterization of LRRFIP1-DBD in the presence of DNA suggests that LRRFIP1 remains in a highly extended random coil conformation when it binds DNA. The aggregation of LRRFIP1 at higher concentrations of DNA suggests that LRRFIP1 oligomerization may be a critical part of its signaling mechanism. Overall, the structure of LRRFIP1-CC constitutes a valuable tool for probing the mechanism of LRRFIP1 signaling by site-directed mutagenesis and for structural studies with larger LRRFIP1 constructs.
Supplementary Material
Acknowledgments
We thank Ian Berke for assistance in SAXS data collection and analysis, and William Eliason for guidance with MALS experiments. This work was supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease award to Y.M., and by NIH grant P01 GM022778-37. J.B.N was supported in part by NIH training grant T32 GM08383.
Abbreviations used
- CC
coiled-coil
- EGS
ethylene glycol bis(succinimidylsuccinate)
- LRR
leucine-rich repeat
- EM
electron microscopy
- LRRFIP1
Leucine Rich Repeat (in Flightless-1) Interacting Protein
- MALS
Multi-angle Light Scattering
- rmsd
root mean square deviation
Footnotes
Accession numbers
The atomic coordinates of LRRFIP1-CC have been deposited in the Protein Data Bank with accession code PDB 4H22.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- Berke IC, Modis Y. Structural basis of innate immune recognition of viral RNA. Cell Microbiol. 2012 doi: 10.1111/cmi.12061. In press. [DOI] [PubMed] [Google Scholar]
- Botos I, Segal DM, Davies DR. The Structural Biology of Toll-like Receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Chang JF, Hall BE, Tanny JC, Moazed D, Filman D, et al. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure. 2003;11:637–649. doi: 10.1016/s0969-2126(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Science signaling. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KS, de Couet HG. Novel proteins interacting with the leucine-rich repeat domain of human flightless-I identified by the yeast two-hybrid system. Genomics. 1999;58:146–157. doi: 10.1006/geno.1999.5817. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR. Interplay of Fli-I and FLAP1 for regulation of beta-catenin dependent transcription. Nucleic Acids Res. 2006;34:5052–5059. doi: 10.1093/nar/gkl652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Yin HL. Identification of the binding partners for flightless I, A novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J Biol Chem. 1998;273:7920–7927. doi: 10.1074/jbc.273.14.7920. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Molinard G, Petty TJ, Broger L, Gabus C, et al. Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1. PLoS genetics. 2012;8:e1002484. doi: 10.1371/journal.pgen.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, et al. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′ Phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Suriano AR, Sanford AN, Kim N, Oh M, Kennedy S, et al. GCF2/LRRFIP1 represses tumor necrosis factor alpha expression. Mol Cell Biol. 2005;25:9073–9081. doi: 10.1128/MCB.25.20.9073-9081.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SA, Brown EC, Kingsman AJ, Kingsman SM. TRIP: a novel double stranded RNA binding protein which interacts with the leucine rich repeat of flightless I. Nucleic Acids Res. 1998;26:3460–3467. doi: 10.1093/nar/26.15.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.