SUMMARY
The monoubiquitylation of histone H2B plays an important role in gene expression by contributing to the regulation of transcription elongation and mRNA processing and export. We explored additional cellular functions of this histone modification by investigating its localization to intergenic regions. H2B ubiquitylation is present in chromatin around origins of DNA replication in budding yeast, and as DNA is replicated its levels are maintained on daughter strands by the Bre1 ubiquitin ligase. In the absence of H2B ubiquitylation, the pre-replication complex is formed and activated, but replication fork progression is slowed down and the replisome becomes unstable in the presence of hydroxyurea. H2B ubiquitylation promotes the assembly or stability of nucleosomes on newly replicated DNA, and this function is postulated to contribute to fork progression and replisome stability.
INTRODUCTION
The monoubiquitylation of histone H2B (H2Bub1) has emerged as an important modification in the RNA polymerase II (Pol II) transcription cycle (Weake and Workman, 2008). The majority of H2Bub1 in chromatin is localized to gene coding regions through the co-transcriptional association of the Bre1 ubiquitin ligase and Rad6 ubiquitin conjugase with elongating Pol II (Fleming et al., 2008; Minsky et al., 2008; Schulze et al., 2009; Xiao et al., 2005). The ubiquitylation of H2B is dynamic, and cycles of ubiquitylation and deubiquitylation that occur during transcription are a prerequisite for optimal gene expression (Henry et al., 2003; Osley et al., 2006). A key role for H2B ubiquitylation is to control Pol II elongation. The absence of H2Bub1 destabilizes elongating Pol II, while the persistence of H2Bub1 inhibits recruitment of the CTK1 kinase that regulates events important for transcription elongation (Fleming et al., 2008; Wyce et al., 2007). Additional transcription-coupled roles for H2Bub1 include mRNA 3’ end formation, processing, and export, along with pre-mRNA splicing (Pirngruber et al., 2009; Shieh et al., 2011; Tomson et al., 2011; Vitaliano-Prunier et al., 2012). Finally, H2Bub1 controls the trans-methylation of histone H3 on lysines 4 and 79, two modifications that also have important roles in various aspects of transcriptional regulation (Nakanishi et al., 2009).
H2Bub1 also performs transcription-independent functions in the cell. These include roles in meiosis, the UV-induced checkpoint pathway, DNA double-strand repair, apoptosis, and the Set1-directed methylation of the kinetochore-associated protein, Dam1 (Chernikova et al., 2010; Game and Chernikova, 2009; Giannattasio et al., 2005; Latham et al., 2011; Nakamura et al., 2011; Walter et al., 2010; Yamashita et al., 2004). This raised the question of whether H2Bub1 has additional cellular functions. The similarities between DNA replication and transcription suggested that H2Bub1 could have roles in DNA replication analogous to its functions in transcription. Both processes involve the recruitment of multi-subunit polymerase complexes to specific genomic sites, followed by the movement of polymerases along the chromatin fiber. Importantly, similar patterns of nucleosome dynamics occur during transcription elongation and DNA replication. Ahead of the advancing polymerases, nucleosomes are displaced from DNA, and behind the polymerases nucleosomes are assembled to restore chromatin structure (Corpet and Almouzni, 2009; Schwabish and Struhl, 2004). Other similarities between the two processes include a functional coupling between nucleosome assembly and polymerase progression. Failure to properly reassemble nucleosomes during transcription elongation leads to destabilIzation of Pol II (Fleming et al., 2008). Similarly, the replisome becomes unstable when nucleosome assembly is defective during DNA replication (Burgess and Zhang, 2010b; Clemente-Ruiz and Prado, 2009; Groth et al., 2007; Groth et al., 2005).
In this study, we report that H2Bub1 is present in chromatin adjacent to several well-characterized origins of replication in yeast. During replication H2Bub1 levels are maintained via the association of Bre1 with newly replicated DNA. In the absence of H2Bub1, cells became sensitive to hydroxyurea, implicating this mark in the cellular response to replication stress. Analysis of the loading of key replisome factors onto origins in an htb-K123R mutant showed that the pre-replication (pre-RC) complex was assembled and activated normally. However, there was decreased association of factors required for DNA synthesis, which corresponded with a defect in progression of the replication fork and destabilization of the replisome. Histone occupancy was also impaired around replicated origins in the absence of H2Bub1, supporting the view that this histone modification promotes the stability and advancement of replication forks by regulating nucleosome assembly or stability on newly replicated DNA.
RESULTS
H2Bub1 is present in chromatin adjacent to replication origins and maintained on duplicated daughter strands
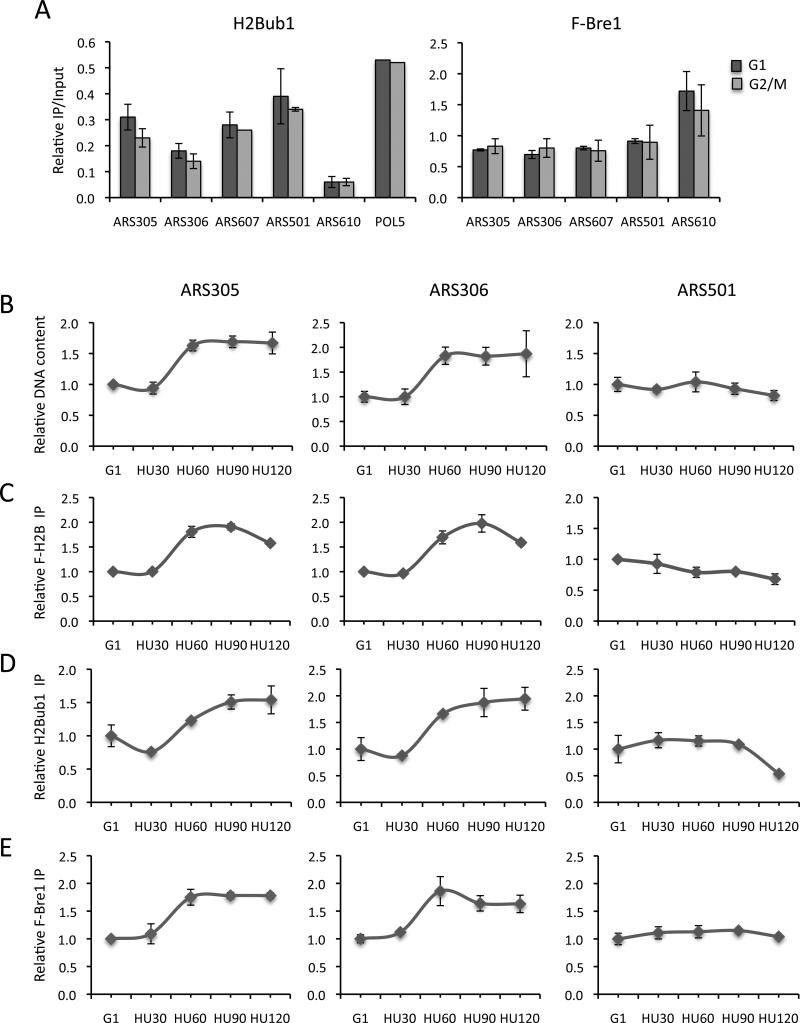
Several large scale studies have reported that H2B is ubiquitylated in nucleosomes that flank replication origins, even though these regions are ORF-free (Unnikrishnan et al., 2010; Schulze et al., 2011). To confirm the presence of H2Bub1 at origins, we used an H2Bub1 antibody (Schulze et al., 2009) in ChIP experiments with several well-characterized early (ARS305, ARS306, ARS607) and late (ARS501, ARS610) origins (Fig. 1A, left panel). H2Bub1 and histones were present at each origin and their occupancy was not significantly different between cells in G2/M and G1 phase (Fig. S1B). Although the levels of H2Bub1 were generally lower at origins than on transcribed genes, they were significantly higher than those at the transcriptionally silent HMRa locus (Fig. 1A, POL5 and Fig. S1A, left panel) (Shieh et al., 2011). The one exception was the telomere-proximal late origin, ARS610, which had very low levels of H2Bub1.
Figure 1. H2Bub1 is maintained on replicating DNA.
ChIP analysis of H2Bub1 and Flag-Bre1 at selected replication origins in log phase cells. A. H2Bub1 (left panel) and Flag-Bre1 (right panel) levels in G2/M and G1 arrested cells. B-E. G1 cells arrested with α-factor were released into 0.2M hydroxyurea (HU). Samples were analyzed by qPCR for the levels of origin-proximal DNA (B) or by ChIP for the accumulation of Flag-H2B (C), H2Bub1 (D) and Flag-Bre1 (E) at two early (ARS305 and ARS306) and one late (ARS501) origin. Error bars indicate the standard deviation from 2-3 independent experiments (A, C, D, E) or three technical replicates (B). In all cases, IP signals at ARS sequences were normalized to IP signals at the ASI1 locus. In panel A, relative IPs were further normalized to input DNA. In panels B-E, IP levels were normalized to G1 IP levels, which were set as 1.
Similar to H2Bub1, Bre1 was present at the same early and late origins in both G2/M and G1 phase cells (Fig. 1A, right panel). The origin-associated levels of Bre1 were also equivalent between the two cell cycle stages and comparable to Bre1 levels at POL5, a moderately transcribed gene (Fig. S1A, right panel). Interestingly, Bre1 levels were highest at ARS610, which had the lowest levels of H2Bub1, suggesting that the ligase might have another function at this origin (Fig. 1A). Together, the data indicate that both Bre1 and H2Bub1 are associated with replication origins.
The similar levels of these factors at origins in G1 and G2/M cells suggested that the modification is maintained after origins are fired. To follow the levels of H2Bub1 at origins during DNA replication, we arrested wild type cells in G1 with α-factor and then released the cells into media containing 0.2M hydroxyurea (HU). Cells enter S phase in the presence of HU and DNA synthesis proceeds from activated early origins at a reduced rate for several kilobases before replication forks stall. Two early (ARS305, ARS306) and one late (ARS501) origins were first analyzed for DNA content and H2B levels during release of cells into HU. The levels of both DNA and H2B increased ~2-fold at the early origins around 60 min after the release of cells into HU, reflecting the duplication of DNA and chromatin at these elements (Fig. 1B, C). There was no change in either DNA or H2B levels at the late origin because of activation of the intra-S phase checkpoint (Fig. 1B, C)(Santocanale and Diffley, 1998).
The levels of H2Bub1 and Bre1 also increased ~2- fold exclusively at early origins, with the increase in Bre1 occupancy slightly preceded the accumulation of H2Bub1 (Fig. 1D, E and Fig. S1D, E; data not shown). Because the levels of H2Bub1 and Bre1 increased in direct proportion to the amount of DNA replicated, H2Bub1 and Bre1 must be present on both daughter strands (Fig. S1C). When cells entered S phase in the presence of HU, Bre1 and H2Bub1 accumulated at regions several Kb distal to origins (Fig. S1D, E). This suggested that H2Bub1 is maintained at origins through the association of Bre1 with replicating chromatin. Bre1 is recruited to regions of active transcription and travels with elongating Pol II (Kim and Roeder, 2009; Xiao et al., 2005). However, the pattern of Bre1 accumulation was not consistent with Bre1 traveling with the replisome.
Cells are sensitive to hydroxyurea in the absence of H2B ubiquitylation
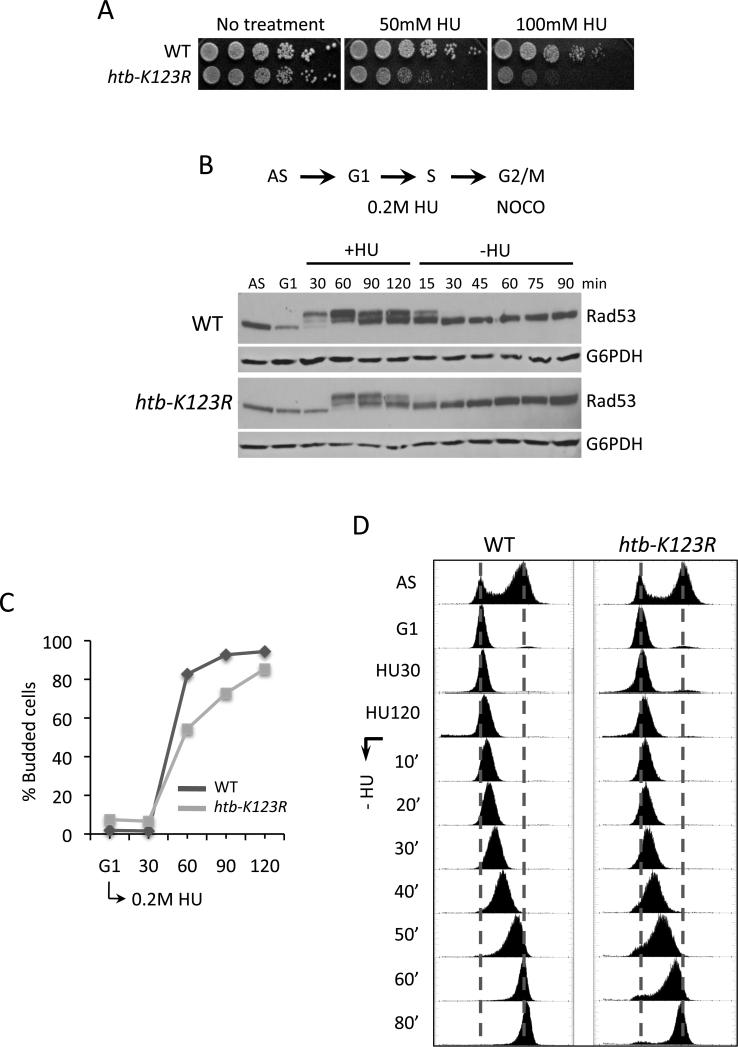
To investigate the functional significance of H2Bub1 at origins, we examined the sensitivity of htb-K123R cells to agents that inhibit DNA replication. An htb-K123R mutant was hypersensitive to HU but mildly sensitive to methyl methane sulfonate or camptothecin (Fig. 2A and Fig. S2A, B). The HU sensitivity conferred by htb-K123R was found in different genetic backgrounds and as well as in a bre1Δ mutant (Fig. S2A). Importantly, hht-K4A and hht-K79A mutants, alone or in combination, were significantly less sensitive to HU than an htb-K123R mutant (Fig. S2A). This indicates that the HU-sensitivity of htb-K123R is not a consequence of the H2Bub1-dependent trans-methylation of H3 by Set1 and Dot1 (Nakanishi et al., 2009).
Figure 2. htb-K123R cells are sensitive to hydroxyurea and show a delay in S phase entry.
A. Wild type (WT) and htb-K123R cells were serially diluted and spotted onto YPD plates containing HU, and photographed after 3 days. B. Rad53 phosphorylation was analyzed by western blot analysis of TCA extracts using a polyclonal Rad53 antibody. G6PDH served as a loading control. C, D. Cells were arrested in G1 with α-factor, released into 0.2M HU for 120 minutes, and then incubated in medium without HU but with 20μg/ml nocodazole. The percentage of budded cells was determined by microscopic examination (C), and DNA content was measured by FACS (D).
We asked if the HU sensitivity of an htb-K123R mutant represented a defect in the intra-S phase checkpoint by monitoring phosphorylation of the Rad53 kinase. Rad53 phosphorylation is triggered by agents that block fork progression to stabilize stalled replisomes adjacent to active origins and prevent the firing of late origins (Branzei and Foiani, 2006; Segurado and Tercero, 2009). We followed Rad53 phosphorylation by western blot analysis after cells were released from G1 into HU (Fig. 2B). In wild type cells phospho-Rad53 appeared ~30 minutes after release into HU, indicating activation of the intra-S checkpoint. In the htb-K123R mutant phospho-Rad53 was not detected until 60 minutes after release. The delayed activation of the checkpoint in these cells likely represents the delayed entry of the mutant into S phase, as discussed below.
When the HU block was removed, phospho-Rad53 was lost in both wild type and htb-K123R cells, indicating deactivation of the checkpoint (Fig. 2B). Phospho-Rad53 disappeared more rapidly in the mutant and the levels of phospho-Rad53 were also lower during HU. The diminished levels of phospho-Rad53 in htb-K123R cells likely reflect the reduced levels of ssDNA and RPA at origins, as shown in later results (Fig. 5D; Fig. S7B). However, there was no evidence for DNA replication at late origins (Fig. 1B and data not shown) and htb-K123R cells did not prematurely release from the HU-induced S phase block as measured by FACS (Fig. 2D). Additionally, the checkpoint responsive genes RNR2, RNR3, and RNR4 were upregulated in both wild type and mutant cells during HU (data not shown). Together, the data indicate that the intra-S phase checkpoint remained fully active in htb-K123R cells throughout the entire HU time-course
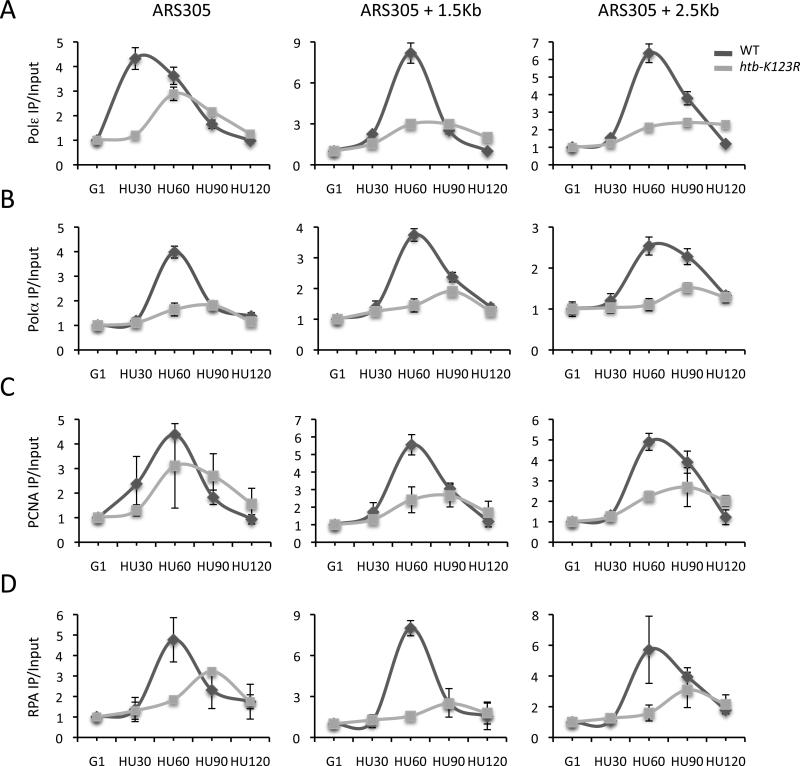
Figure 5. Leading and lagging strand factors have reduced origin association in the absence of H2Bub1.
ChIP analysis of factors mediating leading and lagging strand DNA synthesis as described in Fig. 4 legend. A. DNA Polε; B. DNA Polα; C. PCNA; D. RPA. Error bars represent the standard deviation between two independent experiments (C, D) or from three technical replicates (A, B). IP signals were normalized as described in Fig. 4 legend.
DNA replication is initiated in the absence of H2B ubiquitylation
To investigate the point in the DNA replication cycle that was sensitive to the absence of H2Bub1, we arrested cells in G1, followed by release into HU. S phase entry was monitored by bud formation, which occurs co-incidentally with the initiation of DNA synthesis (Fig. 2C). Multiple experiments showed that both htb-K123R and bre1Δ cells had an approximately 15-20 minute delay in bud formation compared to wild type (Fig. S3B and data not shown). We attributed this delayed entry into S phase to the reduced expression of several cyclin genes (CLN1, CLN2, and CLB5) that are important for the G1 to S phase transition (Fig. S2C)(Bloom and Cross, 2007). A cell cycle delay also occurs in rad6Δ and bre1Δ cells and has been ascribed to a role of Rad6 and Bre1 in transcriptional regulation of cyclin genes (Zimmermann et al., 2011). Despite the transcriptional defect, htb-K123R cells eventually budded and entered S phase, generally with a 15-20 minute delay, consistent with the delayed activation of the intra-S phase checkpoint.
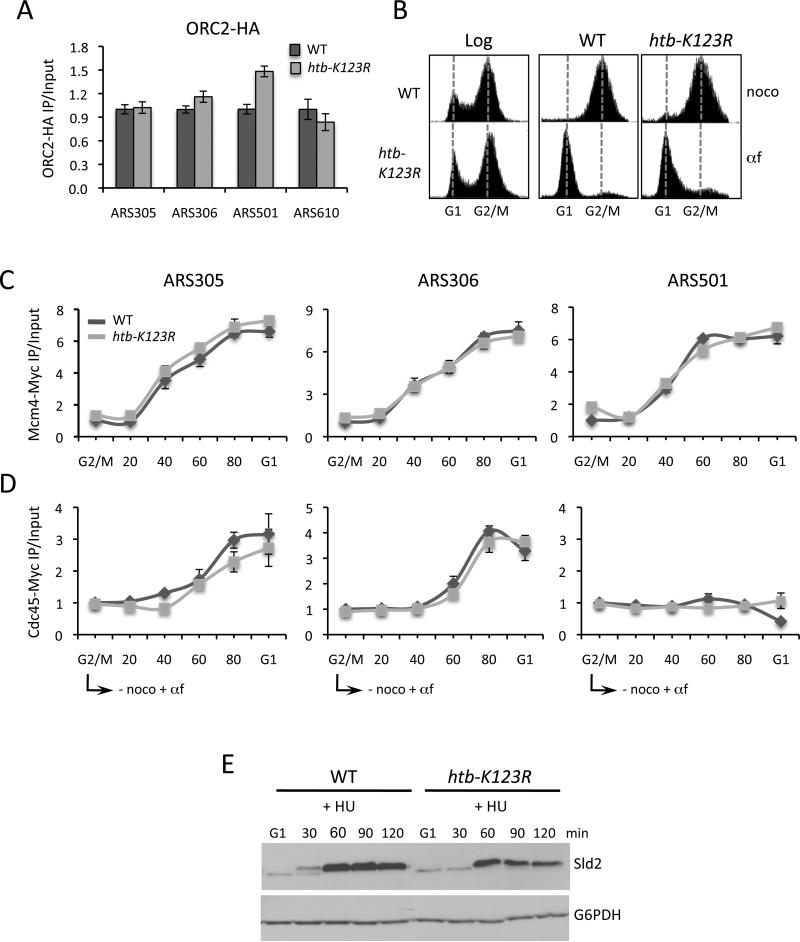
In budding yeast, the Origin Recognition Complex (ORC) is present at both early and late origins throughout the cell cycle (Toone et al., 1997). During G1, Cdc6 and Cdt1 bind to ORC and coordinate the loading of the MCM helicase, which together with ORC forms the pre-replication complex (pre-RC) (Lei and Tye, 2001; Takara and Bell, 2011). MCM binding completes pre-RC licensing, making it competent for firing when cells enter S phase. To investigate if the delayed initiation of S phase in htb-K123R cells was due to a defect in the establishment of the pre-RC, we first measured Orc2-HA occupancy at origins. Orc2 was present at equivalent levels at origins in wild type and htb-K123R cells, indicating that H2Bub1 was dispensable for its loading (Fig. 3A). We then monitored the kinetics of Mcm4-Myc recruitment to origins as cells progressed synchronously from G2/M phase to G1 phase (Fig. 3B). Mcm4 was recruited to both early and late origins in htb-K123R cells with similar kinetics and to the same levels as in wild type cells (Fig. 3C and Fig. S4A).
Figure 3. The pre-RC is assembled and activated in the absence of H2Bub1.
A. ChIP analysis of Orc2-HA levels at four replication origins in log phase cells. Error bars represent the standard deviation between two independent experiments. B. FACS analysis of DNA content in log phase cells, nocodazole-arrested cells (G2/M), or nocodazole-arrested cells released into α-factor for 100 minutes (G1). The FACS data correspond to the ChIP data in C and D. C, D. Mcm4-Myc (C) and Cdc45-Myc (D) occupancy at origins in cells released from nocodazole arrest into media supplemented with α-factor. Error bars represent the standard deviation from three technical replicates. E. CDK-dependent phosphorylation of Sld2 was monitored by western blot analysis of TCA lysates from cells arrested in G1 and released into 0.2M HU. G6PDH was used as a loading control. The ChIP data in panels A, C, D represent IP values for each ARS relative to the IP value at the ASI1 ORF, and then corrected for the ratios of amplification using input DNA. The htb-K123R values are represented relative to WT values in asynchronous cells (A) or G2/M arrested cells (C, D).
The pre-RC is primed for release of MCM at the G1 to S transition after the recruitment of Cdc45 and the CDK-dependent phosphorylation of Sld2 and other factors (Aparicio et al., 2006; Araki, 2010). Cdc45 associates with the MCM complex exclusively at early origins (Heller et al., 2011; Labib, 2010). To investigate if H2Bub1 was required for Cdc45 recruitment to these origins, Cdc45 occupancy was measured by ChIP as cells were released from G2/M into G1 (Fig. 3D and Fig. S4B). Cdc45 was present at equivalent levels at early origins in both wild type and htb-K123R cells, but absent from late origins. We then monitored the phosphorylation of Sld2 in both strains. Sld2 interacts with a complex that includes Dpb11, GINS, and DNA polymerase epsilon (Polε) to form the pre-loading complex (pre-LC)(Araki, 2010). The association of this complex with the pre-RC is thought to complete its activation to initiate DNA synthesis (MacNeill, 2010). Because htb-K123R cells have decreased expression of several G1 cyclin genes whose products form the active CDK, it was possible that reduced CDK activity accounted for the delayed entry of the mutant into S phase. Western blot analysis revealed a shift in the apparent molecular weight of Sld2 in both wild type and htb-K123R cells when they entered S phase in the presence of HU (Fig. 3E). This shift could be reversed by phosphatase treatment and was consistent with Sld2 phosphorylation (Fig. S4C). The ~30 minute delay in the appearance of phospho-Sld2 in htb-K123R cells again correlated with delayed entry of these cells into S phase (Fig. 3E). Thus, the key factors required for the initiation of DNA synthesis are present and activated in the absence of H2Bub1.
Cells lacking H2B ubiquitylation show a defect in S phase progression
The maintenance of H2Bub1 on newly replicated chromatin suggested that the mark plays a role in the progression of S phase. To examine this possibility, cells were arrested in G1, allowed to initiate S-phase in the presence of HU, and then released into media without HU but containing nocodazole. This resulted in the passage of cells through a single S phase and excluded effects on DNA replication resulting from the delayed entry of htb-K123R cells into S phase. FACS analysis revealed a delay in the completion of DNA synthesis in htb-K123R cells after HU was removed (Fig. 2D). Whereas wild type cells completed DNA synthesis ~60 minutes after release from HU, it took about 20 minutes longer for mutant cells to duplicate their genome. We confirmed the S phase kinetics by measuring the global levels of the S phase marker, H3K56ac, and the G2/M marker, Clb2, in the same experiment. In wild type cells maximum levels of H3K56ac occurred ~60 minutes post-HU release (Fig. S3A), coinciding with the completion of DNA synthesis as measured by FACS (Fig. 2D). By 80 minutes, H3K56ac levels were significantly decreased and Clb2 had accumulated, indicating that cells had reached G2 phase (Fig. S3A). In contrast, in htb-K123R cells H3K56ac was present for a longer period of time and not significantly turned over until 100 minutes post-HU release, around the same time that Clb2 accumulated (Fig. S3A). Together, the data indicate that H2Bub1 is important for progression of S phase after removal of HU-induced replication stress.
To investigate whether H2Bub1 played a role in S phase progression in an unperturbed cell cycle, we monitored DNA content by FACS when cells were released from G1 in the absence of HU. Bud counts confirmed that htb-K123R cells again had a slower (~15 min) entry into S phase compared to wild type cells (Fig. S3B). After accounting for the delay in S phase initiation, S phase was ~15 min longer in the mutant (Fig. S3C). Thus, the data suggest that H2Bub1 has a role in normal S phase progression.
H2B ubiquitylation promotes replisome stability during a G1 to HU shift
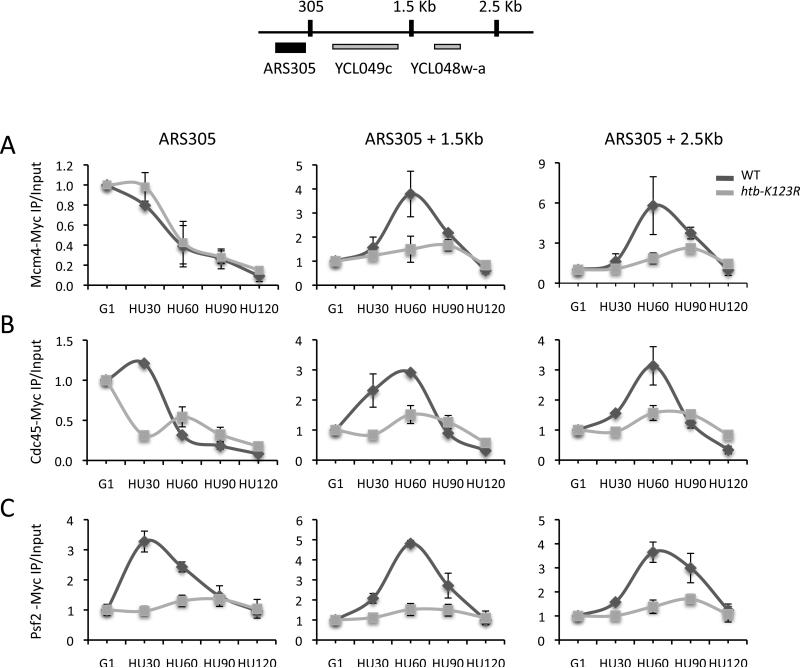
The slower progression of htb-K123R cells through S phase could result from the instability of the replisome at stalled replication forks. To test this hypothesis, we used ChIP to monitor the association of key replisome components with two early origins during a G1 to HU shift. Mcm4 and Cdc45 were recruited to similar levels at these origins in both wild type and htb-K123R cells during G1 (Fig. 3C, D). The amount of Mcm4 and Cdc45 at origins decreased when these cells were released from G1 into HU (Fig. 4A, B and Fig. S5A, B). In wild type cells this decrease was due to the movement of both factors away from the origin to positions several kilobases distal (Fig. 4A, B and Fig. S5A, B). However, in htb-K123R cells only a small fraction of Mcm4 and Cdc45 redistributed downstream of the origin (Fig. 4A, B and Fig. S5A, B). These data suggest that in mutant cells a significant fraction of Mcm4 and Cdc45 dissociated from DNA as cells entered S phase, leading to fewer stable replisomes at origin-distal positions.
Figure 4. H2Bub1 is important for replisome stability in the presence of HU.
ChIP analysis of selected replisome factors at ARS305 and two ARS305-distal positions (1.5 Kb and 2.5 Kb) after the release of cells from G1 into 0.2M HU. A. Mcm4-Myc. B. Cdc45-Myc. C. Psf2-Myc. Error bars represent the standard deviation from two independent experiments (A, B) or from three technical replicates (C). The ChIP data represent IP values for each ARS relative to the IP value at the ASI1 ORF, and then corrected for the ratios of amplification using input DNA. The data were further normalized to G1 values, which were set as 1.
Next, we monitored factors that associate with origins only after activation of the pre-RC (Gambus et al., 2009; MacNeill, 2010). We first measured the occupancy of Psf2, a component of the GINS complex that is important for both the establishment of the replication fork at origins and fork progression (Aparicio et al., 2006; Kanemaki and Labib, 2006; Labib and Gambus, 2007). In wild type cells Psf2 specifically associated with origins as cells entered S phase and then migrated to adjacent sequences with the advancing fork (Fig. 4C and Fig. S5C). In contrast, only very low levels of Psf2 associated with these same origins in htb-K123R cells, consistent with the instability of MCM/Cdc45 in this strain (Fig. 4C and Fig. S5C).
Factors required for leading and lagging strand DNA synthesis, including DNA Polε, DNA Polα, PCNA, and RPA, also associated with origins and then redistributed to origin-distal positions when wild type cells entered S phase (Fig. 5A-D and Fig. S6A-D). However, in htb-K123R cells Polα, Polε, and RPA associated inefficiently with origins and did not accumulate to significant levels at positions downstream of origins (Fig. 5A, B, D and Fig. S6A, B, D). This phenotype appeared to be somewhat origin specific. While Polε was present at low levels at both ARS305 and ARS306 (Fig. 5A), it accumulated to levels similar to those in wild type cells at ARS607 (Fig. S6A). Importantly, these replication factors were not present at the ARS501 late origin in either wild type or mutant cells, indicating that the intra-S phase checkpoint remained functional (Fig. S6A-D). Moreover, the reduced occupancy of replisome factors in htb-K123R cells was not the consequence of the reduced expression of genes that encode these proteins as evidenced by RT-PCR or western blot analysis (Fig. S7A and data not shown).
In contrast to Polα, PCNA occupancy at origins was not significantly lower in the htb-K123 mutant compared to the wild type strain (Fig. 5C and Fig. S6C). This phenotype is consistent with origin firing and subsequent collapse in the absence of H2Bub1 (Papamichos-Chronakis and Peterson, 2008). In support of this conclusion, reduced levels of single-stranded DNA replication intermediates (RI) were present at ARS305 in htb-K123R cells during a G1 to HU shift (Fig. S7B) and RPA occupancy was lower at this origin (Fig. 5D). The collective data suggest that H2Bub1 plays an important role in the progression and integrity of replication forks.
H2B ubiquitylation regulates histone occupancy on replicated DNA
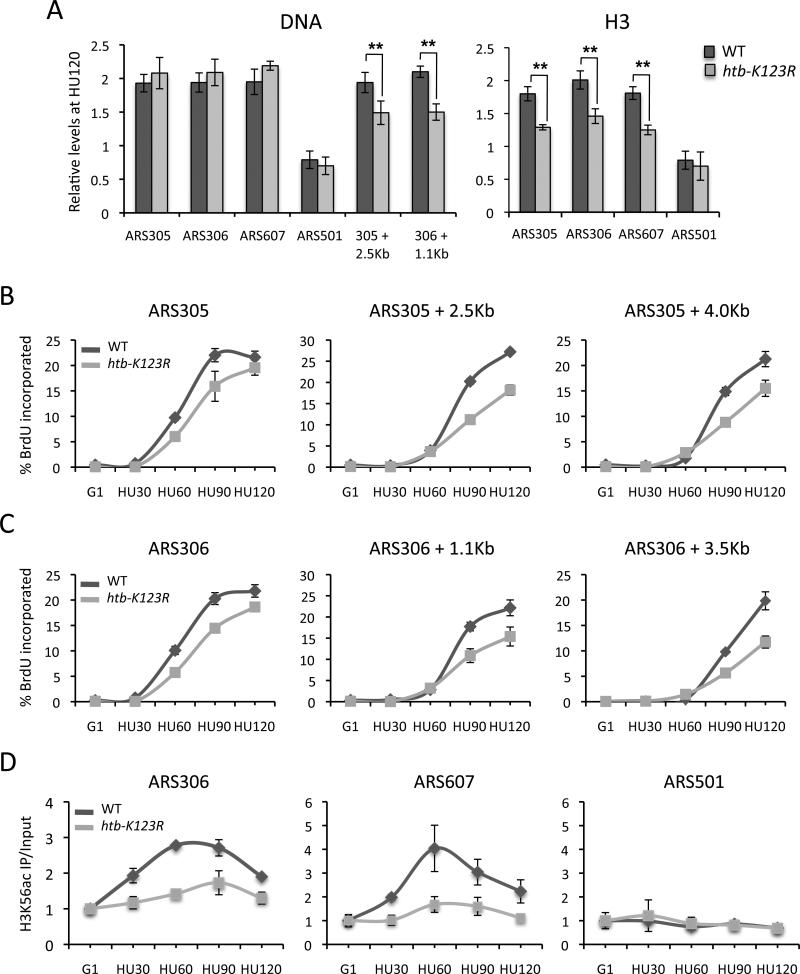
As the replisome progresses, one nucleosome is displaced in front of the fork and two nucleosomes are assembled on newly replicated DNA behind the fork (Corpet and Almouzni, 2009). Newly assembled nucleosomes are then “matured” to restore chromatin organization and stability. To investigate if H2Bub1 regulates nucleosome assembly or stability during replication, we first measured the amount of DNA replicated immediately adjacent to three early origins upon a G1 to HU shift using both a quantitative PCR assay of genomic DNA and BrdU ChIP. By 120 min after the shift, origin-proximal DNA was replicated to the same extent (~2-fold increase over G1) and with similar kinetics in both wild type and htb-K123R cells, while no replication occurred at the ARS501 late origin (Fig. 6A-C and Fig. S8A, C). However, at origin distal positions, there was a significant decrease in the amount of replicated DNA as well as a pronounced kinetic delay in replication in the htb-K123R mutant (Fig. 6A-C and Fig. S8C). These results support our previous observation that htb-K123R cells initiate replication normally but have a defect in replication fork progression that becomes apparent at origin-distal positions. Because origin-proximal sequences were replicated in the mutant, the reduced occupancy of replisome components at origins must therefore reflect the reduced stability of these factors rather than a failure to recruit them.
Figure 6. H2Bub1 plays a role in histone deposition during DNA replication.
A. DNA content at each ARS and two ARS-distal positions was quantitated by qPCR and normalized to the DNA content at ASI1 120 minutes after release of cells from G1 into HU (left panel). ChIP analysis of H3 (right panel). IP signals were normalized as described in Fig. 4 legend. Error bars represent the standard deviation from 4-6 independent experiments, and the asterices indicate p values of <0.01 as determined by Student's T-Test. B, C. Anti-BrdU ChIP at two origins and origin-distal positions. Values represent the % BrdU incorporated relative to input DNA. Error bars represent the standard deviation from three technical replicates. D. H3K56ac levels were measured by ChIP at the ARS305 and ARS607 early origins and ARS501 late origin. IP values were normalized as in Fig. 4 legend. Error bars represent the standard deviation from 3-4 independent experiments.
We then examined histone occupancy on replicated DNA by measuring H3 levels on origin-proximal DNA (Fig. 6A and Fig. S8B). The level of H3 at ARS305, ARS306, and ARS607 almost doubled in wild type cells by 120 min after a shift to HU, but remained unchanged at the ARS501 late origin. In contrast, in htb-K123R cells there was ~25% less H3 at these origins and the kinetics of H3 deposition were slower (Fig. 6A and Fig. S8B). Reduced levels of H3K56 acetylation, a mark of newly synthesized H3, were also found at early origins in mutant cells (Fig. 6D and data not shown). Importantly, total cellular levels of H3 and H3K56ac were not affected by the absence of H2Bub1 (data not shown). These data indicate that despite the duplication of sequences immediately adjacent to active origins in htb-K123R cells, substantially less H3 was present at these origins. These results are consistent with either a defect in nucleosome assembly at origins or a failure to stabilize assembled nucleosomes during chromatin maturation.
The MCM helicase contributes to fork progression by unwinding DNA and displacing nucleosomes ahead of the fork by its association with FACT, a histone chaperone and nucleosome re-organization factor (Abe et al., 2011; Han et al., 2010; Kundu et al., 2011; Tan et al., 2006; Tan et al., 2010). H2Bub1 and Spt16, the histone-binding subunit of yeast FACT, co-operate to restore nucleosome occupancy during transcription elongation. Moreover, H2Bub1 stabilizes Spt16 association with transcribing DNA (Fleming et al., 2008). We asked if H2Bub1 had similar functional interactions with Spt16 during replication. An htb-K123R spt16-197 double mutant showed enhanced HU sensitivity, consistent with overlapping roles in replication (Fig. S8E). We used ChIP to measure recruitment of Spt16 to origins during a G1 to HU shift. Spt16 was recruited to the ARS305 early origin but not to the ARS501 late origin in both the presence and absence of H2Bub1 (Fig. S8D and data not shown). In wild type cells, Spt16 was also present at two ARS305-distal positions in a pattern consistent with its transient association with the advancing replisome (Fig. S8D). However, Spt16 occupancy was significantly reduced at these same distal positions in htb-K123R cells, suggesting that H2Bub1 stabilizes Spt16 on replicating chromatin.
The HR pathway is important for DNA replication in the absence of H2B ubiquitylation
Despite the destabilization of the replisome in htb-K123R cells during exposure to HU, S phase was eventually completed when the inhibitor was removed. The homologous recombination (HR) pathway can promote the recovery of stalled or collapsed forks that lead to DNA double-strand breaks (DSBs) (Branzei and Foiani, 2006). Very low levels of the DSB-induced modification, phospho-H2A, were present at early origins during a HU shift in both wild type and htb1-K123R cells (data not shown). However, in checkpoint proficient yeast HU does not generate DSBs, and single-stranded DNA formed at stalled forks can initiate the HR pathway (Alabert et al., 2009). We investigated the role of HR in HU recovery by examining the phenotypes of wild type and htb-K123R strains in which the RAD52 gene was deleted. RAD52 encodes a key player in the HR pathway and rad52Δ cells are highly sensitive to HU. A rad52Δ htb-K123R double mutant was even more sensitive to HU than rad52Δ alone (Fig. S9A). Moreover, DNA replication was significantly slower in the double mutant compared to a rad52Δ strain when cells were released from an HU block (Fig. S9B). Together, the data suggest that the absence of H2Bub1 triggers HR-dependent processes to complete DNA synthesis as a consequence of replisome instability that occurs during HU treatment.
DISCUSSION
In this study, we present evidence directly linking H2B ubiquitylation to the regulation of DNA replication in yeast. H2Bub1 is present in chromatin adjacent to several well-characterized origins of replication, as is the H2B ubiquitin ligase, Bre1. As the replication fork advances, both H2Bub1 and Bre1 levels are maintained on newly replicated DNA. In the absence of H2Bub1, cells become sensitive to hydroxyurea, which causes replication fork stalling, and the completion of DNA synthesis occurs more slowly after removal of the HU block. This phenotype results from the instability of the MCM helicase and the reduced association of key factors that mediate DNA replication. H2Bub1 regulates the assembly or stability of nucleosomes on newly replicated DNA, and this role is postulated to promote replication fork progression and replisome stability.
Ubiquitylation of H2B at origins during DNA replication
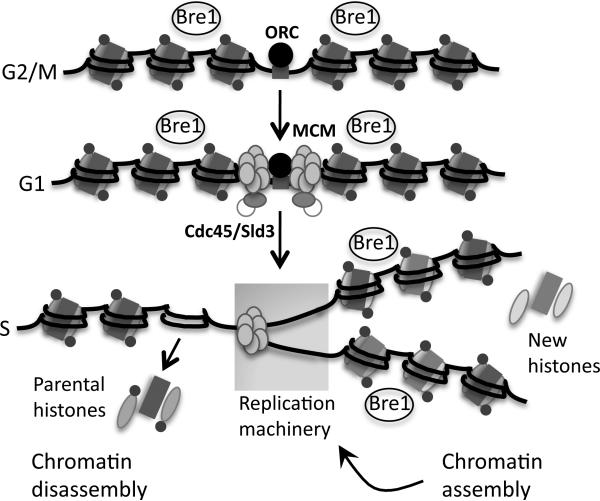
We hypothesize that during replication H2Bub1 levels are maintained on replicated DNA by the recycling of marked parental nucleosomes coupled with Bre1-dependent ubiquitylation of newly synthesized H2B deposited on DNA behind the fork (Fig. 7). Yeast replication origins are present in ORF-free regions that are flanked by well-positioned nucleosomes, raising the question of how Bre1 could be targeted to H2B in this chromatin context (Eaton et al., 2010). The pattern of Bre1 association with replicating DNA is not consistent with Bre1 traveling with the activated replisome, as it does with elongating Pol II during transcription (Xiao et al., 2005). Instead, Bre1 appears to associate with replicated DNA de novo. Bre1 interacts with oligonucleosomes in vitro and has not been reported to directly bind to DNA (Kim and Roeder, 2009). Thus, Bre1 could recognize some special feature of chromatin assembled behind the fork such as H3 or H4 acetylation, which marks newly deposited histones (Falbo and Shen, 2009). These marks turn over as chromatin matures, but Bre1 remains associated with chromatin as the fork advances. This scenario could account for both the recruitment of Bre1 to newly assembled chromatin as well as the observation that Bre1 does not actively move with the fork.
Figure 7. H2Bub1 is maintained on replicating DNA to promote efficient fork progression.
H2Bub1 (small circles) and Bre1 are present around origins throughout the cell cycle. At the onset of S phase, one nucleosome is removed in front of the advancing fork and two nucleosomes are assembled behind the fork on newly synthesized daughter strands. H2Bub1 levels are maintained around origins by the recycling of H2Bub1-marked parental nucleosomes coupled with Bre1-dependent de novo ubiquitylation of newly synthesized H2B assembled into nucleosomes behind the fork. In the absence of H2Bub1 histone occupancy is decreased at origins, reflecting a defect in nucleosome assembly or stability. The absence of H2Bub1 is postulated to impede fork progression driven by the MCM helicase.
H2B ubiquitylation promotes nucleosome assembly or stability during replication
In the absence of H2Bub1 H3 occupancy is decreased on origin-proximal DNA soon after it is replicated. This phenotype is consistent with a role for H2Bub1 in promoting nucleosome assembly during replication. Parallel pathways involving histone acetytransferases and histone chaperones deposit newly synthesized histones onto newly replicated DNA. The ASF1-Rtt109/CAF-I-Rtt106 pathway deposits H3 marked by acetylated lysine 56, while the Gcn5/CAF-I-Rtt106 pathway deposits H3 acetylated on lysine 27 and enhances H3K56ac deposition by CAF-I (Driscoll et al., 2007; Li et al., 2008; Burgess et al., 2010; Burgess and Zhang, 2010a; Burgess and Zhang, 2010b; Li et al., 2011). The reduced occupancy of H3K56ac at origins in htb-K123R cells suggests that H2Bub1 interacts with one or both pathways to regulate the deposition of newly synthesized H3. H2Bub1 might stabilize the interaction of H3K56ac with ASF1 or promote the transfer of H3K56ac to CAF-I. Alternatively, it might affect H3K27 acetylation by Gcn5 or its transfer to CAF-I. In support of a role for H2Bub1 in these assembly pathways, an asf1Δ htb-K123R double mutant is significantly more sensitive to HU than either single mutant (data not shown).
Both the ASF1 and FACT histone chaperones have been implicated in deposition of displaced parental histones behind the fork (Groth et al., 2007; Groth et al., 2005; Jasencakova and Groth, 2010; Abe et al., 2011). The reduced stability of Spt16 on replicated DNA in htb-K123R cells suggests that H2Bub1 might also affect the deposition of parental nucleosomes through similar chaperone interactions. H2Bub1 co-operates with Spt16 (FACT) to assemble nucleosomes in the wake of transcribing Pol II (Fleming et al., 2008). Strikingly, Spt16 promotes the deposition of H3-H4 evicted by Pol II, which is similar to the recycling of parental H3-H4 histones during replication (Jamai et al., 2009). This raises the possibility that H2Bub1 and Spt16 co-operate to assemble histones displaced in front of advancing polymerases.
An alternative role for H2Bub1 at the fork is that it stabilizes newly assembled nucleosomes on replicated DNA, consistent with its global role in promoting chromatin stability (Batta et al., 2011; Chandrasekharan et al., 2009). These two roles need not be mutually exclusive. H2Bub1 could both promote histone deposition and stabilize nucleosomes behind the advancing fork. Finally, H2Bub1 might contribute to nucleosome positioning around origins. Nucleosomes immediately adjacent to origins are well positioned, an arrangement that facilitates origin activation and promotes fork progression (Berbenetz et al., 2010; Eaton et al., 2010). Because several well-studied origins in htb-K123R cells show normal ORC association and pre-RC activation, this suggests that the nucleosomal structure at origins is generally maintained. Global analysis of chromatin structure around origins in the htb-K123R mutant will address this issue.
H2B ubiquitylation promotes efficient replication fork progression and replisome stability
Several lines of evidence suggest that inefficient replication in htb-K123R cells is a consequence of a defect in unwinding DNA in the context of chromatin. First, less ssDNA is formed during a G1 to HU shift. Second, there is reduced RPA association with active origins but fairly normal recruitment of PCNA to the same origins. These data implicate H2Bub1 in the activity of the MCM helicase. MCM promotes fork progression through its contribution to nucleosome displacement ahead of the fork in addition to its role in DNA unwinding, and both activities are required for replisome advancement (Groth et al., 2007). We consider two mechanisms by which H2Bub1 could influence these MCM functions. First, MCM-dependent nucleosome displacement in front of the fork could be coupled to nucleosome assembly behind the fork; in the absence of proper nucleosome assembly or stability in htb-K123R cells, a negative feedback loop would be activated to inhibit the helicase. Alternatively, H2Bub1 could affect the association of MCM with FACT. FACT stimulates the helicase activity of MCM and together the two factors promote replication on chromatin templates (Abe et al., 2011; Han et al., 2010; Kundu et al., 2011; Tan et al., 2006; Tan et al., 2010). Because FACT is not stably associated with origin-adjacent chromatin in the absence of H2Bub1, the lower levels of FACT could in turn affect the activity and stability of MCM, impeding fork progression and leading to replisome destabilization.
The genome is eventually duplicated in htb-K123R cells after replication stress is removed. This is accomplished in part by a pool of stable replisomes that resume replication when HU is removed. Additional licensed origins that fire when the checkpoint is alleviated could also contribute to the completion of DNA replication. However, our studies suggest that the HR pathway plays an important role in restoring replication in mutant cells. While we have not directly examined the effect of htb-K123R on HR, we have not detected increased levels of markers of DSBs in this mutant in either the presence or absence of HU (unpublished data).
In summary, the data have extended the range of H2Bub1 cellular functions to include DNA replication. H2Bub1 is present in chromatin adjacent to replication origins and maintained on chromatin after passage of the replication fork. During replication H2Bub1 promotes the assembly and/or stability of nucleosomes behind the advancing replication fork, in striking similarity to its function in transcription elongation. This function could play a key role in promoting the efficient progression of the replication fork and in stabilizing the replisome upon replication stress.
EXPERIMENTAL PROCEDURES
Yeast strains and growth conditions
Strains are listed in Supplemental Table 1. Cells were grown in YPD medium at 30°C unless otherwise noted. Strains containing tagged or deleted genes were constructed by standard yeast genetic and molecular techniques.
Cell Synchronization
Cells were grown in YPD medium at 30°C to a density of 0.4 O.D. units prior to the addition of 5μg/ml α-factor (United Biochemical Research). Additional α-factor aliquots were added after 45 and 90 min for a total of 135 min. G1 arrested cells were collected for ChIP, western blot analysis, and FACS. The remainder of the culture was centrifuged, washed, and resuspended in pre-warmed YPD medium containing 60μg/ml pronase (Sigma), 5mM CaCl2, and 0.2M hydroxyurea (HU; Sigma) for 2 hours. HU was removed from cells by centrifugation and washing, followed by resuspension in pre-warmed YPD medium containing 20μg/ml nocodazole (Sigma). In experiments investigating pre-RC formation, mid-log phase cells growing in YPD medium were arrested with 20μg/ml nocodazole for 90 minutes, collected and washed with prewarmed medium, and resuspended in YPD medium containing 5μg/ml α-factor. Additional α-factor boosts were given at 45 and 90 minutes post-nocodozole release.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed on formaldehyde fixed, sonicated chromatin using the following antibodies: anti-Flag M2 agarose (Sigma); anti-HA 12CA5 (Roche); anti-Myc 9E10 (Millipore); anti-H3K56ac (Millipore); anti-Pol30 (P. Kaufman); anti-Spt16 (T. Formosa); anti-RPA (W-D. Heyer); anti-H2Bub1 (A. Shitatifard); anti-H3 C-terminus (Active Motif); and anti-BrdU (GE Healthcare). ChIP DNAs were quantified by real-time qPCR using a SYBR-Green PCR mix (Fermentas) and an ABI7900 real time PCR system. ChIP antibody conditions and qPCR primers are available upon request. IP signals at origins were normalized to the IP signal at the ASI1 ORF, which is 40Kb from the nearest origin. Where indicated, data were further normalized to G1 samples and input DNA. For H2Bub1 ChIP, background levels were determined by performing an anti-H2Bub1 ChIP in an htb-K123R strain alongside the wild type strain (Schulze et al., 2009). The background levels were then subtracted from the wild type qPCR values prior to normalization.
Additional experimental procedures are presented in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
H2Bub1 levels are maintained on newly replicated DNA during S-phase
H2Bub1 promotes efficient replication fork progression and replisome stability
H2Bub1 regulates nucleosome assembly or stability on newly replicated DNA
ACKNOWLEDGEMENTS
Ali Shitlatifard, Tim Formosa, Paul Kaufman, and Wolf-Dietrich Heyer are gratefully acknowledged for generous gifts of antibodies against yeast H2Bub1, Spt16, Pol30, and RPA, respectively. Cory Hillyer is thanked for expert technical assistance, Rebecca Sanderson is acknowledged for the construction of Flag-Bre1 strains, and Cheng-Fu Kao is thanked for the p306-BrdU-inc plasmid. Scott Houghtaling is thanked for helpful comments. FACS analysis was performed in the UNM Cancer Center's Flow Cytometry Shared Resource. The research was supported by NIH grants GM40118 and GM40118-S1 to M.A.O. and 1K22 CA163485 to K.M.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011;286:30504–30512. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Bianco JN, Pasero P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009;28:1131–1141. doi: 10.1038/emboj.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio T, Ibarra A, Mendez J. Cdc45-MCM-GINS, a new power player for DNA replication. Cell Div. 2006;1:18. doi: 10.1186/1747-1028-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22:766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbenetz NM, Nislow C, Brown GW. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 2010;6(9):e1001092. doi: 10.1371/journal.pgen.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Exp Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Burgess RJ, Zhang Z. Histones, histone chaperones and nucleosome assembly. Protein Cell. 2010a;1:607–612. doi: 10.1007/s13238-010-0086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RJ, Zhang Z. Roles for Gcn5 in promoting nucleosome assembly and maintaining genome integrity. Cell Cycle. 2010b;9:2979–2985. doi: 10.4161/cc.9.15.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova SB, Dorth JA, Razorenova OV, Game JC, Brown JM. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat Res. 2010;174:558–565. doi: 10.1667/RR2184.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M, Prado F. Chromatin assembly controls replication fork stability. EMBO Rep. 2009;10:790–796. doi: 10.1038/embor.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends in Cell Biology. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre NC, Berger SL. Histone H2B ubiquitylation and deubiquitylation in genomic regulation. Cold Spring Harb Symp Quant Biol. 2004;69:289–299. doi: 10.1101/sqb.2004.69.289. [DOI] [PubMed] [Google Scholar]
- Falbo KB, Shen X. Histone modifications during DNA replication. Mol Cells. 2009;28:149–154. doi: 10.1007/s10059-009-0127-7. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst) 2009;8:470–482. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Han J, Li Q, McCullough L, Kettelkamp C, Formosa T, Zhang Z. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 2010;24:1485–1490. doi: 10.1101/gad.1887310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin- dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Puglisi A, Strubin M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol Cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Groth A. Restoring chromatin after replication: how new and old histone marks come together. Semin Cell Dev Biol. 2010;21:231–237. doi: 10.1016/j.semcdb.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu LR, Seki M, Watanabe N, Murofushi H, Furukohri A, Waga S, Score AJ, Blow JJ, Horikoshi M, Enomoto T, et al. Biphasic chromatin binding of histone chaperone FACT during eukaryotic chromatin DNA replication. Biochim Biophys Acta. 2011;1813:1129–1136. doi: 10.1016/j.bbamcr.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Latham JA, Chosed RJ, Wang S, Dent SY. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146:709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Tye BK. Initiating DNA synthesis: from recruiting to activating the MCM complex. J Cell Sci. 2001;114:1447–1454. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- Li Q, Burgess R, Zhang Z. All roads lead to chromatin: Multiple pathways for histone deposition. Biochim Biophys Acta. 2011;1819:238–246. [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA, Fleming AB, Kao CF. Histone ubiquitylation and the regulation of transcription. Results Probl Cell Differ. 2006;41:47–75. doi: 10.1007/400_006. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3'-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biol Cell. 2009;101:617–627. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- Shieh GS, Pan CH, Wu JH, Sun YJ, Wang CC, Hsiao WC, Lin CY, Tung L, Chang TH, Fleming AB, et al. H2B ubiquitylation is part of chromatin architecture that marks exon-intron structure in budding yeast. BMC Genomics. 2011;12:627. doi: 10.1186/1471-2164-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. EMBO J. 2011;30:4885–4896. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Liu H, Lin CL, Lee SC. Functional cooperation between FACT and MCM is coordinated with cell cycle and differential complex formation. J Biomed Sci. 2010;17:11. doi: 10.1186/1423-0127-17-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson BN, Davis CP, Warner MH, Arndt KM. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3'-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics. 2011;188:273–289. doi: 10.1534/genetics.111.128645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone WM, Aerne BL, Morgan BA, Johnston LH. Getting started: regulating the initiation of DNA replication in yeast. Annu Rev Microbiol. 1997;51:125–149. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Babour A, Herissant L, Apponi L, Margaritis T, Holstege FC, Corbett AH, Gwizdek C, Dargemont C. H2B ubiquitylation controls the formation of export-competent mRNP. Mol Cell. 2012;45:132–139. doi: 10.1016/j.molcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D, Matter A, Fahrenkrog B. Bre1p-mediated histone H2B ubiquitylation regulates apoptosis in Saccharomyces cerevisiae. J Cell Sci. 2010;123:1931–1939. doi: 10.1242/jcs.065938. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci U S A. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C, Chymkowitch P, Eldholm V, Putnam CD, Lindvall JM, Omerzu M, Bjoras M, Kolodner RD, Enserink JM. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc Natl Acad Sci U S A. 2011;108:18748–18753. doi: 10.1073/pnas.1115885108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.