Abstract
Purpose
To improve image registration accuracy in neurodegenerative populations.
Materials and Methods
This study used primary progressive aphasia, aged control, and young control T1-weighted images. Mapping to a template image was performed using single-channel Large Deformation Diffeomorphic Metric Mapping (LDDMM), a dual-channel method with ventricular anatomy in the second channel, and a dual-channel w/appendage method, which utilized a priori knowledge of template ventricular anatomy in the deformable atlas.
Results
Our results indicated substantial improvement in the registration accuracy over single-contrast-based brain mapping, mainly in the lateral ventricles and regions surrounding them. Dual-channel mapping significantly (p<0.001) reduced the number of misclassified lateral ventricle voxels (based on manually-defined reference) over single-channel mapping. Dual-channel (w/appendage) method further reduced (p<0.001) misclassification over dual-channel method, indicating that the appendage provides more accurate anatomical correspondence for deformation.
Conclusion
Brain anatomical mapping by shape normalization is widely used for quantitative anatomical analysis. However, in many geriatric and neurodegenerative disorders, severe tissue atrophy poses a unique challenge for accurate mapping of voxels, especially around the lateral ventricles. In this paper, we demonstrate our ability to improve mapping accuracy by incorporating ventricular anatomy in LDDMM and by utilizing a priori knowledge of ventricular anatomy in the deformable atlas.
Keywords: Atlas, Ventricles, Mapping, Brain, MRI, Diffeomorphic
INTRODUCTION
Normalization-based image analysis, such as voxel-based morphometry, is a powerful tool for the quantitative analysis of MR brain images (see e.g. (1,2)). Algorithms of this type find the appropriate transformations necessary to map all voxels in one brain to anatomically correspondent voxels in another brain. Once the voxel-to-voxel mapping is complete, voxel intensity and morphology can be quantitatively compared (3). In addition, if the template brain is pre-parcellated, the target brain can be automatically parcellated by transferring the boundary definition in a process known as atlas-based parcellation (4–10). While these are powerful approaches for investigating the complex anatomy of the brain, the accuracy of the derived measurements is critically dependent upon the accuracy of the image transformations, necessitating the continual validation and improvement of these methods.
Due to a high degree of variability in brain anatomy, transformation of the shape of one brain to another, a process known as registration, is not always straightforward. Registration algorithms often fail to properly handle certain anatomical changes found in pathological brains, particularly those of older subjects. Mapping failure is especially pronounced in the structures situated around the lateral ventricles due to age-related tissue atrophy (11–13). Large Deformation Diffeomorphic Metric Mapping (LDDMM), part of the class of diffeomorphic approaches (e.g., DARTEL (14), Diffeomorphic Demons (15), and LDDMM (16,17)), attempts to allow a large degree of deformation while simultaneously preserving the topology of the object, thus constraining transformations such that connected tissues remain connected and distinct (different) tissues are not joined. This algorithm achieves highly accurate image registration in normal brains (18–22), but it still faces difficulty accommodating large changes in ventricle morphology often seen in an elderly population.
Diffeomorphic mapping algorithms typically minimize a cost function with two terms: a) an error term, which enforces matching of the images and roughly scales as area of mismatch, and b) a regularization term, which imposes a smoothness constraint on the deformation. In LDDMM the diffeomorphism ϕ matching a template Itemp to an observed image I is calculated by integrating the velocity vector field that is found by minimizing the following equation:
where the first term ensures a smooth transformation and the second term enforces matching of the template and subject image. σ2 is a parameter which controls the weighting of the regularization and data matching terms (17).
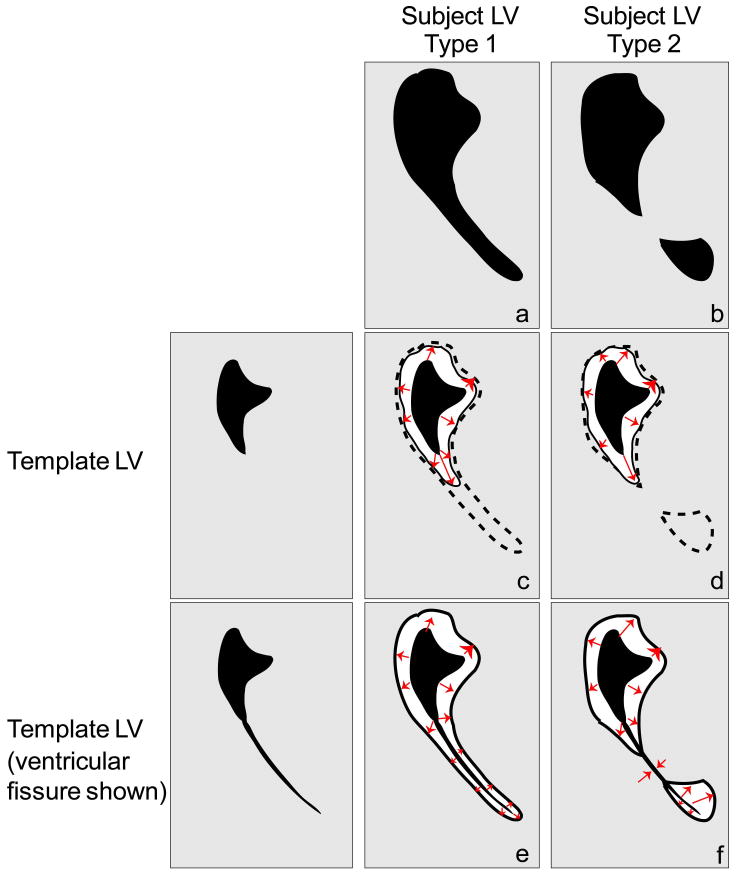
The challenges of accurate mapping using the diffeomorphic approach are illustrated in Fig. 1, which shows deformations between a healthy adult lateral ventricle and two classes of lateral ventricle morphologies seen in dementia patients. The first type of ventricle involves an opening of the posterior horn, which is visible as a thin, long protrusion into the occipital parenchyma (Fig. 1a). Here, the algorithm may fail to map the template to subject properly because the thin protrusion is a small area of mismatch (the error term) and because a drastic protrusion poses a high cost (the regularization term), so the algorithm ignores the protrusion and does not map it to the lateral ventricle (Fig. 1c). In order to perform this matching, the regularization term would need modification by increasing the elasticity constant, α/γ (a parameter explained in (23)), and relaxing the smoothness constraint on the deformation, which would lead to over-fitting in other areas of the brain, such as the cortex. The second type of ventricle deformation involves a topological change where one ventricle is split into two unconnected compartments (Fig. 1b). For a mapping to be a diffeomorphism, it needs to be bijective, continuous, and smooth (differentiable), with a continuous and smooth inverse, which necessitates topological equivalency. Since the template and subject ventricles are not topologically equivalent, a diffeomorphic mapping between them fails (Fig. 1d).
Figure 1.
Schematic of possible deformations of the lateral ventricle (LV). Four mappings shown resulting from two template LVs and two types of subject LV deformations. Gray region = parenchyma, black region = lateral ventricle space, dotted line = desired mapping boundary, solid line = actual mapping boundary, red arrows = hypothetical mapping path. (a) Type 1 = lateral ventricle of patient in which a thin protrusion of the ventricle is observed; (b) Type 2 = lateral ventricle that is composed of two separate compartments; (c) failed mapping of original template LV to subject LV Type 1; (d) failed mapping of original template to subject LV Type 2; (e) and (f) correct mapping for subject LV deformations Type 1 and 2, respectively, resulting from template ventricle with fissure.
The reason for such anatomical changes in the lateral ventricle as aging progresses is rather simple: In healthy young adult brains, a large section of the posterior horn of the lateral ventricle exists as a closed fissure, which is smaller than the spatial resolution of typical MR images, and thus does not exist in normal adult template images. Fig. 1e and 1f show the actual deformation processes that take place if the ventricular fissure is accounted for. Now, the first type of ventricle can be handled by diffeomorphic mapping algorithms because the expansion of the fissure is a smooth transformation (Fig. 1e), and the second type of ventricle can be better approximated by the new template because the ventricle can expand to fill both ventricular compartments (Fig. 1f).
This paper proposes a method for improving the mapping between normal and pathological brains, specifically in the lateral ventricle, by addressing the issue of the ventricular fissure. The method involves incorporation of the ventricular anatomy (closed posterior horn) of the template image as information for determining a more accurate mapping. Because this information cannot be included in the actual T1-weighted images of the template brain, we utilize the dual-channel approach described below.
A multi-channel diffeomorphic mapping method for mapping white matter structures in diffusion tensor imaging data was introduced previously (23). In it, FA (fractional anisotropy) and b0 images are simultaneously used to represent white matter and ventricular anatomy, respectively. This technique was successfully applied to an Alzheimer’s disease population with severe tissue atrophy (20). Diffusion tensor imaging, however, is not as commonly available to researchers as traditional T1-weighted images, and the spatial resolution tends to be much lower. The possibility of using a T1-weighted image-based, two-channel approach in which both image intensity and ventricle segmentation results (without a fissure) are used simultaneously to drive the mapping has been demonstrated previously (24). This method was tested in FSL (http://www.fmrib.ox.ac.uk/fsl), where it visually improved mapping accuracy in an Alzheimer’s disease population (24). Such a dual-channel approach is hypothesized by us to be a natural way to integrate the closed ventricular fissure, which would be an otherwise indiscernible structure in T1-weighted images.
Here, the diffeomorphic multi-channel scheme (23) is utilized, with the second channel containing segmentation of the lateral ventricles in order to better approximate anatomical correspondence between healthy and enlarged ventricles. The ventricles are first segmented, an appendage is included in the segmented template ventricle definition, as a one-time operation, to account for the presence of the ventricular fissure, and mapping is performed using both the raw T1-weighted image and the segmented ventricle image through dual-channel LDDMM. For comparison, we perform the dual-channel mapping using the segmented template ventricle images without the appendage. Through a novel synthesis of ventricular appendages to represent the anatomical principles underlying brain atrophy and multi-channel LDDMM as a vehicle for integrating this information, we demonstrate significant improvement in brain mapping of geriatric and neurodegenerative populations.
MATERIALS AND METHODS
Subjects
In this study, three different populations with a varying degree of brain atrophy and, thus, ventricle enlargement were tested: primary progressive aphasia (PPA) patients (n=12, ages ranging from 56 to 84 years old; mean, 68 years); age-matched controls (n=13, ages ranging from 49 to 76 years old; mean, 56 years); and young controls (n=5, ages ranging from 22 to 49 years old; mean, 33 years). PPA patients and most of the aged controls were observed to have enlarged lateral ventricles, particularly in the atrium and posterior horns, either in one or both hemispheres. The data were obtained at the Johns Hopkins Hospital, US, and at the Kennedy Krieger Institute, US. This study was approved by the Institutional Review Board of each participating site, and written informed consent was obtained.
MRI Data
T1-weighted images (T1-WI) for PPA patients and the age-matched controls were MPRAGE, matrix 256×256, FOV 230mm×230mm, slice thickness 1mm, in-plane voxel size 0.8984mm, TE 3.2ms, and TR 6.85ms. For young controls, the parameters were MPRAGE, matrix 256×256, FOV 240mm×240mm, slice thickness 1.2mm, in-plane voxel size 0.9375mm, TE 3.15ms, and TR 6.837ms.
Image Processing
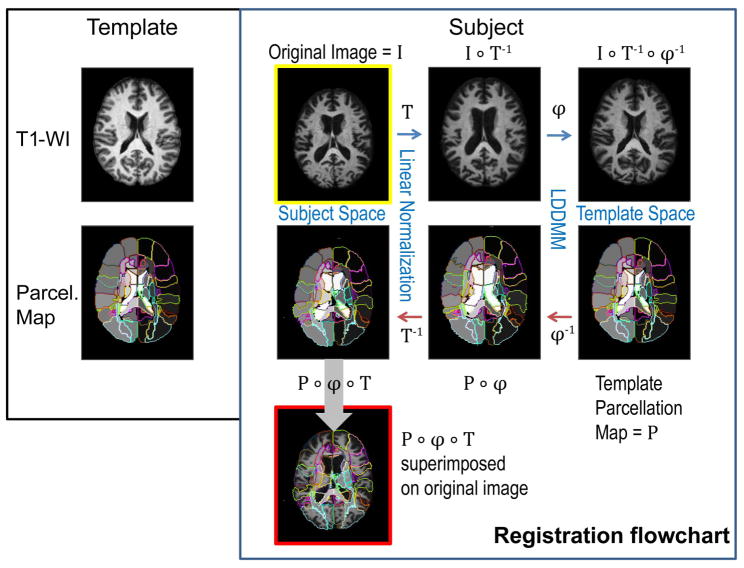
SPM8 (The Wellcome Dept. of Imaging Neuroscience, London; www.fil.ion.ucl.ac.uk/spm) was used to skull-strip the images. The subsequent normalization process, performed using DiffeoMap (Li, X.; Jiang, H.; and Mori, S.; Johns Hopkins University, www.MriStudio.org), is shown in Fig. 2. As the template for normalization, the JHU-DTI-MNI “Eve” atlas (20) was chosen, which is a single-subject template in the ICBM-DTI-81 space and is extensively parcellated and labeled for 159 regions (25). This parcellation will be referred to as the “parcellation map”. The template has an isotropic 1mm voxel size and image size of 181×217×181. The images were first normalized to the ICBM-DTI-81 coordinates (26) using a 12-parameter affine transformation of Automatic Image Registration (AIR) (27) (“Linear Normalization, T” in Fig. 2). The images were further transformed non-linearly to the single-subject template. For this non-linear transformation we used LDDMM with cascading α/γ ratios of 0.01, 0.005, and 0.002 (23). Because of the invertible nature of LDDMM, the transformation results can be used to warp the parcellation map to the original MRI data (red arrows in Fig. 2), thus automatically dividing each brain into sub-regions. We used three different approaches that will be described next: (Approach 1) single-channel LDDMM, based on T1-WIs only; (Approach 2) dual-channel LDDMM, based on the contrast of T1-WI and semi-automatically defined lateral ventricle (LV) maps; and (Approach 3) dual-channel LDDMM as in (Approach 2), but with an additional appendage defined in the LV map and parcellation map of the template (Fig. 3).
Figure 2.
Schematic of typical mapping procedure. The template is a single-subject, T1-weighted image and a parcellation map. The subject image I is first linearly normalized (T = forward linear registration), followed by cascading LDDMM (ϕ = forward nonlinear registration). The resulting image I ∘ T−1 ∘ ϕ−1 is an approximation of the template image. The template parcellation map P is mapped back to the original image by inverse transformations ϕ−1 and T−1. The resulting P ∘ ϕ ∘ T is the automatic parcellation of the subject. Blue arrows = forward transformation, red arrows = inverse transformation, P = parcellation map. A composite function g ∘ f: X→Z is defined by (g ∘ f)(x) = g(f(x)) for all x in X.
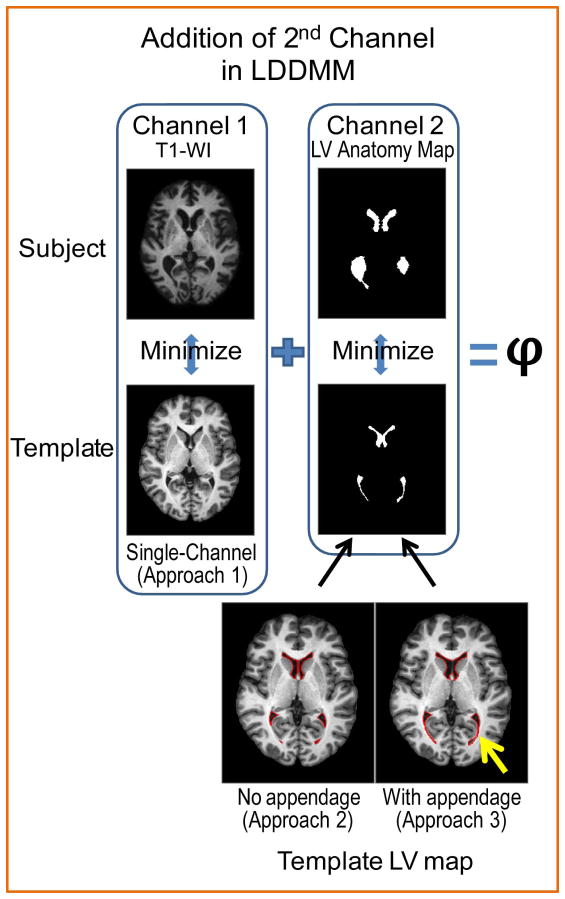
Figure 3.
Approach 1 uses Channel 1 to determine optimal mapping ϕ; Approach 2 and Approach 3 use both Channels 1 and 2. Channel 2 is composed of binary maps of the lateral ventricle (LV) anatomy, which provide the correspondence between the lateral ventricles of the subject and template. While the original Eve atlas lateral ventricle definition is used as the template LV map in Approach 2, for Approach 3 the template LV map with appendage is obtained by manually adding a ventricular appendage (yellow arrow) to account for the existing fissure in the template. This appendage is defined once in the template LV and in the template parcellation map. Subject LV maps are obtained the same way for Approach 2 and 3.
Approach 1: Single-channel Transformation
In the single-channel transformation method, we used LDDMM based on T1-WI contrast as the non-linear transformation step (“LDDMM, ϕ” in Fig. 2) and warped the parcellation map to the original data. The resulting transformed parcellation map was an automatic parcellation of 159 sub-regions for each subject.
Approach 2: Dual-channel Transformation
For this method, we included a second channel, which was composed of the semi-automatically obtained lateral ventricle maps of the subjects and the template. The second channel of LDDMM is utilized in order to guide the registration with more accuracy and, specifically, to account for the deformation of the lateral ventricles. The multi-channel approach is described by Ceritoglu et al. (23). In it, there are C-channel observable images and C-channel template images, and the diffeomorphism ϕ matching the template Itemp to the observed image I is calculated by integrating the velocity vector field that is found by minimizing the following equation:
Index c denotes the contrast images. The parameters , c = 1,…,C control the weights of contrast matching terms and smoothness regularization terms (23). Lateral ventricle anatomy maps were fed into “Channel 2,” as indicated in Fig. 3. For the parcellation of lateral ventricle maps, ROIEditor software (Li, X.; Jiang, H.; Li, Y.; and Mori, S.; Johns Hopkins University, www.MriStudio.org or www.kennedykrieger.org) was used. At each axial slice, a seeded region-growing algorithm was applied to the lateral ventricles, which delineated voxels that were connected and had a value below a specified threshold. This step was followed by a manual correction step, in which the third ventricle and any brain matter were explicitly excluded from the ventricle definition. The manual correction took approximately five minutes for each subject. This 2-D region-growing algorithm was used because a 3-D algorithm was not able to capture the lateral ventricles correctly and because this was a time-conserving alternative. The “Template LV” map was derived from the parcellation map of Eve.
Approach 3: Dual-channel Transformation with an Appendage in the Template
In this approach, the subject LV maps were the same as in Approach 2, but the template LV map was modified. Namely, at ten consecutive axial slices, two appendages were manually included in regions where the ventricles were closed, which extended from the tip of the posterior horn into the occipital portion of the brain (Fig. 3, “Template LV map, with appendage”) and the inferior horn of the ventricles were dilated by two voxels, which is the minimum number of voxels to define a structure. The appendages were approximations of the extension of the mesial portions of the lateral ventricles, which are typically closed in healthy young adult brains. They were added to obtain an optimal mapping that better approximates the actual anatomical deformation. Appendage delineation was performed by a trained radiologist with extensive knowledge of ventricular anatomy and ten slices were chosen based on where the closed ventricles appeared to be visually. This operation needed to be done only once and only on the template image to be used. It added 1900 mm3 (8% of the entire lateral ventricle volume) of extra ventricle space that is under the resolution limit of MRI and, thus, the ventricle volume was erroneously increased by this small amount.
Evaluation Measurements
Our primary interest was to improve and test the registration technique for patients whose lateral ventricles were significantly deformed and enlarged. Since our patients had asymmetric ventricular enlargement, we considered each brain hemisphere, containing the respective lateral ventricle, as one sample. We classified the resulting sixty hemispheres based on the morphology of the posterior horn of the lateral ventricle: open or closed. We performed the following tests for open and closed ventricle samples separately.
To test the accuracy of the transformation, we utilized semi-automatically-derived subject LV anatomy maps as the gold standard to represent the location, size, and shape of the lateral ventricles. It should be noted that the subject LV maps were used both as input into the second channel and as the gold standard. This could pose a bias in evaluating Approach 1 against Approach 2 and 3, which we recognize as a limitation of our study. These maps are the same for Approach 2 and 3 and thus any change in result between Approach 2 and 3 is necessarily due to the different template LV maps (i.e. w/o appendage and with appendage). The warped atlas (parcellation map) was overlaid on the image of the LV map and the number of lateral ventricle voxels that were misclassified as a neighboring region (specifically, as the lingual gyrus, the cuneus, the posterior thalamic radiation, and the hippocampus) were counted. To compare the accuracy of the lateral ventricle definition between single-channel and both dual-channel methods, we compared the degree of misclassification of true lateral ventricles as neighboring regions using the Wilcoxon signed-rank test. All data were analyzed in size-normalized space (i.e. after images were pre-registered with a 12 parameter affine registration).
The preceding evaluation could be readily performed because it only requires accurate definition of the ventricles in each subject. Alternatively, we could define the four brain structures neighboring the ventricles (the lingual gyrus, the cuneus, the posterior thalamic radiation, and the hippocampus) in each subject and measure the LDDMM-derived segmentation accuracy of these structures. This latter approach, however, is not feasible because the first three structures do not lend themselves to accurate definition by manual delineation and the low intra- and inter-rater reproducibility suggest that the resulting segmentation is too low in quality for use as a gold standard.
Image Normalization by FSL
For visual comparison, we performed the same atlas mapping on one PPA subject using FSL (http://www.fmrib.ox.ac.uk/fsl). A template-to-subject transformation was initially calculated using the T1-WIs. Default transformation parameters were used in FSL, with the exception of the sub-sampling scheme, which was changed from [4, 2, 1, 1] to [8, 4, 2, 2] to reduce computational time. The transformation was then applied to the parcellation map, thus creating a warped, subject-specific parcellation.
RESULTS
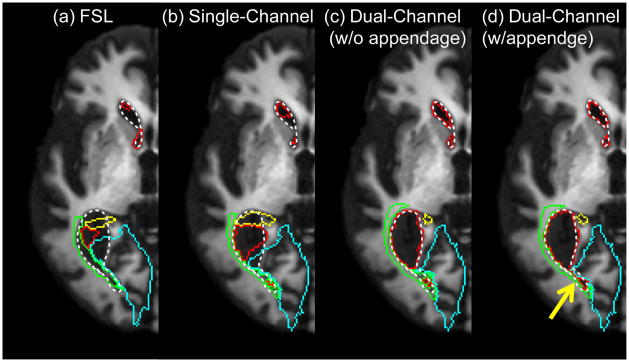
We first visually inspected the change in ventricle mapping resulting from inclusion of the second channel and the appendage in LDDMM. Fig. 4 shows an example of brain mapping from a single-subject atlas to a typical PPA subject with severe parenchymal atrophy and enlarged ventricles. The parcellation results of ventricles, as well as several adjacent structures, are shown. Fig. 4a shows parcellation results using a comparable non-diffeomorphic method (FSL). In Fig. 4b–d, results of the automated parcellation are shown for our three different LDDMM approaches. Note that only the dual-channel w/appendage method was able to give an accurate segmentation of the lateral ventricle and provide the correct segmentation of the surrounding areas. The subtle effect of the appendage can be seen in the area indicated by the yellow arrow in Fig. 4d; the appendage forced a connection between two isolated compartments of the lateral ventricle. Importantly, it should be noted that in Fig. 4c a portion of the posterior thalamic radiation is shown to be medial to the lateral ventricle horn, which violates the topology of this structure, while proper topology is maintained in Fig. 4d.
Figure 4.
Parcellation results based on warping of the parcellation map. Locations of the defined lateral ventricle and surrounding regions are superimposed on the subject T1-WI. (a) FSL-based parcellation is included for visual comparison. (b) The single-channel LDDMM method results show inaccurate registration. The dual-channel LDDMM methods (c) without and (d) with an appendage improve mapping. The addition of the appendage forces the connection of the two seemingly separated compartments of the lateral ventricle (yellow arrow). Color scheme: white dotted line = actual location/size of lateral ventricle; red = mapped lateral ventricle; blue = lingual gyrus/cuneus; green = posterior thalamic radiation; yellow = hippocampus.
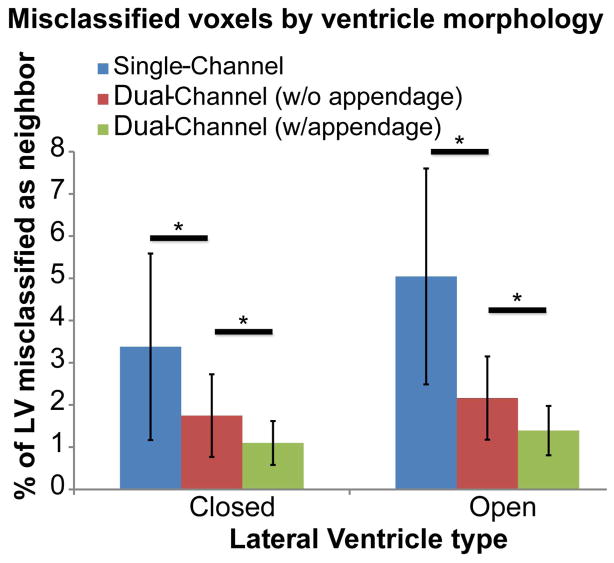
To assess these improvements quantitatively, the topology of the ventricle was first classified into “closed” and “open” types. Closed ventricles were those which had no opening in the posterior horn (n=23); open ventricles were those which had a partial opening in the posterior horn or complete opening resulting in one enlarged space (n=37). Fig. 5 compares the total misclassification of the lateral ventricle as parenchyma using the three different mapping approaches. The bar graph shows the percentage of the lateral ventricles misclassified as regions flanking the posterior horn (lingual gyrus, cuneus, posterior thalamic radiation (PTR), and hippocampus) based on ventricle morphology. The single-channel method yielded the highest percentage of misclassified voxels in both ventricle types (closed: mean = 3%, st. dev. = 2.2%; open: mean = 5%, st. dev = 2.5%). As expected, there was significant improvement in accuracy when adding a second channel in both closed and open ventricle types (Wilcoxon signed-rank test, p<0.001; closed: mean = 1.7%, st. dev = 1%; open: mean = 2.1%, st. dev. = 1%). Further improvement was observed when adding an appendage in the template definition (closed: mean = 1.1%, st. dev = 0.5%; open: mean = 1.4%, st. dev. = 0.6%). This improvement was statistically significant for both closed and open ventricle types (Wilcoxon, p<0.001). Because of the nature of parenchymal atrophy (which leads to ventricle enlargement), the opposite misclassification case (tissue misclassified as a ventricle) made up a negligible percentage of the total brain tissue in the single-channel method (0.1%) and we did not focus on reducing this error with our dual-channel approach.
Figure 5.
Results of parcellation based on different mapping methods. Graph depicts the percentage of lateral ventricle voxels that were misclassified as a neighboring region (the lingual gyrus, the cuneus, the posterior thalamic radiation, and the hippocampus). The highest error occurred for the single-channel method and the lowest error occurred for the dual-channel method with appendage. “Closed” refers to ventricles with unopened fissures at the posterior horn (n=23), “open” refers to partially or completely open ventricles (n=37). * p<0.001, Wilcoxon signed-rank test.
In order to assess how error reduction in individual neighboring regions contributed to the overall improvement, the bar graph from Fig. 5 was broken down into misclassification for individual regions. Tables 1 and 2 enumerate the ventricular volume (mm3) misclassified as a neighboring region, namely lingual gyrus, cuneus, PTR, and hippocampus. Misclassification was significantly reduced in every region for both ventricle types with addition of the second channel. Including the appendage caused a further reduction in misclassification error for the lingual gyrus in open and closed cases (p<0.05), for PTR in the open case (p<0.001) and closed cases (p<0.05), and for the hippocampus in both the open and closed cases (p<0.001), indicating that the appendage provided better mapping primarily of the hippocampus and PTR. The highest significant overall improvement ratio was for the hippocampus.
Table 1.
Closed lateral ventricle volumes misclassified as a neighboring region (mm3).
| Region | Volume of LV misclassified as region | Overall Improvement Ratio |
|---|---|---|
| Mean(SD) Single-channel/Dual-channel/Dual-channel w/app |
|
|
| Lingual Gyrus | 62(84)/31(41)/31(32)a,c | 1.97 |
| Cuneus | 7.5(13.7)/3.1(4.6)/1.5(2)a | 5.06 |
| PTR | 118(156)/50(50)/41(39)a,c | 2.86 |
| Hippocampus | 325(279)/124(67)/71(39)a,b | 4.59 |
p<0.001 between single-channel and dual-channel (w/o appendage)
p<0.001 between w/o appendage and w/appendage
p<0.05 between w/o appendage and w/appendage
PTR = Posterior Thalamic Radiation
Table 2.
Open lateral ventricle volumes misclassified as a neighboring region (mm3).
| Region | Volume of LV misclassified as region | Overall Improvement Ratio |
|---|---|---|
| Mean(SD) Single-channel/Dual-channel/Dual-channel w/app |
|
|
| Lingual Gyrus | 190(227)/72(72)/56(41)a,c | 3.41 |
| Cuneus | 75(119)/12(16)/7(8.5)a | 10.64 |
| PTR | 234(153)/162(124)/102(71)a,b | 2.31 |
| Hippocampus | 720(1085)/171(127)/109(79)a,b | 6.61 |
p<0.001 between single-channel and dual-channel (w/o appendage)
p<0.001 between w/o appendage and w/appendage
p<0.05 between w/o appendage and w/appendage
PTR = Posterior Thalamic Radiation
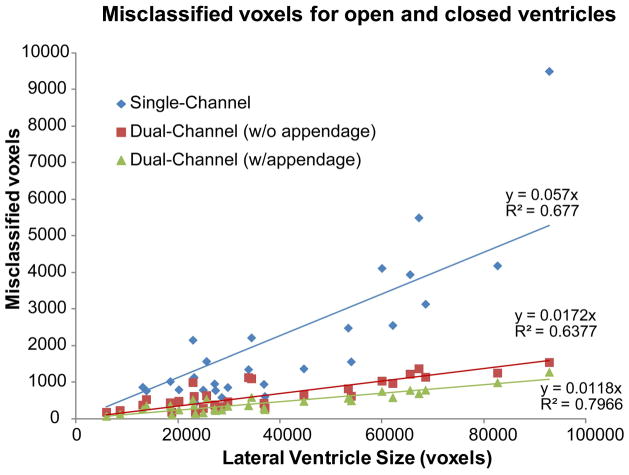
Finally, the relationship between ventricle size and amount of misclassification was examined. A clear correlation was found (see Fig. 6): the larger the ventricle, the less accurate the parcellation of the nearby structures for all mapping methods. Addition of the second channel provided only minor improvement for smaller ventricles, but as the ventricles became larger the improvement imparted by the second channel became more substantial. These large ventricles corresponded to the PPA patients with atrophied parenchyma and problematic anatomies. Furthermore, inclusion of the appendage in the template definition of the second channel caused an additional significant (p<0.001) decrease in error (as compared to dual-channel method w/o appendage) in all subjects, demonstrating that the anatomical constraint provided by the appendage was always beneficial.
Figure 6.
Misclassification of lateral ventricle as a function of ventricle size. The single-channel method has a steeper slope, indicating a higher rate of error as a function of ventricle size. Both dual-channel methods have significantly (p<0.001) lower slopes, indicating the methods are more robust.
The data points in Fig. 6 show the dependence of misclassification on ventricle size. We fitted lines to the data in Fig. 6 and compared the slopes of these regressed lines. Different slopes between methods indicate different rates of error correction as a function of lateral ventricle size. A lower slope indicates that the method is more robust in handling anatomical mismatch between subjects and template. The single-channel method showed that, as ventricle size increased, the error increased rapidly and the method handled error poorly, as indicated by its steep slope (slope = 0.057). In both dual-channel methods, while errors still increased with ventricle size, they did so at significantly (p<0.001) lower rates (w/o appendage slope = 0.017, w/appendage slope = 0.011), attesting to their robustness. We found that appendage inclusion consistently outperforms the regular dual-channel method for every subject and this improvement is significant for the population as a whole (Wilcoxon, p<0.001). We regressed the paired difference between the dual-channel methods against ventricle size and found that the resulting slope is not significantly different from zero (p>0.01). This result indicates that the improvement due to inclusion of the appendage is not strongly dependent on ventricle size.
DISCUSSION
In general, segmentation of the ventricles is relatively straightforward because of the sharp T1-contrast boundary between the CSF and the parenchyma. The ventricles are, however, challenging objects on which to perform registration (24,28), and the seemingly non-diffeomorphic changes of the ventricles could confound the algorithm. As such, warping algorithms often fail to map the normal template brain to a dementia brain and vice versa.
One potential approach to circumvent this issue is to use an age-matched atlas or to generate a study-specific, population-averaged atlas (29). However, the level of atrophy in the geriatric population varies substantially (30); while some subjects have the entire lateral ventricles enlarged and connected as one space, a sizeable number of age-matched control subjects have ventricular anatomy akin to healthy, young individuals, with small ventricles and/or partially closed sections. Furthermore, many individuals’ left and right lateral ventricles are asymmetric and have different topologies. Therefore, using such a study-specific or age-matched atlas would not eliminate the problem.
Diffeomorphic image matching is driven by two competing terms of a cost function: global intensity agreement and local transformation smoothness. Different elasticity values effectively change how much weight is placed on the smoothness term. Unbounded elasticity would allow perfect fitting of arbitrary shapes but would lead to instability of transformation, non-anatomical solutions due to trapping in local minima, and high parameter dependency due to potential multiple solutions (23). In practice, we need to limit the degree of elasticity to obtain stable and anatomically relevant results; however, this might lead to local anatomical mismatch in areas that require a large amount of non-smooth deformation, especially in the posterior horn of the ventricles and in the sucli. Within this framework, addition of the second channel with the ventricle definitions leads to a higher degree of local deformation at the ventricle and tissue boundaries by shifting the balance of the cost function, while still using the same conservative elasticity value. Addition of the appendage further shifts the balance by specifying a target that could be diffeomorphically mapped with a smaller amount of deformation (Fig. 1).
The idea of a multi-channel cost function was introduced in a previous publication (6,23,31). In this paper, we utilize this approach to achieve accurate mapping of the ventricle segments. This approach is relatively simple because the ventricle, as mentioned above, is one of the easiest structures to define semi-automatically or with a fully automated segmentation tool. The downside is the extra step required for subject ventricle segmentation. In this paper, the ventricle was semi-automatically delineated using a simple region-growing tool, followed by manual correction to ensure an accurate definition of the ventricle. In practice, if fully automated brain mapping is required to process large amounts of data, an accurate automated tool would be necessary to employ the proposed method.
Our results raise an important question: When is it advantageous to use the second channel? The line graph in Fig. 6 shows error as a function of ventricle size and can be used as a litmus test to determine which approach to take. At the lower end of the spectrum in ventricle size, both single-channel and dual-channel methods produce comparable and acceptable results, such that the benefits of the second channel may not outweigh the additional work required in preprocessing it. This is true for both PPA and normal subjects, although the majority of PPA subjects will fall on the opposite end of the size spectrum. However, as the size of the ventricles increases, the results between the methods begin to diverge, with the single-channel method yielding more error in registration. At the higher end of the spectrum, the dual-channel approach provides substantial improvement.
The second important idea tested in this study was that the addition of an appendage in the closed ventricle regions of the template image would further improve registration accuracy in subjects. As seen in Figs. 4 and 5 the reduction in error due to addition of the appendage was significant and was not strongly dependent on ventricle size (Fig. 6). Although the appendage adds 1900 mm3 of error in the template ventricle, it improves tissue parcellation even for those subjects with closed ventricles (Fig. 4).
In conclusion, accurate mapping of the brain anatomy by image transformation is a challenging task when two brains have ostensibly different ventricle morphologies. We tested a dual-channel method, in which the binary definition of the ventricle shapes was added to conventional intensity-based images (T1-WI) along with a closed fissure in the template ventricle definition. We observed significant improvement in parcellation accuracy of the ventricles and surrounding structures by employing this approach.
Acknowledgments
Grant Support: UL1RR025005 from NCRR/NIH and NIH Roadmap for Medical Research (AVF), R21AG033774 (KO), P41RR015241 (MIM), RO1AG020012 (SM), P41RR15241 (SM), NS058299 (SM).
The authors thank John Issa for his assistance in editing the manuscript.
References
- 1.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 2.Whitwell JL. Voxel-based morphometry: an automated technique for assessing structural changes in the brain. J Neurosci. 2009;29:9661–9664. doi: 10.1523/JNEUROSCI.2160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Styner M, Gerig G, Lieberman J, Jones D, Weinberger D. Statistical shape analysis of neuroanatomical structures based on medial models. Medical Image Analysis. 2003;7:207–220. doi: 10.1016/s1361-8415(02)00110-x. [DOI] [PubMed] [Google Scholar]
- 4.Iosifescu DV, Shenton ME, Warfield SK, et al. An automated registration algorithm for measuring MRI subcortical brain structures. Neuroimage. 1997;6:13–25. doi: 10.1006/nimg.1997.0274. [DOI] [PubMed] [Google Scholar]
- 5.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 6.Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 7.Granander U, Miller MI. Computational anatomy: an emerging discipline. Statistical computing and graphics newsletter. 1996;7:3–8. [Google Scholar]
- 8.Warfield S, Dengler J, Zaers J, et al. Automatic identification of gray matter structures from MRI to improve the segmentation of white matter lesions. J Image Guid Surg. 1995;1:326–338. doi: 10.1002/(SICI)1522-712X(1995)1:6<326::AID-IGS4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Pitiot A, Delingette H, Thompson PM, Ayache N. Expert knowledge-guided segmentation system for brain MRI. Neuroimage. 2004;23 (Suppl 1):S85–96. doi: 10.1016/j.neuroimage.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 10.Gee JC, Reivich M, Bajcsy R. Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images. Journal of Computer Assisted Tomography. 1993;17:225–236. doi: 10.1097/00004728-199303000-00011. [DOI] [PubMed] [Google Scholar]
- 11.Nestor SM, Rupsingh R, Borrie M, et al. Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain. 2008;131:2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacova C, Peters KR, Beattie BL, et al. Cognitive Impairment No Dementia–Neuropsychological and Neuroimaging Characterization of an Amnestic Subgroup. Dementia and Geriatric Cognitive Disorders. 2008;25:238–247. doi: 10.1159/000115848. [DOI] [PubMed] [Google Scholar]
- 13.Carmichael OT, Thompson PM, Dutton RA, et al. Mapping ventricular changes related to dementia and mild cognitive impairment in a large community-based cohort. 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro; 2006. pp. 315–318. [Google Scholar]
- 14.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009;45:S61–72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Miller MI, Trouve A, Younes L. On the metrics and Euler-Lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- 17.Beg M, Miller M, Trouve A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. International Journal of Computer Vision. 2005;61:139–157. [Google Scholar]
- 18.Faria AV, Zhang JY, Oishi K, et al. Atlas-based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. Neuroimage. 2010;52:415–428. doi: 10.1016/j.neuroimage.2010.04.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faria AV, Hoon A, Stashinko E, et al. Quantitative analysis of brain pathology based on MRI and brain atlases--applications for cerebral palsy. Neuroimage. 2011;54:1854–1861. doi: 10.1016/j.neuroimage.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oishi K, Faria A, Jiang H, et al. Atlas-based whole brain white matter analysis using Large Deformation Diffeomorphic Metric Mapping: Application to normal elderly and Alzheimer’s disease participants. Neuroimage. 2009;46:486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oishi K, Mori S, Donohue PK, et al. Multi-contrast human neonatal brain atlas: application to normal neonate development analysis. NeuroImage. 2011;56:8–20. doi: 10.1016/j.neuroimage.2011.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oishi K, Akhter K, Mielke M, et al. Multi-modal MRI analysis with disease-specific spatial filtering: initial testing to predict mild cognitive impairment patients who convert to Alzheimer’s disease. Frontiers in Neurology. 2011:2. doi: 10.3389/fneur.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceritoglu C, Oishi K, Li X, et al. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. Neuroimage. 2009;47:618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. NeuroImage. 2010;51:221–227. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- 25.Oishi K, Zilles K, Amunts K, et al. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage. 2008;43:447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori S, Oishi K, Jiang H, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40:570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. Journal of computer assisted tomography. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- 28.Reig S, Penedo M, Gispert JD, et al. Impact of ventricular enlargement on the measurement of metabolic activity in spatially normalized PET. NeuroImage. 2007;35:748–758. doi: 10.1016/j.neuroimage.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 29.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23(Supplement 1):S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 30.Schott JM, Price SL, Frost C, Whitwell JL, Rossor MN, Fox NC. Measuring atrophy in Alzheimer disease. Neurology. 2005;65:119–124. doi: 10.1212/01.wnl.0000167542.89697.0f. [DOI] [PubMed] [Google Scholar]
- 31.Park HJ, Kubicki M, Shenton ME, et al. Spatial normalization of diffusion tensor MRI using multiple channels. Neuroimage. 2003;20:1995–2009. doi: 10.1016/j.neuroimage.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]