Abstract
The structure of Panicum Mosaic Virus (PMV) was determined by X-ray diffraction analysis to 2.9 Å resolution. The crystals were of pseudo symmetry F23; the true crystallographic unit cell was of space group P21 with a=411.7 Å, b=403.9 Å and c=412.5 Å, with β=89.7°. The asymmetric unit was two entire T=3 virus particles, or 360 protein subunits. The structure was solved by conventional molecular replacement from two distant homologues, Cocksfoot Mottle Virus (CfMV) and Tobacco Necrosis Virus (TNV), of ~20% sequence identity followed by phase extension. The model was initially refined with exact icosahedral constraints and then with icosahedral restraints. The virus has Ca++ ions octahedrally coordinated by six aspartic acid residues on quasi threefold axes, which is completely different than for either CfMV or TNV. Amino terminal residues 1–53, 1–49 and 1-21 of the A, B and C subunits, respectively, and the four C-terminal residues (239-242) are not visible in electron density maps. The additional ordered residues of the C chain form a prominent “arm” that intertwines with symmetry equivalent “arms” at icosahedral threefold axes, as was seen in both CfMV and TNV. A 17 nucleotide hairpin segment of genomic RNA is icosahedrally ordered and bound at 60 equivalent sites at quasi twofold A–B subunit interfaces at the interior surface of the capsid. This segment of RNA may serve as a conformational switch for coat protein subunits, as has been proposed for similar RNA segments in other viruses.
Keywords: Crystal structure, X-ray, icosahedral, NCS, tombusvirus, refinement, RNA, virus crystallography
1. Introduction
Panicum Mosaic Virus (PMV), a small spherical virus, is a pathogen of the Gramineae family of grasses in which it produces systemic mosaic symptoms or chlorotic mottling (Niblett and Paulsen, 1975; Scholthof 1999). It is the type member of the genus Panicovirus within the family Tombusviridae (Murphy et al., 1995), a family that also includes Tomato Bushy Stunt Virus (TBSV) (Hopper et al., 1984), Turnip Crinkle Virus (TCV) (Hogle et al., 1986), Tobacco Necrosis Virus (TNV) (Oda et al., 2000), and Carnation Mottle Virus (CMV) (Morgunova et al., 1994), all of whose structures have been determined by X-ray crystallography (Carrillo-Tripp et al., 2009). From negative stain electron microscopy (Buzen et al., 1984) and atomic force microscopy (Makino et al., 2005) the PMV virion has a diameter of about 30 nm and sediments in the ultracentrifuge at 109S as a single species. PMV infections are frequently accompanied by the presence of a serologically unrelated satellite virus that sediments at 42S, which is known as Satellite Panicum Mosaic Virus (SPMV). In this sense, it is very similar to TNV and its Satellite Tobacco Necrosis Virus (STNV). The structures of both SPMV and STNV have also been solved by X-ray crystallography (Jones and Liljas, 1984; Ban and McPherson, 1995; Ban et al., 1995). At least two satellite RNAs, satRNAs, are also known to be associated with PMV (Monis et al., 1992; Turina et al., 1998; Cabrera and Scholthof, 1999).
PMV is a T = 3 icosahedral virus (Caspar and Klug, 1962) composed of 180 copies of a single coat protein of 26.4 kDa (Turina et al., 1998; Niblett and Paulsen, 1975; Gasteiger et al., 2005) that adopt three distinct and slightly different conformations in the capsid. Five subunits, designated A subunits, form twelve fivefold symmetric capsomeres, and three pairs of subunits, designated B and C, form 20 quasi sixfold (exact threefold) symmetric capsomeres. Subunits A, B, and C are also related by quasi threefold symmetry. The protein, whose amino acid sequence is known, encapsidates a single positive sense RNA molecule of 4326 nucleotides (Turina et al., 1998). During infection, a subgenomic RNA fragment of 1475 nucleotides is transcribed to encode five proteins, including the coat protein (Turina et al., 1998; Turina et al., 2000).
A number of years ago we determined the structure of SPMV, which is the smallest encapsidated virus known, and refined a model to 1.9 Å resolution (Ban and McPherson, 1995). That was followed by study of other crystal forms that revealed additional features of the capsid protein as well as some segments of RNA (Makino et al., 2006). Because of our interest in the PMV-SPMV system, we pursued the crystallization and structure determination of PMV as well. We readily obtained crystals of PMV under a variety of conditions, but in no case did any crystal diffract to beyond about 8 Å resolution. We attributed this to the observation that PMV appears unstable after purification and spontaneously loses structure and infectivity in a few days time. For a number of small icosahedral viruses, including the related TNV, however, it is also known that swelling and loss of infectivity occurs most rapidly above pH 7.0 (Kaper, 1975). With this in mind, we modified the preparative procedure and crystallization conditions for PMV to maintain a pH of no more than 6.0 throughout. At pH 5.5, we were then able to obtain a crystal form that diffracted beyond 2.9 Å resolution (Makino et al., 2005).
Data was subsequently collected, using synchrotron radiation, from a single flash frozen crystal to 2.9 Å resolution. Preliminary data analysis, combined with an atomic force microscopy analysis, showed that these crystals were pseudo cubic (F23) in symmetry but of actual space group P21, with a= 411.7 Å, b = 403.9 Å, c = 412.5 Å, and β = 89.7° (Makino et al., 2005). The low symmetry required that the asymmetric unit be comprised of two entire virus particles having a combined molecular weight of about 12.3 MDa. Thus, the crystals presented one of the largest asymmetric unit sizes yet addressed by X-ray crystallography, an asymmetric unit composed of 360 protein subunits, or 120 independent trimers plus nucleic acid.
Pertinent to the crystallographic structure determination of PMV reported here is the similarity of PMV to two other viruses, the tombusvirus TNV, as noted above, and Cocksfoot Mottle Virus (CfMV), a sobemovirus, whose structure has also been solved by X-ray crystallography (Tars et al., 2003). Although the sequence identity with PMV after alignment (Barton, 1990) was in the ambiguous zone (Smith and Waterman, 1981) of about 20% for both, these two viruses were considered the most appropriate probes for molecular replacement searches. Indeed, as described below, they did supply models that ultimately led to the PMV structure solution. In particular, searches with the TNV full capsid, along with packing considerations imposed by the pseudo cubic symmetry, combined with self-rotation and translation searches, and all under the constraint of icosahedral symmetry, allowed determination of the relative orientations of all 360 subunits, and the dispositions of the virus particles in the unit cell (Makino et al., 2005).
Here we present the three dimensional structure of PMV and its refinement to 2.9 Å resolution. We also describe the positions of bound calcium ions and the observation of a 17 nucleotide fragment of RNA bound at the interior surface of the capsid. We further analyze the similarities and differences between PMV and other, similar viruses, particularly TNV.
2. Materials and Methods
2.1 Purification, crystallization and data collection
Preparation, purification, and crystallization of PMV were described in detail earlier (Day et al., 1994; Makino et al., 2005), but briefly, the virus was purified from pearl millet, which had been co-infected with PMV and SPMV. Purification principally used PEG fractionation, all carried out at pH 6.0 or lower. Crystals used for data collection were grown from 30% w/v PEG 400 buffered with 0.1 M sodium citrate at pH 5.0 to 5.5. The crystals were grown at room temperature and reached sizes of half a millimeter on edge. Because of the high concentration of PEG 400 in the mother liquor, crystals could be mounted on loops and flash frozen directly in the cryostream. Native data for the structure determination were collected at the Advanced Light Source at the Lawrence Berkeley Laboratories in Berkeley, CA on beamline 5.0.2 using a crystal to detector distance of 380 mm and a wavelength of 1.00 Å. Oscillation angles were 0.5 and exposure times were 20 sec. Remarkably, the crystal, which provided the data used here, showed virtually no indications of decay in the X-ray beam, even after more than 24 hours of exposure. In all, 942 frames of data were collected which yielded a total of 17,212,205 intensity observations to a resolution limit of 2.71 Å. The data were processed using the program d*Trek (Pflugrath, 1999), and in spite of the large unit cell, rather few reflections were eliminated due to overlap. Statistics for the data used in refinement are shown in Table 1. The data set contains reflections significantly above background (greater than 2 esd) to about 2.71 Å resolution, but only reflections within the range 50.1 Å to 2.9 Å resolution were used in the refinement of the model. Redundancy to 2.9 Å resolution was about 4 observations/reflection, and the last shell at that resolution was 24.8% complete.
Table 1. Crystal data and model statistics from both constrained and restrained refinement.
For reflection statistics, the highest resolution shell values are indicated in parenthesis. For B factor statistics, values for particles 1 and 2, respectively, are in parenthesis.
| Crystal data | |
|---|---|
| Space Group | P21 |
| Unit Cell Dimensions (Å, °) | a= 411.74, b= 403.90, c= 412.46, β = 89.65 |
| Z | 4 |
| No. observations | 17,212,205 |
| No. unique reflections | 2,279,719 |
| Crystal mosaicity (°) | 0.373-0.544 |
| Resolution range (Å) | 50.01-2.71 (2.81-2.71) |
| Completeness (%) | 63.0 (4.9) |
| Rmerge | 0.136 (0.346) |
| I/σ(I) | 6.8 (2.5) |
| Redundancy | 7.55 (3.14) |
| Model statistics | constrained model | restrained model |
|---|---|---|
| Resolution Range (Å) | 50 - 3 (3.14 - 3.00) | 50 – 2.9 (3.00-2.90) |
| Sigma cut-off | 0 | 0 |
| R-factor | 0.3395 (0.4675) | 0.2514 (0.4947) |
| R-free | 0.3420 (0.4681) | 0.2850 (0.4988) |
| Number of reflections | ||
| working set | 1,932,101 (108,109) | 1,995,520(66,059) |
| test set | 215,045 (12,121) | 221,945 (7,210) |
| Particle centers-particle 1 | 102.243, 0.636, 1.038 | 102.242, -0.308, 1.048 |
| -particle 2 | 104.217, 203.708, 206.702 | 104.187, 202.789, 206.668 |
| Total number of atoms from | ||
| Protein | ||
| A subunits (aa 54-238) | 1,427 | 171,120 |
| B subunits (aa 50-238) | 1,452 | 174,120 |
| C subunits (aa 22-238) | 1,675 | 200,880 |
| Ca2+ ions | 1 | 120 |
| Nucleotide | 269 | 30,240 |
| Average B factor (Å2) | ||
| Protein | ||
| A subunits | 62.69 | 62.5 (53.1 & 71.9) |
| B subunits | 57.96 | 59.6 (50.9 & 68.2) |
| C subunits | 70.86 | 64.9 (56.5 & 73.2) |
| Ca2+ ions | 16.25 | 97.4 (89.7 & 105.1) |
| Nucleotide | 152.79 (12 nucleotides) | 218.3 (212.8 & 223.8) |
| R.m.s. deviations | ||
| Bonds (Å) | 0.0104 | 0.0087 |
| Angles (°) | 1.746 | 1.409 |
| Ramachandran plot | ||
| Most favored (%) | 359 (72.2) | 48,605 (81.5) |
| Allowed (%) | 129 (25.9) | 11,031 (18.5) |
| Disallowed (%) | 9 (1.8) | 4 (0.00) |
2.2 Heavy atom derivatives
Data to 5.0 Å resolution were also collected on six putative heavy atom derivatives to provide multiple isomorphous replacement phases should that have been needed. Data for the derivatives were processed as described for the native data. Although three of the derivatives indeed gave good substitution and the heavy atom sites could be located unambiguously, in the end, isomorphous phasing was not required to solve the structure.
2.3 Molecular replacement
Preliminary molecular replacement results were presented in an earlier paper (Makino et al., 2005), but to reiterate, self-rotation functions and translation functions, combined with the constraints imposed by the pseudo-symmetry of the crystal provided initial orientations and centers of mass for the two particles within the asymmetric unit. Distinctive pseudo origin peaks found in the native Patterson map along with the Atomic Force Microscopy images of the surfaces of PMV crystals, supported the molecular replacement solutions. The results were initially somewhat ambiguous, however, because the two particles in the asymmetric unit had different orientations within the unit cell. Furthermore, neither center of mass was confined to a crystallographic special position; hence, there was some question as to whether the center of mass locations would prove sufficiently accurate. Ambiguities were resolved to a great extent, however, by the results of molecular replacement searches using TNV and CfMV models as probes. Both gave the same results, and those results were completely consistent with earlier deductions. The orientations of the particles and their exact positions were of crucial importance here, as the phase extension and refinement depended absolutely on the rotation matrices and translation vectors relating the 120 ABC trimers in the two virus particles.
Initial models were poly-alanine backbones of entire TNV and CfMV virus particles, without solvent or accessory ions. Loops at the surfaces of particles were also removed, leaving intact only the cores of the protein subunits. Cross-rotation and translation search routines in CNS v.1.1 (Brünger et al., 1998) were used as in conventional molecular replacement, except that the search model was a virus particle of about 35,000 residues or 175,000 atoms. Observed structure amplitudes within 20 to 8 Å resolution and greater than 2.0 were used. This resulted in two rotation peaks but only one translation solution, centered at approximately (1/4, 0, 0) in the unit cell. The most likely explanation for this, which indeed turned out to be correct, was that the center of mass of the second particle was related to the first particle by an origin shift, thereby relating the two particles by an inter-particle translation vector of (0, ½, ½). This agreed with the appearance of an exceptionally strong native Patterson peak at (0, ½, ½), and the packing arrangement of the particle in the unit cell as deduced by atomic force microscopy (Makino et al., 2005). Thus the second particle was located at approximately (1/4, 1/2, 1/2). The orientation and position of both particles were refined as two independent rigid bodies within various resolution ranges, thus maintaining strict icosahedral symmetry. A trimer consisting of subunits A, B, and C was arbitrarily chosen and then superimposed onto the remaining 119 trimers in the asymmetric unit. This generated a total of 120 pairs of rotation matrices and translation vectors. An important consideration that impacted subsequent model refinement was that these non-crystallographic symmetry (NCS) operators do not constitute a closed set of matrices and vectors. Refinable variables such as particle diameter and overall position of the trimer cannot, therefore, be properly treated, since the other 119 trimers are tied to the defined rotation and translation constraints imposed by the NCS operators. Therefore, it was imperative that all positional refinement variables converged to the most accurate values possible. The refined centers for the two particles were (0.248, 0.002, 0.003) and (0.253, 0.504, 0.501), in fractional coordinates.
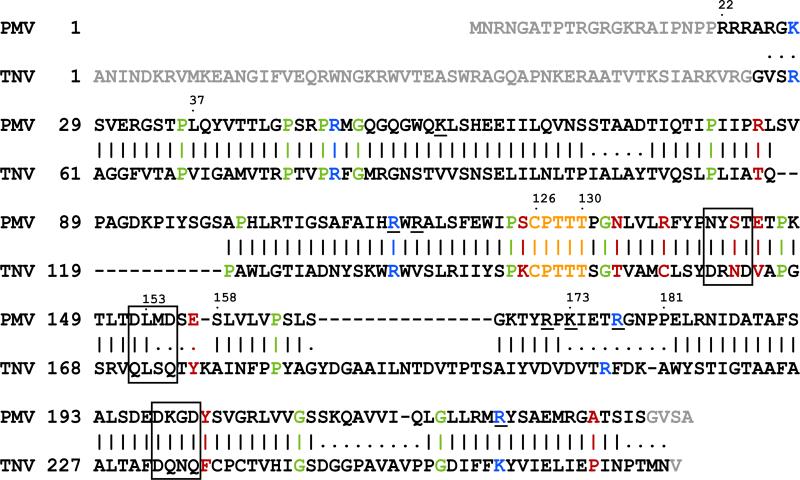
Non-crystallographic symmetry is a powerful asset in the structure determination of icosahedral viruses, and can be utilized to determine and improve phases by incremental extension to increasingly higher resolution (Rossmann and Arnold, 2001). Combined with density modification (Zhang et al., 2001), NCS is now commonly used to refine and extend phases against observed amplitudes beginning with model phases calculated at low, 25 to 20 Å resolutions, and by gradually adding intensities to higher resolution. For PMV, the starting model was a modified TNV model developed from the molecular replacement solution and subsequent side chain rebuilding; TNV, although a member of the same Tombusviridae family, has only 20% sequence identity with PMV. Their structural sequence alignment is shown in Fig. 1, which is consistent with, but not identical to the sequence alignment calculated with the program MULTALIGN (Barton, 1990) using comparison matrix Blosum62 (Henikoff and Henikoff, 1992).
Fig. 1.
Amino acid sequence alignment of the coat protein of PMV against TNV based on structure superposition. Sequence identity is about 20%. A short segment of sequence identity (CPTTT) is highlighted in orange. Positively charged residues located on the interior surface of the capsid that possibly interact with the observed RNA loop are underlined; conserved positively charged residues are colored blue. Residues involved in protein-protein interactions are indicated in red; conserved proline and glycines are in green. Vertical bars indicate good backbone superposition, dots indicate regions not well superposed, and blanks are for the large insertions or deletions. Regions not observed in electron density maps are shown in grey. The DXXD motifs are enclosed in boxes.
In phase extension, it is critical to properly define the boundaries of the solvent and averaging masks, because phases are calculated from averaged and solvent flattened electron density maps. Usually, the same solvent mask is used for both averaging and solvent flattening, and problems with mask overlap, or gaps between them are minimal. Because the 120 NCS operators are improper, and do not constitute a closed symmetry set, the mask must encompass only a unique protomer in order for the averaging procedure to work correctly. Therefore, we designed a mask around the trimer and extended it towards the center of the particle in a wedge shape so that the solvent mask enclosed no more than the volume assigned for averaging. All points outside the boundaries were considered solvent. All masks were generated with the program MAMA (Kleywegt and Jones, 1999). The averaging, density modification and resolution extension procedures of phase extension were performed with CNS v1.1 (see Fig. S.1).
The TNV molecular replacement model was rebuilt with the PMV sequence to the 3 Å averaged map obtained from phase extension as an NCS constrained model composed of a single ABC trimer (see Fig. S.2 for an example of density at 4 Å resolution using these phases). The rebuilt model was refined with the same NCS constraints used in phase extension. Because the R factors were elevated, we expanded the NCS constrained model to the full crystallographic asymmetric unit of two full virions and continued refinement, first by rigid body refinement of 120 protein subunits then by using NCS restraints with the understanding that strict icosahedral symmetry would not be maintained. Eight NCS restraint groups containing 60 molecules each were defined, four groups per particle such that all the icosahedrally equivalent A or B or C subunits comprised a group and the RNA fragments comprised the fourth group. During restrained refinement, it was clearly evident that loop 88-96 extending from many subunits suffered from close inter-particle contacts. Non-averaged 2Fo-Fc electron density maps revealed alternate backbone conformations for these loops, which were rebuilt accordingly (Figs. S.3 and S.4). This loop was consequently removed from the NCS restraints definitions and refinement continued. Other exterior and interior loops as well as the N- and C-termini were removed from the NCS restraints definitions to see what impact they had on the Rfree value. In all cases the Rfree value either increased or remained essentially the same, implying that including them in the NCS restraints definitions was appropriate. Ultimately, the positional and B-factor NCS restraint weights were optimized against Rfree by 3 cycles of positional and B-factor refinement of the same model using various restraint weights. Whereas the CNS default values are 300 and 2 for the positional and B-factor weights, respectively, the optimized values for PMV were 150 and 32. During analysis of the final model, electron density maps suggested that individual side chains within NCS restraint groups may differ in conformation or even have alternate conformations. We concluded that identifying, rebuilding and removing individual residues from the restraint groups would be cumbersome, time-consuming and yield minimal benefit in terms of both information gained and improved R factors. Therefore, we did not pursue additional model rebuilding to account for these side chain differences.
The initial icosahedrally constrained model was built using averaged Fourier maps calculated with the observed structure amplitudes and the phases obtained by phase extension. The difference Fourier maps used for heavy atom determination were also calculated using the phases from phase extension and then averaged using the 120 NCS operators. Rebuilding of the constrained models employed 2Fo-Fc and Fo-Fc Fourier maps obtained from model phases and subsequently averaged. Restrained models were rebuilt to non-averaged Fourier maps.
Subunit superpositions of PMV subunits onto themselves were carried out with CNS v1.3 using the Cα atoms of 171 residues (aa 56-88 and 96-233, which excluded the N-terminal residues, the last five C-terminal residues and the highly variable loop 89-95). The superposition of TNV onto PMV was also performed with CNS v1.3 using 138, 137 and 164 Cα atoms of residues of the A, B and C subunits, respectively, that were identified as equivalent from the structural alignment found previously. In both sets of superpositioning calculations, mean rms differences are reported over all subunits of the PMV model and correspond to the 185 common Cα atoms of the A subunit in the PMV self-comparison and the fitted Cα atoms in the TNV comparison.
Constrained refinement procedures were carried out with the original or modified CNS v1.1 (Brünger et al., 1998) scripts. The final NCS restrained refinements were carried out with scripts for CNS v1.3 (Brünger, 2007). The graphics programs O v8.0 (Jones et al., 1991) and COOT (Emsley and Cowtan, 2004) were used for model building, GRASP (Nicholls et al., 1991) for surface electrostatic potential calculations, and figures were produced with PyMOL v1.0r0 (DeLano, 2002) and Adobe Photoshop and/or Adobe Illustrator.
3. Results
3.1 Structure Determination and Refinement
Poly-alanine models derived from TNV and CfMV representing two PMV particles were positioned and oriented as indicated by molecular replacement results (Makino et al., 2005). The particles were first refined as two rigid bodies against 8 Å resolution data with F/σ > 6.0. Using the correlation coefficient as the initial refinement target, the resulting R-factor value was 0.56. Further constrained refinement was then performed. Starting from an initial phase set calculated from the most current model for data between 25 Å and 50 Å resolution, phases were extended to 3.5 Å employing a combination of NCS averaging (using the previously determined 120 NCS operators) and solvent flattening with appropriate averaging and solvent masks.
A Fourier map based on observed structure amplitudes and phases extended to 3.5 Å resolution showed characteristic densities for many additional side chains, as well as alternate paths for loops where there were either insertions or deletions in PMV relative to TNV. It was at this point that a fragment of RNA was identified and built into the Fourier map. Adjustments of the PMV backbone and final side chain mutations were subsequently carried out along with several cycles of simulated annealing, minimization, and group B-factor refinement with side-chain and main-chain atoms as two separate groups for each residue. Up to this point, strict icosahedral constraints and averaged maps were used for refinement and model building. The R and Rfree values for the constrained refinement were 0.340 and 0.342, respectively, for 3 Å data.
The model was expanded to the full crystallographic asymmetric unit (2 full virions) and refinement was continued using restraints in which each set of equivalent subunits in each particle was a restrained group, resulting in 8 restraint groups (6 protein subunit groups and 2 nucleic acid groups). Initially, the model was refined without using NCS restraints, yielding R values of 0.268 and 0.330 for R and Rfree, respectively, for 2.9 Å data. The restraint weights and B factor sigmas were optimized by varying the values and selecting those that produced the lowest Rfree. Early in the restrained refinement we judged there to be an excess of close contacts. Analyzing these regions of the model against SIGMAA (Read, 1986) weighted 2Fo-Fc and Fo-Fc maps, revealed that indeed the chains were not equivalent even within a restrained group. Loop 88-96 of a number of chains exhibited noteworthy departures from the predominant conformation due to inter-particle contacts (Figs. S.3 and S.4). This loop corresponds to the 13 amino acid insertion in the PMV sequence relative to TNV and is clearly dynamic in character. Rebuilding loop 88-96 of selected chains and removing the loop from the restraint conditions for all restraint groups reduced Rfree to 0.296.
Other regions that might exhibit differences from subunit to subunit were also tested by removing them from the restraint definitions. These regions included the N- and C-termini and every loop that was either on the interior or the exterior, with no spatial constraints. For all the loops tested, the Rfree value increased, while for the N- and C-termini there was essentially no change. Hence, the only region that was not restrained in the refinement of the final model was loop 88-96 of all capsid subunits. The final model with optimized positional and B factor NCS restraint weights gave R=0.2514 and Rfree=0.2850. No attempt was made to model discrete solvent or local disorder. Model statistics for both the constrained model and the restrained model are given in Table 1. Since the constrained model is clearly inadequate due to departures from strict icosahedral symmetry, only the restrained model coordinates have been deposited in the Protein Data Bank (Berman et al., 2000).
3.2 Heavy atoms
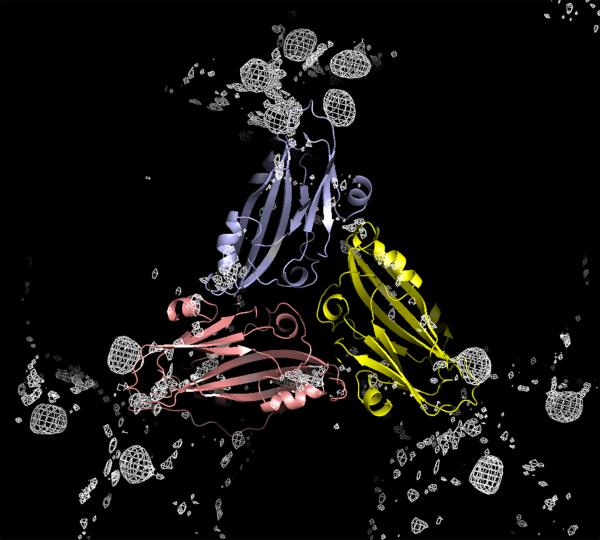
At one point in the crystallographic analysis we were concerned that the orientation matrices and centers of mass might be insufficiently precise to allow accurate model averaging and phase extension. Fortunately, this concern was unwarranted as conventional phase extension and refinement, even with two entire virus particles in the asymmetric unit, was ultimately adequate to phase the structure amplitudes. Nevertheless, to insure eventual success, X-ray intensities were collected to 4 Å resolution from six potential heavy atom derivatives. Using phases calculated from the TNV model placed in the unit cell according to the molecular replacement results (Makino et al., 2005), Fderivative – Fnative difference Fourier syntheses were computed, averaged over all NCS operators, and examined. For three of the putative isomorphous derivatives, OCMP (o-chloromercuriphenol), PCMB (p-chloromercuribenzoic acid), and K2PtCl4, unambiguous peaks, eight times sigma, appeared. The difference electron density for OCMP, for example, is shown in Fig. 2. The set of peaks observed for OCMP and PCMB were virtually identical, which was consistent with expectations based on chemical considerations. Because the PMV coat protein contains a single cysteine residue, we felt it reasonable to assume (as turned out to be the case) that the OCMP and PCMB peaks marked the position of the lone cysteine thiol.
Fig. 2.
Subunits A, B and C about a quasi threefold axis are shown in blue, yellow and pink, respectively, with the difference electron density for the OCMP heavy atom derivative superimposed. Five density peaks appear about the fivefold icosahedral axes, but only three about the quasi sixfold axes, as the mercury atoms are bound by Cys126 of only the A and B subunits.
Equally important to identifying a fiducial point in the structure, there were two additional consequences. First, the heavy atom experiments demonstrated that TNV was a sufficiently good model for PMV to provide reliable initial phases, and second, that the orientation matrices and centers of mass for the two particles in the asymmetric unit, obtained from molecular replacement and used for averaging the electron density maps and extending the phases, were sufficiently precise.
3.3 Virion Properties
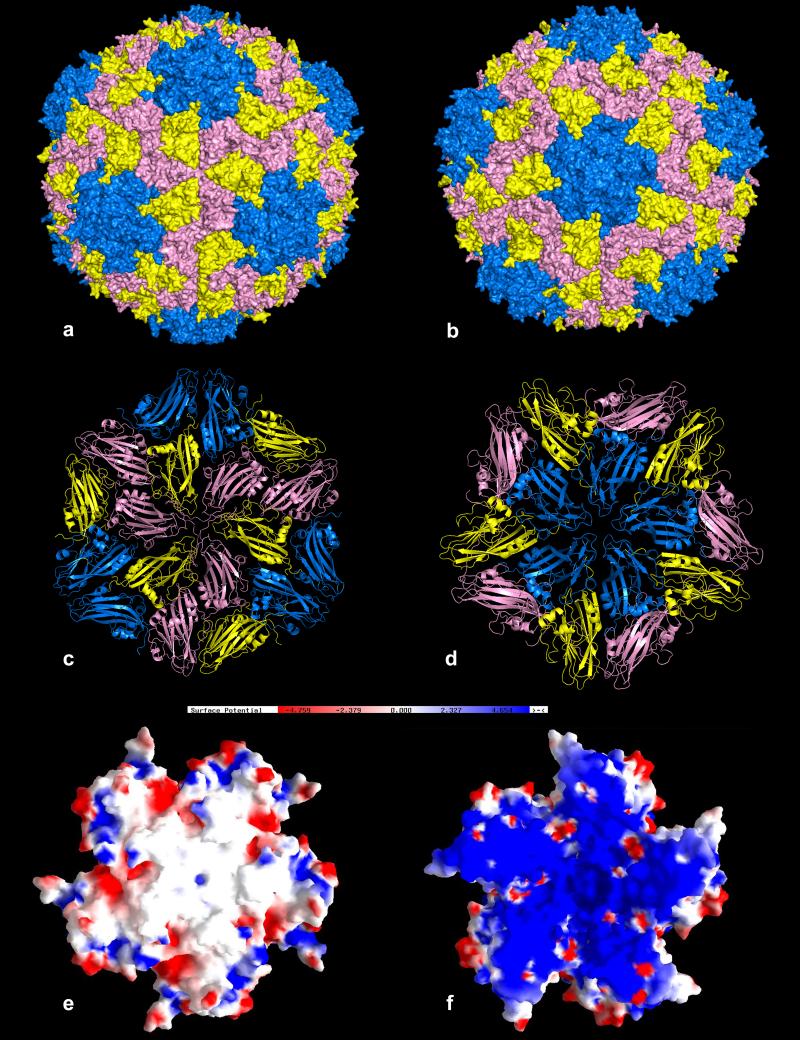
The PMV particle exhibits a relatively smooth external surface with very little protrusion of capsomeres at quasi sixfold (Fig. 3a) and fivefold axes (Fig. 3b). The arrangement of the subunits about the threefold and fivefold axes are illustrated in Figs. 3c and 3d, which show that the B and A subunits have similar dispositions with respect to the proximal symmetry axes whereas the C subunits are slightly rotated when compared to the B subunits (see Fig. S.5). The observed protein shell extends from a minimum radius of 107 Å to a maximum of 159 Å, with a mean radius for all observed protein atoms of 131 Å (Fig. S.6). This is consistent with observations by negative stain electron microscopy (Buzen et al., 1984) and atomic force microscopy (Makino et al., 2005). The interior cavity of a PMV virion has a volume of 5.3 × 106 Å3. This must accommodate not only the 4326 nucleotides of the single stranded genomic RNA, but the unseen 7380 amino acids (equivalent to about 30 capsid protein subunits) of the amino terminal polypeptides of the A, B and C subunits. In any case, the amino terminal amino acids of the coat proteins must occupy somewhere between 16% and 20% of the total interior volume. From this it can be calculated that the RNA volume to mass ratio in the virus, Vm (Matthews, 1968), is about 2.91 Å3/Da, which is more or less consistent with what has been observed for other small icosahedral viruses (Johnson and Rueckert, 1997).
Fig. 3.
In (a) and (b) are van der Waals surface representations of the PMV virion viewed along icosahedral threefold and fivefold axes, respectively. The quasi hexamer of three B and three C subunits of PMV with neighboring subunits, as they are organized about a true threefold axis, is shown in (c), and the pentamer of A subunits with neighbors in (d). The A subunits are in blue, the B subunits in yellow, and the C subunits in pink. In (e) and (f) are the electrostatic potential distributions of the exterior and interior surfaces, respectively, of a PMV pentamer. The kT/e level is 4.7 for both images.
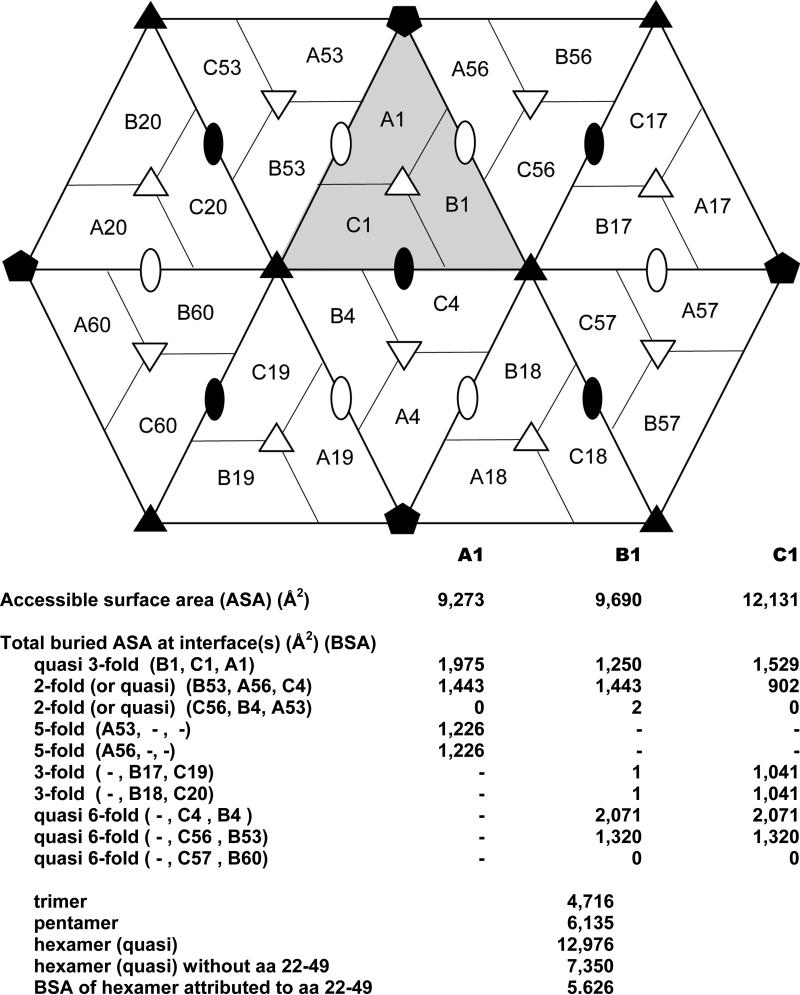
Solvent accessible and buried surface areas were calculated for A, B and C subunits of PMV. The diagram of subunit interfaces in Fig. 4 indicates all possible neighboring subunits around the primary trimer labeled A1, B1, and C1. Inspection of accessible surface areas for individual subunits and inaccessible areas in the presence of neighboring subunits demonstrate that all the major interfaces exceed 1000 Å2 in buried surface area (BSA). The buried surface areas for the B4-C1 (quasi sixfold) and A1-B1 (quasi threefold) interfaces are about 2000 Å2. Interestingly, the interface with the least contacting surface area is observed for subunits C1 and C4 around the exact twofold icosahedral axis (BSA is 902 Å2). The C subunits, however, contribute to the stabilization of the protein capsid via their amino termini that surround the icosahedral threefold axes, as discussed further below. This network of interactions, unique to the C subunits, creates an additional 5626 Å2 of total buried surface area to the quasi hexamers. Equivalent polypeptide segments in A and B subunits are disordered.
Fig. 4.
Accessible and buried surface areas of subunits in the PMV capsid. The diagram shows a primary trimer, labeled as A1, B1, and C1, and the neighboring subunits. Exact icosahedral symmetry axes are indicated by solid symbols and quasi symmetry axes by open symbols. Below the diagram, accessible and buried surface areas are tabulated for each subunit of the primary trimer for each indicated relationship and total buried surface areas are given for subunit assemblies.
For the most part the PMV coat protein structure is unexceptional for plant icosahedral viruses (Johnson and Argos, 1985; Johnson and Rueckert, 1997; Argos and Johnson, 1984; Rossmann and Erickson, 1985). Its principal elements are a canonical Swiss roll beta barrel of eight strands that lies relatively parallel with the overall surface of the particle and a long, largely unstructured amino terminal polypeptide containing a preponderance of positively charged amino acid residues. The A, B, and C subunits, whose organization about the quasi icosahedral threefold axis can be seen in Fig. 2, are essentially identical in structure with a mean rms difference between their common elements (see Methods section) of 0.67 Å (average of 129,240 pairwise superpositions). The greatest divergences involve the additional 30 or so residues observed in the amino terminal polypeptide of the C subunit relative to the A and B subunits. This peptide forms a distinctive arm that is not visible, and is presumably disordered in the other subunits. At residue Gln54 the divergence exceeds 9 Å between B and C subunits (Fig. S.7).
Two other differences between the quasi-equivalent subunits exist. First, the conformation of the backbone at Asp155-Glu157 of the B subunit differs from that of the A and C subunits by 2-2.5 Å, most probably due to the different quaternary interactions discussed below (Fig. S.8). Second, as noted above, the conformations of loop 88-96 differ due to different inter-particle contacts (Figs. S.3 and S.4).
The exterior and interior electrostatic surfaces presented by PMV are shown in Figs. 3e and 3f, respectively. The exterior surface is characterized by a roughly equal number of positive and negative charges interspersed among hydrophobic patches. The interior surface, as in many small icosahedral viruses, is quite different. It is an almost solid sheet of positive charge. This is all the more impressive because the polypeptides included in this rendering do not begin until residues 54 and 50 in the A and B subunits, respectively, and residue 22 in the C subunits. Thus, there is, in addition to the charges reflected in Fig. 3f, many additional positive charges contributed by the unseen amino terminal tails of the A, B, and C subunits. The accumulation of positive charge at and near the interior surface of the capsid undoubtedly reflects the existence of strong protein – nucleic acid electrostatic interactions. It is difficult not to think these important in virus assembly.
3.4 Comparison of PMV with TNV
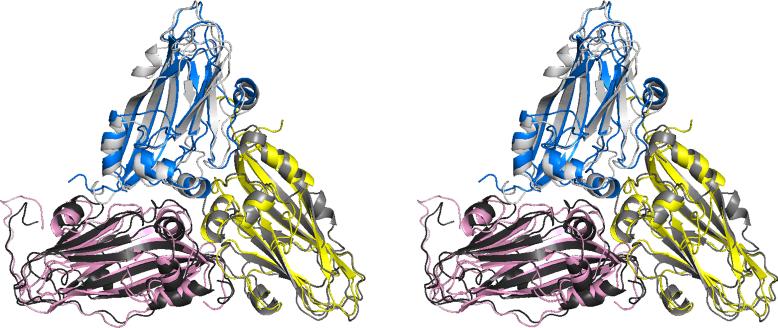
The coat protein of TNV is 276 amino acids in length while that of PMV is 242 and, as is evident in Fig. 1, the sequence identity between PMV and TNV is marginal at best (20%). Nonetheless, aside from the modest insertions and deletions, the secondary and tertiary structures of the two coat proteins are virtually the same. This is in large part because TNV contains a 32 residue extension at its amino terminus, and this polypeptide, along with a substantial number of upstream residues, is not visible in electron density maps of TNV, nor does it appear in the final TNV model (Oda et al., 2000). The polypeptide is disordered in the interior of the virion. The mean rms difference between 539 corresponding Cα atoms of the 120 ABC trimers of PMV and TNV is 2.6 Å (1.6 Å for 439 superposed Cα atoms), but the differences arise more from the very slightly different arrangement of the three subunits about the quasi threefold axis than from actual differences in the courses of their backbones. An ABC trimer of PMV is seen superimposed on a corresponding trimer of TNV in Fig. 5. The individual subunits have mean rms differences of 2.4, 2.4, and 2.9 Å for the A, B, and C subunits respectively (1.6, 1.4, and 1.5 Å, respectively for fitted atoms).
Fig. 5.
Stereoview of the trimer of A, B and C subunits of PMV, shown in blue, yellow, and pink, respectively, superimposed on the corresponding subunits of TNV, which are colored in three shades of gray.
In the sequence alignment in Fig. 1, there is a 15 residue segment in TNV that is absent in PMV following Ser166 in the latter. This segment exists in TNV as a flexible loop lying at the interior surface of the capsid. The loop contains no positively charged side chains, would not appear to be involved in nucleic acid binding, and would likely be structurally dispensable. The principal insertion in PMV with respect to TNV is a 13 amino acid segment between Leu86 and Ala100, which forms an extended loop that protrudes into solvent on the exterior of the capsid.
The loop assumed one discrete conformation when fully exposed to solvent, but when in the proximity of, or in contact with the surfaces of other virus particles in the crystal lattice, the loop took on alternate conformations. The upshot of this is that virus particles in the crystal lattice did not exhibit exact icosahedral symmetry, nor were the two particles within the crystallographic asymmetric unit absolutely identical. The deviation from precise icosahedral symmetry illustrated by this loop likely pervades entire virus particles, though perhaps in less prominent and perceptible ways than revealed by this example.
There are also 2 deletions and 1 insertion of single amino acids in the sequence of PMV with respect to TNV, all of which are in solvent exposed loops. One deletion in PMV, however, is of particular interest because it belongs to a loop, bound by Leu153 and Ser158, located at an interface between trimers. In TNV, this loop contributes to the coordination of a calcium ion. A corresponding calcium ion is absent in PMV. The reduction in length of the loop by one residue and the consequent alteration of the loop conformation in PMV likely accounts for the absence of calcium binding. Another single site deletion in the PMV sequence is within a loop comprised of residues 212 to 220 that is found on the exterior surface of the particle and is unlikely to be of consequence.
The only single amino acid insertion in PMV compared to TNV, is at the end of the segment from Lys173 to Pro181; this alteration appears significant. The segment forms a loop that approaches the quasi threefold axis of the ABC trimer and is located on the interior surface of the capsid. The structural alignment shown in Fig. 1 shows no sequence conservation here between PMV and TNV, which is reflected in a radical difference in loop conformation between PMV and TNV (Fig. S.9). There is nearly a 180° rotation about pivot residues Glu175 and Pro181. As a consequence, the loop in PMV directs the Arg177 side chain towards the interior of the capsid in proximity of the backbone of the RNA segment described below. In contrast, the arginine in the corresponding loop of TNV is not directed inward.
3.5 The Amino Terminal Polypeptides
Amino terminal polypeptides deserve attention because of their role in nucleic acid interactions, and probably in assembly (McPherson, 2005). In PMV, this polypeptide is shorter, but it contains more concentrated positive charge than does the polypeptide in TNV. In the first 36 residues of PMV, there are 9 arginine and 2 lysine residues, which correspond to 30% of the amino acids in the PMV amino terminus. Half of them are concentrated between residues 22 and 28. These basic residues alone are sufficient to neutralize nearly 50% of the negative charges of the encapsidated RNA.
There is also a high incidence of glycine and proline in the first 45 amino acids (9 glycine and 7 proline residues), which probably insures conformational fluidity in the absence of RNA, and allows multiple discrete conformations in its presence. Residues up to position 54 and 50 in chains A and B, respectively, are disordered in PMV. The extremely hydrophilic, flexible character, and unusually positively charged amino terminal tail is consistent with interaction with nucleic acid.
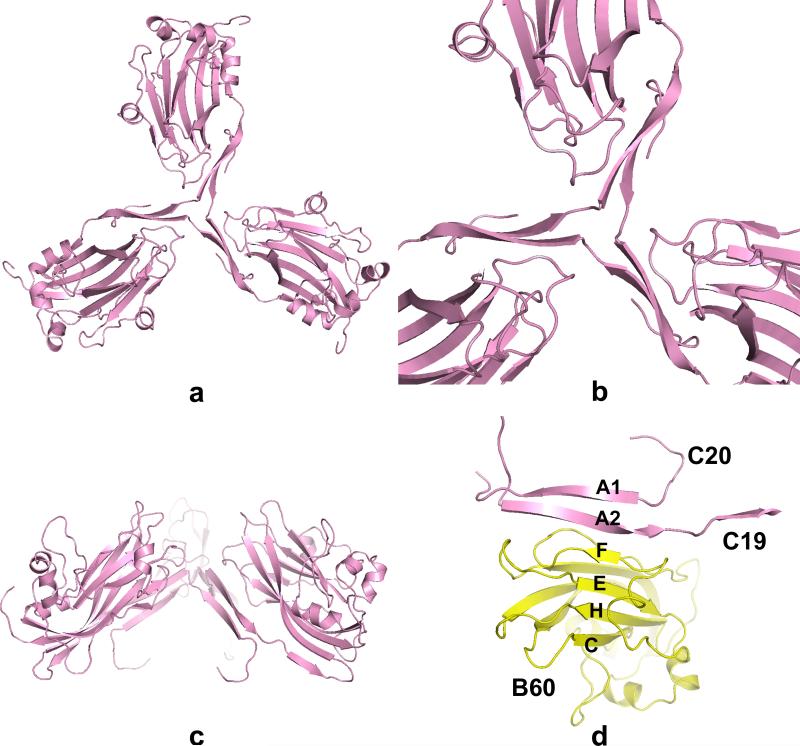
Amino and carboxy terminal polypeptides have frequently been assigned unique roles in virion structure. Amino termini are commonly associated with protein-RNA interactions (McPherson, 2005) while carboxy termini often form beta annuli or other prominent structural elements that maintain the capsid (Speir et al., 1995; Canady et al., 1996; Larson et al., 2000). In PMV the amino termini of the three quasi-equivalent subunits are distinctly different (see Fig. S.7); whereas the amino termini of the A and B subunits are disordered up to Gln54 and Met50, respectively, the amino termini of the C subunits, before being lost in the virus interior, cross over one another at the exact threefold axis (the quasi sixfold axis) and form a tight triangle as shown in Figs. 6a-c. The C subunit forms, as noted above, a unique “arm” made possible by the tight turn at Gly55, which turn is absent in the A and B subunits, and the subsequent inter-strand hydrogen bonding. This turn allows the formation of strand A3 (residues 49-53) that extends the BIDG β sheet of the C subunit to five strands. There are three proline residues (Pro36, Pro45, Pro48), which are the only conserved residues in the visible N-terminal arms up Arg49 of both PMV and TNV, that are located at or near inflection points along the arm and delimit the strand segments A1 (residues 29-35), A2 (residues 37-44) and A3. As illustrated in Fig. 6d, these strands result in an intricate weave of strands that extends four-stranded CHEF β sheets of the B subunits to six strands with strands A2 and A1 (each from a different C subunit) as the fifth and sixth strands, respectively. A similar arrangement was observed for both TNV and CfMV, where it was postulated to participate in conformational switching during assembly of particles (Oda et al., 2000; Tars et al., 2003).
Fig. 6.
In (a), (b) and (c), the concerted interactions of C subunits (C1, C19 and C20 in Fig. 4) around the icosahedral threefold axis through their amino terminal polypeptides are shown. In (d), the amino terminal polypeptides of C19 and C20 pass by the threefold axis to extend the CHEF β-sheet of the B60 subunit to six strands.
3.6 Interactions About the Fivefold and Quasi Sixfold Axes
There is another short segment of somewhat unique composition that is common to TNV, CfMV and PMV. It is also conserved in other tombusviridae and sobemoviruses such as Olive Mild Mosaic Virus (Cardoso et al., 2005) and Southern Bean Mosaic Virus (cowpea strain) (Chen et al., 1989, Silva and Rossmann, 1985). This segment has the sequence Cys – Pro – Thr – Thr – Thr (aa 126 – 130 in PMV). In some other viruses, such as Olive Latent Virus, serine replaces threonine (Felix et al., 2005). The peptide segment is a loop between consecutive strands of the beta sheet. Symmetrical loops create a crown, or cusp, at both the fivefold and quasi sixfold axes of the virion. The conservation of this segment among proteins of such marginal homology suggests that it may indeed be crucial in maintaining virion structure, or in assembly of particles.
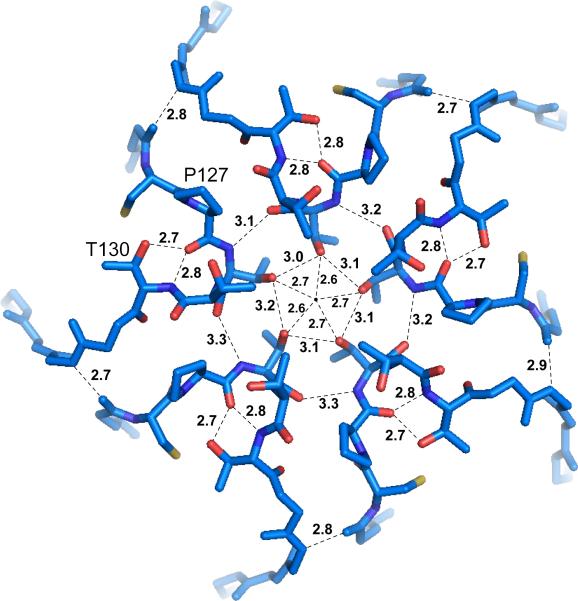
At a fivefold icosahedral axis, shown in Fig. 7, Thr128 from five symmetry related subunits are arranged around the axis such that their side chains, as well as their main chain nitrogen and carbonyl oxygen atoms, form hydrogen bonds with one another to form a closed circle. The average separation between the hydrogen bonded OG1 side chain atoms of these residues is about 3.1 Å, which translates to a circle of radius of 2.64 Å. Subtracting the 1.4 Å van der Waals radius of the oxygen atoms gives a pore radius of about 1.24 Å, large enough only for the smallest of ions without requiring displacement of the Thr128 residues. That constellation is made possible by proline residues 127 and 131 that create a relatively unrestrained loop conformation. Additional hydrogen bonds that contribute to the stability of this pore are formed by the main chain nitrogen and the hydroxyl oxygen of Thr130 and the carbonyl oxygen of Pro127. With the exception of Pro131, all residues are conserved between PMV and TNV.
Fig. 7.
The conserved amino acid sequence motif C(126)-P-T-T-T(130) found in both PMV and TNV are located in a loop, and five of these loops come into close proximity about the icosahedral fivefold axis. The cluster of loops produces a pore radius of 1.24 Å and severely inhibits any entry into the particle along this direction. The distances shown are for the pentamer containing A1 of particle 1.
At the quasi sixfold axes, the arrangement of Thr128 residues is much different. Instead of a contiguously hydrogen bonded threonine arrangement forming a symmetrical hexagon, they form hydrogen bonded pairs at the corners of equilateral triangles with the shortest distance between pairs of 9.7 Å. Compensating for the van der Waals radii of the oxygen atoms, this translates to a pore size of about 4.2 Å radius. However, this sizeable pore is filled by the side chains of the three Leu37 residues on the N-termini of the C subunits associated with the quasi hexamer. The pore radius created by the Leu37 side chains is 1.28 Å, approximately the same as the pore at the fivefold axes. Therefore, only the smallest ions have access to the interior of the particle along either of the fivefold or quasi sixfold axes.
As noted above, the only cysteine (Cys126) in the coat protein, which lies at the amino terminal end of the loop forming segment, binds mercury when exposed to OCMP or PCMB. Not all of the 180 cysteine residues in the capsid react with Hg, however, which would seem to imply some small variation in the disposition of the loop. The cysteine of subunits about the fivefold axis are reactive, but at the quasi sixfold axis, only the B subunits react with mercury, the C subunits do not. This is curious, since the solvent accessibilities to the cysteine residues in both B and C subunits seem to be the same, except that the neighboring backbone residues of this reactive cysteine in C subunits, make hydrogen bonds with the backbone of its own N-terminus. This possibly restricts the loop of the C subunits from undergoing an alteration in conformation that would allow its thiol group to become accessible to the heavy atom reagent. This is the threefold axis where N-termini of the C subunits meet to form beta strand annulus interactions.
3.7 Calcium ion binding
PMV is completely different from both TNV and CfMV in terms of calcium binding. Given the similarities in the three dimensional structures of the three viruses, this seems to suggest that coordination of calcium may play only an ancillary role in the determination of capsid architecture and its stability.
TNV has 5 independent calcium binding sites. Three of these sites are located between the monomers of the quasi-equivalent ABC trimer about 14 Å from the quasi threefold axis and have essentially equivalent coordination. These sites, also found as the only calcium binding sites in CfMV, are characterized in each structure by a pair of aspartate residues (Asp160 and Asp163 in the TNV sequence) of one monomer and the two or three ligands provided by the other monomer. The TNV sites are square planar except for the site between the A and C subunits in which a water molecule was modeled at an apex of a possible octahedron; the Ca++...O distances are in the range of 2.48-3.30 Å, which are long for calcium-oxygen coordination. In contrast, the CfMV sites are square pyramidal with the apex occupied by the terminal carboxylate group and the Ca++...O distances range from 2.29 to 2.51 Å, which are very characteristic of calcium-oxygen coordination. In the PMV structure at these locations, on the other hand, there are possible hydrogen bonds between Ser143 OG and Glu197 O of one subunit and Tyr202 OH and Arg184 NH2 of the other subunit, respectively. No density corresponding to a calcium ion could be seen.
The other two TNV calcium sites are quasi-equivalent also. They have the same coordination but one site is located between each pair of A subunits around the pentamers while the other site is located between C-B pairs of subunits in the quasi hexamer but not between B-C pairs. Based on the model in the PDB, these sites are best described as square planar, are about 28 Å from the quasi threefold axis and have coordination distances in the range of 2.62 –2.90 Å, which again are long for calcium-oxygen coordination. These sites are not present in CfMV and PMV. Within about 6 Å of these locations in PMV there are at least five hydrogen bonds. For a particular A subunit there are hydrogen bonds to the neighboring A subunit in the pentamer and to the B subunit of its ABC trimer (subunits A56 and B1 in Fig. 4 for subunit A1). For a particular C subunit, hydrogen bonds are formed with the neighboring B subunit in the hexamer and the A subunit of its ABC trimer (subunits B53 and A1 in Fig. 4 for subunit C1). These hydrogen bonds are A(C)-Arg137 NH2 to B(A)-Ala234 O, A(C)-Asp155 O to A(B)-Ile64 N, A(C)-Glu157 OEx to B(A)-Ala 234 N, A(C)-Ser158 to B(A)-Arg224 NH2, and for the B-C interaction only, C-Glu157 N to B Glu62 OE2.
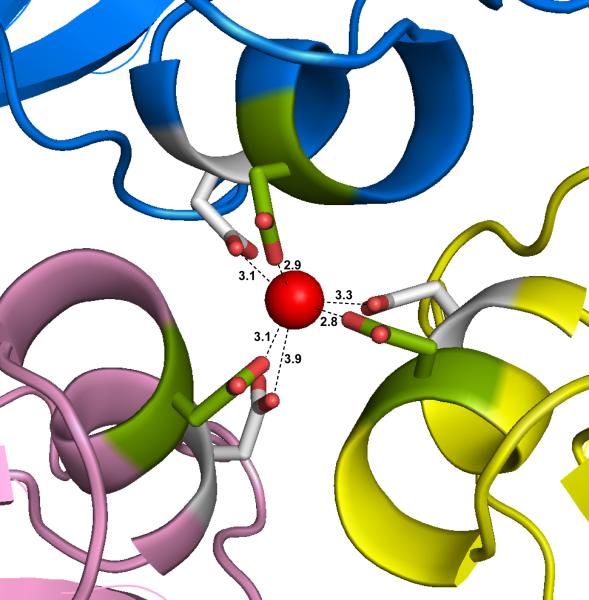
PMV has, however, putative bound calcium ions at another location. As shown in Fig. 8, a calcium ion is found at the quasi threefold axis of symmetry relating the A, B, and C subunits. Three quasi equivalent aspartic acid side chains (Asp198) form a triangle just above the ion and three quasi equivalent Asp201 side chains form a similar triangle just below the ion. The two triangles are rotated slightly with respect to one another and thereby form a distorted octahedral cage about the Ca++ ion. This pair of aspartate residues also form a D-X-X-D configuration of residues though in a completely different location than the D-X-X-D configuration in TNV and CfMV. The distances between the ion and the nearest carboxyl oxygen atoms have a large range of 2.70 – 3.94 Å over the 120 ions in the two particles. Of the six possible calcium ligands, no more than two at any site has a distance less than 3 Å. This suggests that the ions are mobile within the cage as is also suggested by their high B factors. A corresponding ion is absent in TNV and CfMV, but is seen in less closely, but still structurally, related Black Beetle Virus (Hosur et al., 1987) and Sesbania Mosaic Virus (Bhuvaneshwari et al., 1995).
Fig. 8.
The putative calcium ion on the quasi threefold axis is shown as a red sphere, with its six octahedrally arranged carboxylate ligands forming a cage about it. Three of the side chains are provided by symmetry related Asp198 (green) and three by symmetry related Asp201 (white). These were the only calcium ions observed in the PMV capsid. The distances are for the primary quasi trimer A1, B1, and C1 of particle 1.
3.8 Some Additional Tertiary and Quaternary Interactions
Hydrogen bond interactions within and between A, B and C subunits in PMV are too numerous to describe in detail, but some electrostatic interactions, or salt bridges, are worthy of note. These are either intra-subunit bonds that stabilize the tertiary structure of the coat protein, or inter-subunit bonds that appear to be important in the assembly and stability of the capsid.
Using a 4 Å N...O distance criterion, some specific salt bridges can be identified. Possible inter-subunit salt bridges exist between Arg47 and Glu157 of the C and B subunits, respectively, in the quasi hexamers (<d> = 3.99 Å). This interaction does not exist between the B and C subunits because the main chain conformation of the C subunit at Glu157 differs from the B subunit. An equivalent interaction between A subunits around the fivefold axis does not exist for the same reason. Around the quasi threefold axis, there are possible salt bridges between Arg85 and Glu145 of the A and C subunits, respectively (<d>=4.0 Å). However, in the B-A and C-B interfaces, the minimal average distances are 4.51 Å and 6.41 Å and often have positive difference density between the guanidinium and carboxylate groups of the respective amino acids (Fig. S.10).
There are four possible intra-subunit salt bridges but they are not found in every icosahedrally unique subunit. The Arg137 – Glu157 and Arg137 – Asp155 exist in the B subunits (<d> = 3.49 Å and <d> =2.70 Å, respectively), but not in the A and C subunits, a result of differences in backbone conformation between B subunits and the A and C subunits (Fig. S.8). Common to all subunits are the interactions between Arg232 and Glu182 and between Arg85 and Asp187 (<d> = 3.21 Å and <d> = 3.40 Å, respectively).
3.9 RNA
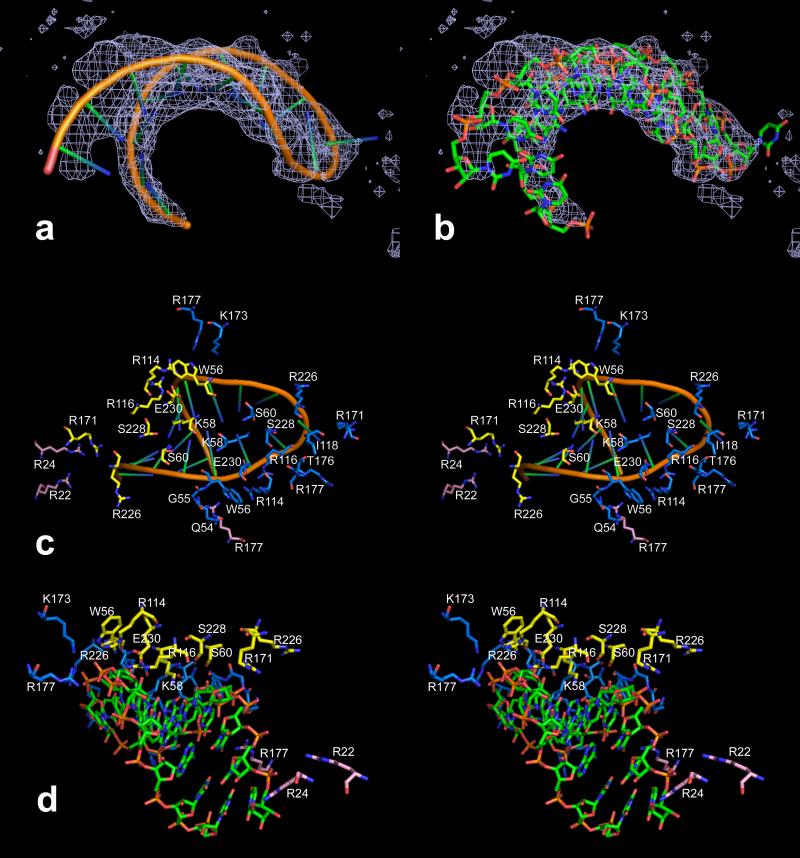
We examined the averaged electron density on the interior of the protein capsid in search of genomic RNA that was consistent with the icosahedral symmetry of the virion (otherwise it would not be seen). Ordered segments of single stranded RNA, as well as loops and double helical stems have now been observed in numerous icosahedral viruses (Johnson and Rueckert, 1997; Larson et al., 1993, 2000, 2005a). In the case of PMV we visualized in 2Fo-Fc and conventional difference electron density maps an RNA hairpin loop of 17 nucleotides. This is shown in Figs. 9a and 9b. The RNA fragment was built as the generic sequence U2A2UAU5AU5 with the four adenine bases paired, based on the best fit to density. The 5’ and 3’ ends were built as uridine because the base density was weak and base pairing appeared to break down as the ends diverge.
Fig. 9.
In (a) and (b), NCS averaged 2Fo–Fc electron density is shown superimposed on the model of the 17 nucleotide hairpin loop of RNA found near the interior surface of the PMV capsid. The map was calculated at 2.9 Å resolution and contoured at 0.4 sigma. In (c) and (d), stereo views of the RNA in two different representations illustrate the possible RNA-protein interactions with capsid residues. All close protein residues are labeled in (c) but not in (d). The A, B and C subunits are colored blue, yellow and pink, respectively.
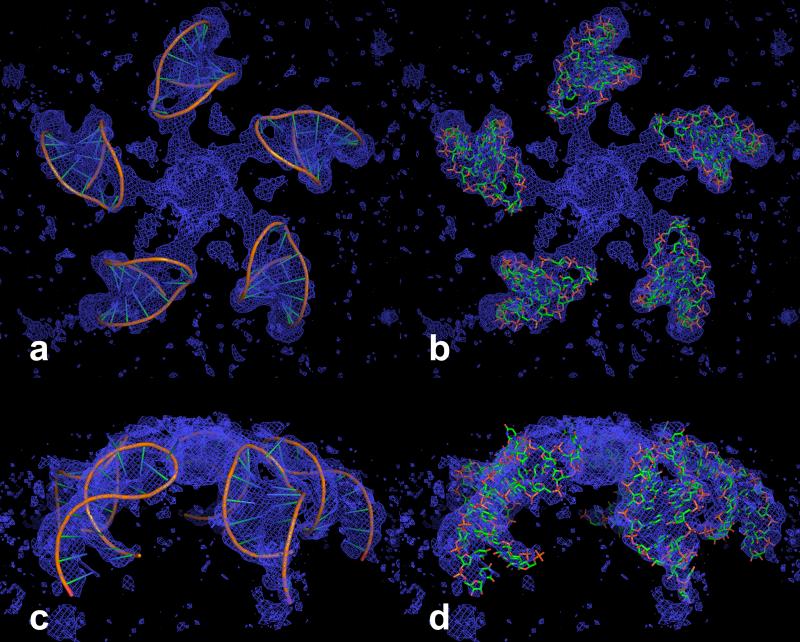
The helical stem is intersected between the U5-A12 and A6-U11 base pairs by the quasi dyad of the A and B subunits (A1 and B53 in Fig. 4). This is illustrated in Figs. 10a, 10c and 10e. As seen in Fig. 11, the RNA loops are disposed about the fivefold axes, with the loop end directed toward the axis and about 18 Å from it. The loops do not invade the pentameric capsomeres as was observed for similar loops seen in Turnip Yellow Mosaic Virus (Larson et al., 2005a) and Desmodium Yellow Mottle Virus (Larson et al., 2000). Upon lowering the contour of the density of an averaged 2Fo-Fc map, however, arms of density appeared leading from the hairpin loop at the phosphate of U9 toward the fivefold axis where the five arms coalesce into a sizeable density mass. The prominent feature on the fivefold axis is about 16 Å long (along the fivefold axis) and about 20 Å in diameter. The arms become continuous between the hairpin loops and the density on the fivefold at a contour level of 0.40 σ. These features are illustrated in Fig. 11.
Fig. 10.
Comparison of the capsid dimer-RNA complex of PMV with the equivalent dimer-RNA complex of STMV. In (a), (c) and (e) are ribbon representations of A (blue) and B (yellow) subunits of PMV (related by an icosahedral quasi twofold axis) and their bound RNA. In (b), (d) and (f) are comparable views of the capsid dimer-RNA complex from STMV. Panels (a-d) are views perpendicular to the twofold axes and panels (e) and (f) are views along the twofold axes from the particle centers.
Fig. 11.
NCS averaged 2Fo-Fc map, calculated at 2.9 Å resolution and contoured at 0.32 sigma, superposed on the five RNA hairpin loops around a fivefold axis. At this contour level, arms of density can be seen connecting the hairpin loops to the mass of density on the fivefold axis. In (a) and (b) the view is down the fivefold axis. In (c) and (d) the view is perpendicular to the fivefold axis.
It should be noted that the B factors for the RNA are elevated (see Table 1) and the RNA density confused in unaveraged electron density maps. However, the density is clear in averaged maps as seen in Figs. 9a, 9b and 11. Since averaging was performed using masks that covered the entire interior of the particle and there is no appreciable density on the interior except for the hairpin and the density leading to the fivefold axes, we feel confident that our model is a fair representation of the visible RNA. Refinement suggests that the occupancy is in the range of 70% to 100%. Large B factors were present throughout the structure, including for capsid proteins and calcium ions, so they were not unexpected for the RNA, particularly given (1) the breakdown of icosahedral symmetry due to the non-repeating RNA sequence (i.e., no 17 nucleotide sequence repeats 60 times through the length of the RNA), (2) disorder created by connection diversity at the ends of the hairpins, and (3) the possibility that not all hairpins are complete loops but may be diverted along the density arms leading to the accumulation at the fivefold axis.
3.10 RNA – Protein Interactions
A number of interactions between the RNA and coat protein are evident, but only involving nucleotides 5-7 and 9-13. These are illustrated in Figs. 9c and 9d. Six hydrogen bonds are formed with main chain atoms (Gln54, Gly55, Trp56, and Leu118 of the A subunit and two bonds from Trp56 of the B subunit). Of these, the interaction of Leu118 would be base-specific since it is in position to form a hydrogen bond with O4 of U9 at the apex of the RNA loop. Cytidine at this position would be sterically unfavorable and, although directionality would be good, the distance to the N7 of purine bases would be greater than 3.5 Å. Ten hydrogen bonds are possible with side chain atoms of Lys58 (3 bonds), Ser60, and Glu230 of the A subunits and Lys58 (2 bonds), Ser60, Arg116 and Glu230 of the B subunits. The quasi dyad symmetry holds well for these interactions. Of these side chain interactions, the pair of Lys58 residues from the A and B subunits are particularly interesting since they protrude into the “minor groove” of the hairpin loop and make hydrogen bonds to O2 of two or three uridine bases that are base-paired in our model (U7, U10 and U11 for the A subunit and U5 and U13 for the B subunit). This insertion is reminiscent of the Val38 side chains in STMV that protrude into the minor groove of the helical segments of that structure (Larson et al., 1993, 1998).
There are also two arginine residues (Arg226 of the A subunits and Arg114 of the B subunits) and one lysine residue (Lys173 of the A subunits) within 4 Å of phosphate oxygen atoms that could be directly or indirectly interacting with the RNA, and another nine arginine residues from all three subunits (residues 114, 116, 171, and 177 of the A subunits; 171 and 226 of the B subunits; and, 22, 24, and 177 of the C subunits) that are directed toward the RNA with their guanidinium groups within 4 to 11 Å of electronegative atoms of the RNA. The average buried surface area of the RNA is 898 Å2, about 26% of the total RNA surface area.
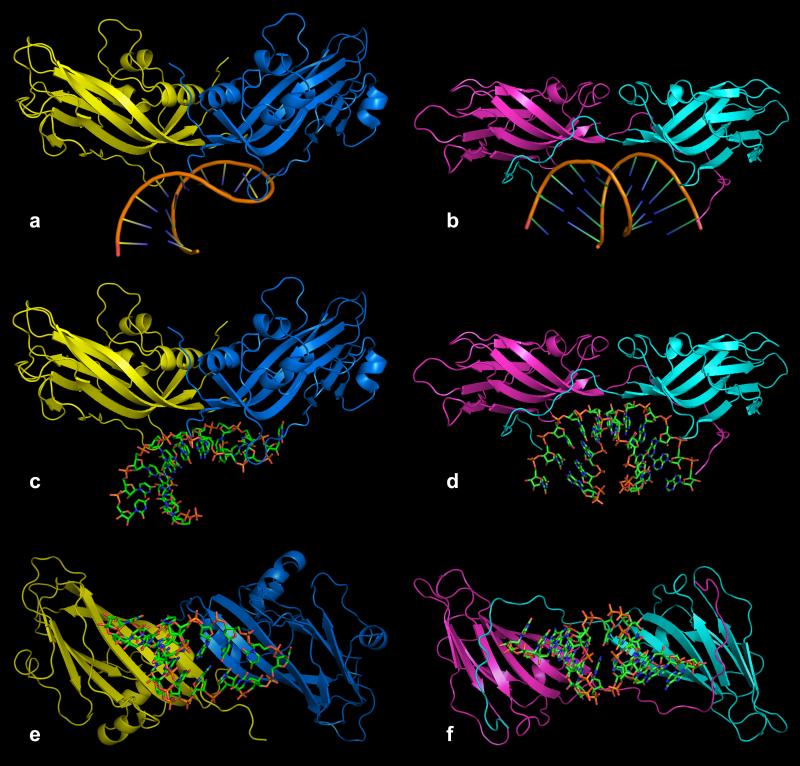
There is a marked resemblance between the RNA fragment and its protein binding site on the interior of the PMV capsid and the RNA and its binding site in satellite tobacco mosaic virus (STMV; Larson et al., 1993, 1998). This is illustrated in Fig. 10. In both viruses the RNA fragment lies at the interface of two protein subunits. In STMV the RNA fragment is bisected by an exact icosahedral twofold axis, which relates two opposing strands of an RNA duplex. In PMV, the RNA fragment is bisected by the quasi twofold axis relating A and B subunits. The overall disposition of the RNA fragment and the orientation of the fragment axis with respect to the beta strands of the protein subunits also appear to be very much the same. There is no known phylogenetic relationship between PMV and STMV, hence we might conclude that the binding site found in the two viruses may represent a more general RNA binding site likely to be found in other viruses.
An RNA duplex residing on an icosahedral twofold axis, but exhibiting no connecting loop, was also observed in the structure of Flock House Virus (FHV; Fisher and Johnson, 1993). They proposed that in Nodaviruses the RNA segment might be an important participant in the assembly of the virus. They further suggested that the RNA segment, along with an associated amino terminal peptide, might serve as a “conformational switch” that determines which of the otherwise identical coat protein polypeptides ultimately become A, B or C subunits. The RNA segment that we see in PMV is compatible with that idea, and we believe it may provide a similar function.
PMV, STMV and FHV are three good examples of helical RNA segments bound to protein dimers, the first located about an icosahedral quasi twofold axis and the latter two located about strict icosahedral twofold axes. Figs.12a-c illustrate these three RNA-protein complexes viewed down the helical axes of the RNA segments. In all three cases the salient feature is the clamp-like binding site, formed by the two protein molecules in each complex, that wrap around a considerable portion of the surface of the RNA helix, regardless of whether the protein dimer has exact or quasi twofold symmetry. Fisher and Johnson (1993) pointed out that the AB quasi twofold dimer in FHV is inwardly bent (see Fig. S.11) with respect to the CC dimer (referred to as “flat”) such that the α-helical features of the protein, which form the clamp arms of the binding site, are closed and, therefore, can't bind a helical RNA segment. In contrast, Figs. 12a and 12d-f illustrate that the CC twofold dimer in PMV, because it is flat, does not form the same clamp-like binding site that is observed in the AB dimer. In each case, the RNA can serve to “select” the dimer relationship (whether quasi twofold or exact twofold), but clearly the protein fold in combination with the “bent” (quasi twofold) or “flat” (exact twofold) relationship of the protein subunits dictate the formation of the binding site.
Fig. 12.
Comparison of binding sites of helical RNA structures with quasi twofold and exact twofold dimers. All views are down the helical axes of the RNA structures. In (a) is a ribbon diagram of the complex found in PMV in which the RNA is bound to the quasi twofold AB dimer (subunits A and B are blue and yellow, respectively). In (b) and (c) are the ribbon diagrams of the helical RNA structures that are bound to icosahedral twofold protein dimers in FHV and STMV, respectively. In each case, the protein dimer forms a clamp-like pocket around the RNA helix. In (d) is a ribbon diagram of the PMV AB dimer without RNA. In (e) the C1-C4 (see Fig. 4) exact twofold dimer (pink) has been superposed on the AB dimer using only Cα atoms of the A and C1 subunits. It is clear that the C4 and B subunits do not align well, illustrating the enormous difference between the quasi twofold and exact twofold dimers in PMV. In (f) the B subunit has been removed, revealing that the C-C dimer does not form a clamp-like structure that could wrap around an RNA helical structure, mainly due to the presence of the N-terminal arms of the two C subunits. These arms seem to simultaneously occupy the potential binding site and spread the dimer into a “flat” dimer versus the “bent” conformation of the AB dimer.
It is tempting to conclude that the RNA driven conformational switch is more general and is operative in all ssRNA viruses where T > 1. Indeed, ordered segments of RNA have now been observed in the virions of numerous small viruses (Carrillo-Tripp et al., 2009). There are, however, examples of T > 1 viruses, TYMV being prominent among them, that appear to assemble completely native capsids with A, B and C subunits in the absence of nucleic acid. Interestingly, however, an appreciable amount of icosahedrally ordered RNA is observed in close association with the interior surface of the capsid of TYMV (Larson et al., 2005a).
4. Discussion
4.1 The Structure Analysis
The crystallographic analysis of PMV represented one of the more challenging in terms of the size of the asymmetric unit and the number of data that had to be recorded, processed, and managed, and because of the number of polypeptide chains and atoms that had to be refined. Because of the low symmetry, with four particles in the monoclinic unit cell, the asymmetric unit of two full virus particles had a molecular weight, protein alone, of 9.5 MDa, or 10 MDa if the observed nucleic acid is included. The number of independent reflections to 2.9 Å was 2,217,465. The ABC protein subunit trimers are 120 fold redundant in the asymmetric unit, thus the number of reflections per atom in the 2.9 Å resolution (restrained) structure is about 3.46, which provides a secure margin even by conventional protein crystallography standards.
The R and R free might be considered elevated in comparison with some well refined proteins; however, we would point out the following. In higher resolution shells of reflections for the PMV crystals, as with most virus crystals, there is a large amount of very weak data. In addition, the crystal from which the data was collected, though frozen and showing no measurable signs of decay, was exposed to the X-ray beam for more than 24 hours. Thus some radiation damage was inevitable. The bulk solvent correction was probably inadequate to the refinement since much of the volume of the crystal, the interiors of the virus particles, are filled with nucleic acid, not low-density water. Finally, there are 7380 amino acids from the unseen amino terminal polypeptides that are present in the crystal, but insufficiently ordered to be modeled.
We refined our model of PMV initially using icosahedral constraints, assuming also that the two particles within the asymmetric unit were identical. It became evident, however, that those assumptions were too stringent in this particular crystallographic problem. Indeed, there are no crystallographic constraints that require the two particles in the asymmetric unit to be identical. We believed them to be so only because they are both PMV virions, and we assume, probably incorrectly, all virions to be identical.
Similarly, there are also no crystallographic constraints that require all ABC subunit trimers to be identical, or even the individual A, B, and C protein subunits throughout a virus particle. We tacitly assume that they are, because we believe all PMV virions exhibit exact icosahedral symmetry, but this is biologically determined, and biology, unlike crystallography, need not be perfect. Many of the subunits in each particle are involved in contacts with neighboring particles in the crystal lattice. Because of the low symmetry, these interactions are different for most subunits. This is demonstrated most explicitly in PMV by the clearly evident variable dispositions assumed by the loop segment formed by amino acids 87 through 99 in response to neighboring virus particle interactions.
It is true that release of strict icosahedral constraints and the requirement for exact particle identity injects parameters into the refinement process. Such an increase alone, it might be argued, could conceivably allow the refinement procedure to drive the working R factor to lower values, that is, for “over refinement” to occur. We note, however, that replacement of rigorous constraints with conservative restraints resulted in a significant reduction in Rfree as well, and the model itself shows, as a consequence of restrained refinement, perceptible improvements in the stereochemistry, and a marked reduction in bad interparticle contacts within the unit cell. There are, in fact, other examples where, for similar reasons, icosahedrally constrained refinement was abandoned in favor of restraints for virus particles (Larson et al., 2005b, Lane et al., 2011).
Comparison of the restrained and constrained models demonstrates that the overall structures are nearly identical and that the former deviates very little from strict icosahedral symmetry. With respect to the overall structures, Table 1 shows that the particle centers changed by almost 1 Å; however, that change is principally in the y direction representing a drift of the origin in the polar space group P21 during refinement. Superposing the particles of the restrained model onto the corresponding particles of the constrained model (Cα atoms of residues 56-88 and 96-233) gave rotation angles of 0.008° and 0.027° and rms deviations of 0.82 Å and 0.91 Å for particles 1 and 2, respectively.
With respect to icosahedral symmetry, we calculated the NCS operators of the two particles in each model in terms of Euler angles (α, β, γ) and compared each of the corresponding sets for particles 1 and 2 of the two models by determining the rms differences between the sets. For particle 1, the rms differences were 0.21°, 0.18° and 0.26° for α, β and γ, respectively. Similarly for particle 2, the deviations were, respectively, 0.21°, 0.18° and 0.25°. After adjustment for the origin shift, the translation vectors (x, y, z) of the NCS operators were, respectively, 0.55 Å, 0.55 Å, and 0.52 Å for particle 1 and 0.76 Å, 0.76 Å, 0.85 Å for particle 2; the centers of mass of the corresponding trimers had rms differences of 0.41 Å and 0.55 Å for particles 1 and 2, respectively, with a maximum difference of 1.01 Å in particle 2.
4.2 Homology With Other Viruses
As suggested by the absence of any sequence identity, and the immunological findings (Masuta et al., 1987; Buzen et al., 1984), there is no obvious structural similarity between the coat proteins of PMV and SPMV other than that seen among virtually all small icosahedral plant viruses. On the other hand, although no longer a source of wonder to most students of protein structure, we continue to be impressed by capsid protein molecules from different viruses having less than 20% sequence identity but exhibiting extraordinary structural congruity. This is what we find for PMV, TNV and CfMV. Structural similarity persists in spite of each virus having a different number and distribution of calcium binding sites, as well as insertions and deletions in their respective sequences. The former suggests that metal binding sites in virus protein shells may, per se, have rather little to do with determining or organizing structure, or promoting assembly, but may be more involved with physiological response to pH and host specific factors, including particle swelling and contraction. It is worthy of note that the major calcium binding locus in TNV, the most closely related homologue, is replaced by a prominent electrostatic association, the Arg85 – Glu145 bridge in PMV. This suggests that some sort of strong structural interaction is required at that point to serve as a “hook and eye” in the tertiary structure, but its exact nature is less important.
4.3 RNA
One of the more unique and intriguing aspects of the PMV structure is the 17 nucleotide hairpin loop seen bound by an A and B subunit near their quasi twofold axis. This was not observed in either TNV or CfMV. Because there are 60 equivalent such sites of binding and if full occupancy is assumed, then the RNA loop accounts for about 17% of the entire single stranded RNA genome. The interactions between the RNA loops and the interior of the capsid are both intimate and extensive, with salt bridges between basic amino acid side chains and negatively charged phosphate groups evident, as well as numerous other close contacts undoubtedly involving hydrogen bonds and hydrophobic associations.
RNA secondary structural elements have now been visualized in a large number of small icosahedral viruses (Johnson and Rueckert, 1997). These elements have included RNA helical duplexes, usually associated with dyad and fivefold axes (Larson et al., 2005a; Makino et al., 2006; Larson et al., 1993; Fisher and Johnson, 1993), and various single stranded loops of diverse conformations that either invade capsomeres (Larson et al., 2005a) or simply make close interactions with the interior of the capsid as we see here (Larson et al., 2000; Chen et al., 1989; Tsao et al., 1991).
The existence of the icosahedrally consistent RNA loop – protein capsid interactions, a distinctive feature of the PMV structure, suggests that the interactions likely play some useful role in either maintaining the integrity of the virion, or in directing its assembly. Both are of course possible, and perhaps likely. By interacting with multiple subunits in an asymmetric manner, the RNA loop may act as a conformational switch to create an AB subunit pair and a quasi dyad axis, rather than allowing formation of a homodimer and an exact axis. A break in symmetry of this kind is essential in the assembly of a T > 1 icosahedral particle. An RNA influence on subunit conformation similar to the one seen here has been previously described for other icosahedral viruses, most notably in Flock House Virus (Fisher and Johnson, 1993).
The presence of unique, icosahedrally ordered protein – RNA interactions is consistent with both kinds of molecules taking part in a cooperative assembly process in which the RNA first recruits protein subunits around it, while the protein, in forming an icosahedral shell, marshals the nucleic acid into its encapsidated conformation (Larson and McPherson, 2001; Larson et al., 1998; McPherson, 2005). It seems implausible that a relatively unstructured and asymmetric mass of free RNA somehow projects 60 icosahedrally symmetric, structurally identical loops to contact the virus capsid at 60 similarly symmetric points. On the other hand, it appears from this, and other studies that encapsidated RNA can exhibit conformations that are more icosahedrally consistent with the protein capsid than might have been anticipated. Increasingly, findings are accumulating that suggest both RNA and amino terminal polypeptides are initially crucial to virus assembly, and that a relatively fluid RNA may ultimately, through interaction with its coat proteins, become more highly structured (McPherson, 2005).
5. PDB accession codes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4FY1, 4FY2, 4FY3, 4FY4, 4FY5, 4FY6, 4FY7, 4FY8, 4FY9, 4FYA.
Supplementary Material
Fig. S.1 A schematic diagram of the phase refinement procedure (with initial and final statistics) used to obtain the Fourier map from which the initial full PMV model was built.
Fig. S.10 Three examples of positive difference density (contoured at 3.0 sigma) found near the carboxyl group of Glu145 between Glu145 and Arg85 of a neighboring subunit. In several cases, the bond lengths in the side chain of Glu145 are elongated as though refinement is compensating for some missing entity, possibly water, that exists between Glu145 and Arg85.
Fig. S.11 A ribbon diagram of the quasi twofold AB dimer in Flock House Virus. The clamp-like binding site of the C-C icosahedral twofold dimer does not exist in this dimer (compare Fig. 12).
Fig. S.2 Icosahedral symmetry averaged Fo electron density map using experimental phases obtained by successive rounds of density modification, averaging, and extension, showing side chain densities of a region of the amino terminal polypeptide (S(29)-VERGST-P(36)) of the C subunit. The TNV poly-alanine model was used for initial phase calculations at 20 Å resolution. The density shown here was computed at 4 Å resolution.
Fig. S.3 Illustration of the difference between loops 88-96 of two capsid subunits. Panels (a-c) are stereo views with loop 88-96 of a C subunit of particle 1 in pink and a C subunit of particle 2 in orange (C44 and F85, respectively, in our working system). The C44 subunit has been superposed on the F85 subunit over 171 Cα atoms excluding terminal residues and residues 89-95. These two loops represent the most extreme difference between any two loops 88-96 with distances between corresponding Cα atoms as large as 9 Å. Panels (a) and (b) show two close-up views of the loop 88-96 superposed on the unaveraged 2Fo-Fc density around subunit F85 contoured at 0.6 sigma that demonstrate the difference between the conformations of these two loops. In (c) these loops are viewed in context of the F85 subunit environment with the proximal B subunit of particle 1 in yellow. Density for the complete F85 subunit is shown in light blue and the darker blue density highlights the density of loop 88-96 of F85.
Fig. S.4 Illustration of the difficulty NCS averaging of the exteriors of the particle can create. In both (a) and (b) the C44 subunit around loop 88-96 is in pink and an A subunit of a neighboring particle is in blue. In (a) is superposed the density of an NCS averaged 2Fo-Fc map (contoured at 0.7 sigma) drawn only around the C44 loop and adjacent residues of C44 and the A subunit. The direction of the loop density is reminiscent of the density of the F85 subunit whose loop has a conformation similar to the majority of the loops (see Fig. S.3). In (b) is superposed the unaveraged 2Fo-Fc density (contoured at 0.6 sigma), which follows the C44 loop and is separated from the density of the proximal A subunit. Building the C44 loop according to the averaged map results in penetration of the loop into the A subunit.
Fig. S.5 There are several observations that identify differences in the B and C subunits. A most likely contributor to these differences is the packing within the capsid. The A, B and C subunits are blue, yellow and pink, respectively. In (a-c) the B subunit has been rotated 60° about the threefold axis toward the C subunit and then translated to bring their centers of mass into coincidence. Clearly, they do not align well with this simple treatment. From the position shown in (a-c), the B subunit was superposed onto the C subunit by a least squares fit of 166 Cα atoms resulting in the rotation axis seen in (a) and a rotation angle of 29.4° shown in (b). The views in (b) and (c) are down the rotation axis. In (c) vectors Vb (B subunit) and Vc (C subunit) are defined by the Cα atoms of residues 76 and 159 in the C and F strands, respectively, of the CHEF sheet. These vectors serve as an internal reference to the dispositions of the two subunits. The angle between these vectors is 29.1°, which is very similar to the angle of rotation required to superpose B onto C. Also, using this vector as a proxy for the plane of the CHEF sheet, we see that Vb is approximately parallel to the threefold axis, whereas Vc is not at all. Thus, within the quasi hexamer the B and C subunits present somewhat different interfaces to the neighboring subunits. In (d-f) the relationships to the proximal symmetry axes are illustrated. The vector L is an internal reference defined by the Cα atoms of residues 218 and 231 of the I strand in the BIDG sheet and is used here to approximate the inclination of each subunit toward its proximal symmetry axis. The A subunit, as is generally the case for T=3 icosahedral viruses, has the steepest inclination with respect to the fivefold axis, followed by the C subunit. The B subunit is the flattest. Although this treatment is fairly crude, it suggests that the B and C subunits differ in inclination toward the threefold axis by nearly 10 .
Fig. S.6 The capsid of PMV is seen in cross section with radial dimensions and directions of icosahedral symmetry axes indicated.
Fig. S.7 Comparison of the N-terminal residues around Gln54, the first residue visible for the A subunits. In (a) and (b) are two views of the N-termini of the A, B and C subunits after superposition of the A and B subunits onto the C subunit. The A subunit is blue, the B is yellow and the C is pink. In (c), (d) and (e) are the A, B and C subunits, respectively, with their N-terminal unaveraged 2Fo-Fc density superposed, contoured at 0.6 sigma.
Fig. S.8 The difference between the conformations of residues Asp155 and Ser156 between the A and B subunits is illustrated. The C subunits are similar to the A subunits around these residues. Each of (a), (b) and (c) are stereo views with essentially the same orientation and shows residues 137 through 157. In (a), a B subunit (B1) has been superposed on an A subunit (A1). The A subunit is blue and the B subunit is yellow. Asp155 and Ser156 are seen to have very distinct conformations between the two subunits. In (b), the A subunit is superposed on its density showing that there is a good fit around Asp155 and Ser156 while in (c) the B subunit is superposed on its density and also shows that Asp155 and Ser156 fit the density well. Arg137 interacts with the main chain carbonyl in the A subunit and with the Asp155 carboxyl in the B subunit.
Fig. S.9 Comparison of the small loop conformation for the non-conserved residues (aa 173-181) between PMV and TNV. In (a) a TNV trimer (in shades of gray) is superposed on the PMV trimer in which the A, B and C subunits are blue, yellow and pink, respectively. The RNA, in cartoon representation, overlays the A subunit. The loop region is circled and lies near the quasi threefold axis in close proximity to the RNA. In (b) is a close up view of the loop. The TNV loop is directed nearly 180° away from the position of the PMV loop. The side chains of the residues in the loop are shown, as is the arginine of TNV.
Acknowledgments
This research was supported by a grant (GM080412) to AM from the NIH. D. Makino was supported by CNPq (Conselho Nacional de Desenvolvimento Cientifico e Technologico), an entity of the Brazilian government for development of science and technology.
Abbreviations
- PMV
Panicum Mosaic Virus
- TNV
Tobacco Necrosis Virus
- CfMV
Cocksfoot Mottle Virus
- RNA
ribonucleic acid
- STMV
Satellite Tobacco Mosaic Virus
- NCS
non-crystallographic symmetry
- FHV
Flock House Virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argos P, Johnson JE. Chemical stability in simple plant viruses. In: Jurnak FA, McPherson A, editors. Biological Macromolecules and Assemblies, Vol. I: Virus Structures. Wiley; New York: 1984. pp. 1–43. [Google Scholar]
- Ban N, Larson SB, McPherson A. Structural comparison of the plant satellite viruses. Virology. 1995;214:571–583. doi: 10.1006/viro.1995.0068. [DOI] [PubMed] [Google Scholar]
- Ban N, McPherson A. The structure of satellite panicum mosaic virus at 1.9 Å resolution. Nat. Struct. Biol. 1995;2:882–890. doi: 10.1038/nsb1095-882. [DOI] [PubMed] [Google Scholar]
- Barton GJ. Protein multiple sequence alignment and flexible pattern matching. Methods Enzymol. 1990;183:403–428. doi: 10.1016/0076-6879(90)83027-7. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvaneshwari M, Subramanya HS, Gopinath K, Savithri HS, Nayudu MV, et al. Structure of sesbania mosaic virus at 3 Å resolution. Structure. 1995;3:1021–1030. doi: 10.1016/s0969-2126(01)00238-6. [DOI] [PubMed] [Google Scholar]
- Brünger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buzen FG, Niblett CL, Hooper GR, Hubbard J, Newman MA. Further characterization of panicum mosaic virus and its associated satellite virus. Phytopathology. 1984;74:313–318. [Google Scholar]
- Cabrera O, Scholthof KBG. The complex viral etiology of St. Augustine decline. Plant Dis. 1999;83:902–904. doi: 10.1094/PDIS.1999.83.10.902. [DOI] [PubMed] [Google Scholar]
- Canady MA, Larson SB, Day J, McPherson A. Crystal structure of turnip yellow mosaic virus. Nat. Struct. Biol. 1996;3:771–781. doi: 10.1038/nsb0996-771. [DOI] [PubMed] [Google Scholar]
- Cardoso JMS, Felix MR, Clara MIE, Oliveira S. The complete genome sequence of a new necrovirus isolated from Olea europaea L. Arch. Virol. 2005;150:815–823. doi: 10.1007/s00705-004-0453-1. [DOI] [PubMed] [Google Scholar]
- Carrillo-Tripp M, Shepherd CM, Borelli AI, Venkataraman S, et al. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acid Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chen Z, Stauffacher C, Li Y, Schmidt T, Bomer W, et al. Protein-RNA interactions in an icosahedral virus at 3.0 Å resolution. Science. 1989;245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- Day J, Ban N, Patel S, Larson SB, McPherson A. Characterization of crystals of satellite panicum mosaic virus. J. Mol. Biol. 1994;238:849–851. doi: 10.1006/jmbi.1994.1339. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PYMOL Molecular Graphics System, v1.0r0. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Felix MR, Cardoso JMS, Oliveira S, Clara MIE. Viral properties, primary structure and phylogenetic analysis of the coat protein of an olive latent virus 1 isolate from Olea europaea L. Virus Res. 2005;108:195–198. doi: 10.1016/j.virusres.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Fisher AJ, Johnson JE. Ordered duplex RNA controls the capsid architecture of an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; New York: 2005. pp. 571–607. [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle JM, Maeda A, Harrison SC. Structure and assembly of turnip crinkle virus 1. X-ray crystallographic structure-analysis at 3.2 Å resolution. J. Mol. Biol. 1986;191:625–638. doi: 10.1016/0022-2836(86)90450-x. [DOI] [PubMed] [Google Scholar]
- Hopper P, Harrison SC, Sauer RT. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J. Mol. Biol. 1984;177:701–713. doi: 10.1016/0022-2836(84)90045-7. [DOI] [PubMed] [Google Scholar]
- Hosur MV, Schmidt T, Tucker RC, Johnson JE, Gallagher TM, et al. Structure of an insect virus at 3.0-Å resolution. Proteins: Struct., Funct., Genet. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Argos P. Virus particle stability and structure. In: Francki RIB, editor. The Plant Viruses: Polyhedral Virions with Tripartite Genomes. Plenum Pres; New York: 1985. pp. 19–56. [Google Scholar]
- Johnson JE, Rueckert RR. Packaging and release of the viral genome. In: Chiu W, Burnett RM, Garcea R, editors. Structural Biology of Viruses. Oxford University Press; Oxford: 1997. pp. 269–287. [Google Scholar]
- Jones TA, Liljas L. Structure of satellite tobacco necrosis virus after crystallographic refinement at 2.5 Å resolution. J. Mol. Biol. 1984;177:735–767. doi: 10.1016/0022-2836(84)90047-0. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A: Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kaper JM. The chemical basis of virus structure, dissociation and reassembly. North-Holland; Amsterdam: 1975. pp. 333–352. [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Software for handling macromolecular envelopes. Acta Crystallogr. D: Biol. Crystallogr. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- Lane SW, Dennis CA, Lane CL, Trinh CH, Rizkallah PJ, et al. Construction and crystal structure of recombinant STNV capsids. J. Mol. Biol. 2011;413:41–50. doi: 10.1016/j.jmb.2011.07.062. [DOI] [PubMed] [Google Scholar]
- Larson SB, Day J, Canady MA, Greenwood A, McPherson A. Refined structure of desmodium yellow mottle tymovirus at 2.7 Å resolution. J. Mol. Biol. 2000;301:625–642. doi: 10.1006/jmbi.2000.3983. [DOI] [PubMed] [Google Scholar]
- Larson SB, Day J, Greenwood A, McPherson A. Refined structure of satellite tobacco mosaic virus at 1.8 Å resolution. J. Mol. Biol. 1998;277:37–59. doi: 10.1006/jmbi.1997.1570. [DOI] [PubMed] [Google Scholar]
- Larson SB, Koszelak S, Day J, Greenwood A, Dodds JA, et al. Double helical RNA in satellite tobacco mosaic virus. Nature. 1993;361:179–182. doi: 10.1038/361179a0. [DOI] [PubMed] [Google Scholar]
- Larson SB, Lucas RW, Greenwood A, McPherson A. The RNA of turnip yellow mosaic virus exhibits icosahedral order. Virology. 2005a;334:245–254. doi: 10.1016/j.virol.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Larson SB, Lucas RW, McPherson A. Crystallographic structure of the T=1 particle of brome mosaic virus. J. Mol. Biol. 2005b;346:815–831. doi: 10.1016/j.jmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Larson SB, McPherson A. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Curr. Opin. Struct. Biol. 2001;11:59–65. doi: 10.1016/s0959-440x(00)00166-4. [DOI] [PubMed] [Google Scholar]
- Makino D, Day JS, Larson SB, McPherson A. Investigation of RNA structure in satellite panicum mosaic virus. Virology. 2006;351:420–431. doi: 10.1016/j.virol.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Makino DL, Larson SB, McPherson A. Preliminary analysis of crystals of panicum mosaic virus (PMV) by X-ray diffraction and atomic force microscopy. Acta Crystallogr. D: Biol. Crystallogr. 2005;61:173–179. doi: 10.1107/S090744490403077X. [DOI] [PubMed] [Google Scholar]
- Masuta C, Zuidema D, Hunter BG, Heaton LA, Sopher DS, et al. Analysis of the genome of satellite panicum mosaic virus. Virology. 1987;159:329–338. doi: 10.1016/0042-6822(87)90471-5. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McPherson A. Micelle formation and crystallization as paradigms for virus assembly. BioEssays. 2005;27:447–458. doi: 10.1002/bies.20196. [DOI] [PubMed] [Google Scholar]
- Monis J, Sopher DS, Jackson AO. Biologically active cDNA clones of panicum mosaic virus satellites. Phytopathology. 1992;82:1175. [Google Scholar]
- Morgunova E.Yu., Dauter Z, Fry E, Stuart DI, Stel'mashchuk V.Ya., et al. The atomic structure of carnation mottle virus capsid protein. FEBS Lett. 1994;338:267–271. doi: 10.1016/0014-5793(94)80281-5. [DOI] [PubMed] [Google Scholar]
- Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, et al. Virus Taxonomy-Classification and nomenclature of viruses. Springer-Verlag; Wien, New York: 1995. [Google Scholar]
- Niblett CL, Paulsen AQ. Purification and further characterization of panicum mosaic virus. Phytopathology. 1975;65:1157–1160. [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct., Func., Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Oda Y, Saeki K, Takahashi Y, Maeda T, Naitow H, et al. Crystal structure of tobacco necrosis virus at 2.25 Å resolution. J. Mol. Biol. 2000;300:153–169. doi: 10.1006/jmbi.2000.3831. [DOI] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. D: Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A: Found. Crystallogr. 1986;42:140–149. [Google Scholar]
- Rossmann MG, Arnold E. Non crystallographic symmetry averaging of electron density for molecular-replacement phase refinement and extension. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. vol. F. Kluwer Acad. Pub.; London, UK: 2001. pp. 279–292. [Google Scholar]
- Rossmann MG, Erickson JW. Structure and assembly of icosahedral shells. In: Casjens S, editor. Virus Structure and Assembly. Jones & Bartlett Publishers; Boston: 1985. pp. 29–73. [Google Scholar]
- Scholthof KBG. A synergism induced by satellite panicum mosaic virus. Mol. Plant-Microbe Interact. 1999;12:163–166. [Google Scholar]
- Silva AM, Rossmann MG. The refinement of southern bean mosaic virus in reciprocal space. Acta Crystallogr. B: Struct. Sci. 1985;41:147–157. [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Speir JA, Munshi S, Wang GJ, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryoelectron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tars K, Zeltins A, Liljas L. The three-dimensional structure of cocksfoot mottle virus at 2.7 Å resolution. Virology. 2003;310:287–297. doi: 10.1016/s0042-6822(03)00148-x. [DOI] [PubMed] [Google Scholar]
- Tsao J, Chapman MS, Agbandje M, Keller W, Smith K, et al. The three dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- Turina M, Desvoyes B, Scholthof KBG. A gene cluster encoded by panicum mosaic virus is associated with virus movement. Virology. 2000;266:120–128. doi: 10.1006/viro.1999.0069. [DOI] [PubMed] [Google Scholar]
- Turina M, Maruoka M, Monis J, Jackson AO, Scholthof KBG. Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology. 1998;241:141–155. doi: 10.1006/viro.1997.8939. [DOI] [PubMed] [Google Scholar]
- Zhang KYJ, Cowtan KD, Main P. Phase improvement by iterative density modification. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. vol. F. Kluwer Acad. Pub.; London, UK: 2001. pp. 321–324. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S.1 A schematic diagram of the phase refinement procedure (with initial and final statistics) used to obtain the Fourier map from which the initial full PMV model was built.
Fig. S.10 Three examples of positive difference density (contoured at 3.0 sigma) found near the carboxyl group of Glu145 between Glu145 and Arg85 of a neighboring subunit. In several cases, the bond lengths in the side chain of Glu145 are elongated as though refinement is compensating for some missing entity, possibly water, that exists between Glu145 and Arg85.
Fig. S.11 A ribbon diagram of the quasi twofold AB dimer in Flock House Virus. The clamp-like binding site of the C-C icosahedral twofold dimer does not exist in this dimer (compare Fig. 12).
Fig. S.2 Icosahedral symmetry averaged Fo electron density map using experimental phases obtained by successive rounds of density modification, averaging, and extension, showing side chain densities of a region of the amino terminal polypeptide (S(29)-VERGST-P(36)) of the C subunit. The TNV poly-alanine model was used for initial phase calculations at 20 Å resolution. The density shown here was computed at 4 Å resolution.
Fig. S.3 Illustration of the difference between loops 88-96 of two capsid subunits. Panels (a-c) are stereo views with loop 88-96 of a C subunit of particle 1 in pink and a C subunit of particle 2 in orange (C44 and F85, respectively, in our working system). The C44 subunit has been superposed on the F85 subunit over 171 Cα atoms excluding terminal residues and residues 89-95. These two loops represent the most extreme difference between any two loops 88-96 with distances between corresponding Cα atoms as large as 9 Å. Panels (a) and (b) show two close-up views of the loop 88-96 superposed on the unaveraged 2Fo-Fc density around subunit F85 contoured at 0.6 sigma that demonstrate the difference between the conformations of these two loops. In (c) these loops are viewed in context of the F85 subunit environment with the proximal B subunit of particle 1 in yellow. Density for the complete F85 subunit is shown in light blue and the darker blue density highlights the density of loop 88-96 of F85.
Fig. S.4 Illustration of the difficulty NCS averaging of the exteriors of the particle can create. In both (a) and (b) the C44 subunit around loop 88-96 is in pink and an A subunit of a neighboring particle is in blue. In (a) is superposed the density of an NCS averaged 2Fo-Fc map (contoured at 0.7 sigma) drawn only around the C44 loop and adjacent residues of C44 and the A subunit. The direction of the loop density is reminiscent of the density of the F85 subunit whose loop has a conformation similar to the majority of the loops (see Fig. S.3). In (b) is superposed the unaveraged 2Fo-Fc density (contoured at 0.6 sigma), which follows the C44 loop and is separated from the density of the proximal A subunit. Building the C44 loop according to the averaged map results in penetration of the loop into the A subunit.
Fig. S.5 There are several observations that identify differences in the B and C subunits. A most likely contributor to these differences is the packing within the capsid. The A, B and C subunits are blue, yellow and pink, respectively. In (a-c) the B subunit has been rotated 60° about the threefold axis toward the C subunit and then translated to bring their centers of mass into coincidence. Clearly, they do not align well with this simple treatment. From the position shown in (a-c), the B subunit was superposed onto the C subunit by a least squares fit of 166 Cα atoms resulting in the rotation axis seen in (a) and a rotation angle of 29.4° shown in (b). The views in (b) and (c) are down the rotation axis. In (c) vectors Vb (B subunit) and Vc (C subunit) are defined by the Cα atoms of residues 76 and 159 in the C and F strands, respectively, of the CHEF sheet. These vectors serve as an internal reference to the dispositions of the two subunits. The angle between these vectors is 29.1°, which is very similar to the angle of rotation required to superpose B onto C. Also, using this vector as a proxy for the plane of the CHEF sheet, we see that Vb is approximately parallel to the threefold axis, whereas Vc is not at all. Thus, within the quasi hexamer the B and C subunits present somewhat different interfaces to the neighboring subunits. In (d-f) the relationships to the proximal symmetry axes are illustrated. The vector L is an internal reference defined by the Cα atoms of residues 218 and 231 of the I strand in the BIDG sheet and is used here to approximate the inclination of each subunit toward its proximal symmetry axis. The A subunit, as is generally the case for T=3 icosahedral viruses, has the steepest inclination with respect to the fivefold axis, followed by the C subunit. The B subunit is the flattest. Although this treatment is fairly crude, it suggests that the B and C subunits differ in inclination toward the threefold axis by nearly 10 .
Fig. S.6 The capsid of PMV is seen in cross section with radial dimensions and directions of icosahedral symmetry axes indicated.
Fig. S.7 Comparison of the N-terminal residues around Gln54, the first residue visible for the A subunits. In (a) and (b) are two views of the N-termini of the A, B and C subunits after superposition of the A and B subunits onto the C subunit. The A subunit is blue, the B is yellow and the C is pink. In (c), (d) and (e) are the A, B and C subunits, respectively, with their N-terminal unaveraged 2Fo-Fc density superposed, contoured at 0.6 sigma.
Fig. S.8 The difference between the conformations of residues Asp155 and Ser156 between the A and B subunits is illustrated. The C subunits are similar to the A subunits around these residues. Each of (a), (b) and (c) are stereo views with essentially the same orientation and shows residues 137 through 157. In (a), a B subunit (B1) has been superposed on an A subunit (A1). The A subunit is blue and the B subunit is yellow. Asp155 and Ser156 are seen to have very distinct conformations between the two subunits. In (b), the A subunit is superposed on its density showing that there is a good fit around Asp155 and Ser156 while in (c) the B subunit is superposed on its density and also shows that Asp155 and Ser156 fit the density well. Arg137 interacts with the main chain carbonyl in the A subunit and with the Asp155 carboxyl in the B subunit.
Fig. S.9 Comparison of the small loop conformation for the non-conserved residues (aa 173-181) between PMV and TNV. In (a) a TNV trimer (in shades of gray) is superposed on the PMV trimer in which the A, B and C subunits are blue, yellow and pink, respectively. The RNA, in cartoon representation, overlays the A subunit. The loop region is circled and lies near the quasi threefold axis in close proximity to the RNA. In (b) is a close up view of the loop. The TNV loop is directed nearly 180° away from the position of the PMV loop. The side chains of the residues in the loop are shown, as is the arginine of TNV.