Abstract
Overexpression of cyclin D1 is believed to endow mammary epithelial cells (MECs) with a proliferative advantage by virtue of its contribution to pRB inactivation. Accordingly, abrogation of the kinase-dependent function of cyclin D1 is sufficient to render mice resistant to breast cancer initiated by ErbB2. Here, we report that mouse cyclin D1KE/KE MECs (deficient in cyclin D1 activity) upregulate an autophagy-like process but fail to implement ErbB2-induced senescence in vivo. In addition, immortalized cyclin D1KE/KE MECs retain high rates of autophagy and reduced ErbB2-mediated transformation in vitro. However, highlighting its dual role during tumorigenesis, downregulation of autophagy led to an increase in senescence in cyclin D1KE/KE MECs. Autophagy upregulation was also confirmed in human mammary epithelial cells (HMECs) subjected to genetic and pharmacological inhibition of cyclin D1 activity and, similar to our murine system, simultaneous inhibition of Cdk4/6 and autophagy in HMECs enhanced the senescence response. Collectively, our findings suggest a previously unrecognized function of cyclin D1 in suppressing autophagy in the mammary epithelium.
Keywords: Cyclin D1, autophagy, senescence, ErbB2, mammary gland
Introduction
Macroautophagy (referred to hereafter as autophagy) is an evolutionarily conserved and highly regulated catabolic process that involves the sequestration and subsequent lysosomal-mediated degradation of organelles and long-lived proteins (1). The morphological hallmark of autophagy is the formation of double-membrane vacuoles, also known as autophagosomes, which transport cytoplasmic cargo to the lysosomes for degradation and substrate recycling (2). Autophagy-mediated recycling of cellular components provides basic biochemical substrates that can be utilized for energy production and in biosynthetic reactions, a function that is critical for cell survival in situations of metabolic stress (2–4). Nonetheless, in the context of a nutrient-rich environment, basal levels of autophagy are still required to ensure a quality control mechanism that prevents the accumulation of aggregates of proteins and damaged organelles (1, 5). In addition to these prosurvival functions, there is also evidence that, under certain conditions, autophagy can also be a mechanism of cell death (4, 6).
Early observations suggested that autophagy acts as a suppressor of tumor formation. For example, Beclin-1/Atg6, a gene essential for the induction of autophagy, was found hemiallelically deleted in 40–75% of sporadic breast, ovary, and prostate cancers (7). In addition, autophagy-deficient Beclin-1+/− mice show an increased frequency of spontaneous malignancies, indicating that Beclin-1 is a haplo-insufficient tumor-suppressor gene (8, 9). Moreover, indirect evidence for autophagy downregulation during tumorigenesis is provided by the observation that human cancers commonly display an activation of the PI3K/Akt/mTOR pathway, which negatively regulates autophagy (5). Among the mechanisms that may explain autophagy’s effects on tumorigenesis, autophagy-mediated attenuation of oxidative stress is one of the best studied (5). Accordingly, autophagy-deficient tissues display a protumorigenic phenotype partially as a result of ROS accumulation and increased DNA damage (10). Recently, a more direct link between autophagy and tumor suppression has been postulated in which autophagy becomes activated during Ras-induced senescence in human fibroblasts (11). Thus, failure to implement senescence upon inhibition of autophagy may result in the acquisition of a proliferative advantage that accelerates tumor formation (12). Despite the connection between autophagy and tumor suppression, there is also significant evidence that autophagy is required for proliferation and therapy resistance in established cancers and cancer cell lines (1). In this scenario, downregulation or inhibition of autophagy leads to an impairment of cell proliferation or enhancement of the therapeutic effect of cancer drugs (13).
A consistent signature of luminal-type breast tumors is the overexpression of cyclin D1, a cell cycle regulator that functionally integrates multiple mitogenic signals, including those derived from tyrosine kinase receptors (14). It is believed that the oncogenic properties of cyclin D1 depend to a large extent on its ability to activate cyclin-dependent kinases 4 or 6 (Cdk4/6) (15, 16). Consistent with this model, we have shown that in vivo abrogation of the kinase-dependent function of cyclin D1 is sufficient to render mice resistant to breast cancer initiated by ErbB2 (17), a phenotype that can be explained, at least in part, by a reduction in the number of cellular targets for ErbB2-mediated transformation (18). These observations suggest that targeting cyclin D1/Cdk(4/6) is, in principle, a reasonable approach to treat breast cancers. Indeed, two recently developed Cdk(4/6) inhibitors have already entered clinical trials (19). However, a better understanding of the metabolic and signaling pathways that are implemented in response to reduced levels of cyclin D1-associated kinase activity will be necessary to expand the therapeutic window and to confront the almost certain development of drug resistance.
Here we describe the interplay between cellular processes that can affect proliferation in mammary epithelial cells (MECs) derived from kinase-deficient cyclin D1KE/KE knock-in mice. We found that cyclin D1KE/KE mammary tissues are able to sustain increased proliferation in response to ErbB2, this despite the insensitivity of mutant MECs to protumorigenic proliferative activity in the long term. These findings correlated with an upregulation of autophagy in cyclin D1KE/KE mammary tissues along with a failure to upregulate markers of premature ErbB2-induced senescence. However, downregulation or blockade of autophagy in vitro had a detrimental effect on proliferation by increasing senescence, suggesting that upregulation of autophagy observed in cyclin D1KE/KE MECs may constitute a protective mechanism in response to reduced levels cyclin D1 activity. Thus, our findings suggest that blockade of autophagy may enhance the therapeutic effects of Cdk4/6 inhibitors in the treatment of breast cancer.
Materials and Methods
Mouse strains
Cyclin D1KE/KE (17) and MMTV-ErbB-2 mice (strain TG. NK (20)) were maintained in a C57BL/6 × 129 mixed genetic background and genotyping was carried out as described elsewhere (17). Ink4a/Arf-deficient animals have been previously described (21). All animals used were treated humanely and in accordance with the Institutional Animal Care and Use Committee at Tufts Medical Center approved protocols.
Cell lines
Mammary glands harvested from Cyclin D1+/+/Ink4a-Arf−/− and Cyclin D1KE/KE/Ink4a-Arf−/− mice were mechanically disaggregated and digested with Collagenase (Sigma) and Hyaluronidase (Sigma). The resultant organoids were further digested in 0.25% trypsin-EDTA and Dispase/DNAse I and then filtered through a 40 μm mesh. Lineage+ cells (CD45+, CD31+ and Ter119+) were immunomagnetically removed from freshly isolated cells using the Easysep mouse mammary stem cell enrichment kit (Stem Cell Technologies). Lineage (−) cells (mammary epithelial cells) were plated in DMEM/F-12 (1:1) culture medium (Invitrogen) supplemented with 2% fetal bovine serum, 10 μg/ml insulin (Sigma), 5 ng/ml mouse EGF (Sigma), and 500 ng/ml hydrocortisone (Sigma). Cells were periodically passaged until the appearance of immortalized clones. Immortalized mouse mammary epithelial cells generated in this way were characterized using luminal- and myoepithelial-cell markers as described in Fig. S4 and have not been utilized in previous publications. Human mammary epithelial cell lines (MCF10A and HMECs) were kindly provided by Dr. Charlotte Kuperwasser (Tufts University). Since their original generation and description (22–24), these cell lines have been extensively characterized and tested for their lineage and transformability phenotypes using multiple mammary epithelial cell markers (25, 26).
Immunostaining
Mammary glands were fixed overnight in 4% paraformaldehyde/PBS. Sections (5 μm) were de-waxed, re-hydrated, washed in PBS, and subjected to antigen retrieval by boiling in 10mM citrate buffer (pH 6.0). For frozen sections, tissues were fixed in 4% paraformaldehyde/PBS for 2 hours, followed by alternate washes in 50mM Glycine and PBS. Tissues were then incubated in 20% sucrose/PBS overnight at 4°C and in 30% sucrose/PBS for 1 hour at room temperature before being embedded in OCT compound. For immunostaining, tissue sections were incubated overnight at 4 °C with primary antibodies diluted in 2% BSA/PBS. Secondary antibodies were applied for 1 hr at room temperature. Immune complex formation was detected through immunofluorescence using Alexa Fluor 488 and/or Alexa Fluor 546 conjugated antibodies (Invitrogen). Nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI) whenever necessary (Sigma). Alternatively, sections were incubated with horseradish peroxidase (HP)-conjugated antibodies (Jackson) and immune complex formation visualized with 3,3′-diaminobenzidine (DAB) method according to the manufacturer’s instructions (Vector). Nuclei were counterstained with Mayer’s hematoxylin (Sigma). Antibodies used for immunostaining included are listed in the Supplemental Materials and Methods.
Transformation assays
Anchorage-independent growth of ErbB2/NeuT infected cells (20×103) was assayed by scoring the number of colonies formed in 0.4% agarose (with a 0.8% agarose underlay) after three weeks. When necessary, ErbB2/NeuT infected cells (1×106) were injected subcutaneously into immunodeficient (NOD/SCID) mice. Cells of different genotypes were injected into contralateral flanks of the same mouse. After 25 days, tumor-harboring mice were euthanized, tumors removed and weighed.
SA-β-Galactosidase activity
Detection of senescence-associated (SA)-β-Galactosidase activity on tissue sections was carried out essentially as described (27, 28). Briefly, cryosections (7μm) were re-hydrated and then incubated overnight at 37°C in staining solution (1 mg/ml 5-bromo-4-chloro-3-inolyl-β-D-galactoside (X-gal); 40 mM citric acid/sodium phosphate buffer, pH 6.0; 5 mM potassium ferrocyanide; 5 mM potassium ferricyanide; 150 mM NaCl; 2 mM MgCl2). Next day, sections were washed twice in PBS and mounted in glycerol. Alternatively, sections were co-stained with antibodies directed against Ki-67 antigen.
Protein preparation and immunoblotting
Lin(−) MECs and organoid suspensions were lysed in ELB buffer (250mM NaCl, 50mM HEPES pH 7.0, 5 mM EDTA, 0.1% NP40, 1μg/mL aprotinin, 50μg/mL Pefabloc (Roche), 10μg/mL leupeptin, 1mM DTT, 100μM sodium orthovanadate, 10mM β-glycerophosphate, 10mM sodium fluoride, and 2mM sodium pyrophosphate). Immunoblotting was carried out essentially as described (17). Antibodies used in this work are listed in the supplemental Materials and Methods. Immunoblot analysis was carried out using horseradish peroxidase conjugated donkey anti-rabbit or donkey anti-mouse antibodies (Jackson ImmunoResearch Laboratories). For cytokine antibody arrays (Mouse Cytokine Antibody Array, RayBiotech, Inc.), support membranes were incubated with protein lysates derived from organoids (300μg) essentially as described in the manufacturer’s instructions.
Statistical analysis
Numerical data are presented as mean ± S.D or mean ± SEM of at least three independent experiments, and statistical significance between groups was identified using the two-tailed Student’s t test.
Results
Cyclin D1KE/KE mammary tissues sustain high, but temporally limited, proliferation in response to ErbB2i
We have previously shown that cyclin D1 kinase activity is required for ErbB2-mediated breast cancer and the maintenance of mammary progenitors (17, 18). While proliferation is very low in both cyclin D1KE/KE (kinase activity deficient) and cyclin D1+/+ mammary epithelia in the absence of active ErbB2 (not shown), the question remained as to whether ErbB2-derived signals could lead to a proliferative response in the absence of cyclin D1’s function. Examination of ErbB2-expressing cyclin D1KE/KE mammary tissues (cyclin D1KE/KE/MMTV-ErbB2) from aged females (~6 months of age) revealed morphological changes in both the epithelium and the stroma. Ducts from mutant glands appeared greater in diameter, with a higher number of epithelial nuclei per cross section (Fig. 1A, panels and graph). In addition, staining with the myoepithelial cell marker α-Smooth Muscle Actin (SMA) revealed a more prominent and disorganized layer of myoepithelium, composed of densely packed cells (Fig. 1A, right panels). Stromal changes included prominent fibrosis surrounding the ducts (Fig. 1A, trichrome staining, middle panels). Of note, these morphological changes were not observed in cyclin D1KE/KE mammary tissues in the absence of ErbB2 (data not shown). Despite increased cellularity, staining with the proliferation marker Ki67 revealed few residual positive cells in ErbB2-expressing cyclin D1KE/KE mammary ducts. Interestingly, Ki67+ cells co-localized extensively with the myoepithelial marker SMA, a reflection of the differentiation phenotype previously described (18) (Fig. 1B). Collectively, these findings were indicative of an early expansion of ErbB2-expressing cyclin D1KE/KE mammary epithelial cells (MECs), which nonetheless were unable to sustain proliferation late in life, probably due to aberrant differentiation into cells with myoepithelial features. Consistent with this, younger (3-month old) ErbB2-expressing cyclin D1KE/KE mammary tissues displayed high levels of proliferation, similar to levels observed in ErbB2-expressing cyclin D1+/+ mammary tissues but much higher than the levels observed in mammary tissues lacking the ErbB2 transgene (Fig. 1C and not shown). Importantly, ectopic proliferation observed in mutant mammary glands was accompanied by reduced levels of phosphorylation at Serine-780 of pRb, a cyclin D-dependent phosphorylation event (17), as well as reduced levels of the E2F targets cyclin A and cyclin E (Fig. S1A and B). These findings indicate that pRb-independent proliferation, although temporally limited, can still take place in mutant mammary tissues in response to ErbB2.
Figure 1. ErbB2-expressing cyclin D1KE/KE mammary tissues can sustain high but temporally limited proliferation.
(A) Panels: whole mounts, trichrome staining, and SMA/pERM co-immunofluorescence of six-month old mammary glands. Graph: number of nuclei per duct section. pERM (phospho-Ezrin/Radixin/Moesin) was used as an apical marker of luminal cells and SMA (α-smooth muscle actin) identifies myoepithelial cells. Error bars indicate mean± SD (n=3). Scale bars, 500μm (whole mounts) and 50μm (sections). (B) Mammary glands of the indicated genotypes (age 6 months) were co-stained for the proliferation marker Ki67 (red) and SMA (green). Right panels show a higher magnification of insets. (C) Ki67 (green) and ErbB2 (red) co-immunofluorescence mammary glands (age 3 months). Arrows indicate basally localized Ki67 positive nuclei. Nuclei were counterstained with DAPI (blue). Graph: percentage of Ki67 positive cells per duct section (black bars) and percentage of Ki67 positive cells that co-localized with SMA (white bars). ErbB2(−) wild-type mammary tissues were also analyzed as negative controls. Error bars indicate mean± SD (n=3). Scale bars, 50μm.
Upregulation of autophagy in cyclin D1KE/KE mammary tissues
Given the central role of cyclin D1 as an integrator of growth-promoting signals that can also have an impact on autophagy (5), we speculated that this process might be deregulated in mutant MECs, perhaps explaining the proliferative changes in ErbB2-expressing mammary tissues described above. To test this hypothesis, we took advantage of an antibody that recognizes the cleaved form of LC3 (microtubule-associated light chain 3), which has been used as a reliable marker for autophagy using immunohistochemistry on a variety of tissues (29–31). Cleavage of LC3 is the first step in a process that allows the protein to be lipidated and incorporated into the membrane of autophagosomes (2). As shown in Fig. 2A, levels of cleaved LC3 were upregulated in cyclin D1KE/KE mammary tissues compared with cyclin D1+/+ controls, and the intensity of staining was further increased in ErbB2-expressing cyclin D1KE/KE mammary tissues. A similar upregulation of this marker was observed in protein lysates obtained from freshly isolated mutant MECs (Lin(−) fraction) (Fig. 2B). To further support our findings, we studied mammary tissues at the ultrastructural level using transmission electron microscopy (TEM). Although visualization of classical autophagosomes was challenging, this analysis revealed signs of prominent intracellular digestion activity, including the presence of abundant vesicles with variable degrees of electron-density consistent with lysosomes, autolysosomes and residual bodies in cyclin D1KE/KE but not cyclin D1+/+ mammary tissues (Fig. S2 and data not shown). Taken together, our findings indicate that mammary tissues deficient in cyclin D1 activity show signs of increased autophagic activity.
Figure 2. Upregulation of autophagy in cyclin D1KE/KE mammary tissues.
(A) Activated autophagy was assessed on mammary tissues by immunohistochemistry (red) for cleaved LC3. Nuclei were counterstained with hematoxylin (blue). Scale bars, 50μm. (B) Lysates derived from lineage negative (Lin(−)) mammary epithelial cell fractions from the indicated genotypes were immunoblotted for cleaved LC3. GAPDH was used as a loading control.
Cyclin D1KE/KE mammary tissues display reduced ErbB2-mediated senescence
Premature senescence is a potent barrier against unscheduled proliferation of early transformed cells (12, 32). Recently, Young et al. (2009) have shown that autophagy can be induced as an effector mechanism during Ras-induced senescence in vitro. Inhibition of autophagy in this setting resulted in a failure to activate a senescence program (11). Given the observation that ErbB2-expressing cyclin D1KE/KE mammary tissues display an upregulation of autophagy markers and are unable to develop tumors, we speculated that increased autophagy might reflect or facilitate ErbB2-induced senescence in mutant tissues. Surprisingly, ErbB2-expressing cyclin D1KE/KE mammary tissues failed to upregulate senescence associated- (SA)-β-Galactosidase (β-Gal) activity compared to pre-neoplastic ErbB2-expressing cyclin D1+/+ controls (Fig. 3A). Of note, robust β-Gal staining was also detected in cyclin D1+/+/MMTV-ErbB2 tumor lesions (Fig. 3A, middle panels). As expected, however, β-Gal+ regions and Ki67 staining did not co-localize in ErbB2-derived tumors (Fig. S3A), perhaps indicating that proliferative regions evolved following escape from senescence. We also confirmed that senescence observed in ErbB2-expressing cyclin D1+/+ pre-neoplastic tissues was restricted to the ductal epithelium (Fig. S3B).
Figure 3. Cyclin D1KE/KE mammary tissues display reduced ErbB2-mediated senescence.
(A) SA-β-Galactosidase (blue, upper panels) and ErbB2 staining (red, lower panels) of mammary glands and ErbB2-derived tumors. Scale bars, 50μm. (B) Immunostaining for histone 3 tri-methylated at lysine 9 (H3K9me3) (red). Nuclei were counterstained with DAPI (blue). Scale bars, 50μm. (C) Quantitative reverse transcriptase (qRT)-PCR was used to analyze the expression of the senescence-associated secreted factors MMP-2, MMP-3, and PAI-1. Results from whole mammary glands (upper panel) and epithelial-enriched Lin(−) populations (lower panel) are shown. Error bars indicate mean± SD from three different experiments.
The failure to undergo senescence observed in ErbB2-expressing cyclin D1KE/KE mammary tissues was corroborated by staining for histone 3 tri-methylated at lysine 9 (H3K9me3), a marker for heterochromatin formation and a hallmark of senescence (Fig. 3B). As an additional confirmation of our findings, we analyzed the expression of factors that are produced and secreted by senescent cells (32–35). Quantitative real time (RT)-PCR using mammary tissues or purified mammary epithelial cells (Lin(−) fractions) from ErbB2-expressing cyclin D1KE/KE mammary glands, revealed a failure to upregulate the expression of selected factors (Fig. 3C and Fig. S3C). Moreover, cytokine array analyses of protein lysates from ErbB2-expressing cyclin D1KE/KE mammary tissues revealed reduced levels of interleukin-6 (IL-6), another senescence-associated secreted factor (36) (Fig. S3D). Furthermore, although senescence observed in ErbB2-expressing cyclin D1+/+ mammary tissues was not accompanied by an induction of p16Ink4a or p19Arf as observed in other systems (37, 38), we did observe a relative increase in the levels of p18INK4c and p27Kip1 in ErbB2-expressing cyclin D1+/+ mammary tissues compared to the ErbB2-expressing cyclin D1KE/KE counterparts (Fig. S3E). Importantly, we were unable to detect differences in apoptosis in tissues derived from ErbB2-expressing cyclin D1+/+ or cyclin D1KE/KE mice (Fig. S3F). Taken together, despite evidence that autophagy upregulation may be required for the proper orchestration of oncogene-induced senescence, our findings indicate that these processes can occur in a mutually exclusive manner in mammary tissues that are deficient in cyclin D1 function.
KE/Ink4(−) MECs show reduced ErbB2/NeuT-mediated transformation
Despite the inability of ErbB2-expressing cyclin D1KE/KE cells to undergo senescence in vivo, we were unable to propagate purified cyclin D1KE/KE MECs in culture beyond passage 8 (data not shown). Since growth arrest of primary cells in vitro depends on the induction of p16Ink4a and/or p19Arf (39), we reasoned that we could generate an appropriate in vitro model by immortalizing cyclin D1KE/KE/Ink4a-Arf−/− MECs. As shown in Fig. S4A, immortalized MECs retain the expression of the epithelial markers Keratin 18 (K18, a luminal epithelial cell marker) and α-Smooth-Muscle-Actin (SMA, a myoepithelial cell marker). Of note, cyclin D1KE/KE/Ink4a-Arf−/− (KE/Ink4(−)) MECs grew more slowly than cyclin D1+/+/Ink4a-Arf−/− (WT/Ink4(−)) MECs (Fig. S4B), with BrdU incorporation and cell cycle profile analyses revealing a small albeit significant reduction of KE/Ink4(−) cells undergoing cell division (Fig. S4C). Importantly, we were unable to detect any difference in the proportion of cells undergoing apoptosis and, as expected, senescent cells were only sporadically detectable in both cell lines (not shown). The proliferation defect observed in KE/Ink4(−) MECs was improved but did not reach wild-type levels following transduction of ErbB2/NeuT (Fig. S4D), a fact that was also reflected in their poor colony formation ability (Fig. S4E). These findings stand in contrast to the robust proliferation of ErbB2-expressing cyclin D1KE/KE cells in vivo (Fig. 1C) and may reflect contributions from the microenvironment that promote aberrant proliferation in the presence of ErbB2. Nevertheless, even taking the proliferation deficiency into account, ErbB2/NeuT-transduced KE/Ink4(−) MECs showed a disproportionately reduced ability to form colonies in soft agar (Fig. S4F) or tumors after subcutaneous injection into NOD/SCID mice (Fig. S4G). Thus, similar to our in vivo observations, the KE mutant profoundly impairs transformation in MECs without overtly altering senescence or apoptosis, making this system suitable for analyzing the dynamics of autophagy in mutant MECs.
KE/Ink4(−) MECs retain increased rates of autophagy
As a first approach to study autophagy in vitro, we determined LC3 protein levels in immortalized MECs. Following cleavage, LC3 becomes lipidated (LC3-II) and localizes to autophagosome membranes, a modification that allows it to be distinguished from the soluble form (LC3-I) (40). As shown in Fig. 4A, LC3-II and to a lesser extent Beclin-1 protein levels, were increased in KE/Ink4(−) MECs. However, LC3-II did not increase further following expression of ErbB2/NeuT (Fig. S5A).
Figure 4. KE/Ink4(−) MECs retain increased rates of autophagy.
(A) Protein lysates from immortalized MECs were immunoblotted for LC3B (I and II), Beclin-1 and p62/SQSTM1. (B) Immunoblotting for LC3B and p62/SQSTM1. Increased and unimpeded autophagic flux in KE/Ink4(−) MECs was revealed following chloroquine (CQ) treatment (30 μM of CQ for 24 hours). Densitometric ratios of LC3-II over GAPDH signals are shown (graph, average of three independent blots, error bars indicate mean± SD). (C, D) MECs were transiently transfected with the LC3-mCherry-EGFP construct and fluorescent vesicles visualized 24 (C) or 72 (D) hours later. (C) The proportion of autophagosome-containing cells was similar between cell lines (upper graph), but the number of double-positive autophagosomes per cell was increased in KE/Ink4(−) cells (lower graph). Representative images are shown. Scale bars, 25μm and 20μm for KE and WT MECs, respectively. Error bars indicate mean± SD from three independent experiments. (D) Representative images obtained 72 hours post-transfection. A quantification of the proportion of cells containing autolysosomes or autophagosomes is shown. Error bars indicate mean± SD from three independent experiments. Scale bars, 20μm.
Since LC3-II accumulation could also reflect a defect in autophagosome-lysosome fusion (40), we also checked the levels of p62/SQSTM1, a protein that is normally degraded through autophagy (10). As expected, levels of p62/SQSTM1 were reduced in KE/Ink4(−) MECs (Fig. 4A), an indication of increased autophagic activity. In addition, blockade of autophagosome-lysosome fusion using chloroquine (CQ) led to the simultaneous accumulation of LC3-II and p62/SQSTM1 in both cell lines, although LC3-II reached higher levels in KE/Ink4(−) MECs (Fig. 4B), a further indication of increased rates of autophagy. In order to visualize the autophagic process, we took advantage of a doubly tagged LC3 construct (LC3-mCherry-EGFP) in which LC3 has been tandemly fused to a red fluorescent protein (mCherry) and to an enhanced green fluorescent protein (EGFP) (41). Unlike mCherry, the fluorescent signal of EGFP is highly susceptible to the acidic/proteolytic environment of lysosomes. Therefore, autophagosomes that have incorporated tagged LC3 show both fluorescent signals before fusion with lysosomes, but only the red signal following maturation of autophagosomes into autolysosomes. Immortalized MECs were transiently transfected with LC3-mCherry-EGFP and fluorescent structures were examined 24 or 72 hours later. Importantly, transfection efficiencies were comparable between cell lines (31.33±14.5% versus 32.82±11.5%, n=3, for WT and KE cells, respectively). Double-positive autophagosomes were first identified 24 hours post-transfection. While the proportion of autophagosome-containing cells was not significantly different between cell lines (Fig. 4C, upper graph), the number of double-positive autophagosomes per cell was dramatically increased in KE/Ink4(−) MECs (Fig. 4C, lower graph). As expected from normal autophagosome maturation, the number of autophagosome-containing cells and the number of autophagosomes per cell decreased over time, with the concomitant increase in red-only autolysosomes. However, the proportion of KE/Ink4(−) MECs containing autolysosomes was still higher than WT/Ink4(−) MECs (Fig. 4D). Importantly, autolysosomes were invariably positive for endocytosed dextran that labels lysosomes (Fig. S5B), confirming autophagosome-lysosome fusion. Taken together, these data are consistent with a basal upregulation of autophagy in immortalized cyclin D1KE/KE MECs.
Knockdown of ATG5 exacerbates senescence in KE/Ink4(−) MECs
We next studied the proliferative and tumorigenic effects of autophagy downregulation in MECs. Unexpectedly, shRNA-mediated knockdown of Beclin-1 did not lead to a reduction of LC3-II or changes in proliferation (Fig. S6A, B). Further, Beclin-1-depleted KE/Ink4(−) cells were still able to upregulate LC3-II in response to mTORC1 inhibition (Fig. S6C). Thus, in contrast to other systems reported in the literature, these experiments reveal a surprising ability of MECs to induce autophagosome formation even in the context of profoundly reduced levels of Beclin-1.
Unlike Beclin-1, shRNA-mediated downregulation of ATG5 resulted in a dramatic inhibition of autophagy, reflected in the almost undetectable levels of LC3-II and concomitant accumulation of p62/SQSTM1 (Fig. 5A). Surprisingly, disrupting autophagosome formation led to impaired proliferation (Fig. 5B). However, while knocking down ATG5 in WT/Ink4(−) MECs resulted in a modest increase in both apoptosis and senescence, ATG5-deficient KE/Ink4(−) MECs showed a pronounced senescence response (Fig. 5C and D). Accordingly, ATG5 depletion in ErbB2/NeuT-expressing MECs further reduced their ability to be transformed (Fig. 5E). These data suggest that the increased autophagy observed in mutant MECs is required to sustain proliferation and survival in the context of reduced cyclin D1 function.
Figure 5. Downregulation of autophagy increases senescence in KE/Ink4(−) MECs.
(A) shRNA-mediated knockdown of ATG5 in immortalized MECs. Immunoblots for ATG5, LC3B (I and II) and p62/SQSTM1 are shown. (B) Proliferation curves of MECs following shRNA-mediated knockdown of ATG5. (C) Annexin V analysis (apoptosis) of cells shown in (B). Fold increase in the % of Annexin V positive cells (shRNA over ctrl) is shown. Error bars indicate mean± SD (n=3). (D) Quantification of the proportion of SA-β-Gal+ cells. Error bars indicate mean± SD (n=3). (E) MECs carrying control (Ctrl) or ATG5 shRNAs were transduced with ErbB2/NeuT or vector, and then assayed for their ability to form colonies in soft agar. Error bars indicate mean± SD (n=3).
Simultaneous inhibition of Cdk4/6 activity and autophagy enhances senescence in HMECs
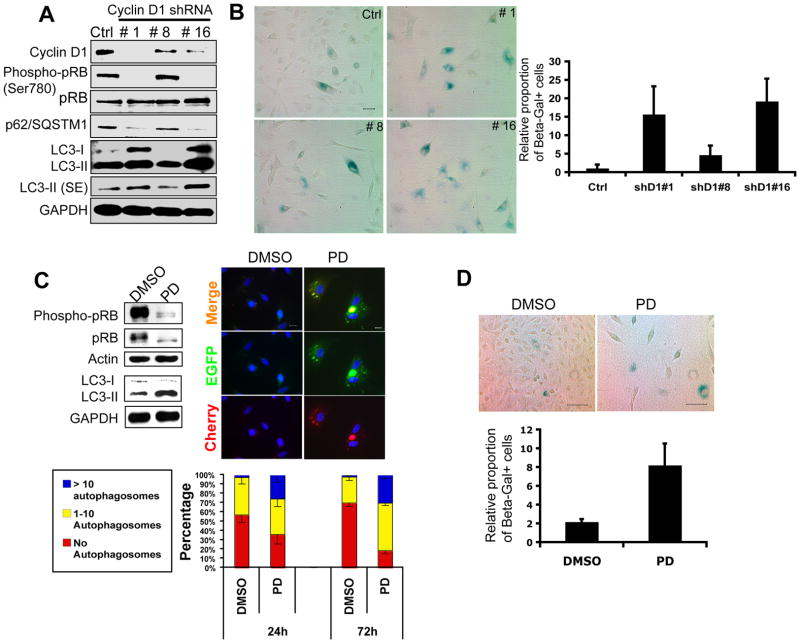
In order to extend our findings to more therapeutically relevant human models, we first set out to reduce the expression of cyclin D1 in telomerase-immortalized human mammary epithelial cells (HMECs) (22). Knockdown of cyclin D1 using three different shRNAs led to dramatic proliferation impairment (Fig. S7A) that correlated with the degree of cyclin D1 reduction and hypophosphorylation of pRB at Ser-780 (Fig. 6A, see shRNAs #1 and 16). Importantly, cyclin D1 reduction was also accompanied by an upregulation of LC3-II and a reduction of p62/SQSTM1 (Fig. 6A). Unlike KE/Ink4(−) MECs, however, the proportion of senescent cells was greater in cyclin D1-depleted HMECs (Fig. 6B). Since differences in senescence between KE/Ink4(−) and D1-depleted HMECs could reflect the ability of kinase independent functions of cyclin D1 to repress induction of senescence, we next wanted to know whether the pharmacological inhibition of Cdk4/6 in HMECs could better reproduce the phenotypes observed in KE/Ink4(−) cells. To this end, we took advantage of the highly specific Cdk4/6 inhibitor PD0332991 (19). Although PD0332991 treatment did not elicit a significant apoptotic response (not shown), the proportion of HMECs in S-G2-M was significantly reduced by the treatment (Fig. S7B). In order to visualize autophagy, HMECs were first transfected with the LC3-Cherry-EGFP construct and then treated with 0.5 μM PD0332991 or vehicle (DMSO) for 24 or 72h. As shown in Fig. 6C (right panels), the number of autophagosomes (vesicles exhibiting both red and green signals) per cell increased significantly following drug treatment. We confirmed these observations by comparing the endogenous levels of LC3-II in drug- or vehicle-treated cells (Fig. 6C, left panels). The proportion of senescent cells also increased after PD0332991 treatment, although to a much lesser extent compared to cyclin D1-depleted HMECs (Fig. 6D). Thus, proliferation arrest achieved through either genetic depletion of cyclin D1 or inhibition of Cdk4/6 activity is accompanied by an early induction of autophagy in HMECs.
Figure 6. Downregulation of cyclin D1 or Cdk4/6 inhibition induce autophagy in HMECs.
(A) Immunoblot analysis of cyclin D1, phosphorylated pRB (Ser-780), total pRB, p62/SQSTM1 and LC3B (I and II) in HMECs transduced with control (ctrl) or cyclin D1 shRNAs (#1, #8 and #16). GAPDH was used as a loading control. SE: short exposure. (B) SA-β-Galactosidase staining of shRNA-transduced HMECs. The relative proportion of positive cells per 20x field is shown. Error bars indicate mean± SD (n=3). (C) Left panels: Immunoblot analysis of phosphorylated pRB (Ser-780), total pRB and LC3B (I and II) in HMECs treated with 0.5 μM PD0332991 for 24 hours. α-Tubulin and GAPDH were used as loading controls. Right panels: HMECs transfected with the LC3-mCherry-EGFP construct were treated with PD-0332991 (PD) or DMSO for 24 or 72h. Representative images are shown. Graphs represent the number of autophagosomes per transfected cell. Error bars indicate mean± SE (n=3). Scale bars, 10 μm. (D) SA-β-Galactosidase staining of PD0332991-treated HMECs. The proportion of positive cells per 20x field is shown. Error bars indicate mean± SD (n=3).
We next tested the combined effects of autophagy downregulation and Cdk4/6 inhibition in HMECs. Similar to what we observed in KE/Ink4(−) MECs (Fig. 5), knockdown of ATG5 in HMECs increased the proportion of senescent cells, a response that was further exacerbated by PD0332991 treatment (Fig. 7A and B). Thus, contrary to recent reports describing autophagy as an effector mechanism of senescence in fibroblasts (11), autophagy reduction in MECs can activate the senescence program. Moreover, these findings lend further support to the concept that autophagy functions primarily as a pro-proliferative process in MECs, and also suggest that the combined use of inhibitors of autophagy and Cdk4/6 might represent a promising therapeutic approach to treat breast cancers. In order to explore this idea, immortalized HMECs and ErbB2/NeuT-expressing MCF10A, a tumorigenic cell line, were treated with chloroquine (CQ), PD0332991 (PD), or a combination of both compounds. Importantly, concentrations of single drugs used in these experiments were kept at a minimum in order to detect enhancing effects when used in combination. As shown in Fig. 7C, combined use of CQ and PD0332991 led to a reduction in proliferation compared with cells that were untreated or treated with a single drug. Furthermore, the ability of ErbB2/NeuT-expressing MCF10A cells to form colonies in soft agar was significantly reduced following CQ or PD0332991 treatment, an effect that was further exacerbated when cells were treated with both compounds (Fig. 7D).
Figure 7. Simultaneous inhibition of Cdk4/6 activity and autophagy enhances senescence in HMECs.
(A) shRNA-mediated knockdown of ATG5 in HMECs. Immunoblots for ATG5, LC3B, and p62/SQSTM1 are shown. (B) SA-β-Galactosidase staining of ATG5-depleted HMECs, with or without PD0332991 treatment (0.5 μM, 72h). The proportion of positive cells per 20x field is shown. Error bars indicate mean± SD (n=3). (C) Proliferation curves of HMECs and MCF10A/ErbB2(NeuT) cells treated with chloroquine (CQ), PD0332991, or both compounds. Error bars indicate mean± SD (n=3). (D) Soft-agar transformation assay for MCF10A/ErbB2(NeuT) cells treated with chloroquine (CQ), PD0332991, or both compounds. Error bars indicate mean± SD (n=3). Scale bar, 250 μM.
Discussion
In the present work, we sought to identify and characterize cellular processes that are altered in MECs displaying reduced cyclin D1 activity in an effort to better understand the biological effects of pharmacological Cdk4/6 inhibition. To this end, we used the cyclin D1KE/KE knockin mouse as our primary model (17, 18). Surprisingly, cyclin D1KE/KE mammary tissues were initially able to sustain aberrant proliferation in response to ErbB2, despite impaired pRb phosphorylation. Since many growth-promoting signals (including those dependent on ErbB2) can increase the levels or activity of cyclin D1 (14, 42) and, at the same time, inhibit autophagy (5, 43), we reasoned that autophagy might be upregulated in cyclin D1KE/KE mammary tissues in order to compensate for reduced cyclin D1 activity. Indeed, using several approaches, we show here that cyclin D1KE/KE mammary tissues display signs of increased autophagic activity. This increase in autophagy in the presence of impaired pRb phosphorylation is consistent with a recent study linking pRb’s E2F binding function to autophagy induction (44). However, other studies have linked the pRb-E2F interaction to autophagy suppression (45, 46), indicating that these functions may be context dependent.
Importantly, unlike recent reports indicating that autophagy upregulation is required for oncogene-induced senescence (11), ErbB2-expressing cyclin D1KE/KE mammary epithelium displayed a significant reduction in senescence while retaining increased levels of autophagy, suggesting that these processes may be regulated differently in mutant mammary glands. Given the high proliferative fraction and the lack of senescence and apoptosis in ErbB2-expressing cyclin D1KE/KE mammary tissues, it is surprising that mutant animals display a profound lack of transformation as they age. Based on the observed increase in myoepithelial cells in mutant mammary glands and the reduction in progenitor cells (18), we surmise that mutant MECs lacking cyclin D1 activity undergo terminal, albeit aberrant, differentiation even in the presence of activated ErbB2, thereby avoiding neoplastic conversion. However, we cannot exclude stromal defects or paracrine effects resulting from, for example, senescence-associated secreted factors, which are consistently reduced in mutant glands. Future studies using transplantation of MECs and stromal cells, alone or in combination, will be required to fully understand the fate of ErbB2-expressing cyclin D1KE/KE mammary MECs.
To better understand the role of autophagy in cyclin D1KE/KE MECs, we established a cell culture system that mimicked many of the in vivo properties of these cells. Consistent with the in vivo data, high rates of autophagy were retained in immortalized cyclin D1KE/KE MECs. In keeping with its pro-survival function, downregulation of autophagy in these cells worsened cell proliferation through an exacerbation of senescence. It is important to note that, despite initial observations pointing to a role for autophagy in tumor suppression (3, 10, 47, 48), evidence from more refined animal models indicates that abrogation of autophagy in selected tissues can impair tumor initiation (49).
How do we reconcile these seemingly opposing functions of autophagy during tumorigenesis? One possibility is that upregulation of autophagy does have a long-term tumor suppressive effect in ErbB2-expressing mammary epithelia by way of preventing the accumulation of protein aggregates, damaged organelles, and reactive oxygen species (ROS) (10). Accordingly, abrogation of autophagy in vivo may favor a mutagenic environment that would allow the acquisition of additional genetic events necessary to escape ErbB2-induced senescence and ensure tumor evolution. In contrast, acute disruption of autophagy in vitro, with the consequent rapid accumulation of cellular damage, would be expected to activate senescence unless other tumor suppressive pathways are also disrupted.
The data presented here might also point to unappreciated relationships between cyclin D1 function, autophagy and metabolism. The co-existence of proliferation and autophagy in ErbB2-expressing cyclin D1KE/KE mammary tissues suggests that despite the ability of the mutant protein to sustain a ErbB2-dependent proliferative response, the non-catalytic functions of cyclin D1 are not sufficient to suppress autophagy in vivo. In this scenario, functional cyclin D1/Cdk(4/6) complexes may act as a nexus to indicate both growth factor and nutrient proficiencies appropriate for a proliferative response. In the context of inactive cyclin D1/Cdk(4/6) complexes, induction of autophagy and proliferation may represent an attempt to respond to growth promoting signals in the perceived absence of metabolic substrates. Further work will be necessary to test this hypothesis.
In summary, highlighting the crucial position of cyclin D1 downstream of many growth-promoting signals, abrogation of cyclin D1 activity triggers the activation of autophagy in both murine and human MECs. Moreover, suppression of autophagy together with cyclin D1-dependent kinase inactivation in immortal and transformed human MECs leads to increased senescence or reduced proliferation and transformation. Therefore, the functional connection between cyclin D1 and autophagy may be exploited through combinatorial pharmacological modulation of Cdk4/6 activity and the autophagy response to more efficaciously treat breast cancer.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant CA104322 (PWH). None of the materials in this manuscript has been published or is under consideration elsewhere.
The authors thank Dr. Terje Johansen for providing with the pDest-mCherry-GFP-LC3B construct and the members of the Hinds lab for their helpful comments.
Footnotes
None of the authors has a conflicting financial interest.
References
- 1.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuchihara K, Fujii S, Esumi H. Autophagy and cancer: dynamism of the metabolism of tumor cells and tissues. Cancer Lett. 2009;278:130–8. doi: 10.1016/j.canlet.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 3.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 5.Chen N, Debnath J. Autophagy and tumorigenesis. FEBS letters. 2010;584:1427–35. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 7.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 8.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of clinical investigation. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. The New England journal of medicine. 2006;355:1037–46. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 13.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. The Journal of clinical investigation. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215–24. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 15.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nature reviews. 2001;1:222–31. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 16.Malumbres M, Barbacid M. Is Cyclin D1-CDK4 kinase a bona fide cancer target? Cancer cell. 2006;9:2–4. doi: 10.1016/j.ccr.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, et al. Cyclin D1 Kinase Activity Is Required for the Self-Renewal of Mammary Stem and Progenitor Cells that Are Targets of MMTV-ErbB2 Tumorigenesis. Cancer cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–32. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 20.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–15. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 21.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 22.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer research. 1990;50:6075–86. [PubMed] [Google Scholar]
- 24.Tait L, Soule HD, Russo J. Ultrastructural and immunocytochemical characterization of an immortalized human breast epithelial cell line, MCF-10. Cancer research. 1990;50:6087–94. [PubMed] [Google Scholar]
- 25.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast cancer research : BCR. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, et al. Defining the cellular precursors to human breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2772–7. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severino J, Allen RG, Balin S, Balin A, Cristofalo VJ. Is beta-galactosidase staining a marker of senescence in vitro and in vivo? Experimental cell research. 2000;257:162–71. doi: 10.1006/excr.2000.4875. [DOI] [PubMed] [Google Scholar]
- 29.Ketteler R, Seed B. Quantitation of autophagy by luciferase release assay. Autophagy. 2008;4:801–6. doi: 10.4161/auto.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sivridis E, Koukourakis MI, Zois CE, Ledaki I, Ferguson DJ, Harris AL, et al. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. The American journal of pathology. 2010;176:2477–89. doi: 10.2353/ajpath.2010.090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nature reviews. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 33.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nature cell biology. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 34.Schnabl B, Purbeck CA, Choi YH, Hagedorn CH, Brenner D. Replicative senescence of activated human hepatic stellate cells is accompanied by a pronounced inflammatory but less fibrogenic phenotype. Hepatology (Baltimore, Md. 2003;37:653–64. doi: 10.1053/jhep.2003.50097. [DOI] [PubMed] [Google Scholar]
- 35.Shelton DN, Chang E, Whittier PS, Choi D, Funk WD. Microarray analysis of replicative senescence. Curr Biol. 1999;9:939–45. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 36.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 37.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 38.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nature reviews. 2002;2:331–41. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 40.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods in enzymology. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 41.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 42.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nature reviews. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 43.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nature reviews. 2007;7:961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, et al. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–93. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 46.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–42. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–27. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.