Abstract
Aims/hypothesis
Recent functional characterisations of genome-wide association study (GWAS) loci suggest that cis-regulatory variation may be a common paradigm for complex disease susceptibility. Several studies point to a similar mechanism at the transcription factor 7-like 2 (TCF7L2) GWAS locus for type 2 diabetes. To address this possibility, we carried out an in vitro scan of this diabetes-associated locus to fine-map cis-regulatory sequences within this genomic interval.
Methods
A systematic cell-based enhancer strategy was employed to interrogate all sequences within the 92 kb type-2-diabetes-association interval for cis-regulatory activity in a panel of cell lines (HCT-116, Neuro-2a, C2C12, U2OS, MIN6 and HepG2). We further evaluated chromatin state at a subset of these regions in HCT-116 and U2OS cells and examined allelic-specific enhancer properties at the type-2-diabetes-associated single nucleotide polymorphism (SNP) rs7903146.
Results
In total, we assigned cis-regulatory activity to approximately 30% (9/28) of constructs tested. Notably, a subset of enhancers was active across multiple cell lines and overlapped with key epigenetic markers suggestive of cis-regulatory sequences. We further replicated the allelic-specific properties for SNP rs7903146 in pancreatic beta cells and additionally demonstrate identical allelic-specific enhancer effects in other cell lines.
Conclusions
These results provide a detailed map of cis-regulatory elements within the TCF7L2 GWAS locus and support the hypothesis of cis-regulatory variation leading to type 2 diabetes predisposition. The detection of allelic-specific effects for SNP rs7903146 in multiple cell lines further alludes to the likelihood of a peripheral defect in disease aetiology.
Keywords: GWAS, cis-Regulatory elements, TCF7L2, Type 2 diabetes
Introduction
Genetic variation in non-coding sequences has been associated with increased risk in a number of common human diseases [1]. The experimental elucidation of the molecular mechanisms underlying these associations often reveals that common variants disrupt long-range cis-regulatory sequences responsible for the spatial, temporal and quantitative aspects of transcription of neighbouring genes.
Consistent with these observations, genetic variation within introns of TCF7L2 constitutes the strongest genetic risk factor for type 2 diabetes across diverse human populations [2]. TCF7L2 is expressed in broad domains, including the pancreas, gut and liver. Notably, both the overexpression [3–5] and ablation [6–8] of TCF7L2 have been described as modifiers of glucose tolerance in humans and animal models. Recent studies have demonstrated allelic-specific enhancer properties for sequences spanning single nucleotide polymorphism (SNP) rs7903146, the variant with the strongest association with disease risk [4, 9]. Our analyses lend support to this conclusion, as we demonstrated, in vivo, that the 92 kb diabetes-associated region represents the primary domain governing regulatory activity in a 500 kb window spanning TCF7L2 [3].
We recently performed an in vivo analysis of enhancers within this genome-wide association study (GWAS) interval, focusing on a subset of regions exhibiting strong sequence conservation [10]. Here, we carried out a comprehensive enhancer screen that assessed the regulatory potential of all sequences spanning the entire 92 kb association interval, using luciferase assays in diverse cell lines representing tissues involved in glucose homeostasis and expressing transcription factor 7-like 2 (TCF7L2). We validated enhancers through chromatin-state analysis and further examined allelic-specific enhancer properties at SNP rs7903146 using this cell panel.
Methods
Generation of reporter library
Human sequences (28 total) tiling the 92 kb associated region were cloned (Fig. 1a and electronic supplementary material [ESM] Table 1) through Gateway technology (Invitrogen, Carlsbad, CA, USA) in pGL4.23 luciferase vectors for cell-based assays.
Fig. 1.
Cell-based enhancer screen of the TCF7L2 association interval. (a) TCF7L2 gene (NM_001146274.1) exon numbers are given; the 92 kb associated interval is highlighted in red. The alternative exon 3b is shown as exon 4. A red asterisk marks the type-2-diabetes-associated SNP rs7903146. The 28 regions analysed for regulatory activity are numbered above and delineated by blue dotted lines. Sequence conservation between human and mouse (VISTA Genome Browser) is shown. ENCODE H3K4me1 is also shown. The results of the enhancer scan are summarised and regions exhibiting enhancer activity are coloured in black. Construct numbers are also given again below the summary. Relative luciferase activity in HCT-116 (b), neuro-2a (c), C2C12 (d), U2OS (e), MIN6 (f) and HepG2 (g) cells is shown. A dotted red line marks the threshold for enhancer activity. In each graph, the construct number is given on the x-axis. The numbers of all constructs exhibiting enhancer activity are displayed
Cell culture and reporter assay of enhancer activity
We examined enhancer activity in human HCT-116 colorectal carcinoma cells, U2OS osteosarcoma cells and HepG2 hepatocellular carcinoma cells as well as murine neuro-2a neuroblastoma cells, C2C12 myoblasts and MIN6 insulinoma cells. Luciferase assays were conducted in 96 well plates. Transfection with 200ng construct DNA using Lipofectamine 2000 (Invitrogen) was performed in triplicate.
Chromatin-state analysis
We evaluated a subset of regions harbouring regulatory activity in HCT-116 and U2OS cells, as these represent the only human cell lines used in the enhancer screen. Immunoprecipitation was conducted on sonicated chromatin using antibodies against histone 3, lysine 27 acetylation (H3K27ac; abcam-ab4729, Abcam, Cambridge, MA, USA) and histone 3, lysine 4 tri-methylation (H3K4me3; active motif-39159, Active Motif, Carlsbad, CA, USA). Quantitative PCR was conducted in triplicates of biological replicates from each experiment.
Statistics
A two-sided Student’s t test was used to assess allelic-specific regulatory effects.
Results
Cell-based enhancer scan of TCF7L2 diabetes-associated locus
Sequences spanning the entire GWAS association interval were cloned and tested using cell-based luciferase assays. We generated 28 constructs spanning 93.2 kb of TCF7L2 intronic sequence (Fig. 1a). Region 7, spanning SNP rs7903146, harboured the protective C allele. For these assays, we used HCT-116, neuro-2a, C2C12, U2OS and MIN6 cells; HepG2 cells served as a negative control, as this interval was devoid of liver enhancer activity in our previous in vivo work [3]. We chose these cell types as they are representative of tissues that express TCF7L2 and are further involved in glucose metabolism [3, 10].
We uncovered cis-regulatory elements in colorectal (HCT-116, Fig. 1b), neuronal (neuro-2a, Fig. 1c), skeletal muscle (C2C12, Fig. 1d), osteosarcoma (U2OS, Fig. 1e) and pancreatic beta cell (MIN6, Fig. 1f) lines (also see ESM Table 2). As expected [3], we did not identify enhancer activity in HepG2 cells (Fig. 1g). In total, we assigned enhancer function to approximately 30% of constructs (9/28). Several of the regions displayed regulatory activity across multiple cell lines. Most enhancer elements span regions of marked sequence conservation between human and mouse. A subset of our identified enhancer also overlap with pronounced H3K4me1 peaks and nearly all span sequences predicted to be strong enhancers from chromatin-state predictions across diverse cell lines, strengthening the notion that these regions indeed represent cis-regulatory elements (Fig. 1a).
Evaluation of chromatin state at HCT-116 and U2OS regulatory elements
We assessed H3K27ac levels, a chromatin signature commonly found at enhancers, in a subset of our regulatory sequences. We also characterised H3K4me3 levels, a modification that is prevalent in active promoters, given recent reports of a cryptic alternative promoter activity in many mouse intronic enhancers. We assayed regulatory sequences uncovered in HCT-116 and U2OS human cell lines (Fig. 1). Supporting our luciferase data, all five sequences exhibited enrichment of H3K27ac in HCT-116 and U2OS cells (ESM Fig. 1). Interestingly, region 25 also displayed high H3K4me3 enrichment in U2OS cells. However, this region contains no evidence of transcription in humans or any other species. Together, these results posit that region 25 likely represents an enhancer sequence in U2OS cells, although we cannot exclude its role as an alternative promoter of an as yet unidentified TCF7L2 isoform.
In vitro functional analysis of SNP rs7903146
We also tested short sequences spanning SNP rs7903146 for potential allelic differences in our cell panel (Fig. 2). We generated the identical 239 bp constructs centred on both the protective (C) and risk (T) allele of SNP rs7903146 that were previously reported to possess allelic-specific regulatory activity [4]. As the detailed studies demonstrating the allelic-specific regulatory properties for this variant were largely limited to pancreatic beta cells [4, 9], our analysis allows for a more systematic interrogation of regulatory potential across multiple cell types.
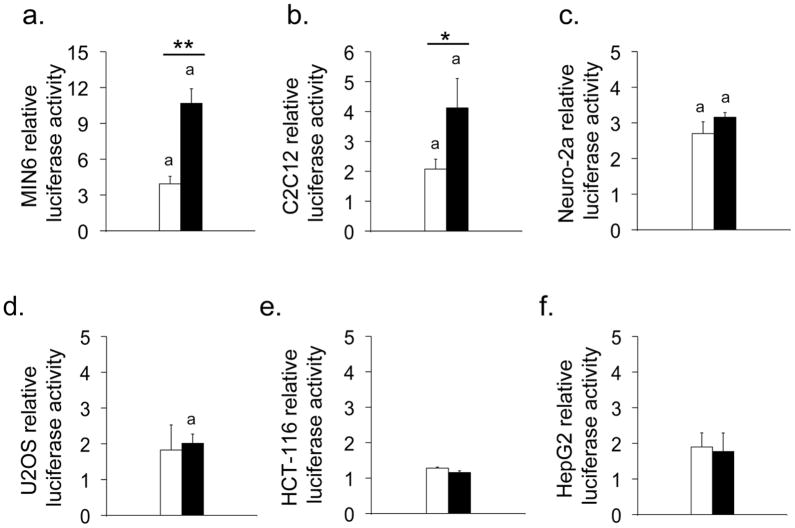
Fig. 2.
In vitro functional analysis of sequences spanning SNP rs7903146. Relative luciferase activity from the 239 bp regions surrounding the protective C (white bars) and risk T (black bars) alleles of SNP rs7903146 in MIN6 (a), C2C12 (b), neuro-2a (c), U2OS (d), HCT-116 (e) and HepG2 (f) cells. aConstructs that exhibited enhancer activity. Neuro-2a cells exhibited a trend (p=0.09) in allelic-specific activity. *p<0.05 and **p<0.01 for the difference in activity between protective and risk allele constructs
We replicated the results generated from previous studies [4, 9] using pancreatic MIN6 cells (Fig. 2a). Interestingly, this sequence also exhibited enhancer activity in myoblast (Fig. 2b), neuronal (Fig. 2c) and bone (Fig. 2d) cell lines. We next determined if any allelic-specific properties were present in these three additional cell lines and identified a significant difference in myoblasts (Fig. 2b), while neuronal cells displayed a strong trend (Fig. 2c). In agreement with previous analyses [4, 9], in both cases the risk T allele maintained stronger regulatory activity than the protective C allele. These results support the notion that the disease risk allele of rs7903146 leads to an increased enhancer activity in multiple tissues, not only pancreatic beta cells.
Discussion
Understanding the regulatory architecture of GWAS loci is critical, as the recent characterisations of these intervals have consistently uncovered functional variants within regulatory elements [1]. To this end, we performed an enhancer scan on the TCF7L2 diabetes-associated interval and generated a detailed fine-scale regulatory map of this region. The 92 kb interval that we tested has been further narrowed using association data in ethnically diverse populations [11]. Nevertheless, recent studies reported associations of variants within TCF7L2 and cardiovascular diseases [12], schizophrenia [13], and colorectal cancer [14]. Besides common diabetes-associated variants, we believe that our analysis can also be used to focus next-generation sequencing efforts to identify rare variants in regulatory sequences. Hence, we believe that a broader and systematic analysis of this locus would be more widely beneficial.
We find that 32% (9/28) of our constructs exhibited regulatory expression in myoblast, neuronal, colorectal, bone and pancreatic beta cell lines. We further validated five of these regions that were found in human cell lines through chromatin mapping. Notably, over 50% of the uncovered enhancer elements within this GWAS interval further generated activity across distinct cell lines. Our analyses carry a number of intrinsic limitations that need to be addressed. For instance, the enhancers identified in this GWAS interval are not exhaustive as they are limited to the cell types used. Nonetheless, our results reflect a considerable degree of cis-regulatory complexity at this locus and a broader panel of cell lines may have uncovered additional regulatory activity. The use of different-sized fragments in our reporter assay may further generate disparate results, as longer sequences allow for the co-interrogation of neighbouring repressor sequences. This may explain differences between region 7 and the shorter 239 bp region harbouring SNP rs7903146 situated within region 7. Alternatively, the use of smaller sequences may not reflect the true function of an element in its endogenous sequence. Consequently, determining the most appropriate approach is difficult.
A subset of these enhancers also exhibited regulatory activity in our recent in vivo analysis [10]. However, differences between studies are also apparent. These discrepancies possibly reflect differences in assessing enhancer activity across diverse platforms. This includes variations in sensitivity using quantitative and qualitative assays, the use of immortalised cancer cell lines for reporter assays as well as spatial and temporal effects on regulatory expression. Nonetheless, these results stress the advantages of integrating diverse strategies to obtain a comprehensive map of enhancer activities.
We further replicated findings on the allelic-specific properties of the diabetes-associated SNP rs7903146 in pancreatic beta cells [4, 9]. Importantly, we expand on these results by identifying allelic-specific properties for this variant in cells outside the pancreas. As pancreatic-specific roles have become a prime focus to understanding diabetes aetiology [15], our results stress the need for analyses of TCF7L2 function in other tissues. Indeed, the complexity of this transcription factor is illustrated from the further association of this interval with colorectal cancer [14], schizophrenia [13] and coronary artery disease [12], all of which had rs7903146 as a disease-associated SNP.
Collectively, our results illustrate the potential involvement of cis-regulatory sequence variations in diabetes susceptibility at this GWAS locus. The localisation of regulatory elements that control expression within distinct cellular domains as well as the expansion of the allelic-specific properties of SNP rs7903146 outside pancreatic beta cells further points to the potential involvement of peripheral tissues in the pathophysiology of type 2 diabetes.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH grants DK078871 and HG004428 (M. A. Nobrega), DK020595 (G. I. Bell) and T32 GM007197 (D. Savic). We also thank the Diabetes Research and Training Center (DRTC) at the University of Chicago and Kovler Family Foundation for their gift.
Abbreviations
- GWAS
Genome-wide association study
- H3K27ac
Histone 3, lysine 27 acetylation
- H3K4me3
Histone 3, lysine 4 tri-methylation
- SNP
Single nucleotide polymorphism
- TCF7L2
Transcription factor 7-like 2
Footnotes
Contribution statement
DS, GIB and MAN contributed to the concept and design of the study. DS, SYP and KAB contributed to the analysis and interpretation of data. DS, SYP, KAB, GIB and MAN contributed to drafting of the manuscript and revisions. The final version of the manuscript was approved by all authors.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
References
- 1.Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cauchi S, El Achhab Y, Choquet H, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med. 2007;85:777–782. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- 3.Savic D, Ye H, Aneas I, Park SY, Bell GI, Nobrega MA. Alterations in TCF7L2 expression define its role as a key regulator of glucose metabolism. Genome Res. 2011;21:1417–1425. doi: 10.1101/gr.123745.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu L, Sauter NS, Schulthess FT, Matveyenko AV, Oberholzer J, Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57:645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 7.Loder MK, da Silva Xavier G, McDonald A, Rutter GA. TCF7L2 controls insulin gene expression and insulin secretion in mature pancreatic beta-cells. Biochem Soc Trans. 2008;36:357–359. doi: 10.1042/BST0360357. [DOI] [PubMed] [Google Scholar]
- 8.Figeac F, Uzan B, Faro M, Chelali N, Portha B, Movassat J. Neonatal growth and regeneration of beta-cells are regulated by the Wnt/beta-catenin signaling in normal and diabetic rats. Am J Physiol Endocrinol Metab. 2010;298:E245–256. doi: 10.1152/ajpendo.00538.2009. [DOI] [PubMed] [Google Scholar]
- 9.Stitzel ML, Sethupathy P, Pearson DS, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savic D, Bell GI, Nobrega MA. An in vivo cis-regulatory screen at the type 2 diabetes associated TCF7L2 locus identifies multiple tissue-specific enhancers. PLoS One. 2012;7:e36501. doi: 10.1371/journal.pone.0036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgason A, Palsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 12.Sousa AG, Marquezine GF, Lemos PA, et al. TCF7L2 polymorphism rs7903146 is associated with coronary artery disease severity and mortality. PLoS One. 2009;4:e7697. doi: 10.1371/journal.pone.0007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen T, Ingason A, Djurovic S, et al. At-Risk Variant in TCF7L2 for type ii diabetes increases risk of schizophrenia. Biol Psychiatry. 2011;70:59–63. doi: 10.1016/j.biopsych.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 14.Folsom AR, Pankow JS, Peacock JM, Bielinski SJ, Heiss G, Boerwinkle E. Variation in TCF7L2 and increased risk of colon cancer: the Atherosclerosis Risk in Communities (ARIC) Study. Diabetes Care. 2008;31:905–909. doi: 10.2337/dc07-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia. 2008;51:1100–1110. doi: 10.1007/s00125-008-1025-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.