Abstract
The k-turn is a commonly occurring structural motif that introduces a tight kink into duplex RNA. In free solution, it can exist in an extended form, or by folding into the kinked structure. Binding of proteins including the L7Ae family can induce the formation of the kinked geometry, raising the question of whether this occurs by passive selection of the kinked structure, or a more active process in which the protein manipulates the RNA structure. We have devised a single-molecule experiment whereby immobilized L7Ae protein binds Cy3-Cy5-labeled RNA from free solution. We find that all bound RNA is in the kinked geometry, with no evidence for transitions to an extended form at the millisecond timescale of the camera. Furthermore, real-time binding experiments provide no evidence for a more extended intermediate even at the earliest times, at a time resolution of 16 ms. The data support a passive conformational selection model by which the protein selects a fraction of RNA that is already in the kinked conformation, thereby drawing the equilibrium into this form.
Introduction
The kink-turn (k-turn) is a widespread structural motif found in many functional double-stranded RNA species that introduces a tight kink into the duplex axis with an included angle of close to 60° (1). Most k-turn sequences comprise a three-nucleotide bulge flanked on the 3′ side by G⋅A and A⋅G pairs, and frequently further nonWatson-Crick basepairs (termed the NC-helix; see Fig. 1). The G⋅A pairs are trans sugar-edge/Hoogsteen-edge basepairs that direct the minor groove edges of the two adenine bases into the minor groove of the opposing duplex (termed the C-helix). The kinked geometry is stabilized by A-minor interactions in the core of the k-turn, resulting in the formation of a number of well-conserved hydrogen bonds (2,3).
Figure 1.

The sequence of Kt-7. (Upper part) Sequence and basepairing in Kt-7, with the nucleotide positions labeled according to the standard k-turn nomenclature. (Lower part) Sequence of the RNA duplex containing a central Kt-7 sequence (boxed) used in the FRET experiments. The RNA is 5′-labeled with Cy-5 and Cy-3.
Many k-turns act as specific binding sites for proteins. For example, most k-turns found in both ribosomal subunits are bound to different proteins (4,5). These include L7Ae, which is bound to Kt-15 in the Haloarcula marismortui 50S ribosomal subunit (4). This is a member of a family of related proteins, including the ribosomal protein L30e, yeast Snu13p, and human 15.5 kDa proteins (6,7) as well as a bacterial homolog YbxF (8). In addition to the role in the ribosome, protein binding to k-turns occurs as a critical initial step in the formation of the box C/D and H/ACA complexes that mediate site-specific modification of RNA in archaea and eukaryotes (9,10), and the 15.5 kDa protein binds U4 snRNA in the U4-U6.U5 tri-snRNP involved in the biogenesis of the spliceosome (7). Crystal structures are available for the complexes of Archaeoglobus fulgidus and Methanococcus jannaschii L7Ae and box C/D RNA (11,12), M. jannaschii L7Ae and box H/ACA RNA (13) as well as that of the human 15.5 kDa protein and the U4 snRNA (14).
In free solution, the k-turn can exist in two alternative conformations. One is the kinked structure that has been observed in a number of crystal structures of ribosomes, riboswitches, and other species. However, in free solution in the absence of proteins or added metal ions, the RNA adopts a less-kinked structure, which is typical of that normally found for a three-nucleotide bulge (15,16). We have found three ways in which the formation of the kinked conformation occurs.
First, for many k-turn sequences, particularly where the sequence approximates that of the consensus k-turn, addition of metal ions such as 100 μM Mg2+ induces the population to adopt the well-characterized kinked structure (16).
Second, we have recently shown that tertiary interactions within a more complex RNA molecule can stabilize a k-turn structure that is unable to fold in isolation by the addition of divalent metal ions (17).
Third, the binding of proteins such as L7Ae can also induce the formation of k-turn conformation in the population (3,18,19).
We have found that A. fulgidus L7Ae binds to H. marismortui Kt-7 with very high affinity, with Kd = 10 pM, inducing the formation of the kinked geometry of the k-turn (3). This is a clear example of a protein-induced structural transition, but the observation raises the question of how this occurs at the molecular level. Two possible mechanisms can be envisaged: one dynamic (induced fit (20)) and the other static (conformational selection (21)). This has been discussed previously for macromolecule-ligand, protein-protein, and protein-nucleic acid interactions (22–27). Most of those cases involve relatively small-scale accommodation of nucleic acid structure, as exemplified by the binding of U1A RNA to the U1A protein (28,29). By contrast, the protein-mediated folding of the k-turn occurs on a rather larger scale. In the induced-fit mechanism, the protein is envisaged to interact with the extended form, somehow mechanically inducing the kinked conformation in situ. In conformational selection, the protein would bind selectively to RNA in a preformed kinked conformation, so displacing the equilibrium to that structure. This requires an equilibrium between an extended and a folded k-turn structure to exist in solution, as previously indicated in our measurement of fluorescent lifetimes (16). In other systems these possibilities have been analyzed by computational methods (27,30,31), and by NMR relaxation (32–36) approaches. In the k-turn folding process, it is difficult to distinguish these alternative mechanisms using ensemble methods in solution, and we have therefore studied L7Ae protein binding events by fluorescence resonance energy transfer (FRET) in single RNA molecules, using what is to the best of our knowledge novel experimental design. These experiments favor the conformational selection model of k-turn folding on protein binding.
Materials and Methods
RNA synthesis
RNA oligonucleotides were made by chemical synthesis using t-BDMS phosphoramidite chemistry (37), as described in Wilson et al. (38). Oligonucleotides were made with 5′ amino linkers, and Cy3 and Cy5 fluorophores were conjugated as n-hydroxysuccinimide esters (Amersham Biosciences, Chandler, AZ). All oligonucleotides were purified by gel electrophoresis in polyacrylamide, and recovered from gel fragments by electroelution followed by ethanol precipitation. Fluorescently labeled RNA was subjected to further purification by reversed-phase HPLC on a C18 column (MicroBondapak; Waters, Milford, MA) using an acetonitrile gradient with an aqueous phase of 100 mM triethylammonium acetate pH 7.0.
The RNA species used in this study were constructed by hybridization of two oligonucleotides of sequence (written 5′ to 3′):
Cy3- CCUGACAUGGGGAGCCACUGACUGG
Cy5- CCAGUCAGUGGCGAAGAACCAUGUCAGG
These were purified by electrophoresis in polyacrylamide gels under nondenaturing conditions.
Synthesis of a gene encoding a U1A - L7Ae fusion, and expression, and purification of the protein
An in-frame fusion of the genes for U1A and L7Ae was constructed by ligation of PCR products generated from genes for Homo sapiens U1A and A. fulgidus L7Ae. The ligated product was subjected to a further round of PCR, and after restriction cleavage was ligated into the vector pET15b. The ligated plasmid was transformed into competent XL1-Blue Escherichia coli cells (Stratagene, La Jolla, CA) and plated onto Luria broth agar containing 0.1 mg/mL ampicillin. Individual colonies were checked by growing in LB medium containing 0.1 mg/mL ampicillin. pET15b containing the complete gene for the U1A-L7Ae fusion protein with a hexahistidine tag was transformed into E. coli BL21-Gold (DE3) pLysS cells (Stratagene), and protein expression was induced by the addition of IPTG to 1 mM. Cleared cell lysate was applied to a Ni2+-chelated HiTrap column (GE Healthcare, Waukesha, WI) installed on an ÄKTA Purifier (GE Healthcare), and eluted using 500 mM imidazole. After dilution, the protein was applied to a HiTrap heparin HP column (GE Healthcare) and eluted using a NaCl gradient. Pooled fractions were concentrated and applied to a Superdex 75 column (GE Healthcare). The U1A-L7Ae fusion protein was confirmed by mass spectrometry to comprise both H. sapiens U1A and A. fulgidus L7Ae proteins. Full experimental details are provided in the Supporting Material.
Total internal reflection single-molecule microscopy
Fluorescence intensities at donor and acceptor wavelengths were acquired from Cy3-Cy5-labeled RNA bound to U1A-L7Ae fusion protein. The slides were mounted on the stage of an inverted microscope (model No. IX70, Olympus, Melville, NY) and excited via the evanescent field generated by the total internal reflection of light from a solid-state 532-nm laser (Crystalaser, Reno, NV) via a quartz prism. Fluorescent emission was collected by a 1.2 numerical aperture 60× water immersion objective lens (Olympus), and separated by a 645-nm dichroic mirror (Chroma Technology, Bellows Falls, VT) into the donor and the acceptor fluorescence. These were focused side-by-side into a back-illuminated, electron multiplying charge-coupled device camera (iXON EMCCD; Andor Technology, South Windsor, CT) (39). Hundreds of molecules could be recorded simultaneously using an image area of 8.2 × 8.2 mm (512 × 512 active pixels). Data were acquired using software written in Visual C++ (Microsoft, Redmond, WA), where each frame had a duration of 33 ms for the population histograms and 16 ms for the real-time binding experiments.
Measurements were performed at room temperature. Single-molecule fluorescence resonance energy transfer (FRET) efficiency after background correction was approximated by EFRET = IA/(IA + ID), where IA and ID are the fluorescence intensities of the acceptor and donor, respectively. Because the quantum yields and detection efficiencies of Cy3 and Cy5 are very close, EFRET closely matches the true efficiency of energy transfer. However, the spectral overlap separation of Cy3 and Cy5 is not absolute and the 645-nm separation led to ∼10% of Cy3 leakage into the Cy5 channel. This gives an apparent EFRET of ∼0.1 for a single active Cy3 fluorophore in the absence of Cy5 acceptor.
Data analysis was carried out using laboratory-written analysis routines developed in MATLAB (The MathWorks, Natick, MA). Single-molecule FRET histograms were obtained using the whole FRET trace, while the population histograms were obtained by averaging frames 11–20 for every individual molecule after manually filtering photobleaching and blinking effects. Filtering involved removal of time-traces in which:
-
1.
the fluorescence intensity of the acceptor was lower than 50–55 (caused by the absence of Cy5 fluorescence);
-
2.
the total intensity was higher than 550–700 (indicating the presence of multiple fluorophores);
-
3.
absence of anticorrelation between both fluorescence intensities during a blinking, dynamic, or a photobleaching event;
-
4.
the total intensity was irregular over the length of the time-trace; and
-
5.
multiple photobleaching event from the same fluorophore.
Cross-correlation analysis was performed using a program implemented in MATLAB. Records of donor and acceptor intensities as a function of time for single junction molecules were analyzed using the equation
| (1) |
where and are the mean donor and acceptor intensities, respectively, over the whole data record, normalized by the number of points summed (N). The function G(τ) correlates the donor intensity at time t with acceptor intensity at time increment τ later.
Observation of single k-turn RNA molecules bound to immobilized L7Ae U1A-L7Ae fusion protein
A cell was constructed using an ultraclean quartz microscope slide drilled with two holes and a coverslip separated by double-sided adhesive tape to leave a volume for the sample. This was then washed by injection of water, followed by 10 mM Tris-HCl (pH 8.0), 5 mM NaCl (T5 buffer). Successive additions were made of 100-μL volumes of (1) 1 mg/mL of a 1:4 mixture of biotinylated and unmodified bovine serum albumin (BSA; Sigma Aldrich, St. Louis, MO), (2) 0.2 mg/mL neutravidin (Thermo Fisher Scientific, Waltham, MA), (3) 3 nM 5′-biotinylated U1A RNA (biotin-dGCAGCGAAUCCAUUGCACUCCGGAUUCGCUGdC), and (4) 4 nM U1A-L7Ae protein, with 5-min incubations at room temperature followed by washing with T5 buffer at each stage. The k-turn RNA was prepared by mixing 20 μL of 80 nM Cy3, Cy5-labeled k-turn RNA with 80 μL imaging solution comprising 60 mg/mL glucose (Sigma Aldrich), 0.08 mg/mL catalase (Roche, Basel, Switzerland), 2 mg/mL glucose-oxidase (Sigma Aldrich), and 0.25 mg/mL Trolox (Sigma Aldrich) in 10 mM Tris-HCl (pH 8.0). This was introduced into the cell located on the microscope stage in two successive injections of 50 μM.
Real-time observation of single k-turn RNA molecules in the act of binding to immobilized U1A-L7Ae fusion protein
The cell was constructed as above, and the same procedure was used to attach the RNA and protein components to the surface as in Steps 1–4. Plastic tubing was attached to the drilled holes in the slide to allow injection of Cy3, Cy5-labeled k-turn RNA during illumination, and collection of fluorescence by the objective lens of the microscope. Data acquisition was initiated simultaneously with injection of fluorescent RNA. Acquired pictures were digitally postprocessed in real-time to perform background subtraction followed by data-scaling to increase image contrast. To capture all binding events, each frame was searched for individual molecules by applying cluster analysis on a rolling average of ∼50 frames. Coordinates of individual molecules were recorded, sorted, and tested for uniqueness. Fluorescence intensity time-traces were later extracted and manually filtered by applying the common criteria (see above).
Results and Discussion
Attachment of L7Ae protein to the surface of a quartz slide
The normal procedure for studying nucleic acid-protein interactions by single-molecule FRET would be to tether the donor-acceptor-labeled nucleic acid to the surface of the slide, and then introduce the protein in solution and allow it to bind, observed in total-internal-reflection microscopy using an EMCCD camera. However, this was complicated for the k-turn, since this is essentially a duplex, where both 5′-termini are required for fluorophore attachment. We therefore opted to immobilize the protein, and deliver the fluorescent k-turn RNA in solution. This has the additional merit that the only fluorescent spots observed must correspond to RNA molecules that are bound by the protein. The RNA used in these experiments consisted of a 25-bp duplex 5′-terminally labeled with Cy3 (acceptor) and Cy5 (donor) and the central sequence of Kt-7 (Fig. 1).
To reduce the possibility of surface-induced denaturation of the protein we adopted the strategy shown in Fig. 2 A. This is based on the use of a U1A-binding RNA hairpin-loop, and a fusion of U1A and L7Ae proteins. The latter was generated by making an in-frame fusion of the required genes, and expressing a fusion protein consisting of U1A and L7Ae at the N- and C-termini, respectively, linked by a GGGGGGEF peptide. The fusion retained the ability to induce the folding of Kt-7 in solution, demonstrated as an increase in FRET efficiency in the Cy3-Cy5-labeled k-turn as the kinking shortens the distance between the termini (Fig. 2 B). In the single-molecule procedure, a 5′-biotinylated RNA strand with a U1A-binding terminal loop is attached to biotinylated BSA on the quartz slide via neutravidin in the normal manner. The U1A-L7Ae fusion protein is then bound to the hairpin via its U1A end, leaving the L7Ae-end free to bind its target k-turn RNA. Thus, the final step is to inject in the fluorescent Kt-7 RNA and allow it to bind to the immobilized L7Ae; this is the only stage involving a fluorescent component. Altogether the surface-attached complex comprises quartz-BSA-neutravidin-RNA hairpin-(U1A-L7Ae fusion)-(Cy3-Cy5-Kt-7 RNA).
Figure 2.
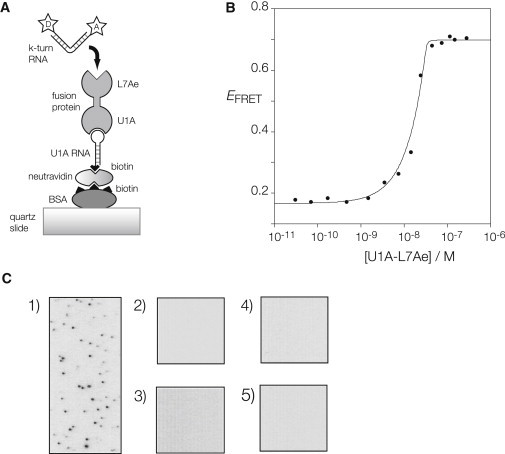
Binding of fluorescent k-turn-containing RNA to L7Ae protein immobilized on a microscope slide. (A) Schematic of the immobilization scheme. U1A-L7Ae fusion binds the loop of an RNA hairpin with a 5′-biotin that is bound to biotinylated BSA via neutravidin. Cy3-Cy5-labeled k-turn RNA flows into the cell, and binds to the L7Ae component of the fusion protein. (B) Induction of folding of Kt-7 upon binding of the U1A-L7Ae fusion protein. Protein-induced folding has been measured in bulk by means of FRET, using the same Cy3-Cy5-labeled RNA used in the single-molecule experiments. Folding kinks the RNA, leading to an increase in the efficiency of energy transfer between the 5′-terminally fluorophores. The plot shows FRET efficiency of Kt-7 as a function of protein concentration. The data have been fitted to a two-state model for L7Ae binding (line) using a stoichiometric binding model. The fitting indicates Kd < 10 pM, although such high affinity cannot be reliably estimated by this technique. The increase in FRET efficiency is ΔEFRET = 0.53, i.e., there is a large increase in FRET efficiency on binding the fusion protein. (C) When all the components are in place, individual bound RNA molecules can be visualized as single points of light (shown inverted as black) at Cy5 emission wavelength. (1) RNA bound to the complete set of components. No bound molecules are observed when the fluorescent RNA is added to (2) BSA only; (3) BSA + neutravidin only; (4) BSA, neutravidin, and U1A RNA only; or (5) BSA, neutravidin, U1A RNA, and U1A protein in place of the fusion.
In principle, the fluorescent Kt-7 RNA should not be able to bind to the surface unless all the components of the assembly are bound correctly. This is demonstrated by the controls presented in Fig. 2 C, which shows a number of frames from the total-internal-reflection microscope. When all the components are assembled together, we observe a number of immobilized single-fluorescent RNA molecules, visualized as single points of light, shown here inverted as black points on a white background. However, if we leave out any of the components (while always adding the fluorescent RNA), or replace the fusion protein with U1A protein alone, we no longer observe immobilization of k-turn RNA molecules. When the full assembly is in place, all the observed points of light correspond to RNA molecules immobilized by binding to the L7Ae protein.
Kt-7 RNA bound to L7Ae exists in a tightly kinked geometry
Fig. 3 A shows an example of the intensity of donor and acceptor emission from one bound k-turn molecule in the presence of 2 mM Mg2+ions, recorded at a 33-ms frame rate. Initially IA > ID, i.e., the bound RNA adopts the kinked geometry (high EFRET). At 125 s, there is a clear Cy5 photobleaching event where the Cy5 emission falls to zero, while the Cy3 increases because it is no longer quenched by FRET; this shows that before this point, this molecule has single functional Cy3 and Cy5 fluorophores, and we have selected such molecules for further analysis. Values of EFRET for 1185 molecules are plotted as a histogram in Fig. 3 B. Most molecules are in a high FRET state, forming a distribution with a mean EFRET = 0.83; thus, the k-turns are bound to L7Ae in a predominantly folded conformation. Close examination of the time-trace (Fig. 3 A) reveals no evidence of transitions to an unfolded (i.e., low EFRET) state, and this was the case for many similar traces examined (further examples given in the Supporting Material). Furthermore, cross-correlation analysis of the data provides no evidence for fast transitions (Fig. 3 C).
Figure 3.
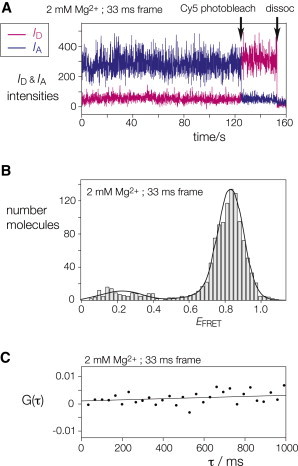
Kt-7 molecules are bound to L7Ae in a predominantly folded conformation in the presence of divalent metal ions. (A) ID and IA versus time for one representative molecule in the presence of 2 mM Mg2+ ions, at 33-ms frame rate. Cy5 was photobleached at 125 s. Near the end of the trace (labeled dissoc) the intensities of both fluorophores fall to background levels; this is most likely due to dissociation of the complex from the surface, although it could result from Cy3 photobleaching. (B) Histogram of EFRET values for Kt-7 molecules bound to L7Ae in 2 mM Mg2+ ions (1185 molecules). Gaussian mixture parameters of data distribution were estimated using an expectation maximization algorithm and maximum likelihood estimator in MATLAB, giving a mean value of EFRET = 0.83. (C) Cross-correlation analysis of a time-trace of ID and IA for a single k-turn molecule in 2 Mg2+, calculated from data recorded at 33-ms frame rate. The fit is linear and almost horizontal with an intercept that is very close to zero, indicating the absence of cross-correlation between donor and acceptor emission down to the timescale of the data collection.
It could be argued that Kt-7 will be predominantly folded in the kinked geometry in the presence of 2 mM Mg2+ even in the absence of bound protein. We therefore repeated the analysis in the absence of added divalent cations, where the structure is predominantly unfolded for protein-free k-turns. A typical time-trace recorded for Kt-7 in 0 mM Mg2+ is shown in Fig. 4 A. Analysis of 1312 molecules generated the histogram of EFRET values shown in Fig. 4 B. Although the mean FRET efficiency is a little lower (EFRET = 0.72) than in the presence of 2 mM Mg2+, it is clear that the RNA molecules are folded, in contrast to their largely unfolded state when not bound to protein under these low-salt conditions. And despite the absence of divalent metal ions, upon performing close examination of the time-trace (and many other similar ones not shown; see the Supporting Material), we could find no evidence for transitions to an unfolded state, as supported by cross-correlation analysis (Fig. 4 C). This remains true at our fastest frame rate; there is no evidence for transient excursions of the bound molecules into the extended conformation.
Figure 4.
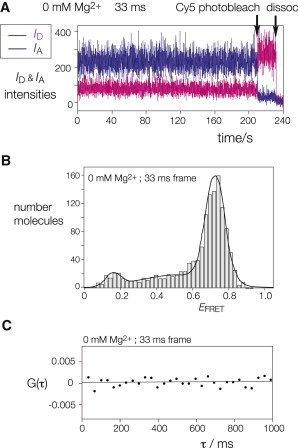
Kt-7 molecules are bound to L7Ae in a predominantly folded conformation in the absence of divalent metal ions. (A) ID and IA versus time for one representative molecule in the presence of 0 mM Mg2+ ions, at 33-ms frame rate. Cy5 was photobleached at 210 s. (B) Histogram of EFRET values for Kt-7 molecules bound to L7Ae in 0 mM Mg2+ ions (1312 molecules). The mean value of EFRET = 0.72. (C) Cross-correlation analysis of a time-trace of ID and IA for a single k-turn molecule in 0 Mg2+, calculated from data recorded at 33-ms frame rate. The fit is linear and horizontal with an intercept at zero, indicating the absence of cross-correlation between donor and acceptor emission down to the timescale of the data collection.
Real-time observation of L7Ae binding to Kt-7
Although we observe no evidence for L7Ae-bound RNA molecules undergoing transitions to the extended state, it is possible that they might initially bind in a less-kinked conformation before becoming converted to a stable kinked state. We therefore probed the possibility that the RNA initially binds in an extended state, but is rapidly induced to switch to the kinked geometry within the binding site of the protein. To this end, we injected fluorescent RNA into the cell while simultaneously collecting the emitted fluorescent light. In this way, we could observe binding events in real-time and measure EFRET for a given RNA molecule at the earliest time within the resolution of our data collection. These experiments were carried out in the presence of 0 mM Mg2+, so that before binding the RNA was predominantly unfolded in free solution, and were studied with the camera operating at a 16-ms frame rate. One such time-trace is shown in Fig. 5 (with further examples in the Supporting Material). At the earliest time there is no bound RNA, and thus ID and IA are at background levels. During the binding event both signals rise, with IA > ID consistent with the tightly kinked conformation. The high FRET state is achieved within a single frame, i.e., 16 ms. This has been observed in many similar time-traces. We have also failed to observe any transient states of lower EFRET at the highest time-resolution possible using our EM-CCD camera (8-ms frame), although the noise level for these data precludes accurate measurement.
Figure 5.
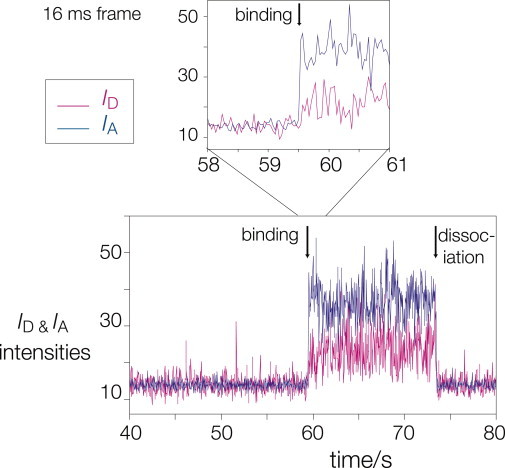
Observation of a Kt-7 molecule binding to L7Ae. A time-trace of ID and IA showing the increase in both signals on binding of the RNA to the fusion protein (at 59.5 s) at 16-ms frame rate. The expansion of the binding region shows that both intensities rise within a single frame. At 74 s, both ID and IA fall back to their initial levels. This could be due to dissociation of the complex or photobleaching of Cy3. Further examples are shown in the Supporting Material.
We can estimate rates of association and dissociation from the single-molecule data (Fig. 6). Measurement of the latter is relatively straightforward, as association and dissociation events are clearly assigned in many traces (see Fig. 5 and Fig. S3 in the Supporting Material). The number of fluorescent complexes remaining bound after a given elapsed time is plotted, and fitted to a single exponential function, giving a dissociation rate of koff = 0.0076 min−1. We have previously measured the rate of dissociation of unfused L7Ae from Kt-7 using ensemble FRET measurements, giving koff = 0.002 s−1 (3). The rate measured here should be the sum of rates of dissociation of Kt-7 from the U1A-L7Ae fusion, the dissociation of the U1A-L7Ae from the U1A RNA, and any photobleaching of Cy3, so a higher rate would be expected, as observed. Moreover, the ensemble measurements were made at lower temperature. The association rate has a greater uncertainty due to imprecision in the timing of the injection and the timing required for mixing. Nevertheless, a plot of the number of bound Kt-7 molecules as a function of the time interval between injection and observation of binding (Fig. 6 A) is well fitted by a single exponential function, giving an observed rate of kobs = 0.002 s−1. The final Kt-7 concentration in the cell was 0.25 nM, giving a bimolecular rate of association of kon = 8.4 × 106 M−1 s−1. This is slower than obtained by ensemble measurements, where rates of kon = 5.4 × 107 and 2 × 108 M−1 s−1 were calculated. The major source of discrepancy is likely to result from the difference between surface binding in the single-molecule experiments, and binding in free solution.
Figure 6.
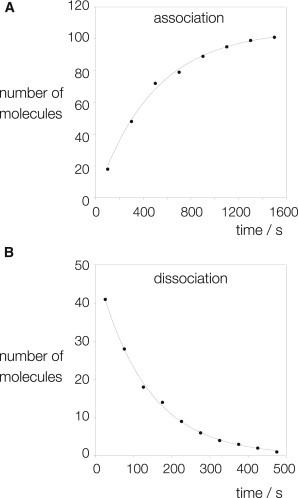
Estimation of binding and dissociation rates from single-molecule data. The times of association were measured as the interval after injection, before a given bound molecule with active Cy3 and Cy5 fluorophores was observed. If a clear dissociation event (such as the example in Fig. 5) was seen, this time was recorded and the lifetime of the bound Kt-7 calculated. (A) Plot of the cumulative number of bound molecules as a function of the time interval, postinjection. The data have been fitted to a single exponential rise. (B) Plot of the cumulative number of bound molecules remaining observable on the surface as a function of the time interval after the initial binding event. The data have been fitted to a single exponential decay.
Conclusion
These experiments have shown that L7Ae-bound k-turn molecules are only observed in their kinked geometry irrespective of the solution conditions. No transient formation of a more extended state was detectable over long periods of observation (a total of 326-min duration of the folded state, summed over 1948 molecules). Furthermore, no extended intermediate structure was observed in the initial binding process. If extended forms exist transiently, they have a lifetime shorter than 16 ms. These data are consistent with the conformational selection mechanism, whereby the protein selectively binds and stabilizes k-turns that already exist in the kinked conformation, and thus the formation of the kinked geometry in the population occurs by drawing the equilibrium toward this state. However, it remains possible that some conformational readjustment could occur at the time of binding, if that happens to be faster than 16 ms.
Acknowledgments
We thank Scott McPhee for RNA synthesis, and David Rueda and Xin Sheng Zhao for technical advice concerning slide preparation.
We thank Cancer Research UK, the Wellcome Trust, and the Human Frontier Science Program for financial support.
Footnotes
Kersten T. Schroeder’s current address is LECOM-Bradenton, 5000 Lakewood Ranch Blvd., Bradenton, FL 34211.
Jonathan Ouellet’s current address is Institut Armand-Frappier, 531 Boulevarde des Prairies, Laval, Québec H7V 1B7 Canada.
Supporting Material
References
- 1.Klein D.J., Schmeing T.M., Steitz T.A. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J., Lilley D.M.J. The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA. 2007;13:200–210. doi: 10.1261/rna.285707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner B., Lilley D.M. The importance of G⋅A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J. Mol. Biol. 2008;381:431–442. doi: 10.1016/j.jmb.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 4.Ban N., Nissen P., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Wimberly B.T., Brodersen D.E., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 6.Koonin E.V., Bork P., Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nottrott S., Hartmuth K., Lührmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird N.J., Zhang J., Ferré-D’Amaré A.R. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA. 2012;18:759–770. doi: 10.1261/rna.031518.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiss-László Z., Henry Y., Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 10.Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 11.Moore T., Zhang Y., Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 12.Suryadi J., Tran E.J., Brown B.A., 2nd The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry. 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- 13.Hamma T., Ferré-D’Amaré A.R. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Vidovic I., Nottrott S., Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 15.Matsumura S., Ikawa Y., Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goody T.A., Melcher S.E., Lilley D.M. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder K.T., Daldrop P., Lilley D.M.J. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure. 2011;19:1233–1240. doi: 10.1016/j.str.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner B., Melcher S.E., Lilley D.M. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woźniak A.K., Nottrott S., Oesterhelt F. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA. 2005;11:1545–1554. doi: 10.1261/rna.2950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshland D.E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai C.J., Ma B., Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44:418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- 22.Pitici F., Beveridge D.L., Baranger A.M. Molecular dynamics simulation studies of induced fit and conformational capture in U1A-RNA binding: do molecular substates code for specificity? Biopolymers. 2002;65:424–435. doi: 10.1002/bip.10251. [DOI] [PubMed] [Google Scholar]
- 23.Okazaki K., Takada S. Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms. Proc. Natl. Acad. Sci. USA. 2008;105:11182–11187. doi: 10.1073/pnas.0802524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weikl T.R., von Deuster C. Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins. 2009;75:104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 25.Hammes G.G., Chang Y.C., Oas T.G. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csermely P., Palotai R., Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem. Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H.X. From induced fit to conformational selection: a continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys. J. 2010;98:L15–L17. doi: 10.1016/j.bpj.2009.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Kormos B.L., Baranger A.M. Molecular dynamics simulation studies of a protein-RNA complex with a selectively modified binding interface. Biopolymers. 2006;81:256–269. doi: 10.1002/bip.20408. [DOI] [PubMed] [Google Scholar]
- 29.Qin F., Chen Y., Chen H.F. Induced fit or conformational selection for RNA/U1A folding. RNA. 2010;16:1053–1061. doi: 10.1261/rna.2008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailor M.H., Sun X., Al-Hashimi H.M. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 31.Silva D.A., Bowman G.R., Huang X. A role for both conformational selection and induced fit in ligand binding by the LAO protein. PLOS Comput. Biol. 2011;7:e1002054. doi: 10.1371/journal.pcbi.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noeske J., Buck J., Wöhnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange O.F., Lakomek N.A., de Groot B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 34.Tang C., Schwieters C.D., Clore G.M. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 35.Wlodarski T., Zagrovic B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. USA. 2009;106:19346–19351. doi: 10.1073/pnas.0906966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boehr D.D., McElheny D., Wright P.E. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc. Natl. Acad. Sci. USA. 2010;107:1373–1378. doi: 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaucage S.L., Caruthers M.H. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 38.Wilson T.J., Zhao Z.-Y., Lilley D.M. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- 39.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


