Abstract
DNA methylation is an important epigenetic mark that is known to induce chromatin condensation and gene silencing. We used a time-domain fluorescence lifetime measurement to quantify the effects of DNA hypermethylation on the conformation and dynamics of a nucleosome. Nucleosomes reconstituted on an unmethylated and a methylated DNA both exhibit dynamic conformations under physiological conditions. The DNA end breathing motion and the H2A-H2B dimer destabilization dominate the dynamic behavior of nucleosomes at low to medium ionic strength. Extensive DNA CpG methylation, surprisingly, does not help to restrain the DNA breathing motion, but facilitates the formation of a more open nucleosome conformation. The presence of the divalent cation, Mg2+, essential for chromatin compaction, and the methyl donor molecule SAM, required for DNA methyltransferase reaction, facilitate the compaction of both types of nucleosomes. The difference between the unmethylated and the methylated nucleosome persists within a broad range of salt concentrations, but vanishes under high magnesium concentrations. Reduced DNA backbone rigidity due to the presence of methyl groups is believed to contribute to the observed structural and dynamic differences. The observation of this study suggests that DNA methylation alone does not compact chromatin at the nucleosomal level and provides molecular details to understand the regulatory role of DNA methylation in gene expression.
Introduction
DNA in eukaryotic cells is folded into a compact form by wrapping around a protein complex, i.e., histone octamer (1,2). This structural assembly, known as chromatin, plays an important role in regulating gene expression by controlling the accessibility of DNA to the transcriptional machinery. The nucleosome is the building block of a chromatin fiber. The high-resolution crystal structure of a nucleosome has revealed the atomic details of this basic unit as composed of 147 bp DNA wrapped around a histone octamer (1). Instead of assuming a static conformation, the nucleosome complex is highly dynamic (3,4). These dynamic features, originating from different types of histone variants, histone posttranslational modifications, and DNA modifications, also known as epigenetic modifications, can have profound effects in gene regulation and cell growth (3–5).
DNA methylation is the most common epigenetic mark that occurs at the DNA level. A healthy mammalian genome is normally filled with >107 methyl groups (6). An aberrant DNA methylation pattern, especially in the promoter region, is closely correlated with the development and the progression of cancer and neurological diseases (7,8). DNA hypermethylation has been shown to lead to the formation of heterochromatin and gene silencing in various cell studies (7,9). However, how DNA methylation regulates gene expression in eukaryotic cells is still an open question. The prevailing opinion suggests that DNA methylation provides preferential binding sites for specific proteins, i.e., MeCP2, which helps to recruit other nucleosome-binding proteins and synergistically promotes the compaction of methylated chromatin (10,11). Meanwhile, there are other studies suggesting that DNA methylation can directly regulate the accessibility of chromatin, by affecting the conformation and dynamics of the formed nucleosome complexes (12–14). The existing experimental evidence accounting for the role of DNA methylation in modulating chromosome conformation does not align well with each other. Whether DNA methylation can directly modulate chromosome conformation remains controversial.
Our study aims to address this issue by evaluating nucleosome conformation and stability using a time-domain fluorescence lifetime spectroscopy approach. Using the DNA sequence with the highest known binding affinity to a nucleosome, i.e., the Widom-601 sequence (15), we reconstituted nucleosomes with different DNA methylation features. We quantified the effects of DNA methylation on the conformational and dynamic features of the reconstituted nucleosomes under a broad range of buffer conditions. Our results suggest that nucleosomes assume a dynamic conformation independent of the DNA methylation level, similar as suggested by existing literature (16–18). However, under various buffer conditions examined in this study, DNA methylation alone does not directly compact a nucleosome, but leads to a more open nucleosome conformation with enhanced DNA end breathing motion. This finding is different from what has been reported previously in the literature (12).
Materials and Methods
Preparation of fluorescently labeled nucleosome samples
We used a 157 bp DNA fragment to reconstitute nucleosomes in this study. The detailed DNA sequence is shown below:
ACTCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCAGT.
This sequence consists of a 147 bp Widom-601 (15) sequence with a 5 bp linker DNA (italics) on both ends. The sequence contains 13 CG sites (underlined). Nucleosome samples are prepared by mixing fluorescently labeled DNA fragments with refolded recombinant histone octamers.
The labeled DNA fragments are produced by polymerase chain reaction (PCR), using primers that have a fluorescent label on their 5′ ends. The PCR products are then purified using a PCR purification kit to remove unincorporated nucleotides and primers. The purified DNA fragment has a comparable purity to the same DNA sequence purified using an ion-exchange HPLC approach. Two types of fluorescently labeled DNAs, i.e., a FAM-labeled and a FAM-TAMRA dual-labeled DNA, are prepared in this study. The fluorescence labeling efficiencies of the purified sample are characterized using their absorption spectrums. The labeling efficiency of a TAMRA dye to a DNA fragment is found to be around 99% (see the Supporting Material). The purity of the DNA fragment is examined using a 10% polyacrylamide gel with 0.5X Tris-borate-EDTA (TBE) buffer at room temperature (Fig. S1).
DNA cytosine methylation is introduced to all the CpG sites flanking the DNA sequence by a methyltransferase reaction. A CpG methyltransferase, i.e., M.SssI (New England Biolabs, Ipswich, MA), is used in this study. The reaction is carried out using standard buffer at 37°C overnight. The methyltransferase is then deactivated by heating to 65°C for 20 min and removed using phenol. The completion of the methylation reaction is verified using BstUI restriction enzyme, whose cleavage ability is blocked by CpG methylation. A typical BstUI digestion pattern of the prepared DNA fragments is shown in Fig. 1. The methylation reaction does not affect the labeling efficiency of the conjugated fluorescence dyes, as verified using absorption spectrums. The methylated DNA fragments exhibit complete resistance to BstUI digestion suggesting that all CpG sites of DNA are completely methylated. The completely methylated DNA fragments are later reconstituted into a nucleosome. Unmethylated and methylated DNA templates are prepared multiple times (>3), and are reconstituted into nucleosomes in multiple batches (>5).
Figure 1.
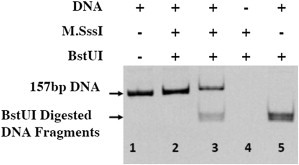
BstUI digestion pattern of different DNA fragments. Lane 1: PCR synthesized DNA fragments; lane 2: DNA fragments reacted with M.SssI for 2 h; lane 3: DNA fragments reacted with M.SssI for 1 h; and lanes 4 and 5: control lanes.
Histone octamers are refolded using four types of recombinant Xenopus laevis histone proteins, i.e., H2A, H2B, H3, and H4. These four types of proteins are individually expressed and purified using an established protocol, as described in previous literature (19). The four types of histone proteins are then mixed at a stoichiometric ratio and refolded into an octamer complex. The refolded histone octamers are purified and then mixed with the appropriately labeled DNA fragments to reconstitute a nucleosome core particle using a salt gradient. All nucleosomes are incubated at 40°C for 2 h to facilitate the positioning of a histone octamer on the central location of the DNA template (19). The quality of each reconstituted sample is examined using 5% (Fig. 2) and 8% (Fig. S2) polyacrylamide gels. The polyacrylamide gels are run in 0.25X TBE buffer at 4°C. These polyacrylamide gels are commonly used to resolve multiple translational settings of nucleosomes (19,20). Due to the strong positioning effects of the Widom-601 sequence, the reconstituted nucleosomes exhibit a single band on both polyacrylamide gels, suggesting that the formed nucleosome assumes a single DNA translational setting. During the assembly of a nucleosome particle, we also adjust the stoichiometric ratio between DNA and histone octamer to eliminate the existence of unbound DNA fragments (free DNA) in the nucleosome sample. Unbound DNA fragments are not observed in our PAGE results (Fig. 2). Free DNA, therefore, is not expected to contribute to the measured fluorescence signals.
Figure 2.
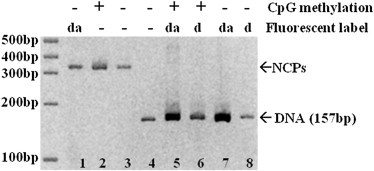
A typical 5% polyacrylamide gel of DNA and reconstituted nucleosomes. Lanes 1–3: Reconstituted nucleosomes; lanes 4–8: 157 bp DNA fragments. da: dual-labeled sample. d: FAM-only labeled sample. The gel was run for 3 h in 0.25X TBE buffer at 4°C and 150 V.
We also monitored the solubility of nucleosomes under various salt concentrations being explored in this study using a similar sedimentation assay as described in (21). Nucleosomes remain largely soluble within the various salt concentrations being explored in this study.
Time-domain fluorescence lifetime measurements
Time-domain fluorescence measurement is a widely used spectroscopy approach to explore the structure and dynamics of biological molecules (22–24). Compared with steady-state experiments, time domain measurements are more informative by providing more insights into the subpopulation of fluorescence species that coexist in the sample. The time-resolved fluorescence decay curve can routinely resolve the relative intensity of two fluorescent species (see the Supporting Material). In this case, it can provide us with detailed information about the breathing of DNA ends entering and exiting a histone octamer surface, which accounts for the dynamic feature of a nucleosome (Fig. 3). Although this approach cannot provide details of individual molecules as in a single-molecule experiment, it can be performed at relatively high nucleosome concentrations, which prevents the occurrence of dilution-induced nucleosome dissociation events (25,26).
Figure 3.
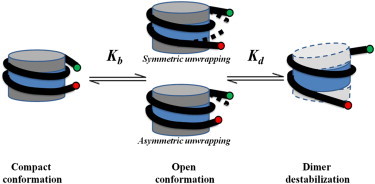
Schematic drawing of the DNA breathing motion and dimer destabilization of a nucleosome.
Time domain fluorescence decay curves are collected using fluorescently labeled nucleosome samples. The sample concentration is kept above 1 μM by titrating in unlabeled nucleosomes (reconstituted in vitro using recombinant histone proteins). The sample integrity during the fluorescence measurements is monitored by a 5% polyacrylamide gel. All time-domain fluorescence decay curves are collected at room temperature with a ChronosBH lifetime spectrometer (ISS, Champaign, IL). The samples are excited with 440 nm, 20 MHz laser pulses. A narrow emission filter (505–545 nm) is applied to separate the donor dye fluorescence emission from the direct excitation beam as well as to prevent collection of fluorescence emission from the acceptor dye. The decay curve is calculated using the equipped time-correlated single photon counting card. The decay curves are analyzed using the Vinci Fluorescence Spectroscopy Analysis software (ISS, Champaign, IL). The detailed equation used to analyze the decay curves (I(t)) is listed in Eq. 1:
| (1) |
In this expression, I is the collected decay curves decoupled with the instrument response function, is the decay time, represents the amplitudes of each component at time t = 0, and n is the number of decay times (27,28). The value of n is dependent on the number of distinctive fluorescence species that coexist in the sample (the detailed approach to identify n is included in the Supporting Material). In addition to the multiple-component model as shown in Eq. 1, a fluorescence decay curve can also be analyzed using a distribution function. However, we selected the multiple-component model over the distribution model based on existing single-molecule experimental evidence suggesting that a nucleosome can assume a close and an open state as illustrated in Fig. 3 (16–18, and see 30).
The fluorescence decay curves of both the donor-only and the donor-acceptor labeled nucleosomes are collected in this study. Typical time-domain fluorescence decay curves collected using nucleosome samples containing either a donor molecule or a fluorescence resonance energy transfer pair are presented in the Supporting Material (Fig. S3). The fluorescence lifetime of the donor (FAM) molecules collected in the absence (τd) and the presence (τda) of acceptor molecules (TAMRA) can be obtained from model analysis. These two values can provide the detailed spatial information about the ends of DNA that surround a histone octamer following Eq. 2:
| (2) |
In this expression E is the energy transfer efficiency, r is the distance between the donor and the acceptor labels, i.e., the distance between the DNA entry/exit sites to a nucleosome, and R0 is the Förster distance, which corresponds to 5 nm based on the calibration experiments performed using a short 17 bp DNA fragment (Fig. S4) and theoretical calculations (29). Different ionic strengths do not affect the Förster distance, as monitored by anisotropy and Förster distance calibration experiments (Figs. S5–S10).
Results
Reconstituted nucleosomes exhibit significant DNA breathing motion under physiological conditions
It has been long postulated that a nucleosome will assume a dynamic structure under physiological conditions (30). This dynamic feature can be reflected as the two DNA ends that enter and exit a nucleosome structure assume different spatial organizations. This type of variation in nucleosome conformation is commonly known as the DNA breathing motion and has been previously reported in both steady-state and single-molecule experiments (16–18,30). Time-domain fluorescence spectroscopy is capable of revealing the subpopulations of different fluorescence species and can be performed at relatively high concentrations (μM) so that nucleosomes retain the same dynamic features as under a typical cell nucleus condition. The analysis of the time-domain fluorescence decay curves of our study unambiguously suggests the existence of the DNA breathing motions (Fig. 3) over a broad range of salt concentrations.
Fig. 4 illustrates a typical time-domain fluorescence decay curve collected using a nucleosome sample containing both donor and acceptor labels. The decay curves are consistently best fitted using a two-component model, which suggests the existence of two distinctive fluorescence species. The χ2 values of a one- and two-component fitting, as shown in Fig. 4 are found to be 4.82 and 1.02, respectively. The two nucleosomal conformations observed in this study have DNA end-to-end distances that vary by >2.5 nm. This distance variation, if arising from different DNA translational settings, would require at least 10 bp shifting in histone octamer location, which should appear as a second band on an 8% native polyacrylamide gel. The reconstituted nucleosome only exhibits a single band on the polyacrylamide gel as shown in Fig. 2 and Fig. S2. Furthermore, the nucleosome samples do not contain any free DNA. Combining all of these, we expect that the two fluorescence species we observed in our experiments correspond to two distinctive nucleosome conformations that coexist in the measured samples. These two conformations are expected to reflect the dynamic structural feature of a nucleosome, i.e., DNA breathing motions as illustrated in Fig. 3, although the relative abundance of these two conformations reveals the equilibrium between the two nucleosomal states, i.e., the compact and the open state of a nucleosome.
Figure 4.
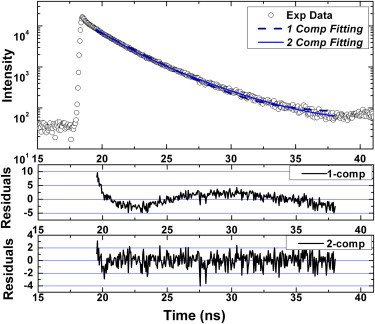
A typical time-domain fluorescence decay curve collected using dual-labeled nucleosome samples. The curvy residual pattern from the one-component fitting indicates the existence of a secondary fluorescence species in the sample. The values of χ2 in one-component and two-component fitting are 4.82 and 1.02, respectively. The one-component fitting gives a single lifetime (τ) of 2.62 ns. The two-component fitting gives two lifetimes of 1.52 ns (τ1) and 3.50 (τ2), and a fraction of the short lifetime (f1) at 0.432.
Based on the measured fluorescence lifetime, we can quantify the distance between DNA ends using their individual energy transfer efficiencies. The two distinctive distances observed under low salt concentrations are summarized in Fig. 5. At 120 mM KCl, the DNA end-to-end distance of a nucleosome in compact and open state is found to be 4.5 and 6.5 nm, respectively. Comparing with the DNA end-to-end distance measured within a crystal structure (4.9 nm, PDB 1ZBB), we find that the distance of the compact conformation (4.5 nm) is close to the one measured in the crystal structure. The open-state of a nucleosome, primarily due to DNA end breathing motion, cannot be observed in a crystal structure due to the close packing of molecules within the crystal lattice. The average DNA end-to-end distance that we report here (5.3 nm) is comparable or slightly smaller than the average DNA end-to-end distance values reported in literature (31). The variation in DNA end-to-end distance between a compact and an open nucleosome conformation is around 2-3 nm for nucleosomes reconstituted using both unmethylated and methylated DNA. This suggests that roughly 8–12 DNA basepairs are involved in the temporary binding and unbinding to a histone octamer surface either on one linker DNA or distributed between both linker DNAs. The fraction (f1) corresponds to the relative abundance of nucleosomes in the compact conformation. This number can therefore be used to measure the equilibrium of the DNA breathing motion. Nucleosomes with different DNA methylation level exhibit similar two-state conformations originating from the DNA breathing motion.
Figure 5.
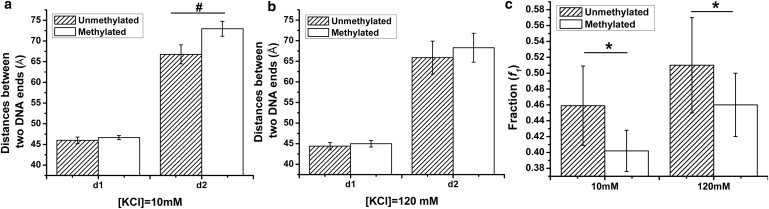
The distances between DNA ends enter/exiting a nucleosome (d1: compact nucleosome; d2: open nucleosome) measured under (a) 10 mM and (b) 120 mM KCl concentrations. (c) The fraction of the compact nucleosome within the whole nucleosome population. Data points: mean ± 1σ, n = 21. ∗ represents p-value < 0.02, # represents p-value < 0.002.
The effect of monovalent counterions on the DNA breathing and nucleosome dissociation dynamics
The dynamic feature of nucleosomes, i.e., partial and full dissociation of DNA from the histone octamer surface, is closely related to many cellular events that require the participation of DNA fragments. One of the major forces that stabilize a nucleosome is the electrostatic interaction. Modulating the buffer ionic strength is, therefore, expected to affect the nucleosome stability and reveal the dynamic behavior of nucleosomes when they participate in different enzymatic reactions such as replication and transcription.
Fig. 6 summarizes the dependence of nucleosome conformations under different ionic strengths, adjusted by different potassium chloride concentrations. Fig. 6 a describes the dependence of the averaged fluorescence lifetime calculated based on the fraction-weighted fluorescence lifetimes. With increasing ionic strengths, the average fluorescence lifetimes of dual-labeled nucleosomes increase. This trend indicates that nucleosomes assume a less compact conformation with two DNA ends further away from each other. A detailed analysis of the respective compact and open conformations further reveals the structural details of the dynamic nucleosome conformations, as illustrated in Fig. 6, b and c. At low ionic strength, i.e., [KCl] < 200 mM, the DNA end-to-end distances corresponding to two different conformations, remain almost independent of salt concentrations. The fraction of the compact nucleosomes fluctuates within this concentration range, as suggested in Fig. 6 c. The slight increase in compact nucleosome population as observed on the leftmost part of this concentration range originates from the surface charge neutralization due to increasing counterion concentrations. The equilibrium constant between these two conformations can be calculated using Eq. 3, where f1 is the fraction of the compact nucleosomes:
| (3) |
Figure 6.
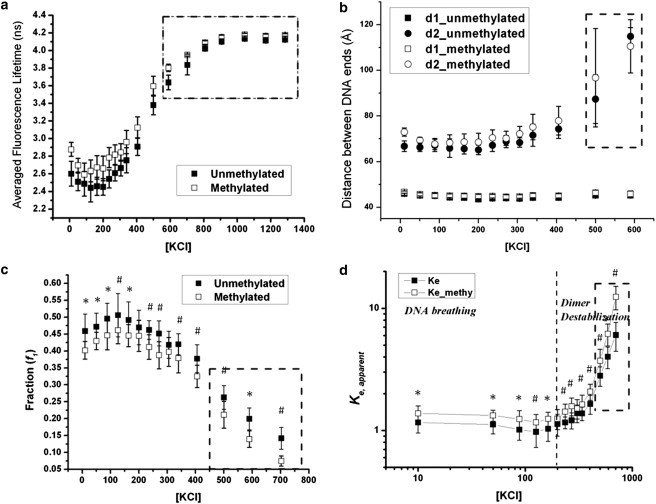
The effect of different monovalent counterion on (a) the fraction weighted averaged fluorescence lifetime; (b) the DNA end-to-end distances of the compact and the open nucleosome conformation; (c) the fraction of the compact nucleosome; and (d) the equilibrium constant between the open and the compact nucleosome conformation. All KCl concentrations are in mM. DNA starts to dissociate from the nucleosome complex in the dotted regions. Data points: mean ± 1σ, n = 21. ∗ represents p-value < 0.02, # represents p-value < 0.002.
These calculated apparent equilibrium constants are plotted versus salt concentrations, as in Fig. 6 d. Similar to the fraction numbers, the equilibrium constant remains almost a constant at low salt concentrations. The predominant nucleosome dynamics originate from the breathing motion of DNA ends. The nucleosome complex remains stable without any significant protein or DNA dissociation, agreeing with previous nucleosome dynamic studies (30,32).
As salt concentration increases further, the accumulation of K+ counterions starts to screen out the attraction forces that hold a nucleosome together. As a result, the averaged fluorescence lifetime of the nucleosome sample starts to increase at salt concentrations above 200 mM. This salt concentration is similar to the previously reported value of the onset of temporary H2A-H2B dimer dissociation from a histone octamer interface (32). The compact conformation remains unaffected within these moderately high salt concentrations. The open nucleosome conformation, on the other hand, shows that the DNA ends move further away from each other. Meanwhile, the relative abundance of compact nucleosomes gradually decreases, and the apparent equilibrium constant measured between the compact and the open nucleosome conformation increases abruptly with increasing salt concentrations.
As salt concentration increases further to above 600 mM, nucleosome destabilization becomes apparent. DNA end breathing, H2A-H2B dimer dissociation, and DNA dissociation can happen simultaneously. The detailed analysis of each individual nucleosome conformation is therefore rendered infeasible using current experimental techniques. Because this concentration deviates significantly from physiological conditions, the related nucleosome dynamics will not be discussed in this work.
Based on the previous observations, the nucleosome exhibits distinctive conformational fluctuations at low to medium ionic strength. DNA breathing motion dominates in low KCl concentration below 200 mM. When salt concentration increases further, the apparent equilibrium constants exhibit a distinctive transition, indicating the onset of a different dynamic behavior in nucleosomes. Existing knowledge of nucleosome stability under different ionic strengths indicates that conformational transitions related to dimer destabilization start at monovalent concentrations just above 200 mM (32). Similar transition is observed in our data.
Although the unmethylated and the methylated samples deviate from each other in their detailed conformations during the disassembling process, the general dissociation pathways remain similar to each other.
The effect of Mg2+ ions on the conformational dynamics of a nucleosome
Divalent counterions, e.g., Mg2+ are commonly used to induce the compactness of nucleosomes and nucleosome arrays (33,34). High Mg2+ concentration above 5 mM is known to promote the self-association of nucleosomes and lead to the formation of nucleosome aggregates. However, there is little literature evidence examining the role of divalent counterions on the dynamics of a nucleosome. This work will quantify the effects of low Mg2+ concentration (<2 mM) on the DNA breathing dynamics as observed at low ionic strength.
The existence of a low concentration of Mg2+ lowers the measured averaged fluorescence lifetime, as illustrated in Fig. 7. This indicates that the nucleosomes assume more compact conformations with an excessive amount of Mg2+ counterions. Detailed analysis of the two nucleosomal subpopulations further reveals that the conformations of the compact nucleosome are mostly affected by increasing Mg2+ concentrations. The existence of divalent counterions brings the two labeled DNA ends closer to each other by around 0.5 nm. The compaction saturates ∼1 mM, and the compact nucleosomes assume a steady conformation thereafter. A similar trend is observed for the fraction (f1) of the compact nucleosomes. The open nucleosome conformation, on the other hand, remains unaffected. However, as the divalent counterion concentration further increases, its ionic effect will dominate and an increase in the end-to-end distance of the open nucleosome conformation is observed at MgCl2 concentrations above 1.5 mM. Similar effects of Mg2+ are observed for both unmethylated and methylated nucleosomes.
Figure 7.
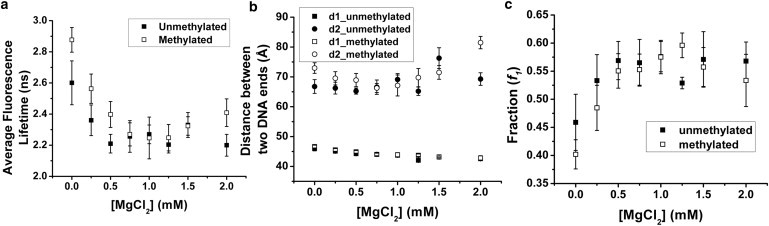
The effect of different divalent counterion on (a) the fraction weighted averaged fluorescence lifetime; (b) the DNA end-to-end distances of the compact and the open nucleosome conformation; and (c) the fraction of the compact nucleosome (the averaged fraction of the unmethylated and methylated samples are statistically different from each other with p-value < 0.05 up to [MgCl2] = 0.5 mM using a t-test). Data points: mean ± 1σ, n = 11.
The effect of a methyl-group donor (SAM) on nucleosome dynamics
S-Adenosyl methionine (SAM) is a common methyl donor used in all de novo methyltransferase reactions. In addition to participating in the enzymatic reactions, this molecule also behaves as a polycounterion under physiological conditions. The concentration of SAM molecules is conventionally used as a methylation index in the cell environment, suggesting the on and off state of chromatin (35). However, how the local enrichment of this methyl donor molecule can directly affect the conformation and dynamics of nucleosomes has not been well characterized. In this study, we evaluate the conformational transitions of fluorescently labeled nucleosome particles under different SAM concentrations. The results are summarized in Fig. 8.
Figure 8.
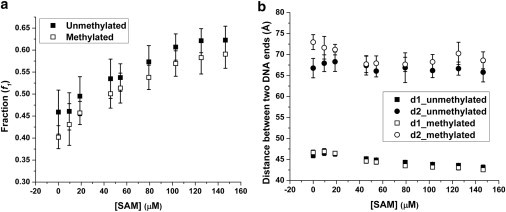
The effect of different SAM concentrations on (a) the fraction of the compact nucleosome state (the fractions of the unmethylated and methylated samples are statistically different from each other with p-value < 0.02 using a t-test); and (b) the conformations of the two nucleosomal states. Data points: mean ± 1σ, nunmet = 28, nmet = 21. The sample contains 10 mM KCl and 0 mM MgCl2.
SAM concentration in a typical cell nucleus environment can vary from 10 μM to several hundreds of micromolars. A typical in vitro DNA methylation requires the presence of SAM molecules at a concentration around 160 μM. Under similar or even lower SAM concentrations, we observed that the nucleosome conformation undergoes a compaction process, resulting from the presence of excessive SAM molecules. Existence of SAM molecules promotes the prevalence of the compact nucleosome conformation. Meanwhile, the compact conformation further closes, with the two DNA ends getting closer to each other. This phenomenon is similar to the previously observed effects of divalent counterions (Mg2+). Surprisingly, the presence of methyl donor molecules by itself can affect the compaction of nucleosomes.
Unmethylated and methylated nucleosomes show a similar compaction trend under the SAM gradient.
Discussion
Dynamics of nucleosomes under different buffer conditions
The dynamic features of a nucleosome have been studied previously using steady-state and single-molecule fluorescence experiments (12,16–18,30,31). The fluorescence lifetime spectroscopy used in this study is unique in its capability of resolving the existence of multiple fluorescence species at a relatively high nucleosome concentration (22–24,27). The experimental findings unambiguously confirm the existence of two nucleosome conformations in equilibrium, covering a broad range of salt concentrations, including the physiological condition. At low salt concentrations, the coexistence of an open and a compact conformation of a nucleosome is believed to originate from the breathing motion of DNA ends. This dynamic feature makes the nucleosomal DNA sequence more accessible and generates kinetic binding sites for external protein factors. The equilibrium between these two conformations can be affected by different buffer compositions, suggesting that chromatin compactness can be further modulated by the activity of counterions and small molecules transported across the nuclear membrane.
To gain more insights into the thermodynamic feature of these two conformations, we calculate the apparent equilibrium constants between the open and the compact conformation. The apparent equilibrium constants obtained at low salt concentrations fluctuate around a constant value (1.1 and 1.3 for unmethylated and methylated samples, respectively) and show a weak dependence on the salt concentrations, as shown in Fig. 9 a. The slightly curved feature at low ionic strength reflects the initial charge neutralization due to the presence of excessive amounts of counterions followed by the electrostatic screening that compromises the attraction forces between DNA ends and a histone octamer surface. Within these low salt concentrations, the histone octamer remains almost intact (32,36). The transient peeling off of DNA end fragments from a histone octamer surface, therefore, dominates the dynamic motion observed in this concentration range. The variations of DNA end-to-end distances as measured in the open and compact nucleosome conformations indicate that there are a few DNA bases (∼8–12 bp) contributing to the DNA end breathing motions.
Figure 9.
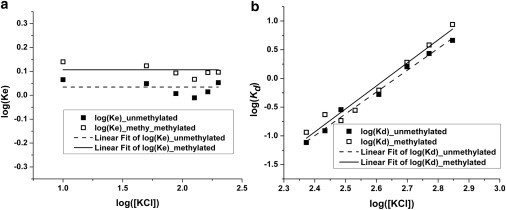
The equilibrium constants of (a) the DNA breathing motion and (b) the dimer destabilization fitted using linear functions.
Two possible DNA breathing motion mechanisms can account for the open state of the nucleosomes, as indicated in Fig. 3. In those mechanisms, the DNA ends can peel off the histone octamer surface in a symmetric or asymmetric way as suggested in previous works (30,37–39). Our current labeling scheme cannot differentiate between symmetric or asymmetric unwrapping. However, the conformational and thermodynamic comparison between the unmethylated and methylated samples should still be valid.
Introducing CpG methylation to 13 distinctive CpG sites on the DNA backbone can affect the equilibrium constants between these two states. The equilibrium constants of DNA breathing motions (Kb), equivalent to the measured apparent equilibrium constant within a low salt concentration range, are found to be 1.1 ± 0.1 and 1.3 ± 0.1 for unmethylated and methylated nucleosomes, respectively. The difference between these equilibrium constants is ∼20%. A t-test for comparison of means showed that these equilibrium constants are statistically different from each other at a 99.9% confidence level.
Single-molecule experiments have shown that at low salt concentrations the equilibrium constant, calculated as Keq = [open nucleosome]/[compact nucleosome], similar to our definition in this study, can vary between 0.1 and 1.45 (16,18). The broad range of equilibrium constants arises from nucleosome dissociation due to low sample concentrations used in single-molecule experiments. In bulk experiments, i.e., steady-state fluorescence, the equilibrium constant values are reported to be in the order of 0.1 (30). However, these experiments are based on an important assumption that at low salt concentration, i.e., 10 mM, the nucleosome assumes a fully compact conformation without any DNA end breathing (Keq,10 mM = 0) (30). This assumption is not supported by our experimental findings in this study. The equilibrium constants that we report in this study agree well with the findings of single molecule experiments.
With a further increase of ionic strength, the apparent equilibrium constants exhibit an abrupt transition, and the relative ratio of the open conformation starts to increase. Meanwhile, the distances between the two ends of the open conformation become even larger. This transition is observed at salt concentrations of around 200–250 mM, similar to a previous observation (32). It suggests the onset of new dynamic behaviors of nucleosomes within this concentration region. Different from the observation at low ionic strength, the apparent equilibrium constant exhibits a significant dependence on salt concentrations. This new dynamic feature can be captured by decoupling the effects of DNA breathing motion from the measured apparent equilibrium constant. The equilibrium constant of the DNA breathing motion remains relatively independent of salt concentrations, and is, therefore, considered as a constant. This new equilibrium constant (Kd) associated with further dissociation of a nucleosome (as illustrated in Fig. 3), can be calculated using Eq. 4:
| (4) |
The calculated dissociation constants exhibit a power law dependence on the salt concentrations, as illustrated in Fig. 9 b. The slope of the linear fitting function can be used to reveal the number of ion pairs that are involved in this dissociation process (32,40). The number of ion pairs found for unmethylated and methylated nucleosomes are 4.2 ± 0.2 and 4.4 ± 0.3, respectively. These numbers are consistent with the number of ion pairs that have been previously reported to be associated with histone H2A-H2B dimer destabilization (32). The observed open nucleosome conformation is expected to include contributions from both the DNA end breathing and the histone H2A-H2B dimer dissociation. The octamer stability, H2A-H2B dimer partial dissociation in particular, is most likely to dominate the nucleosome dynamics at this moderately high ionic strength. DNA methylation does not affect H2A-H2B dimer destabilization.
As salt concentrations increase further (>600 mM), in addition to the already observed dynamic features, H2A-H2B dimer will start to dissociate from the nucleosome complex, and DNA will start to peel off from a histone octamer surface. Multiple fluorescence species (>4) are expected to coexist in the nucleosome samples, which will be impractical to differentiate using time-domain fluorescence spectroscopy. The high ionic strength bears little similarity to in vivo conditions; it will therefore not be discussed in this work.
The presence of divalent counterions can bridge the interactions between DNA molecules, in addition to affecting the ionic strength of the solution (41,42). In this study, the presence of Mg2+ has been shown to affect the equilibrium between the two nucleosome conformations originating from the DNA breathing motion. It favors the formation of compact nucleosomes by restraining the DNA end breathing motion. Furthermore, the presence of the divalent counterion leads to a tighter nucleosome conformation. This more compact structure potentially originates from the attractive forces between spatially close DNA fragments mediated by Mg2+ counterions (42). The compaction process leads to the orientational change of the DNA end segments and is expected to be reflected in the spatial arrangement of linker DNA fragments within a chromatin structure. This finding suggests that divalent counterions, i.e., Mg2+, can affect chromatin folding by modulating the detailed nucleosome conformation in addition to bridging the interactions between neighboring nucleosomes.
The effect of DNA methylation on the conformation and stability of a nucleosome
It has long been postulated that DNA methylation might be able to lead directly to the compaction of a chromatin fiber, without help from DNA methylation-specific binding proteins. However, the current experimental evidence has been highly controversial.
Our results, obtained using nucleosomes with methylated and unmethylated Widom-601 DNA sequence suggest that the presence of DNA methylation does not directly compact a nucleosome. Instead, it leads to the prevalence of a more open nucleosome structure. The same results have been observed using steady-state and time-resolved fluorescence measurements with a different fluorescence labeling strategy (Fig. S11). Detailed analysis of the nucleosome conformations near physiological conditions reveals that the nucleosome reconstituted on a methylated DNA assumes a looser, more open conformation, as compared with the unmethylated one. The two nucleosomal conformations (compact and open conformation) that coexist in equilibrium show a different dependence on the DNA methylation level. The compact nucleosome is unaffected by the prevalence of methyl groups on the DNA. The open nucleosome conformation, on the other hand, exhibits differences. These differences persist within a broad range of monovalent salt concentrations. The significance of the observed difference between unmethylated and methylated samples is tested using the unpaired t-test with equal variances. The calculated p-values of nucleosomal features exhibiting dependence on the DNA methylation level are reported in the captions of Figs. 5–7.
The DNA methylation level also affects the dynamic feature of a nucleosome, particularly at low monovalent salt concentrations. Nucleosomes with methylated DNA backbones exhibit enhanced DNA end breathing motion (Kb = 1.3 ± 0.1) as compared with unmethylated ones (Kb = 1.1 ± 0.1). In other words, the DNA sequences located within a methylated nucleosome tends to be 20% more accessible than those in unmethylated nucleosomes, particularly on the DNA fragments entering/existing a nucleosome. Because a chromatin fiber consists of hundreds of thousands of nucleosomes, we expect that small changes observed at the nucleosome level can be enhanced at the level of chromatin fiber and become more relevant to different biological processes.
The presence of the DNA methylation level, however does not affect the stability of histone octamer. The destabilization of nucleosomes at high monovalent salt concentrations (>200 mM), due to H2A-H2B dissociation, occurs at the same salt concentration with similar strength independent of DNA methylation level.
Similarly, methylated nucleosomes exhibit a less compact conformation at low Mg2+ concentrations. However, as divalent concentration increases, the difference between the unmethylated and the methylated nucleosome samples completely diminishes, and these two types of nucleosomes behave similarly both structurally (the open and the compact conformation) and dynamically (the apparent equilibrium constant).
Unexpectedly, these observations suggest that the presence of DNA methylation can enhance the DNA breathing motion and cause more DNA nucleotides to be involved in this transient opening up of the compact nucleosome structure. This difference is likely to arise from the stiffness of various DNA, which is determined by DNA methylation patterns. With extensive cytosine methylation, the DNA backbone is known to assume decreased bending flexibility (43) and therefore has to overcome larger energetic barriers to comply with the surface curvature of a histone octamer. As a result, the formed nucleosome is less thermodynamically stable and can assume a more open structure with enhanced DNA end breathing motion, as observed in this study. The reduction in DNA flexibility originates largely from the spatial confinement between neighboring bases. The existence of divalent cations can bridge the intramolecular contacts and enhance the elastic properties of a DNA strand (44). High Mg2+ concentrations are therefore capable of mitigating the confinement in molecular contacts introduced by methyl groups. It also explains our experimental observation that the structural and dynamic differences between differently methylated DNA templates diminish at high Mg2+ concentrations.
These findings suggest that DNA methylation does not directly facilitate the compaction of a chromatin fiber at the nucleosomal level. The different conformations observed for unmethylated and methylated nucleosome at low ionic strengths can potentially change the linker DNA entry-exit angle and lead to the change of orientation of neighboring nucleosomes and affect the overall compactness of a chromatin fiber. Additionally, the observed enhanced DNA breathing motion of methylated nucleosome can potentially facilitate the binding of DNA methylation-specific proteins, e.g., MeCP2, on the DNA fragments entering/existing a nucleosome. This enhanced binding event can facilitate the recruitment of other protein factors, e.g., MeCP2, that synergistically accelerate the chromatin compaction process.
The DNA sequence used in this study, i.e., the Widom-601 sequence, is the DNA sequence with the highest known binding affinity to a histone octamer surface. A similar trend, i.e., a larger distance between DNA ends favoring open nucleosome conformation with an increasing DNA methylation level, is also observed in nucleosomes with other DNA sequences (Fig. S12). The effects of DNA methylation on nucleosome structure is therefore expected to be representative of the general role of DNA methylation. However, it is noteworthy that changes in DNA sequence by itself may have a more profound effect than DNA methylation in nucleosomal conformation.
Nucleosome dynamics during a typical DNA methylation reaction
A de novo DNA methylation pattern is introduced to genomic DNA using different DNA methyltransferase. Independent of the DNA methyltransferase type, all methylation reactions require a universal methyl donor, SAM, as a cofactor. This study reveals that the simple presence of SAM molecules can have a significant effect on the observed conformation and thermodynamics of nucleosomes. Due to the zwitterionic nature of SAM, SAM molecules facilitate the compaction of a nucleosome and suppress the DNA breathing motion. These observations can explain the previously reported nucleosome compaction when undergoing an in vitro DNA methylation reaction as reported by Choy et. al. (12). The nucleosomes with unmethylated and methylated DNA respond similarly to increasing SAM concentrations. Throughout the SAM concentration explored in this study, methylated nucleosomes assume a less compact conformation. This unexpected role of SAM molecules in nucleosome dynamics suggests that small molecules present in the cell nucleus, e.g., Mg2+ and SAM, can potentially modulate nucleosome conformations by different concentration gradients.
Conclusions
In summary, our data suggest that a nucleosome assumes a dynamic structure under the physiological condition. The DNA breathing motion dominates under low ionic strengths, followed by a similar dimer destabilization as suggested in the literature (32). Contrary to our expectation, the existence of extensive cytosine methylation on the DNA template is found to slightly open the nucleosome structure and enhance the DNA breathing motion. However, the difference between unmethylated and methylated nucleosomes diminishes with increasing concentration of Mg2+. The existence of Mg2+, as well as SAM molecules, facilitates the compaction of nucleosomes and suppresses the DNA breathing motion. Furthermore, being the essential cofactor required in DNA methylation reactions, this surprising role of SAM molecules in nucleosome stability can potentially shed light on understanding DNA methylation-mediated gene silencing and the cross talk between DNA methylation and histone methylation modifications. Our results suggest that DNA methylation alone does not compact a chromatin at nucleosome level, but may modulate chromatin compaction via the change of the linker DNA entry/exit angle and modulate the nucleosome accessibility to DNA methylation-specific proteins.
Acknowledgments
We gratefully acknowledge Prof. Tim Richmond in ETH, Zürich for generously sharing with us the expression plasmids of histone proteins. We also want to acknowledge Prof. Richard Kuhn at Purdue University for access to the spectromax fluorescence plate reader and Mr. Nathan Nurse for critical reading of this manuscript.
This work was supported by the Purdue Engineering School faculty start-up package and the American Chemical Society (grant No. PRF# 50918-DNI7).
Supporting Material
References
- 1.Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg R.D., Lorch Y.L. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Luger K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 4.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T.H., Tycko B. Creation of genomic methylation patterns. Nat. Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 7.Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 8.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 9.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 11.Kass S.U., Pruss D., Wolffe A.P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 12.Choy J.S., Wei S., Lee T.H. DNA methylation increases nucleosome compaction and rigidity. J. Am. Chem. Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey C.S., Pennings S., Allan J. A determining influence for CpG dinucleotides on nucleosome positioning in vitro. Nucleic Acids Res. 2004;32:4322–4331. doi: 10.1093/nar/gkh749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godde J.S., Kass S.U., Wolffe A.P. Nucleosome assembly on methylated CGG triplet repeats in the fragile X mental retardation gene 1 promoter. J. Biol. Chem. 1996;271:24325–24328. doi: 10.1074/jbc.271.40.24325. [DOI] [PubMed] [Google Scholar]
- 15.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 16.Gansen A., Valeri A., Seidel C.A. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelbauskas L., Chan N., Lohr D. Sequence-dependent variations associated with H2A/H2B depletion of nucleosomes. Biophys. J. 2008;94:147–158. doi: 10.1529/biophysj.107.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koopmans W.J.A., Buning R., van Noort J. spFRET using alternating excitation and FCS reveals progressive DNA unwrapping in nucleosomes. Biophys. J. 2009;97:195–204. doi: 10.1016/j.bpj.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luger K., Rechsteiner T.J., Richmond T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 20.Dorigo B., Schalch T., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 21.Bertin A., Mangenot S., Livolant F. Structure and phase diagram of nucleosome core particles aggregated by multivalent cations. Biophys. J. 2007;93:3652–3663. doi: 10.1529/biophysj.107.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassi G.S., Murchie A.I.H., Lilley D.M. Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J. 1997;16:7481–7489. doi: 10.1093/emboj/16.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klostermeier D., Millar D.P. Time-resolved fluorescence resonance energy transfer: a versatile tool for the analysis of nucleic acids. Biopolymers. 2001-2002;61:159–179. doi: 10.1002/bip.10146. [DOI] [PubMed] [Google Scholar]
- 24.Walter N.G. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods. 2001;25:19–30. doi: 10.1006/meth.2001.1212. [DOI] [PubMed] [Google Scholar]
- 25.Claudet C., Angelov D., Bednar J. Histone octamer instability under single molecule experiment conditions. J. Biol. Chem. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- 26.Godde J.S., Wolffe A.P. Disruption of reconstituted nucleosomes. The effect of particle concentration, MgCl2 and KCl concentration, the histone tails, and temperature. J. Biol. Chem. 1995;270:27399–27402. doi: 10.1074/jbc.270.46.27399. [DOI] [PubMed] [Google Scholar]
- 27.Chabbert M., Hillen W., Bousquet J.A. Structural analysis of the operator binding domain of Tn10-encoded Tet repressor: a time-resolved fluorescence and anisotropy study. Biochemistry. 1992;31:1951–1960. doi: 10.1021/bi00122a008. [DOI] [PubMed] [Google Scholar]
- 28.Grinvald A., Steinberg I.Z. On the analysis of fluorescence decay kinetics by the method of least-squares. Anal. Biochem. 1974;59:583–598. doi: 10.1016/0003-2697(74)90312-1. [DOI] [PubMed] [Google Scholar]
- 29.Yuan C.L., Rhoades E., Archer L.A. Mismatch-induced DNA unbending upon duplex opening. Biophys. J. 2005;89:2564–2573. doi: 10.1529/biophysj.105.065722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li G., Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 31.Tóth K., Brun N., Langowski J. Chromatin compaction at the mononucleosome level. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 32.Böhm V., Hieb A.R., Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirier M.G., Oh E., Widom J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz P.M., Felthauser A., Hansen J.C. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 35.Waterland R.A. Assessing the effects of high methionine intake on DNA methylation. J. Nutr. 2006;136(6, Suppl):1706S–1710S. doi: 10.1093/jn/136.6.1706S. [DOI] [PubMed] [Google Scholar]
- 36.Park Y.J., Dyer P.N., Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 37.Miyagi A., Ando T., Lyubchenko Y.L. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50:7901–7908. doi: 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 38.Shlyakhtenko L.S., Lushnikov A.Y., Lyubchenko Y.L. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voltz K., Trylska J., Langowski J. Unwrapping of nucleosomal DNA ends: a multiscale molecular dynamics study. Biophys. J. 2012;102:849–858. doi: 10.1016/j.bpj.2011.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Record M.T., Jr., Lohman M.L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 41.Luan B., Aksimentiev A. DNA attraction in monovalent and divalent electrolytes. J. Am. Chem. Soc. 2008;130:15754–15755. doi: 10.1021/ja804802u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X.Y., Andresen K., Pollack L. Inter-DNA attraction mediated by divalent counterions. Phys. Rev. Lett. 2007;99:038104. doi: 10.1103/PhysRevLett.99.038104. [DOI] [PubMed] [Google Scholar]
- 43.Nathan D., Crothers D.M. Bending and flexibility of methylated and unmethylated EcoRI DNA. J. Mol. Biol. 2002;316:7–17. doi: 10.1006/jmbi.2001.5247. [DOI] [PubMed] [Google Scholar]
- 44.Baumann C.G., Smith S.B., Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelman L.M., Cheong R., Kahn J.D. Fluorescence resonance energy transfer over approximately 130 basepairs in hyperstable lac repressor-DNA loops. Biophys. J. 2003;84:1131–1145. doi: 10.1016/S0006-3495(03)74929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You Y., Tataurov A.V., Owczarzy R. Measuring thermodynamic details of DNA hybridization using fluorescence. Biopolymers. 2011;95:472–486. doi: 10.1002/bip.21615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haas E., Katchalski-Katzir E., Steinberg I.Z. Effect of the orientation of donor and acceptor on the probability of energy transfer involving electronic transitions of mixed polarization. Biochemistry. 1978;17:5064–5070. doi: 10.1021/bi00616a032. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; NY: 2001. Methods in Molecular Cloning. [Google Scholar]
- 49.Stühmeier F., Welch J.B., Clegg R.M. Global structure of three-way DNA junctions with and without additional unpaired bases: a fluorescence resonance energy transfer analysis. Biochemistry. 1997;36:13530–13538. doi: 10.1021/bi9702445. [DOI] [PubMed] [Google Scholar]
- 50.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


