Abstract
Aims: Uridine diphosphate-glucuronosyltransferase 2B (UGT2B) enzymes conjugate testosterone metabolites to enable their excretion in humans. The functional significance of the UGT2B genetic variants has never been described in humans. We evaluated UGT2B variants in relation to plasma androstane-3α,17β-diol-glucuronide (AAG) levels and the prostate cancer risk. Results: AAG levels were measured in sera from 150 controls and compared to the polymorphisms of UGT2B17, UGT2B15, and UGT2B7. Genomic DNA from controls (301) and cases (148) was genotyped for the polymorphisms, and odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated using unconditional logistic regression analyses. Having two copies of UGT2B17 was associated with higher AAG levels in controls among Whites (p=0.02), but not Blacks (p=0.82). Logistic regression models adjusting for age and race revealed that homozygosity for the G allele of the UGT2B15D85Y polymorphism was directly associated with the prostate cancer risk (OR=2.70, 95% CI=1.28, 5.55). Conclusions: While the small sample size limits inference, our findings suggest that an association between the UGT2B17 copy number variant (CNV) and serum AAG levels in Whites, but unexpectedly not in Blacks. This novel observation suggests that genetic determinants of AAG levels in Blacks are unrelated to the UGT2B17 CNV. This study replicates the results that show an association of UGT215D85Y with an increased prostate cancer risk.
Introduction
Prostate cancer is the second leading cause of cancer death behind lung cancer in men in the United States (U.S. Cancer Statistics Working Group, 2010). In 2012, it is estimated that there will be 241,740 new cases and ∼28,000 deaths from prostate cancer in the United States (Siegel et al., 2012). The fact that androgen ablation is extremely effective at reducing tumor burden in men with prostate cancer has led to the hypothesis that androgens may play a role in prostate cancer etiology.
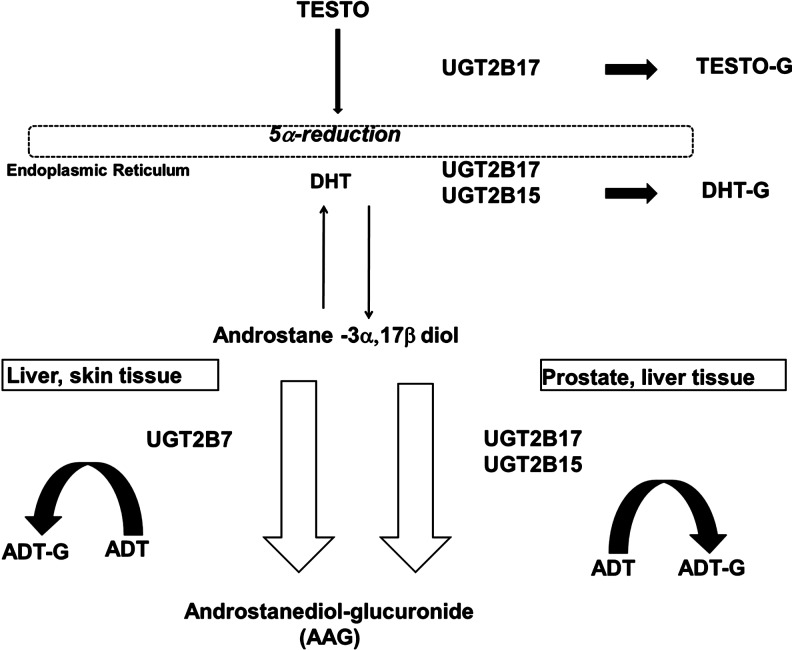
The uridine diphosphate-glucuronosyltransferase 2B (UGT2B) enzymes, UGT2B7, UGT2B15, and UGT2B17, catalyze the glucuronidation of multiple substrates. Through this process, endobiotic or xenobiotic substances, including hormones, are conjugated, and eliminated from the body through excretion in the urine. Of relevance, these enzymes exhibit specificity for androgen metabolites, such as testosterone, dihydrotestosterone, androsterone, and androstane-3α,17β-diol (3α-diol; Fig. 1) (Turgeon et al., 2001). The UGT2B enzymes each target different androgen metabolites, and their conjugation of one metabolite in particular, androstane 3α,17β diol, contributes to the circulating serum levels of its glucuronide, androstanediol-glucuronide (also known as androstane-3α,17β-diol-glucuronide [AAG]). UGT2B15 and UGT2B17 conjugate AAG in the lumen and basal epithelial tissue of the prostate, respectively, and UGT2B7 also conjugates AAG in the skin (Fig. 1) (Turgeon et al., 2001; Chouinard et al., 2007; Valentini et al., 2007). A common genetic polymorphism in the UGT2B17 gene is gene deletion, and evidence shows that individuals who have two deleted UGT2B17 alleles secrete little-to-no urinary testosterone compared to individuals with at least one copy of the gene (Jakobsson et al., 2006). In addition, a polymorphism in the regulatory region of the UGT2B17 gene has been associated with altered levels of AAG (Hu et al., 2010). While both the UGT2B17 and UGT2B15 polymorphisms are significantly associated with AAG levels in European populations, no observations have been made in individuals of African descent (Hsing, 2001; Swanson et al., 2007; Olsson et al., 2011).
FIG. 1.
Glucuronidation targets for UGT2B7, UGT2B17, and UGT2B15 enzymes in the liver, skin, and prostate tissue (Turgeon et al., 2001; Chouinard et al., 2007; Valentini et al., 2007). Narrow black arrows indicate reversible catabolism of DHT; thick black arrows indicate irreversible production of testosterone- and DHT-G; gray arrows (on left and right) indicate irreversible production of ADT-G; and parallel black outlined arrows indicate irreversible production of AAG. Testo, testosterone; DHT, dihydrotestosterone; ADT, androsterone; G, glucuronide.
The UGT2B17 and UGT2B15 genes have variants that have been characterized and evaluated in population-based association studies. UGT2B17 has a copy number variant (CNV) that is present in 0, 1, or 2 copies (Wilson et al., 2004; Jakobsson et al., 2006), while an UGT2B15 variant, UGT2B15D85Y, has a missense polymorphism at codon 85 that changes an aspartic acid residue (D allele) to a tyrosine residue (Y allele), resulting in an increased Vmax. The resulting phenotype of the enzyme leads to quicker androgen metabolite clearance, while the wild type confers lower clearance, possibly raising the effective amount of steroids within the prostate (Levesque et al., 1997; Chouinard et al., 2008). While several UGT2B7 variants have been described, they have not been examined in relation to prostate cancer (Turgeon et al., 2001; Menard et al., 2011). However, the expression of all three genes has been measured in prostate cancer cell lines (Valentini et al., 2007). The association of the UGT2B15D85Y polymorphism with prostate cancer has been examined with conflicting results (MacLeod et al., 2000; Gsur et al., 2002; Hajdinjak and Zagradisnik, 2004; Park et al., 2004; Cunningham et al., 2007). Similarly, numerous studies have explored the association of the UGT2B17 CNV and prostate cancer also with inconsistent conclusions (Park et al., 2006, 2007; Gallagher et al., 2007; Karypidis et al., 2007; Olsson et al., 2008; Setlur et al., 2010). Some of the inconsistencies in these findings could be explained by differences in the composition of the study participants. To date, an assessment of the risk for prostate cancer by single-nucleotide polymorphisms (SNPs) in linkage with UGT2B17 and UGT2B7 has not been accomplished.
In this report, UGT2B genetic variants were assessed for their relationship with serum AAG levels in healthy men and for their association with the risk for prostate cancer in a case–control analysis.
Materials and Methods
Study populations
The details of the case–control study have been previously reported (Antonelli et al., 2009). In brief, male patients from the Durham Veterans Affairs Medical Center (DVAMC) in Durham, North Carolina, who were undergoing a prostate needle biopsy between January 2007 and July 2010, were recruited to enroll in a hospital-based, prostate cancer case–control study. Eligibility criteria for cases included age >18 years, undergoing a prostate biopsy for concerns of potential prostate cancer after presentation with elevated prostate specific antigen (PSA), and/or abnormal digital rectal examination and diagnosed with prostate cancer after pathological review of biopsy tissue. A control population was recruited from the VA Internal Medicine Clinic who fit the criteria of age >18 and had a PSA test performed, but were not recommended to undergo biopsy. Of the 768 eligible control men, 377 signed consent forms (768/377=49% participation rate) by July 2010. Questionnaires were administered to prostate cancer cases before biopsy and to controls to assess risk factors, including race and age. Of the 759 men with a biopsy indication and who were screened for eligibility, 539 (759/539=71% participation rate) provided written consent to participate by July 2010. Of these 539, 517 underwent a biopsy, of which 202 were biopsy positive (cases). This report is limited to the first 150 cases for CNV analysis and 100 cases for genotyping based on available funding. The healthy controls were selected for the genotype (n=297), CNV (n=201), and (n=150) AAG analyses. Institutional Review Board approval was obtained at the Duke University, North Carolina Central University, and the DVAMC, and all patients signed an informed consent at the DVAMC before enrollment.
AAG measurements
AAG was measured on only the first 150 healthy control serum samples due to availability of funding. An enzyme immunoassay (EIA; ALPCO Diagnostics) at the Children's Hospital Boston, Department of Laboratory Medicine, Clinical and Epidemiologic Research Laboratory, was employed for determination of concentration. The EIA follows the basic principle of competitive binding assays where there is competition between an unlabeled antigen and an enzyme-labeled antigen for a fixed number of antibody-binding sites. A 96-well microtiter plate is coated with a polyclonal antibody to AAG. AAG in the samples competes with an AAG/horseradish peroxidase conjugate for binding sites. After incubation, unbound materials are removed by aspirating and washing the wells. The substrate tetramethylbenzidine is added, and a color is generated that is indirectly proportional to the amount of AAG in the sample. An acidic stopping solution is added, and the degree of enzymatic turnover of the substrate is determined by dual-wavelength absorbance measurement. The assay possesses a sensitivity of 0.1 ng/mL and a run-to-run imprecision at AAG concentrations of 0.98, 7.05, and 20.92 ng/mL of 10.4%, 6.5%, and 10.8%, respectively. The samples were from healthy controls with available sera and completed questionnaires. Only samples from Whites and Blacks were included in the final analysis, which reduced the number to 147.
Genotyping
Major UGT2B variants, UGT2B17 CNV and UGT2B15D85Y (rs1902023), were selected for genotyping as well as SNP Tags for UGT2B17 (rs7434408) and UGT2B7 (7435335). Candidate SNP Tags were selected from the UGT2B17 and UGT2B7 genes that were in high linkage disequilibrium (R2≥0.8), and had minor alleles at frequencies that were at least 50% different in the CEU (European/White) and YRI (African/Black) HapMap populations. DNA was isolated from peripheral blood by standard DNA isolation (Qiagen, Inc.) and quantified by ultraviolet spectrophotometry. Before genotyping, DNA concentration was determined using PicoGreen assay (Life Technologies) and measured using the fluorescence intensity measurements plotted against a standard curve that was generated from the average fluorescence intensity of standards run in the replicate. Based on the PicoGreen quantification, 10 ng of genomic DNA from each sample was used in the iPlex assay for Sequenom-iPlex Genotyping (Sequenom, Inc.). The Sequenom MassArray (Sequenom, Inc.) was used, and the assays for UGT2B17 (rs7434408), UGT2B15D85Y (rs1902023), and UGT2B7 (rs7435335) were designed by the Sequenom online assay tools (Assay Designer 4.0) at the David H. Murdock Research Institute (DHMRI) Genomics Laboratory. The data were analyzed by Sequenom-Typer 4.0. The Sequenom-iPlex genotyping and analysis were validated with CEPH gDNA controls by performing the iPlex assay and scanning on the MALDI-TOF mass spectrometer. At the time of this analysis, 364 samples were submitted, and 358 successfully produced good spectra for genotyping at a failure rate of <2.2%. The post-QC (call rate) of UGT2B17 (rs7434408), UGT2B15D85Y (rs1902023), and UGT2B7 (rs7435335) was 100%. The assays included DHMRI control DNA (CEPH) on each plate in replicates that were checked for concordance for each SNP. Samples that were analyzed for the UGT2B17 CNV were quantified with PicoGreen as described above and genotyped using the ABI 7900HT (Applied Biosystems) platform and commercially available TaqMan CNV assays. Each sample was assayed in quadruplicate and compared with controls previously genotyped (Wilson et al., 2004). Data analysis was accomplished using SDS v2.4 and CopyCaller v1.0 (Applied Biosystems), and the confidence level was ≥0.99.
Statistical analyses
All analyses were done using Stata Software. Quartiles of AAG levels in White and Black healthy controls were determined for UGT2B CNV, and genotypes and p-values were calculated by Kruskal-Wallis equality-of-population rank test. Chi-square tests were used to compare risk factors such as age and race with tumor grade based on low (Gleason sum <7) or high grade (Gleason sum >7). Unconditional logistic regression analyses were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for prostate cancer risk. Models were adjusted for age and race separately for CNV and SNPs, and analyses were completed assuming an ordinal, recessive, or dominant model. We estimated ORs for the association between the minor allele for SNPs, rs7434408 and rs7435335, and prostate cancer risk, adjusting for age and race. These analyses were repeated stratified by race and tumor grade. All p-values<0.05 were considered significant.
Results
The distribution of the UGT2B17 CNV genotypes was significantly associated with median serum AAG levels in Whites (p=0.02), but unexpectedly not in Blacks (Table 1). Specifically, a higher number of CNVs was associated with higher AAG levels. Conversely, there was a weak suggestion that the UGT2B15D85Y genotypes may be associated with AAG levels in Whites, although the association did not reach significance (p=0.15). No other significant associations between SNPs and AAG levels were observed in Whites. Notably, no significant association of genotypes and AAG levels was observed in Blacks. We did not observe any significant racial differences in AAG levels in the present study.
Table 1.
Median Androstane-3α,17β-Diol-Glucuronide Levels Among Healthy Controls and Quartiles and Associated UGT2B Sequence Variants
| |
Whites |
Blacks |
||
|---|---|---|---|---|
| Genotype | Individuals (%) | Median (25th, 75th percentile) | Individuals (%) | Median (25th, 75th percentile) |
| UGT2B17 | ||||
| CNV 0 | 6 (6.4) | 4.34 (3.3, 6.19) | 2 (4.2) | 8.8 (5.64, 12) |
| 1 | 41 (44.1) | 6.51 (4.08, 9.24) | 22 (45.8) | 6.9 (3.91, 10.57) |
| 2 | 46 (49.5) | 7.68 (5.69, 13.69) | 24 (50.0) | 6.75 (4.14, 8.8) |
| p Value | 0.02 | 0.82 | ||
| UGT2B17 (rs7434408) | ||||
| A | 28 (30.8) | 6.71 (4.63, 9.33) | 36 (76.6) | 6.71 (4.28, 10.78) |
| A/G | 41 (45.0) | 6.84 (4.47, 12.14) | 2 (4.2) | 7.39 (4.07, 10.16) |
| G | 22 (24.2) | 7.64 (5.44, 13.71) | 9 (19.1) | 5.07 (3.38, 6.77) |
| p Value | 0.77 | 0.67 | ||
| UGT2B15D85Y(rs1902023) | ||||
| G | 17 (18.7) | 8.75 (5.36, 14.14) | 17 (36.2) | 5.52 (3.74, 10.61) |
| G/T | 46 (50.5) | 6.85 (4.83, 11.44) | 16 (34.0) | 7.62 (4.41, 11.12) |
| T | 28 (30.8) | 6.49 (3.54, 9.33) | 14 (29.8) | 7.21 (4.62, 10.37) |
| p Value | 0.15 | 0.44 | ||
| UGT2B7 (rs7435335) | ||||
| G | 65 (72.2) | 6.71 (4.63, 11.52) | 26 (55.3) | 6.01 (3.75, 9.85) |
| G/A | 24 (26.7) | 7.12 (4.65, 11.02) | 19 (40.4) | 7.39 (4.26, 11.86) |
| A | 1 (1.1) | 10.07 (10.07) | 2 (4.2) | 5.55 (4.71, 6.4) |
| p Value | 0.73 | 0.59 | ||
UGT2B, uridine diphosphate-glucuronosyltransferase 2B; CNV, copy number variant.
Demographic characteristics of the study population were compared among controls, men with low-grade tumors (Gleason <7), and those with high-grade tumors (Gleason ≥7) (Table 2). Cases were significantly older than controls (p=0.004). Black men comprised over half of the cases (61.6% vs. 38.4%) when compared to Whites and had the majority of both high-grade tumors (60.3% vs. 39.7%) and low-grade tumors (63% vs. 37%).
Table 2.
Characteristics of Study Participants
| Age | Controls (%) | Cases 1 (low grade) | Cases 2 (high grade) | Significance p value |
|---|---|---|---|---|
| <50 | 18 (6) | 2 (2.7) | 1 (1.4) | |
| 50–59 | 80 (26.6) | 21 (28.8) | 14 (19.2) | |
| 60–69 | 162 (53.8) | 38 (52.0) | 43 (59) | |
| ≥70 | 41 (13.6) | 12 (16.4) | 15 (20.5) | |
| Mean age [SD] | 61.4 [7.5] | 62.2 [7] | 64.6 [6.4] | 0.004 |
| Race | ||||
| Whites | 192 (63.8) | 27 (37) | 29 (39.7) | |
| Blacks | 109 (36.2) | 46 (63.0) | 44 (60.3) | <0.001 |
Dominant, heterozygous, and recessive genotypes for the UGT2B17 CNV, UGT2B17 SNP rs7434408, UGT2B15D85Y, and the UGT2B7 SNP 7435335 were evaluated in relation to the prostate cancer risk (Table 3). Crude analyses of the homozygous dominant allele (G) of the UGT2B15 missense polymorphism, UGT2B1585D (rs1902023), showed a significant association with increased risk for prostate cancer (OR=3.22, 95% CI=1.59, 6.67). Adjusting for age and race did not alter these findings (OR=2.70, 95% CI=1.28, 5.55). No other significant associations between SNPs and prostate cancers were observed. Analyses stratified by race were done to determine if the association of the UGT2B17 CNV and other UGT2B SNPs with prostate cancer differed between Whites and Blacks, though when this was done, the power to detect associations was reduced compared to the whole-population analyses (data not shown).
Table 3.
Odds Ratios for the Association Between Genetic Variants of UGT2B17 CNV, UGT2B17 A/G (rs7434408), UGT2B15D85Y G/T (rs1902023), and UGT2B7 A/G (rs7435335) and prostate cancer risk
| Genotype | Cases (%) | Controls (%) | Crude OR (95% CI) | Adjusted OR (age, race) |
|---|---|---|---|---|
| UGT2B17 CNV | ||||
| 2 | 79 (53.4) | 101 (50.2) | Reference | Reference |
| 1 | 58 (39.2) | 91 (45.3) | 0.81 (0.52, 1.27) | 0.83 (0.53, 1.31) |
| 0 | 11 (7.4) | 9 (4.5) | 1.56 (0.62, 3.95) | 1.64 (0.63, 4.28) |
| UGT2B17 (rs7434408) | ||||
| A | 60 (66.7) | 136 (45.8) | Reference | Reference |
| A/G | 15 (16.7) | 66 (22.2) | 0.58 (0.32, 1.07) | 0.85 (0.44, 1.67) |
| G | 17 (18.9) | 95 (32.0) | 0.36 (0.19, 0.67) | 0.64 (0.32, 1.31) |
| A/G+G vs. A | 32 (34.8) | 136 (45.8) | 0.45 (0.28, 0.73) | 0.75 (0.42, 1.31) |
| UGT2B15D85Y (rs1902023) | ||||
| T | 13 (14.1) | 90 (30.3) | Reference | Reference |
| G/T | 47 (51.1) | 139 (46.8) | 1.39 (0.81, 2.38) | 1.19 (0.68, 2.13) |
| G | 32 (34.8) | 68 (22.9) | 3.22 (1.59, 6.67) | 2.70 (1.28, 5.55) |
| G/T+G vs. T | 60 (65.2) | 229 (77.1) | 1.78 (1.09, 3.03) | 1.51 (0.89, 2.56) |
| UGT2B7 (rs7435335) | ||||
| G | 67 (72.8) | 208 (70.2) | Reference | Reference |
| G/A | 20 (21.7) | 83 (28.0) | 0.75 (0.43, 1.31) | 0.59 (0.32, 1.06) |
| A | 5 (5.4) | 5 (1.7) | 3.10 (0.87, 11.05) | 2.16 (0.56, 8.30) |
| G/A+A vs. G | 25 (27.1) | 88 (29.7) | 0.88 (0.52, 1.49) | 0.68 (0.39, 1.12) |
OR, odds ratio; CI, confidence interval.
Discussion
UGT2B enzymes are important in the biotransformation of steroid hormones, though their associations with serum steroid levels remain unclear. Moreover, though some studies have suggested their association with prostate cancer risk (MacLeod et al., 2000; Hajdinjak and Zagradisnik, 2004; Park et al., 2004, 2006, 2007; Karypidis et al., 2007), others have not (Gsur et al., 2002; Cunningham et al., 2007; Gallagher et al., 2007; Olsson et al., 2008; Setlur et al., 2010), making it difficult to draw any firm conclusions. In the current study, among U.S. veterans, the UGT2B17 CNV genotypes were significantly correlated with AAG levels in White healthy controls, but showed no association with AAG levels in Black healthy controls. The results suggested the novel observation that AAG levels in Blacks may not be associated with UGT2B17 CNV and may be determined by other UGT2B variants. This study also confirmed the results of some previous studies that showed the association of the UGT2B15D85Y polymorphism with prostate cancer risk. Individuals homozygous for the UGT2B1585G (G) allele had a significantly increased risk for prostate cancer on both crude and adjusted analyses.
A study investigating the relationship of the UGT2B genes and hormones showed that both the UGT2B15D85Y polymorphism and the UGT2B17 CNV were associated with serum levels of AAG (p<0.001), but not with levels of another steroid glucuronide, 3α-diol-3-glucuronide, in a population-based cohort of young adult (n=1068; mean age=18.9 years) and elderly (n=1001; mean age=75.3 years) Swedish men (Swanson et al., 2007). We did not determine 3α-diol-3-glucuronide levels in our study as AAG represents 80% of the combined isoforms of 3α-diol, and thus we felt measuring AAG, which gave a good overall measure of glucorinidation activity. Other studies showed that UGT2B17 CNV was associated with AAG levels in subjects of European descent. Olsson et al. (2011) showed that AAG levels were significantly associated with the UGT2B17 CNV, suggesting that the UGT2B17 CNV genotypes are significantly associated with AAG levels. Thus, the relationship we observed between UGT2B17 CNV and AAG levels in Whites is consistent with the two previous reports. Importantly, the levels of AAG measured in this study are similar to levels of AAG previously reported by an NHANES III study, suggesting that the measurements of AAG are stable and reliable (Rohrmann et al., 2007). A key surprising finding from our study, which has not been previously examined, was the absence of an association between UGT2B17 CNV and AAG among Black healthy controls. If confirmed, this suggests that the clearance of intraprostatic steroids may be influenced by different genetic determinants among Black men.
We replicated the findings of previous studies that have shown an association of the UGT2B15D85Y polymorphism and prostate cancer risk (Table 4). However, in the literature, both significant and null associations have been observed. The results from this study showed a significant increase in the risk for prostate cancer in individuals homozygous for the G allele, consistent with previous reports (Table 4) (MacLeod et al., 2000; Hajdinjak and Zagradisnik, 2004; Park et al., 2004). However, null associations of the UGT2B15D85Y polymorphism have also been described. The study by Gsur et al. (2002) showed that the homozygous UGT2B1585Y (T) allele was not associated (OR=1.2; 95% CI=0.65, 2.22) with prostate cancer. The authors of that study stated that a potential limitation to their study could be the use of a control population of men with benign prostate hyperplasia. In addition, a case–control study by Cunningham et al. also found no significant association with prostate cancer risk for the UGT2B15D85 (G) allele, although the association with familial prostate cancer showed borderline significance (OR=1.4, 95% CI=1.0, 1.9) (Cunningham et al., 2007). The results from previous reports assessing the association of the UGT2B17 CNV and prostate cancer risk have been inconsistent (Table 4). We showed no association with prostate cancer risk consistent with some previous reports (Gallagher et al., 2007; Olsson et al., 2008; Setlur et al., 2010). However, other studies reported that individuals homozygous for the UGT2B17 deletion were at increased risk for prostate cancer, while a report showed that only carriers of the UGT2B17 deletion had increased risk (Park et al., 2006, 2007; Karypidis et al., 2007). These differences across studies could be the result of interindividual variability in the UGT2B gene expression and haplotype structure of the UGT2B genes within each study population (Izukawa et al., 2009; Menard et al., 2009). This variability may be more apparent in individuals that have the UGT2B17 gene deleted. Recent evidence however points to the relevance of UGT2B15 to prostate cancer etiology. Expression analysis of UGT2B15 showed that decreased levels of UGT2B15 mRNA and proteins were associated with hormone-naïve and castration-resistant prostate tumors when compared to benign prostatic hyperplasia (Paquet et al., 2011). It would be important in future association studies to investigate the impact of functional polymorphisms in high linkage with the UGT2B15D85Y polymorphism, given its effect on enzyme kinetics.
Table 4.
Summary of Studies Evaluating the Association Between UGT2B Variants and Prostate Cancer
| UGT2B variant | Genotype | Study | Controls | Cases | Blacks | OR | 95% CI | p | References |
|---|---|---|---|---|---|---|---|---|---|
| UGT2B17 CNV | Del/Del | Incident Pca | 487 | 420 | 247 | Cau 1.9 | (1.2–3.0) | 0.006 | Park et al. (2006) |
| AA 1.3 | (0.6–2.7) | 0.32 | |||||||
| UGT2B17 CNV | Del/Del | Incident Pca | 363 | 356 | 34 | 1.7 | (1.03–2.9) | n.d. | Park et al. (2007) |
| UGT2B17 CNV | Del/Del | Incident Pca | 161 | 176 | 0 | 1.63 | (0.37–7.15) | n.d. | Karypidis et al. (2007) |
| Del/Del+Del/Ins | 2.07 | (1.32–3.25) | |||||||
| UGT2B17 CNV | Del/Del | Incident Pca | 397 | 411 | 0 | 0.89 | (0.55–1.45) | n.d. | Gallagher et al. (2007) |
| UGt2B17 CNV | Del/Del | Incident Pca | 1722 | 2779 | 1.01 | (0.83–1.23) | 0.91 | Olsson et al. (2008) | |
| UGT2B17 CNV | Del/Del | Incident Pca | 205 | 221 | 0 | 0.88 | (0.51–1.16) | 0.71 | Setlur et al. (2010) |
| UGTB15D85Y | D85/D85 | Incident Pca | 64 | 64 | 6 | 2.96 | (χ2=7.34) | <0.01 | Macleod et al. (2000) |
| UGTB15D85Y | Y85/Y85 | Incident Pca | 190 | 190 | 0 | 1.20 | (0.65–2.22) | 0.54 | Gsur et al. (2002) |
| UGTB15D85Y | D85/D85 | Incident Pca | 155 | 155 | 0 | 2.70 | (1.1–6.6) | n.d. | Park et al. (2004) |
| UGTB15D85Y | D85/D85 | Sporadic Gleason scores | 178 | 206 | 2.04 | 0.032 | Hajdinjak and Zagradisnik (2004) | ||
| UGTB15D85Y | D85/Y85 | Familial and sporadic | 493 | 438 | 0 | 1.4 | (1.0, 1.9) | n.d. | Cunningham et al. (2007) |
| 493 | 499 | 1.1 | (0.8, 1.4) | n.d. |
Pca, prostate cancer; n.d., no data.
The strengths of this study are that while hospital based, the setting is the DVAMC, where there is equal access to care regardless of race, providing a pool of Blacks in the dataset compared to other studies. Also, the DVAMC more readily provides access to health care for diverse populations, allowing enrollment of a large number of Black men. As with any case–control study, especially those studies with patients with prostate cancer, it is possible that some controls may have actually had prostate cancer. However, this misclassification would tend to bias the results toward the null, and thus may underestimate the strength of the true significant associations between the genotype and prostate cancer risk. The main limitation of our study is the small sample size for prostate cancer cases and controls, which limited our inability to detect modest, but potentially important, associations. However, the results which show differences in the relationships between the UGT2B17 CNV genotypes and AAG levels in White and Black healthy controls point to a need to include diverse populations in future studies to determine the association of molecular markers of steroid metabolism with serum metabolites across racial groups. The importance of including diverse racial groups is further accentuated by results from a recent study that found that UGT2B CNVs had different associations with a risk for biochemical recurrence after surgery and levels of androgen glucuronides in Whites compared to Asians who underwent radical prostatectomies (Nadeau et al., 2011).
In summary, the inclusion of Blacks in this study population, although limited by sample size, revealed the novel observation that the UGT2B17 genotype was associated with AAG levels, but that the association is limited to Whites. This observation points to a need to develop hypotheses about the genetic determinants of AAG clearance in Black populations given the disparity in prostate cancer incidence and mortality. We also found a direct association between the UGT2B15D85 polymorphism and prostate cancer, an association that persisted after adjustment for age and race. Future research that evaluates the association of the UGT2B sequence variants with survival and prostate cancer risk should assess the importance of these genes as future biomarkers and therapeutic targets.
Acknowledgments
We would like to thank Gary Bradwin of the Children's Hospital of Boston; Allen Zillmer of the David H. Murdock Research Institute, Kannapolis, NC; and Megan Wilson of North Carolina Central University and Loretta Taylor and Kathryn Newman of Duke University for assistance. Financial support: Department of Defense PC060233, NIH P20 MD000175, S06-GM008049-33, Department of Veterans Affairs, and the AUA Foundation/Astellas Rising Star in Urology.
Author Disclosure Statement
No competing financial interests exist for any author.
References
- Antonelli JA. Jones LW. Banez LL, et al. Exercise and prostate cancer risk in a cohort of veterans undergoing prostate needle biopsy. J Urol. 2009;182:2226–2231. doi: 10.1016/j.juro.2009.07.028. [DOI] [PubMed] [Google Scholar]
- Chouinard S. Barbier O. Belanger A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J Biol Chem. 2007;282:33466–33474. doi: 10.1074/jbc.M703370200. [DOI] [PubMed] [Google Scholar]
- Chouinard S. Yueh MF. Tukey RH, et al. Inactivation by UDP-glucuronosyltransferase enzymes: the end of androgen signaling. J Steroid Biochem Mol Biol. 2008;109:247–253. doi: 10.1016/j.jsbmb.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Cunningham JM. Hebbring SJ. McDonnell SK, et al. Evaluation of genetic variations in the androgen and estrogen metabolic pathways as risk factors for sporadic and familial prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:969–978. doi: 10.1158/1055-9965.EPI-06-0767. [DOI] [PubMed] [Google Scholar]
- Gallagher CJ. Kadlubar FF. Muscat JE, et al. The UGT2B17 gene deletion polymorphism and risk of prostate cancer A case-control study in Caucasians. Cancer Detect Prev. 2007;31:310–315. doi: 10.1016/j.cdp.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsur A. Preyer M. Haidinger G, et al. A polymorphism in the UDP-glucuronosyltransferase 2B15 gene (D85Y) is not associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:497–498. [PubMed] [Google Scholar]
- Hajdinjak T. Zagradisnik B. Prostate cancer and polymorphism D85Y in gene for dihydrotestosterone degrading enzyme UGT2B15: frequency of DD homozygotes increases with Gleason Score. Prostate. 2004;59:436–439. doi: 10.1002/pros.20024. [DOI] [PubMed] [Google Scholar]
- Hsing AW. Hormones and prostate cancer: what's next? Epidemiol Rev. 2001;23:42–58. doi: 10.1093/oxfordjournals.epirev.a000795. [DOI] [PubMed] [Google Scholar]
- Hu DG. Gardner-Stephen D. Severi G, et al. A novel polymorphism in a forkhead box A1 (FOXA1) binding site of the human UDP glucuronosyltransferase 2B17 gene modulates promoter activity and is associated with altered levels of circulating androstane-3alpha,17beta-diol glucuronide. Mol Pharmacol. 2010;78:714–722. doi: 10.1124/mol.110.065953. [DOI] [PubMed] [Google Scholar]
- Izukawa T. Nakajima M. Fujiwara R, et al. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug metabolism and disposition: the biological fate of chemicals. 2009;37:1759–1768. doi: 10.1124/dmd.109.027227. [DOI] [PubMed] [Google Scholar]
- Jakobsson J. Ekstrom L. Inotsume N, et al. Large differences in testosterone excretion in Korean and Swedish men are strongly associated with a UDP-glucuronosyl transferase 2B17 polymorphism. J Clin Endocrinol Metab. 2006;91:687–693. doi: 10.1210/jc.2005-1643. [DOI] [PubMed] [Google Scholar]
- Karypidis AH. Olsson M. Andersson SO, et al. Deletion polymorphism of the UGT2B17 gene is associated with increased risk for prostate cancer and correlated to gene expression in the prostate. Pharmacogenomics J. 2007;8:147–151. doi: 10.1038/sj.tpj.6500449. [DOI] [PubMed] [Google Scholar]
- Levesque E. Beaulieu M. Green MD, et al. Isolation and characterization of UGT2B15(Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 1997;7:317–325. doi: 10.1097/00008571-199708000-00007. [DOI] [PubMed] [Google Scholar]
- MacLeod SL. Nowell S. Plaxco J, et al. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann Surg Oncol. 2000;7:777–782. doi: 10.1007/s10434-000-0777-3. [DOI] [PubMed] [Google Scholar]
- Menard V. Eap O. Harvey M, et al. Copy-number variations (CNVs) of the human sex steroid metabolizing genes UGT2B17 and UGT2B28 and their associations with a UGT2B15 functional polymorphism. Human Mutat. 2009;30:1310–1319. doi: 10.1002/humu.21054. [DOI] [PubMed] [Google Scholar]
- Menard V. Eap O. Roberge J, et al. Transcriptional diversity at the UGT2B7 locus is dictated by extensive pre-mRNA splicing mechanisms that give rise to multiple mRNA splice variants. Pharmacogenet Genomics. 2011;21:631–641. doi: 10.1097/FPC.0b013e3283498147. [DOI] [PubMed] [Google Scholar]
- Nadeau G. Bellemare J. Audet-Walsh E, et al. Deletions of the androgen-metabolizing UGT2B genes have an effect on circulating steroid levels and biochemical recurrence after radical prostatectomy in localized prostate cancer. J Clin Endocrinol Metab. 2011;96:E1550–1557. doi: 10.1210/jc.2011-1049. [DOI] [PubMed] [Google Scholar]
- Olsson M. Ekstrom L. Guillemette C, et al. Correlation between circulatory, local prostatic, and intra-prostatic androgen levels. Prostate. 2011;71:909–914. doi: 10.1002/pros.21307. [DOI] [PubMed] [Google Scholar]
- Olsson M. Lindstrom S. Haggkvist B, et al. The UGT2B17 gene deletion is not associated with prostate cancer risk. Prostate. 2008;68:571–575. doi: 10.1002/pros.20700. [DOI] [PubMed] [Google Scholar]
- Paquet S. Fazli L. Grosse L, et al. Differential expression of the androgen-conjugating UGT2B15 and UGT2B17 enzymes in prostate tumor cells during cancer progression. J Clin Endocrin Metab. 2011;97:E428–E432. doi: 10.1210/jc.2011-2064. [DOI] [PubMed] [Google Scholar]
- Park J. Chen L. Ratnashinge L, et al. Deletion polymorphism of UDP-glucuronosyltransferase 2B17 and risk of prostate cancer in African American and Caucasian men. Cancer Epidemiol Biomarkers Prev. 2006;15:1473–1478. doi: 10.1158/1055-9965.EPI-06-0141. [DOI] [PubMed] [Google Scholar]
- Park J. Chen L. Shade K, et al. Asp85tyr polymorphism in the udp-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J Urol. 2004;171:2484–2488. doi: 10.1097/01.ju.0000117748.44313.43. [DOI] [PubMed] [Google Scholar]
- Park JY. Tanner JP. Sellers TA, et al. Association between polymorphisms in HSD3B1 and UGT2B17 and prostate cancer risk. Urology. 2007;70:374–379. doi: 10.1016/j.urology.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Rohrmann S. Nelson WG. Rifai N, et al. Serum sex steroid hormones and lower urinary tract symptoms in Third National Health and Nutrition Examination Survey (NHANES III) Urology. 2007;69:708–713. doi: 10.1016/j.urology.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Setlur SR. Chen CX. Hossain RR, et al. Genetic variation of genes involved in dihydrotestosterone metabolism and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:229–239. doi: 10.1158/1055-9965.EPI-09-1018. [DOI] [PubMed] [Google Scholar]
- Siegel R. Naishadham D. Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Swanson C. Mellstrom D. Lorentzon M, et al. The uridine diphosphate glucuronosyltransferase 2B15 D85Y and 2B17 deletion polymorphisms predict the glucuronidation pattern of androgens and fat mass in men. J Clin Endocrinol Metab. 2007;92:4878–4882. doi: 10.1210/jc.2007-0359. [DOI] [PubMed] [Google Scholar]
- Turgeon D. Carrier JS. Levesque E, et al. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787. doi: 10.1210/endo.142.2.7958. [DOI] [PubMed] [Google Scholar]
- U.S. Cancer Statistics Working Group. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; Atlanta: 2010. United States Cancer Statistics: 1999–2007 Incidence and Mortality Web-based Report. [Google Scholar]
- Valentini A. Biancolella M. Amati F, et al. Valproic acid induces neuroendocrine differentiation and UGT2B7 up-regulation in human prostate carcinoma cell line. Drug metabolism and disposition: the biological fate of chemicals. 2007;35:968–972. doi: 10.1124/dmd.107.014662. [DOI] [PubMed] [Google Scholar]
- Wilson W. Pardo-Manuel de Villena F. Lyn-Cook BD, 3rd, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–714. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]