Abstract
Parkinson's disease genes PINK1 and parkin encode kinase and ubiquitin ligase, respectively. The gene products PINK1 and Parkin are implicated in mitochondrial autophagy, or mitophagy. Upon the loss of mitochondrial membrane potential (ΔΨm), cytosolic Parkin is recruited to the mitochondria by PINK1 through an uncharacterised mechanism – an initial step triggering sequential events in mitophagy. This study reports that Ser65 in the ubiquitin-like domain (Ubl) of Parkin is phosphorylated in a PINK1-dependent manner upon depolarisation of ΔΨm. The introduction of mutations at Ser65 suggests that phosphorylation of Ser65 is required not only for the efficient translocation of Parkin, but also for the degradation of mitochondrial proteins in mitophagy. Phosphorylation analysis of Parkin pathogenic mutants also suggests Ser65 phosphorylation is not sufficient for Parkin translocation. Our study partly uncovers the molecular mechanism underlying the PINK1-dependent mitochondrial translocation and activation of Parkin as an initial step of mitophagy.
Mutations of the PINK1 gene cause selective degeneration of the midbrain dopaminergic neurons in autosomal recessive juvenile Parkinson's disease (PD)1. The PINK1 gene encodes a serine/threonine kinase with a predicted mitochondrial target sequence and a putative transmembrane domain at the N-terminus2,3,4,5. Loss of the PINK1 gene in Drosophila results in the degeneration of mitochondria in cells with high energy demands, such as muscle and sperm cells, which is suppressed by the introduction of the parkin gene, another gene responsible for autosomal recessive juvenile PD6,7,8. The gene product Parkin encodes a RING-finger type ubiquitin ligase (E3) with a Ubl domain at the N-terminus9,10,11,12.
A series of cell biological studies have provided strong evidence that there are important roles for PINK1 and Parkin in regulating mitochondrial homeostasis. PINK1 is constitutively proteolysed by the mitochondrial rhomboid protease, PARL, at the mitochondrial membrane of healthy mitochondria, resulting in processed forms of PINK113,14,15,16. The processed PINK1 is rapidly degraded by the proteasome2,17. The reduction of ΔΨm leads to the accumulation and activation of PINK1 in the mitochondria17,18,19 through a currently unresolved mechanism20. The accumulation of PINK1 recruits Parkin from the cytosol to the mitochondria with decreased membrane potential, which stimulates Parkin E3 activity, promoting mitochondrial degradation via an autophagic event known as mitophagy17,21,22,23,24. The recruitment of cytosolic Parkin to the mitochondria upon disruption of ΔΨm is believed to be the first step of mitophagy for the removal of damaged mitochondria. This recruitment is required for the kinase activity of PINK117,21,22,23,24,25. Although two separate studies have proposed that Parkin is directly phosphorylated by PINK126,27, others have failed to detect Parkin phosphorylation by PINK121, suggesting that the kinase activity of PINK1 itself is relatively low. One reason biochemical analysis has been unable to obtain direct evidence is that recombinant human PINK1 purified from mammalian cultured cells or bacteria easily loses kinase activity, while insect PINK1 has significant autophosphorylation activity28,29.
Very recently, Kondapalli, C. et al. reported that PINK1 directly phosphorylates Parkin at Ser65 in the Ubl domain18. However, the extent and consequences of Parkin phosphorylation by PINK1 in mitochondrial regulation are still not fully understood.
To address this issue, we attempted to independently monitor and compare the phosphorylation status of Parkin in wild-type and PINK1-deficient cells, thereby excluding the possibility of phosphorylations by uncharacterised kinases other than PINK130. Here, we also report that Parkin is demonstrably phosphorylated at Ser65 in a PINK1-dependent manner. Furthermore, we show that this phosphorylation event is implicated in the regulation of mitochondrial translocation of Parkin and the subsequent degradation of mitochondrial surface proteins during mitophagy.
Results
Parkin is phosphorylated upon depolarisation in ΔΨm
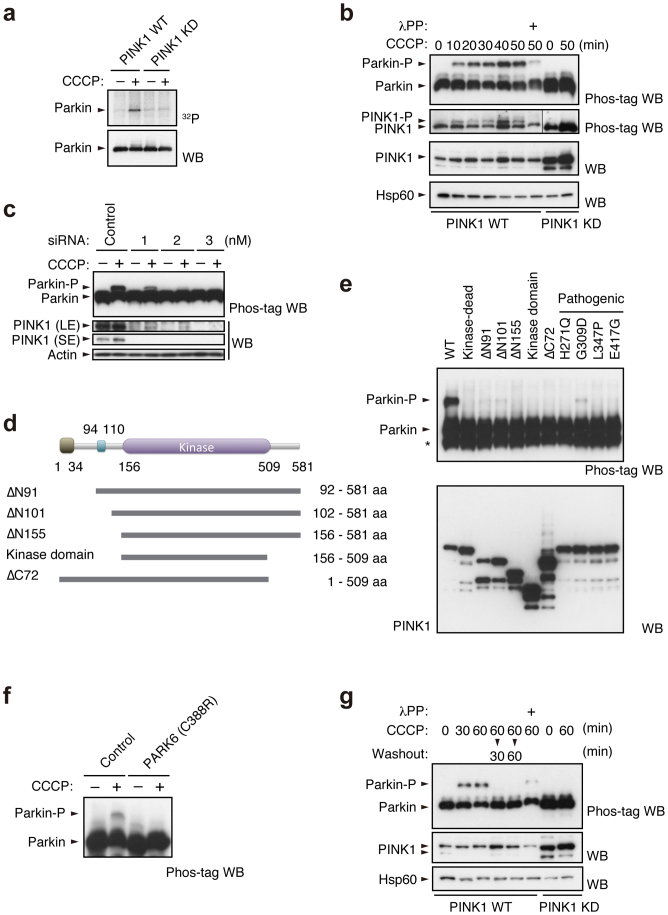
We used [32P] orthophosphate to metabolically label mouse embryonic fibroblasts (MEFs) derived from PINK1 deficient mice, in which HA-tagged Parkin together with FLAG-tagged wild-type or kinase-dead forms (triple mutant with K219A, D362A and D384A) of PINK1 were virally introduced (hereafter referred to as “PINK1-FLAG WT” or “KD/HA-Parkin/PINK1−/−” MEFs) and then induced Parkin-mediated mitophagy via treatment with the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP). As shown in Figure 1a, Parkin was specifically phosphorylated in CCCP-treated PINK1-FLAG WT/HA-Parkin/PINK1−/− MEFs, but not in PINK1-FLAG KD/HA-Parkin/PINK1−/− MEFs. Phos-tag Western blotting, in which phosphorylated proteins appear as slower migrating bands28, revealed that Parkin was phosphorylated within 10 min following CCCP treatment (Fig. 1b). Phosphorylation of Parkin reached its maximum level approximately 40 min after CCCP treatment and was sustained at least until 6 hr (Supplementary Fig. S1). Under these conditions, slower migrating bands of PINK1 also appeared, which very likely reflects the autophosphorylation of PINK1 when activated (Fig. 1b)18. The suppression of PINK1 accumulation by RNA interference suggested thatΔΨm depolarisation-dependent activation of PINK1 along with PINK1 accumulation is a key element for Parkin phosphorylation (Fig. 1c). Every PINK1 deletion and pathogenic mutant we tested failed to stimulate Parkin phosphorylation effectively, strongly suggesting that intact PINK1 is required for this action (Fig. 1d and e). Importantly, human fibroblasts from a patient with PINK1-linked parkinsonism also lacked the activity to phosphorylate Parkin (Fig. 1f). The phosphorylated Parkin disappeared within 30 min during the recovery of ΔΨm depolarisation by the removal of CCCP from the culture medium (Fig. 1g). Further analysis using phosphatase and proteasome inhibitors suggested that phosphorylated Parkin is at least partly degraded by proteasomal activity in the mitochondria (Supplementary Fig. S2).
Figure 1. PINK1-dependent phosphorylation of Parkin in vivo.
(a) PINK1-FLAG WT or KD/HA-Parkin/PINK1−/− MEFs were labelled with [32P] orthophosphate and treated with 30 µM CCCP for 1.5 hr. Phosphorylated Parkin was detected by autoradiography (32P). Immunoprecipitated HA-Parkin was detected by Western blotting (WB) with anti-Parkin. (b) PINK1-FLAG WT or KD/HA-Parkin/PINK1−/− MEFs were treated with or without 30 µM CCCP for the indicated periods of time. Cell lysate was subsequently separated on a Phos-tag gel, followed by WB with anti-PINK1 or anti-Parkin antibodies (Phos-tag WB). Phosphorylated bands of Parkin and PINK1 were confirmed by their disappearance with lambda protein phosphatase (λPP) treatment. Mitochondrial Hsp60 was used as a loading control. (c) Suppression of endogenous PINK1 expression inhibits Parkin phosphorylation. HeLa cells stably expressing non-tagged Parkin were treated with the indicated concentrations of stealth siRNA duplex against PINK1 (Invitrogen) with or without 10 µM CCCP for 1 hr. Long- (LE) and short-exposure (SE) blot signals for PINK1 were shown. Actin was used as a loading control. (d) Truncated PINK1 mutants used in this study. Putative mitochondria-targeting sequence, 1–34 aa; transmembrane domain, 94–110 aa; kinase domain, 156–509 aa. (e) Full-length PINK1 is required for Parkin phosphorylation. PINK1−/− MEFs stably expressing non-tagged Parkin were transfected with various PINK1 constructs with C-terminal FLAG-tags. PINK1 expression was confirmed with anti-FLAG-HRP. (f) Human fibroblasts from a normal control and a PARK6 case with a homozygous C388R mutation44 were transfected with Parkin and were treated with or without 30 µM CCCP for 1 hr. (g) Cells treated with CCCP up to 60 min as in (b) were further incubated with fresh culture medium without CCCP for the indicated periods of time (Washout).
Phosphorylation of Ser65 in the Parkin Ubl domain primes the mitochondrial translocation of Parkin
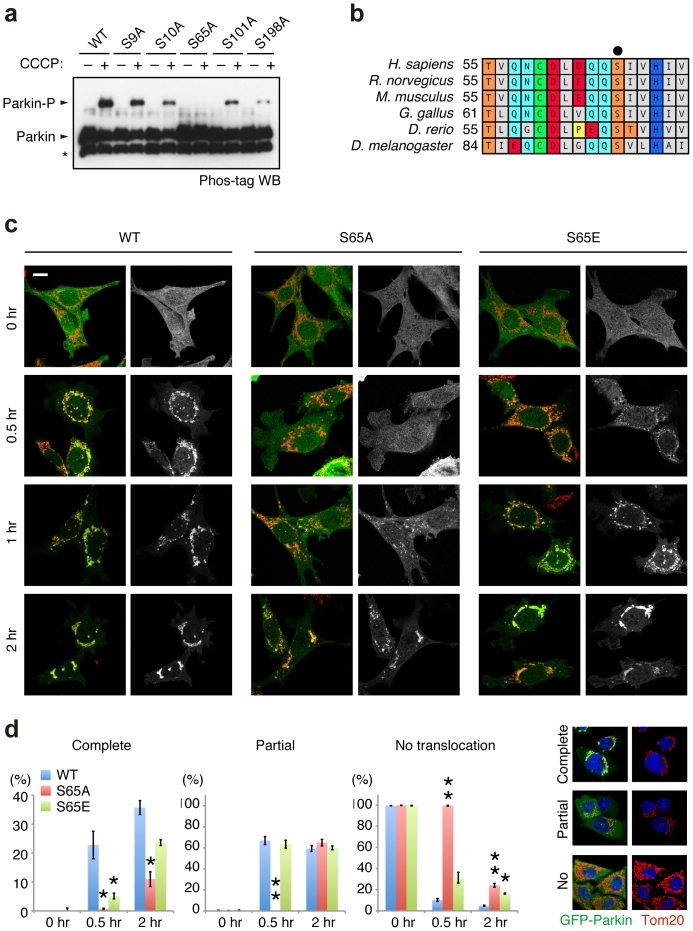
To determine which residue(s) of Parkin are phosphorylated, we immunopurified HA-tagged Parkin from PINK1-FLAG WT or KD/HA-Parkin/PINK1−/− MEFs treated with or without CCCP and performed mass spectrometric analysis for phospho-peptides (Supplementary Fig. S3). Although Phos-tag Western blotting of Parkin mainly detected a single band shift, which represents a single phospho-modification, the mass spectrometric analysis identified Ser9 or Ser10 and Ser65, Ser101 and Ser198 as phosphorylated residues of Parkin. Among these residues, only Ser65 phosphorylation increased (33-fold) in CCCP-treated PINK1-FLAG WT/HA-Parkin/PINK1−/− MEFs (Supplementary Fig. S3). Phos-tag Western blotting with mutant forms of Parkin, in which the identified phospho-serine residues are replaced with alanine, revealed that the band shift represents Ser65 phosphorylation (Fig. 2a). An in vitro kinase assay with recombinant insect PINK1, which has marked kinase activity28, strongly suggested that PINK1 directly phosphorylates Parkin at Ser65 (Supplementary Fig. S4). The Ser65 residue lies in the Ubl domain and is highly conserved from human to Drosophila (Fig. 2b). We next examined whether phosphorylation of Ser65 is required for Parkin-mediated mitophagy. GFP-tagged Parkin WT, which was localised both in the cytoplasm and in the nuclei of mock (DMSO)-treated cells (0 hr, Fig. 2c and d), was translocated to the mitochondria and induced the perinuclear aggregation of mitochondria 2 hr after CCCP treatment, as previously reported (2 hr, Fig. 2c and d)17,23. Replacement of Ser65 with alanine (S65A) did not affect the subcellular localisation of Parkin in mock-treated cells when compared with that of GFP-Parkin WT (0 hr, Fig. 2c and d). However, GFP-Parkin S65A almost completely inhibited the mitochondrial translocation of Parkin and the perinuclear rearrangement of mitochondria 0.5 hr after CCCP treatment (0.5 hr, Fig. 2c and d) and showed delayed translocation in 2 hr (2 hr, Fig. 2c and d). The expression of a putative phospho-mimetic Parkin S65E also showed a subcellular localisation similar to that of GFP-Parkin WT in both DMSO- and CCCP-treated cells (Fig. 2c). However, GFP-Parkin S65E exhibited a mild translocation defect, suggesting that S65E does not fully mimic the phosphorylated Ser65 (Fig. 2d).
Figure 2. Ser65 in the Ubl domain of Parkin is phosphorylated upon depolarisation of ΔΨm.
(a) Phos-tag Western blotting detected phosphorylation of Ser65. HeLa cells were transiently transfected with Parkin WT and a series of alanine mutants for the candidate phospho-residues followed by treatment with or without 20 µM CCCP for 1 hr. Cell lysates were analysed by Phos-tag Western blotting. An asterisk indicates degraded Parkin. (b) Alignment of the amino acid sequences surrounding Ser65 (marked by a black dot) from a variety of animal species. The numbers on the left correspond to the residue numbers of Parkin proteins. (c) Introduction of the S65A mutation delayed Parkin translocation to the depolarised mitochondria in PINK1 WT/GFP-Parkin/PINK1−/− MEFs. Cells retrovirally introduced with GFP-Parkin WT or its phospho-mutants (S65A and S65E) were treated with or without 30 µM CCCP for the indicated periods of time. GFP-Parkin and mitochondria were visualised with anti-GFP (green) and anti-Tom20 (red), respectively. Parkin signals are also shown as monochrome images. Scale bar = 10 µm. (d) Mitochondrial translocation efficiency of Parkin mutants. PINK1 WT/PINK1−/− MEFs stably expressing GFP-Parkin WT, S65A or S65E were treated as in (c). Cells expressing GFP-Parkin perfectly overlapped (Complete, examples are shown on the right), partially overlapped (Partial) or non-overlapped (No) with the Tom20 signal were counted. The data represent means ± SE from three experiments (n = 99–143 cells in each). ** p < 0.01, * p < 0.05 vs. WT at each time point.
Parkin Ser65 phosphorylation is not sufficient for mitochondrial translocation upon depolarisation of ΔΨm
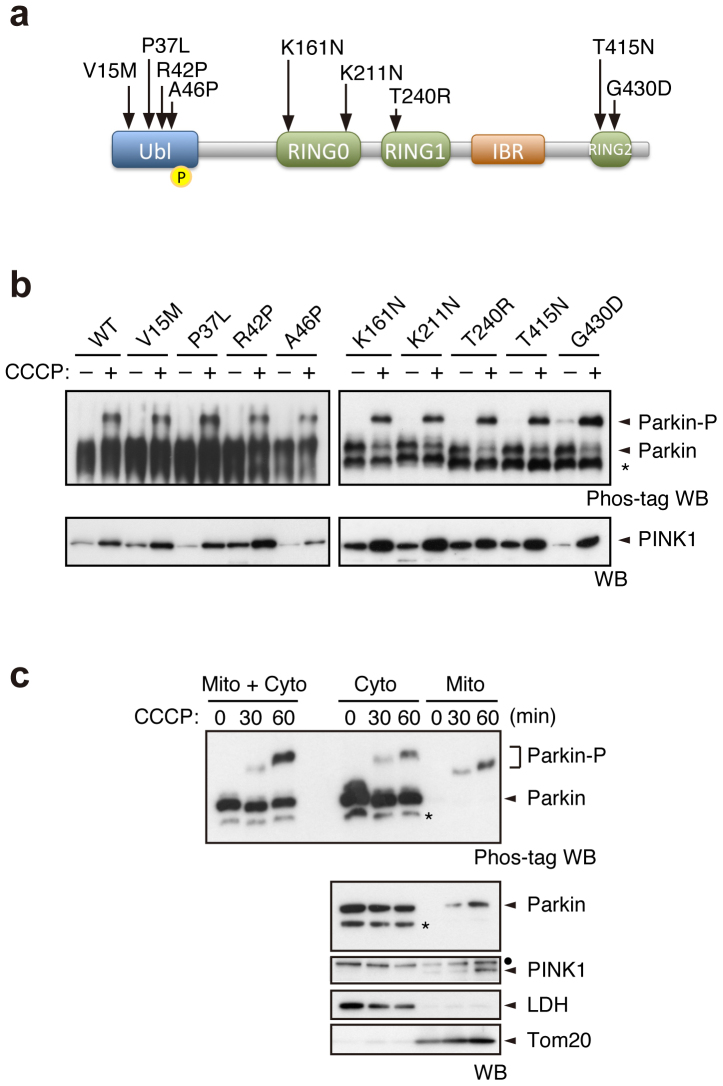
As PINK1-mediated Ser65 phosphorylation appeared to be required for efficient translocation of Parkin, we next examined whether well-characterised pathogenic Parkin mutants were subjected to phosphorylation upon CCCP treatment. In this experiment, we used three kinds of Parkin mutants based on the previous and current studies (Supplementary Fig. S5)17,22,23. The first group, V15M, P37L, R42P and A46P, had intact or weakly impaired mitochondrial translocation activity. The second group, T415N and G430D, had mildly impaired translocation activity. The third group, K161N, K221N and T240R, almost completely lacked translocation activity (Fig. 3a). Surprisingly, all of the mutants possessed comparable phosphorylation efficiencies to those of WT (Fig. 3b). This result suggests that Ser65 phosphorylation is not sufficient for the mitochondrial translocation of Parkin.
Figure 3. Pathogenic mutants of Parkin are subjected to Ser65 phosphorylation.
(a) Diagram of Parkin protein illustrating the pathogenic mutants used in this study. The Ser65 residue in the Ubl domain is shown as a yellow circle. RING, Ring-finger motif; IBR, in-between-Ring fingers domain. (b) Phos-tag Western blotting for Parkin and Western blotting for PINK1 were performed using Parkin WT and a series of pathogenic mutants as shown in Figure 2a. (c) Endogenous Parkin was also phosphorylated in SH-SY5Y cells after CCCP treatment. Post-nuclear cell lysates from SH-SY5Y cells treated with or without 10 µM CCCP for 30 and 60 min were fractionated into mitochondria-rich (Mito) and cytosolic (Cyto) fractions. These two fractions and their combination (Mito + Cyto) were subjected to Phos-tag or normal Western blotting analyses. Endogenous PINK1 was fractionated in the Mito fraction, as previously reported45. Lactate dehydrogenase (LDH) and Tom20 were used as cytosolic and mitochondrial marker proteins, respectively. Asterisks: putative cleaved Parkin; dots: non-specific bands.
Biochemical fractionation of endogenous Parkin from SH-SY5Y cells detected only the phosphorylated form of Parkin in the mitochondrial fraction upon CCCP treatment (Fig. 3c), which strongly suggests that phosphorylation of Parkin is required for mitochondrial translocation. There was a slight difference in the gel mobility of phosphorylated Parkin between the cytosolic and the mitochondrial fractions and between CCCP-treated periods of time. These differences very likely reflect differences in the complexity of the contents of each fraction rather than in the phosphorylation status of Parkin because a single shifted band appears in the mixed fractions (Mito + Cyto in Fig. 3c; CCCP 30 min + 60 min in Supplementary Fig. S6).
Effect of Parkin Ser65 phosphorylation on the autophagic reaction
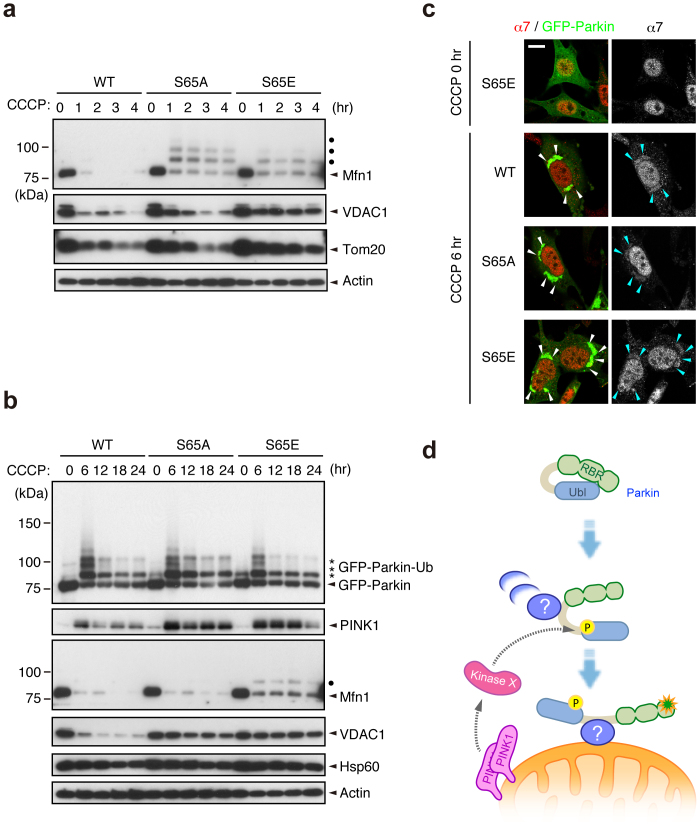
We next examined whether Ser65 phosphorylation is required for the subsequent autophagic reaction, in which various ubiquitin-proteasome- and autophagy-related proteins are involved, including the 26S proteasome, p97/VCP, p62/SQSTM1, LC3, ATG5 and ATG722,23,31,32,33,34,35. Parkin has been reported to be involved in the ubiquitin-proteasome-dependent degradation of a variety of mitochondrial outer membrane proteins, including Mitofusin1 (Mfn1)32, Mfn232, Miro136,37, Miro237, VDAC122 and Tom2031. Degradation of Mfn1, VDAC1 and Tom20 at the mitochondrial outer membrane was observed in PINK1 WT/GFP-Parkin/PINK1−/− MEFs 1 to 4 hr after CCCP treatment (Fig. 4a). While GFP-Parkin harbouring S65A or S65E mutations was also capable of inducing Mfn1, VDAC1 and Tom20 degradation, the efficiency was impaired, especially in Mfn1 and VDAC1 (Fig. 4a). Long-term time course analysis revealed that in cells expressing Parkin with S65A or S65E mutations, Mfn1 and VDAC1 cannot be degraded effectively, and the mitochondrial outer membrane was likely more intact as indicated by the sustained accumulation of PINK1 (Fig. 4b). The impaired degradation cannot be explained simply by the delayed translocation of Parkin mutants because both mutants completed the mitochondrial translocation by the 6 hr time-point (data not shown and see Fig. 4c). In contrast, the profiles of Parkin expression and autoubiquitination in Parkin S65A- or S65E-expressing cells were comparable with those of WT (Fig. 4b). We also examined whether Ser65 mutations affect the accumulation of proteasome (Fig. 4c) and p62 (Supplementary Fig. S7) at the mitochondria during mitophagy via the immunostaining of the proteasome subunit alpha type 7 (α7) and p62. However, there was no evidence that Ser65 mutations inhibit or delay the recruitment of proteasome and p62 to the mitochondria. Finally, we tested whether the Parkin Ubl domain itself is indispensable for the mitochondrial translocation and the substrate degradation (Supplementary Fig. S8). Interestingly, Parkin mutant lacking the Ubl domain (ΔUbl) showed a mild delay in the mitochondrial translocation, slowed the mitochondrial reorganization to the perinuclear region (Supplementary Fig. S8b and c) and impaired the degradation of mitochondrial outer membrane proteins (Supplementary Fig. S8d). These results suggest that proper regulation of the Parkin Ubl domain through the Ser65 phosphorylation is required not only for efficient translocation to mitochondria as an initial step of mitophagy, but also for the degradation of mitochondrial outer membrane proteins during mitophagy through an as yet unknown mechanism.
Figure 4. Ser65 phosphorylation affects the subsequent autophagy reaction.
(a) CCCP-dependent degradation of mitochondrial outer membrane proteins in PINK1 WT/PINK1−/− MEFs expressing WT or mutant forms of GFP-Parkin. Mfn1, VDAC1 and Tom20 were used as markers of mitochondrial outer membrane proteins. Actin: a loading control. Dots: ubiquitinated Mfn1. (b) Long-term time-course analysis of CCCP-dependent mitochondrial protein degradation. The degradation of outer membrane proteins was impaired in cells expressing GFP-Parkin S65A or S65E mutations. Hsp60 was used as a marker of mitochondrial matrix proteins. (c) S65A and S65E mutations do not affect proteasome recruitment to the mitochondria during mitophagy. PINK1 WT/PINK1−/− MEFs expressing WT or mutant forms of GFP-Parkin (green) were treated with 30 µM CCCP for 3 or 6 hr. Cells were stained with anti-proteasome subunit alpha type 7 (α7, red). α7-immunoreactivity was enriched in the nuclei of all three cell genotypes under normal conditions, as displayed in the representative image of S65E (CCCP 0 hr), and overlapped with the aggregated mitochondria (arrowheads) 6 hr after CCCP treatment irrespective of genotype. Similar results were obtained 3 hr after CCCP treatment. Scale bar = 10 µm. (d) Model for Parkin translocation and activation. The Parkin Ubl domain masks C-terminal RING-IBR-RING (RBR) domains for E3 activity46. A Parkin phosphorylation event at Ser65 (P), combined with unknown factor(s) (?), stimulates the mitochondrial translocation of Parkin, releasing the RBR domains from autoinhibition by the Ubl domain.
Discussion
A series of Drosophila genetic and cell biological studies have clearly demonstrated that PINK1 is required for Parkin-mediated mitochondrial maintenance. The mitophagy of damaged mitochondria is a well-characterised event in which PINK1 and Parkin are involved. However, how PINK1 regulates Parkin is largely unclear. This study has shown that Ser65 in the Ubl domain of endogenous Parkin is phosphorylated in an activated PINK1-dependent manner. In addition to mitochondrial accumulation of PINK1, ΔΨm depolarisation-dependent PINK1 autophosphorylation has been reported to be an important element for PINK1 activation and Parkin recruitment19,29. Consistent with these observations, our investigation of PINK1 siRNA suggests that a lower level of PINK1 is able to phosphorylate Parkin afterΔΨm depolarisation (Fig. 1c, compare lanes 1 and 4). Our domain analysis of PINK1 demonstrates that intact PINK1 is required for CCCP-dependent Parkin phosphorylation, and the lack of phosphorylation in fibroblasts from a PARK6 patient implies relevance to the pathogenesis of PD.
The biological significance of this phosphorylation event is suggested by the fact that replacement of Ser65 with alanine or glutamic acid impairs the mitochondrial translocation of Parkin and/or the subsequent mitophagy process. Our observation that maximal phosphorylation of Parkin occurs within 1 hr of CCCP treatment supports the idea that Ser65 phosphorylation is required for the early step of Parkin translocation. In contrast, PINK1 accumulation appears to last at least 6 hr (Fig. 4c and Supplementary Fig. S1b). The difference in time course between PINK1 accumulation and Parkin phosphorylation could be explained by the observation that phosphorylated Parkin is degraded by proteasomal activity. The biochemical evidence that only the phosphorylated form of endogenous Parkin is present in the mitochondrial fraction also implies that Parkin phosphorylation is an essential event for its mitochondrial translocation and subsequent activation (Fig. 3c and Supplementary Fig. S6). Overexpression of PINK1 and Parkin itself leads to mitochondrial translocation of Parkin independently of ΔΨm depolarization, which suggests that excessive amounts of PINK1 and Parkin do not faithfully reflect endogenous reactions. Our study using PINK1−/− MEFs stably co-expressing PINK1 and GFP-Parkin might also be saddled with such a problem. We believe that the endogenous observation in which phosphorylated Parkin is accumulated in mitochondria is a more reliable proposal as a molecular model. The delay of exogenous GFP-Parkin S65A in the mitochondrial translocation would indicate that modification of Ser65 is important for Parkin translocation at least. At the same time, another important finding is that pathogenic mutants that lose their translocation activity are also phosphorylated (Fig. 3b), raising the possibility that phosphorylation of Parkin at Ser65 is insufficient for translocation. Thus, Ser65 phosphorylation likely leads to other events in mitochondrial translocation, such as the association or dissociation of protein(s) involved in the mitochondrial translocation of Parkin or the modification of Parkin itself for activation at a different site(s).
Both the S65A and S65E Parkin mutants cannot undergo efficient mitophagy, as indicated by the incomplete degradation of the mitochondrial outer membrane proteins. Because inhibition of the degradation of the mitochondrial outer membrane proteins by proteasome inhibitors is reported to block mitophagy32,35, it may be that the modification of Parkin Ser65 has a greater than expected impact on the mitophagy process. Although our study does not demonstrate that the S65E mutant behaves exactly like the phosphorylated form of Parkin, the S65E mutant does translocate to the mitochondria in a similar way to WT, although with slightly impaired efficiency, suggesting that S65E has at least some properties that are similar to phosphorylated Parkin. Currently, it is unknown why S65E also inhibits the later processes of mitophagy. One possible explanation is that rapid degradation of phosphorylated Parkin is required for the proper progression of mitophagy, and S65E may not be degraded effectively. However, there is no evidence that S65E is more stable than WT, as shown in Figure 4c.
Very recently, Kondapalli et al. proposed a model to explain the biological significance of Ser65 phosphorylation, in which Ser65 phosphorylation relieves autoinhibition of Parkin E3 activity by the Ubl domain18. This model may explain the depolarised ΔΨm-dependent activation of Parkin. However, our data indicated that the Parkin S65A mutant is also autoubiquitinated (Fig. 4b) and that the ΔUbl mutant showed mild translocation defect and impaired substrate degradation (Supplementary Fig. S8). Moreover, if this is the case, the E3 activity of Parkin pathogenic mutants lacking mitochondrial translocational activity but harbouring intact E3 activity in vitro (such as K161N and K211N, which are subjected to the Ser65 phosphorylation) should be activated in the cytosol38. However, our previous data indicate that K161N and K211N are not activated by CCCP treatment23. Thus, it is conceivable that another step is required for depolarised ΔΨm-dependent activation of Parkin E3. In addition, the Ubl domain might not only autoinhibit its E3 activity but also contribute to the mitochondrial translocation and the substrate degradation through an as yet unknown mechanism. We believe that an appropriate way to estimate Parkin E3 activity in the context of mitophagy is to evaluate the ubiquitination and degradation of substrates in cells with depolarised ΔΨm. Mfn1 is a well-characterised direct substrate of Parkin32, and Parkin-dependent poly-ubiquitination modification of Mfn1 can be detected by Western blotting upon ΔΨm depolarisation32,39,40. Parkin S65A and S65E appear to ubiquitinate Mfn1, as poly-ubiquitinated forms of Mfn1 were observed (Fig. 4b). However, they cannot degrade it effectively, which suggests that the process of substrate degradation is also impaired in these mutants.
Kondapalli et al. have also shown that T. castaneum PINK1 (TcPINK1) directly phosphorylates human Parkin at Ser6518. We confirmed their finding using recombinant TcPINK1 produced from the same construct (Supplementary Fig. S4). The replacement of MBP-Parkin Ser65 with alanine completely abolished PINK1-mediated phosphorylation, indicating that Ser65 is the sole phosphorylation site in vitro. However, experiments in cultured cells showed that the replacement of Ser9, Ser10, Ser101 and Ser198 with alanine affects the Ser65 phosphorylation efficiency (Ser9, ~35% reduction; Ser10, ~76% reduction; Ser101, ~65% reduction; Ser198, ~92% reduction) (Fig. 2a). These residues might be priming phosphorylation sites for Ser65 phosphorylation.
Because PINK1 is believed to be activated in the mitochondria, a topological inconsistency arises from our cell-based data that cytosolic Parkin lacking the mitochondrial translocation activity is phosphorylated. Therefore, it is possible that PINK1 indirectly regulates Parkin phosphorylation. One possible explanation for this is the presence of another cytosolic kinase(s) regulated by PINK1 (Fig. 4d). Alternatively, because mitochondria are a dynamic organelle, cytosolic Parkin adjacent to the moving and fragmented mitochondria with depolarised ΔΨm might be phosphorylated incidentally. The issue as to whether or not PINK1 directly phosphorylates Parkin in cells remains to be solved.
In conclusion, this study has suggested that PINK1-dependent Parkin phosphorylation at Ser65 accelerates the mitochondrial translocation of Parkin and showed that the introduction of mutations at this site also affects subsequent mitophagy processes. Concurrently, our data provide the possibility that there is an elaborate multi-step mechanism for the mitochondrial translocation of Parkin upon the loss of ΔΨm (Fig. 4d), the clarification of which awaits further study.
Methods
Antibodies, plasmids and cell lines
Antibodies used in Western blot analysis were as follows: anti-Parkin (1 : 1,000 and 1 : 5,000 dilution for endogenous and exogenous Parkin, respectively; Cell Signaling Technology, clone PRK8), anti-PINK1 (1 : 1,000 dilution; Novus, BC100-494 or 1 : 1,000 dilution; Cell Signaling Technology, clone D8G3), anti-Mfn1 (1 : 1,000 dilution; Abnova, clone 3C9), anti-VDAC1 (1 : 1,000 dilution; Abcam, Ab15895), anti-Tom20 (1 : 500 dilution; Santa Cruz Biotechnology, FL-145), anti-FLAG-HRP (1 : 2,000 dilution; Sigma-Aldrich, clone M2), anti-GFP (1 : 5,000 dilution; Abcam, ab290), anti-Actin (1 : 10,000 dilution; Millipore, MAb1501), anti-LDH (1 : 1,000 dilution; Abcam, ab7639-1), anti-phospho-GSK3ß (1 : 1,000 dilution; Cell Signaling Technology, clone 5B3), anti-GSK3ß (1 : 1,000 dilution; Cell Signaling Technology, clone 27C10), and anti-Hsp60 (1 : 10,000 dilution; BD Biosciences, clone 24/Hsp60). Antibodies used in immunocytochemistry were as follows: FITC-conjugated anti-GFP (1 : 1,000 dilution; Abcam, ab6662), anti-Tom20 (1 : 1,000 dilution; Santa Cruz Biotechnology, FL-145), anti-Myc (1 : 500 dilution; Millipore, clone 4A6), anti-p62 (1: 500 dilution; Progen Biotechnik, GP62-C), anti-Parkin (1 : 1,000 dilution; Cell Signaling Technology, clone PRK8) and anti-proteasome α7 (1 : 250; a kind gift of Dr S. Murata at the University of Tokyo). cDNAs for human Parkin, PINK1 and its pathogenic and engineered mutants are as described in previous studies23,41. Parkin phospho-mutants were generated by PCR-based mutagenesis followed by sequencing confirmation of the entire gene. PINK1−/− MEFs, cultured as previously described23, were retrovirally transfected with pMXs-puro harbouring non-tagged PINK1, PINK1-FLAG, non-tagged Parkin, HA-Parkin, GFP-Parkin and related cDNA; transfected cells were then selected with 1 µg/ml puromycin. HeLa cells maintained at 37°C in a 5% CO2 atmosphere in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FCS and 1x non-essential amino acids (GIBCO) were retrovirally transfected with pMXs-puro harbouring non-tagged Parkin along with pcDNA3Hyg-mSlc7a1-VSVG and pcDNA3Hyg-mSlc7a1-FLAG (a kind gift of Dr N. Fujita at UCSD). Stable cell lines were selected with 1 µg/ml puromycin and cloned. Transient transfections of cultured cells were performed using Lipofectamine 2000 (Invitrogen) for plasmids and Lipofectamine RNAiMAX (Invitrogen) for stealth siRNA duplexes (Invitrogen), which were used according to the manufacturer's instructions.
Tissue culture
Skin biopsies were obtained from a PARK6 case and a control without mutations in any known PD genes. The study was approved by the ethics committee of Juntendo University, and all participants gave written, informed consent. Dermal primary fibroblasts established from biopsies were cultured in high glucose DMEM supplemented with 10% foetal bovine serum, 1x non-essential amino acids, 1 mM sodium pyruvate (GIBCO), 100 µM 2-mercaptoethanol, and 1% penicillin–streptomycin at 37°C in a 5% CO2 atmosphere.
Mapping of Parkin phosphorylation sites
PINK1−/− MEFs (6.0 x 107) expressing HA-Parkin and PINK1-FLAG were treated with or without 30 µM CCCP for 30 min. HA-Parkin (~500 ng in each) immunopurified with anti-HA-conjugated agarose beads was eluted with 8 M urea buffered with 50 mM Tris-HCl at pH 9.0. Samples from two independent experiments were digested with trypsin or chymotrypsin and analysed by nano-scale liquid chromatography-tandem mass spectrometry (Dionex Ultimate3000 RSLCnano and ABSciex TripleTOF 5600) followed by MASCOT searching and Mass Navigator/PhosPepAnalyzer processing for identification and label-free quantitation, respectively42. Determination of phosphosite localisation was performed based on the presence of site-determining ions43.
Phosphorylation assay and mitochondrial fractionation
PINK1−/− MEFs harbouring HA-Parkin along with wild-type or a kinase-dead form of PINK1-FLAG were metabolically labelled with 175 µCi/ml of [32P] orthophosphate in phosphate-free DMEM (GIBCO) with 10% FBS at 37°C for 3 hr. The medium was then replaced with fresh DMEM containing 10% FBS. Cells were treated with CCCP for 1.5 hr and were lysed on ice with lysis buffer containing 0.2% NP-40, 50 mM Tris (pH 7.4), 150 mM NaCl and 10% glycerol supplemented with protease inhibitor (Roche Diagnostics) and phosphatase inhibitor (Pierce) cocktails, and HA-Parkin and PINK1-FLAG were immunoprecipitated with anti-HA (Wako Pure Chemical, clone 4B2)- or anti-FLAG (Sigma-Aldrich, clone M2)-conjugated agarose beads. Immunoprecipitates were separated by SDS-PAGE and transferred onto a PVDF membrane. Autoradiography and Western blotting were performed to visualise proteins. Phos-tag Western blotting was performed as previously described28. Briefly, phospho-Parkin and phospho-PINK1 were separated on 8% gels containing 50 µM Phos-tag. Mitochondrial and cytosolic fractionations were performed as previously described, with some modifications20. The cytosolic fractions were further clarified by a second centrifugation at 105,000 g for 60 min to remove residual organelle membranes.
Immunocytochemical analysis
Cells plated on 3.5 mm glass-bottom dishes (MatTek) were fixed with 4% paraformaldehyde in PBS and permeabilised with 50 µg/ml digitonin for anti-Tom20 and anti-p62 staining or with 0.1% NP-40 for anti-α7 staining in PBS. Cells were stained with anti-Tom20 or anti-α7 antibodies in combination with FITC-conjugated anti-GFP antibody and were counterstained with DAPI for nuclei. Cells were imaged using laser-scanning microscope systems (TCS-SP5, Leica or LSM510 META, Carl Zeiss).
Statistical analysis
A one-way repeated measures ANOVA was used to determine significant differences between multiple groups unless otherwise indicated. If a significant result was achieved (p < 0.05), the means of the control and the specific test group were analysed using the Tukey-Kramer test.
Author Contributions
K.S., Y. Imai and N.H. designed the research; K.S., Y. Imai, S.Y., T.K. and Y. Ishihama performed the experiments; S.S. contributed new reagents/analytic tools; K.S. and Y. Imai analysed the data; and Y. Imai and N.H. wrote the paper. K.S. and Y. Imai contributed equally to this work.
Supplementary Material
Supplementary Information
Acknowledgments
We thank: Drs K. Tanaka, N. Matsuda, K. Okatsu, T. Kitamura, S. Murata, N. Fujita, N. Furuya, M.M.K. Muqit and R.J. Youle for their generous supply of materials; T. Hasegawa and Y. Imaizumi for the preparation of human fibroblasts; and T. Imura for her technical help. This study was supported by the Naito Foundation, the Novartis Foundation, the Grant-in-Aid for Young Scientists (B) from MEXT in Japan (SK-F, YI), the CREST program of JST (NH) and Grant-in-Aid for Scientific Research on Innovative Areas (NH).
References
- Valente E. M. et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 (2004). [DOI] [PubMed] [Google Scholar]
- Takatori S., Ito G. & Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett 430, 13–17 (2008). [DOI] [PubMed] [Google Scholar]
- Beilina A. et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A 102, 5703–5708 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri L. et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet 14, 3477–3492 (2005). [DOI] [PubMed] [Google Scholar]
- Sim C. H. et al. C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum Mol Genet 15, 3251–3262 (2006). [DOI] [PubMed] [Google Scholar]
- Clark I. E. et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006). [DOI] [PubMed] [Google Scholar]
- Park J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006). [DOI] [PubMed] [Google Scholar]
- Yang Y. et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A 103, 10793–10798 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). [DOI] [PubMed] [Google Scholar]
- Imai Y., Soda M. & Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem 275, 35661–35664 (2000). [DOI] [PubMed] [Google Scholar]
- Shimura H. et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25, 302–305 (2000). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A 97, 13354–13359 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E. et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20, 867–879 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. M. et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191, 933–942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C., Lorenz H., Weihofen A., Selkoe D. J. & Lemberg M. K. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem 117, 856–867 (2011). [DOI] [PubMed] [Google Scholar]
- Whitworth A. J. et al. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis Model Mech 1, 168–174; discussion 173 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8, e1000298 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2, 120080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K. et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun 3, 1016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Jin S. M., Kane L. A. & Youle R. J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell 22, 320–333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C. et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107, 378–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12, 119–131 (2010). [DOI] [PubMed] [Google Scholar]
- Matsuda N. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189, 211–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri S. et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 584, 1073–1079 (2010). [DOI] [PubMed] [Google Scholar]
- Ziviani E., Tao R. N. & Whitworth A. J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A 107, 5018–5023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha D., Chin L. S. & Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet 19, 352–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun 377, 975–980 (2008). [DOI] [PubMed] [Google Scholar]
- Imai Y. et al. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet 6, e1001229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodroof H. I. et al. Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol 1, 110012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A. et al. Parkin phosphorylation and modulation of its E3 ubiquitin ligase activity. J Biol Chem 280, 3390–3399 (2005). [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D. F. & Youle R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183, 795–803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A. et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191, 1367–1380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K. et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15, 887–900 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Kane L. A., Hauser D. N., Fearnley I. M. & Youle R. J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. C. et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 20, 1726–1737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. PINK1 and Parkin Target Miro for Phosphorylation and Degradation to Arrest Mitochondrial Motility. Cell 147, 893–906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet 8, e1002537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N. et al. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem 281, 3204–3209 (2006). [DOI] [PubMed] [Google Scholar]
- Gegg M. E. et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet 19, 4861–4870 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A. et al. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One 6, e16746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K. et al. Parkin stabilizes PINK1 through direct interaction. Biochem Biophys Res Commun 383, 331–335 (2009). [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Sugiyama N., Tanaka N. & Ishihama Y. Human proteome analysis by using reversed phase monolithic silica capillary columns with enhanced sensitivity. J Chromatogr A 1228, 292–297 (2012). [DOI] [PubMed] [Google Scholar]
- Beausoleil S. A., Villen J., Gerber S. A., Rush J. & Gygi S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24, 1285–1292 (2006). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Clinicogenetic study of PINK1 mutations in autosomal recessive early-onset parkinsonism. Neurology 64, 1955–1957 (2005). [DOI] [PubMed] [Google Scholar]
- Zhou C. et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A 105, 12022–12027 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J 30, 2853–2867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information