Abstract
β-Carotene oxygenase 2 cleaves β-carotene asymmetrically at non-central double bonds of the polyene chain, yielding apocarotenal molecules. The hypothesis tested was that apocarotenoids are able to stimulate transcription by activating retinoic acid receptors (RARs). The effects of long- and short-chain apocarotenals and apocarotenoic acids on the activation of RARα and RARβ transfected into monkey kidney fibroblast cells (CV-1) were investigated. We synthesized or purified β-apo-8′-carotenoic acid (apo-8′-CA), β-apo-14′-carotenoic acid (apo-14′-CA), β-cyclocitral (BCL), β-cyclogernanic acid (BCA), β-ionone (BI), β-ionylideneacetaldehyde (BIA) β-ionylideneacetic acid (BIAA) and a C13 ketone, β-apo-13-carotenone (C13). None of the apocarotenoids tested showed significant transactivation activity for the RARs when compared with all-trans retinoic acid (RA). The results suggest that biological effects of these apocarotenoids are through mechanisms other than activation of RARα and β.
Keywords: β-carotene, retinoids, vitamin A, carotenoid metabolism, RARβ, RARα
Introduction
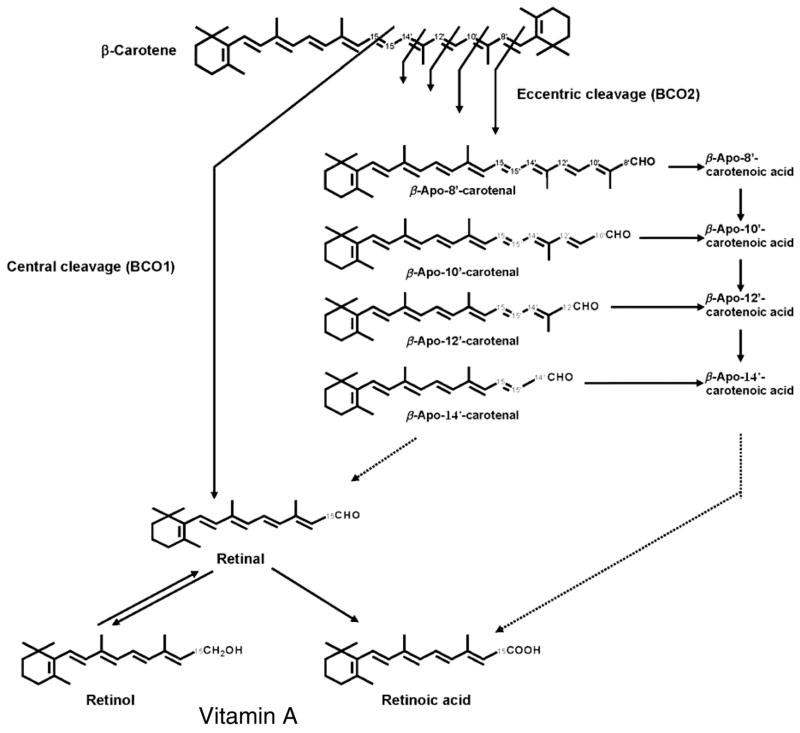
Bacteria, algae, fungi and plants have the ability to synthesize carotenoids; however, animals, including humans, do not. Of the more than 600 different carotenoids characterized from natural sources,1,2 only 60 are present in the human diet.3 The prominent carotenoids found in human plasma include β-carotene, α-carotene, lycopene, lutein, zeaxanthin and β-cryptoxanthin.3 β-Carotene can serve as an important dietary source of vitamin A. It can be cleaved either centrally or at non-central double bonds (eccentrically) (Figure 1). When cleaved centrally by β,β-carotene-15,15′-oxygenase [EC 1.13.11.31] (BCO1), β-carotene yields two molecules of retinal.4 In the eccentric cleavage pathway, β-carotene oxygenase 2 (BCO2), also called β,β-carotene-9′,10′-oxygenase, catalyzes the cleavage of β-carotene at non-central double bonds of the polyene chain yielding apocarotenals like β-apo-8′-, β-apo-10′- and β-apo-12′-carotenals, which may be further metabolized to the corresponding acids and alcohols.5 β-Carotene and some of its derivatives possess vitamin A activity.6 Besides their functions as a precursor of vitamin A and as antioxidants, carotenoids recently attracted research interest because of the possible ability of carotenoid metabolites to modulate retinoid receptors that are responsible for cell differentiation and some aspects of the immune response.7,8 While several studies have shown the possibility of apocarotenoids serving as signaling mediators in the cell, information remains limited. The biological roles and the potential regulation of retinoic acid receptors (RARs) by apocarotenoids are not clear. This research tested the hypothesis that apocarotenoids are able to stimulate transcription by activation of RARs.
Figure 1.
Metabolism of β-carotene. The products formed when cleaved centrally by β,β-carotene 15,15′-oxygenase (BCO1) and eccentrically by β-carotene 9′10′-oxygenase (BCO2).1 BCO1 catalyzes the main pathway of β-carotene metabolism, which cleaves the central 15,15′-double bond of β-carotene to retinal. Retinal as a vitamin A precursor is converted to retinol or further oxidized to retinoic acid in vivo. In comparison, BCO2 catalyzes the eccentric cleavage which cleaves at the double bonds other than the central 15, 15′-double bond of the polyene chain of β-carotene. β-Apocarotenals may be further oxidized to β-apocarotenoic acids18
Materials and methods
Retinoic acid and apocarotenoid preparation
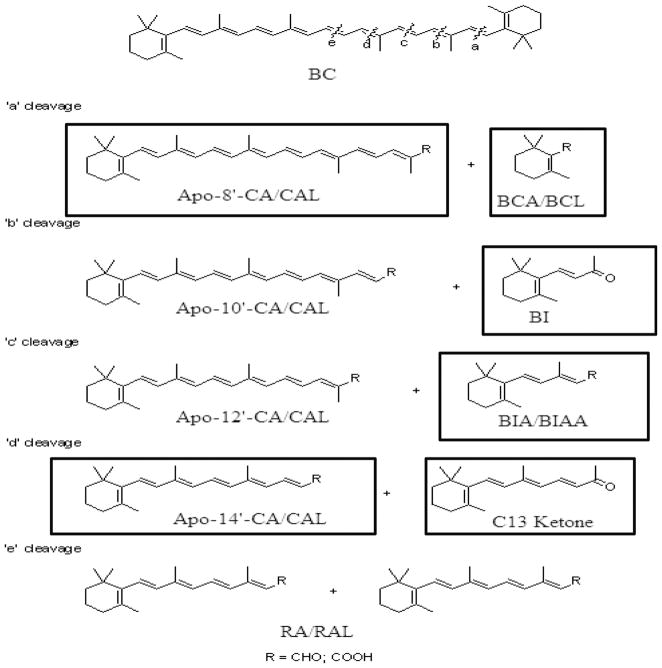
All-trans retinoic acid (RA) was obtained from Sigma (St Louis, MO, USA). Ethyl β-apo-8′-carotenoate was purchased from CaroteNature (Lupsingen, Switzerland). β-Apo-8′-CA was synthesized by saponification of the commercially obtained ethyl ester and purified by preparative thin-layer chromatography (TLC). The β-cyclocitral (BCL) and β-ionone (BI) used were purchased from Sigma-Aldrich (Milwaukee, WI, USA) and further purified by preparative TLC. β-Cyclogeranic acid (BCA) was prepared by air oxidation of BCL and crystallization of the product. β-ionylideneacetic acid (BIAA) was synthesized by Wadsworth-Emmons reaction (triethylphosphonoacetate) with BI and saponification and chromatography. β-Ionylideneacetaldehyde (BIA) was prepared from BIAA by reduction of BIAA and chromatography. The C13 ketone was made by Wittig reaction (acetonyltriphenylphosphonium chloride) with BIA and chromatography. Finally, β-apo-14′-carotenoic acid (apo-14′-CA) was prepared by reaction of triethylphosphonoacetate with retinal (Sigma) and saponification and crystallization. Stock solutions of these compounds were prepared by dissolving in absolute ethanol and stored at −80°C, while serial working dilutions were kept at −20°C. The concentrations of the stock solutions were determined either gravimetrically or by spectrophotometry using known or determined molar extinction coefficients. Figure 2 depicts possible β-carotene metabolites that result from eccentric cleavage and boxed compounds were tested for their abilities to transactivate RARα and RARβ.
Figure 2.
Possible metabolites of β-carotene when eccentrically cleaved. Boxed compounds were tested to see if they could transactivate retinoic acid receptor α or β
Cell culture and DNA preparation
CV-1 cells (male African green monkey kidney fibroblast cell line, ATCC #CCL-70) were purchased from the American Type Cell Culture Collection (Manassas, VA, USA). Cells were kept at 37°C with 5% CO2 in a basic medium, Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS), 100 μg/mL sodium pyruvate, 100 μg/mL penicillin–streptomycin sulfate and 0.5 μg/mL Fungizone (Invitrogen). Plasmids carrying the ampicillin-resistance gene and a reporter construct with an RARE-driven firefly luciferase gene (pRARE-Luciferase), a construct with renilla luciferase driven by the thymidine kinase promoter (pRL-tk Renilla), control vector (pSG5), and expression vectors for RARα and RARβ (pSG5-RARα or β) were gifts from Dr Dianne Soprano, Temple University, School of Medicine (Philadelphia, PA, USA). Like firefly luciferase, renilla luciferase does not require post-translational modification to be activated. Therefore, the renilla luciferase plasmid was used in conjunction with a reporter vector containing the firefly luciferase gene as the transfection normalizer. The constitutive promoter for renilla luciferase was the herpes simplex virus thymidine kinase (HSV-TK). The promoter for the firefly luciferase, a DR5 retinoic acid response element (DR5 RARE), served as the target of the RAR protein produced from the RAR expression plasmids after DNA transfection.
Glycerol stocks of Escherichia coli containing these plasmids were stored at −80°C. Bacteria were cultured on ampicillin containing LB agar plates or liquid media (50 μg/mL) at 37°C. DNA was purified using the Qiagen Qiafilter Midi kit (Qiagen) following the manufacturer’s instructions. DNAs were analyzed by agarose gel electrophoresis while DNA concentration was determined by measuring absorbance at 260 nm. Purity was determined by comparing the absorbance at 260 and 280 nm.
Cotransfection/transactivation assays
Transactivation assays were performed with CV-1 cells. Cells at 85–90% confluency in six-well plates were cotransfected with 2 μg/well pRARE-Luciferase, 0.025 μg/well pRL-tk Renilla, 5 μg/well pSG5-RAR and 10 μL/well Lipofectamine 2000™ (Invitrogen), a cationic lipid-based transfection reagent. Before transfection, medium was changed to serum-free DMEM and cells were transfected with plasmids. After four hours, medium was changed to DMEM containing 10% charcoal-stripped FBS, 100 μg/mL sodium pyruvate, 100 μg/mL penicillin–streptomycin sulfate and 0.5 μg/mL Fungizone. After 24 h, cells were treated with all-trans-RA and apocarotenoids at varying concentrations or with 0.25% ethanol as the control. In each experiment six wells were treated with ethanol alone and the results were averaged to determine the background (uninduced) luciferase expression. Each concentration of each compound was tested in triplicate wells. Multiple, independent experiments were conducted for retinoic acid, β-apo-8′-carotenoic acid and β-apo-14′-carotenoic acid.
Dual luciferase assay
Cells were harvested 50–52 h post-transfection. Luciferase activities (firefly and renilla) were measured sequentially using the Dual-Luciferase reporter (DLR) Assay System (Promega, Madison, WI, USA) following the manufacturer’s instructions. Briefly, cell monolayers were incubated in the presence of Passive Lysis buffer II for 25 min. Lysates were transferred to microcentrifuge tubes and centrifuged at 14,000 rpm at 4°C for 30 s. Twenty microliters of the supernatant was transferred to a 96-well microtiter plate for the assay. DLR assays were run on a luminometer (Promega). The substrate, Luciferase Assay Reagent II (LAR II), for firefly luciferase was autoinjected into each well containing lysate. Firefly luciferase activity was immediately measured by the luminometer, and the luminescence emission over 10 s was recorded. Stop & Glo™ Reagent (Promega) containing renilla luciferase substrate was then autoinjected into the same well, simultaneously quenching the firefly luciferase reaction and activating the renilla luciferase reaction. The renilla luciferase activity was quantified immediately over a 10-s period by the luminometer.
Data analysis
Each transfection was performed in triplicate and repeated multiple times. The firefly luciferase activity was normalized to renilla luciferase levels. Fold induction of gene expression was obtained by comparison with the ethanol control group.
Results
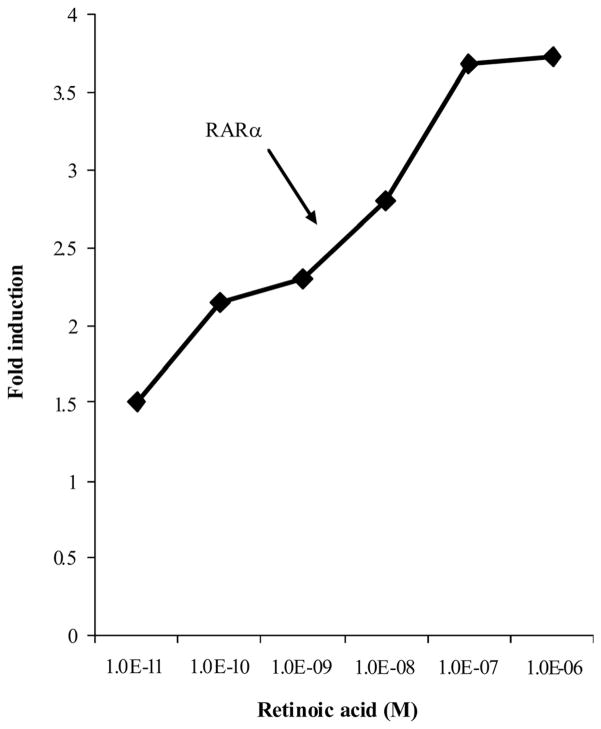
All-trans RA transactivated RAR and the promoter of the firefly luciferase reporter in a dose-dependent manner. Fold induction was taken by the comparison of corrected firefly luciferase activity in RA-treated wells at various concentrations with those in the ethanol-treated control wells. The results revealed that the fold induction for RARα increased with increasing all-trans RA concentration (Figure 3). The induction of firefly/renilla luciferase by all-trans RA compared with ethanol increased from one- to 3.5-fold for RARα with increasing concentrations of RA (10−11 to 10−5 mol/L). The maximum fold induction was achieved at 10 μmol/L all-trans RA, and the half maximal fold induction was achieved at 10−8 mol/L, which is the physiological concentration of RA in human plasma. Similar results were obtained in experiments using RARβ (data not shown).
Figure 3.
The transactivation of RARE-luciferase receptor in monkey kidney fibroblast cells transfected with retinoic acid receptor (RAR)α treated with varying concentrations of retinoic acid
Transactivation of RARα and RARβ
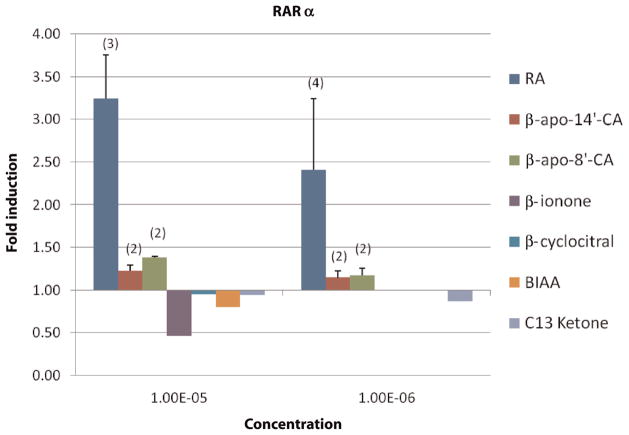
Cells transfected with RARα receptor vector were treated with RA (10−10–10−5 mol/L), β-apo-8′-carotenoic acid (apo-8′-CA), apo-14′-CA and C13 ketone (1 and 10 μmol/L), BI, BCL and BIAA (10 μmol/L) for 22–24 h. Luciferase activities were measured by the DLR system mentioned above and were expressed as fold induction in treated cells. Data are shown in Figure 4. Generally, the transactivation activity of all short- or long-chain apocarotenoic acids was negligible in comparison to all-trans RA, although there was variation among the compounds. The fold inductions by apo-8′-CA and apo-14′-CA were positively correlated with the compound’s concentration, suggesting that apo-8′-CA and apo-14′-CA may be weak RARα ligands. On average, the fold induction by apo-14′-CA was lower than apo-8′-CA, which is consistent with the results that more apo-14′-CA than apo-8′-CA was needed to compete effectively with all-trans RA for binding RARα.9
Figure 4.
Retinoic acid receptor (RAR)α transactivation response to retinoic acid (RA) and apocarotenoids in monkey kidney fibroblasts. Cells transfected with RARα receptor vector were treated for 22–24 h with an apocarotenoid concentration of 10−5 mol/L; the bars from left to right indicate the effects on RARα of RA, apo-14′-CA, apo-8′-CA, β-ionone, β-cyclocitral, β-ionylideneacetic acid and C13 ketone, respectively. At concentrations of 10−6 mol/L, the effects on RARα of RA, apo-8′-CA, apo-14′-CA and C13 ketone were shown by the bars from left to right, respectively. Luciferase activities were measured by dual-luciferase reporter system and were expressed as fold induction in treated cells (see Materials and methods). Data are expressed as the mean ± SD or range from multiple experiments as indicated (n) with three independent observations per experiment. If no ‘n’ is indicated the experiment was conducted once in triplicate
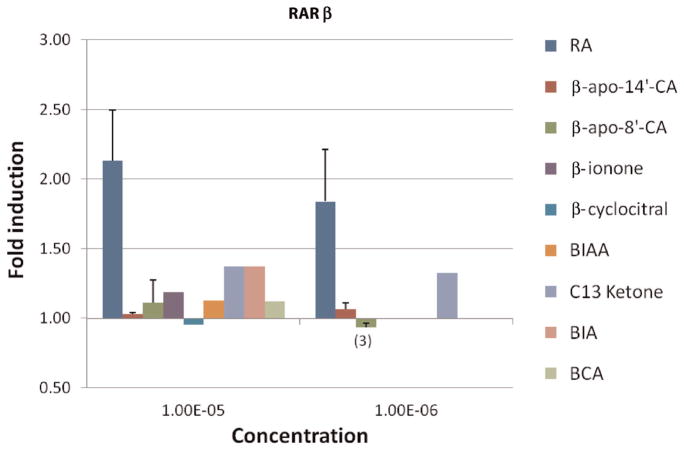
To test the transactivation activity of β-carotene metabolites from eccentric cleavage on RARβ, CV-1 cells were incubated with the following compounds: apo-8′-CA, apo-14′ CA, BI, BIA, BIAA, BCA, BCL and C13 ketone. All-trans RA served as a positive control for all compounds in testing their transactivation of RARβ. Cells transfected with the RARβ receptor vector were treated with 10−10–10−5 mol/L RA, 1 μmol/L and 10 μmol/L apo-8′-CA, apo-14′-CA and C13 ketone and 10 μmol/L BI, BCL and BIAA for 22–24 h in multiple experiments. Luciferase activities are reported as above and data are shown in Figure 5. The fold induction by β-apo-8′-CA and β-apo-14′-CA compared with all-trans RA was negligible indicating no transactivation of RARβ. Short-chain apocarotenoids, BI, BCL, BCA and BIAA did not transactivate RARβ at 10 μmol/L. RARβ was weakly activated by 1 and 10 μmol/L C13 ketone as well as 10 μmol/L BIA.
Figure 5.
Response of retinoic acid receptor (RAR)β transactivation by retinoic acid (RA) and β-carotene metabolites in monkey kidney fibroblast. Cells transfected with RARβ receptor vector were treated for 22–24 h with a concentration of 10−5 mol/L, the bars from left to right indicate the effects on RARβ of RA, apo-14′-CA, apo-8′-CA, β-ionone, β-cyclocitral, β-ionylideneacetic acid, C13 ketone, β-ionylideneacetaldehyde and β-cyclogeranic acid, respectively. At concentrations of 10−6 mol/L, the effects on RARβ of RA, apo-8′-CA, apo-14′-CA and C13 ketone were shown by the bars from left to right, respectively. Luciferase activities were measured by dual-luciferase reporter system and were expressed as fold induction in treated cells (see Materials and methods). Data are expressed as the mean ± SD or range from multiple experiments as indicated (n) with three independent observations per experiment. If no ‘n’ is indicated the experiment was conducted once in triplicate
Discussion
There are two pathways by which β-carotene can be cleaved at double bonds: central and eccentric. The major pathway, the central cleavage, has been more extensively studied. In comparison, the eccentric cleavage pathway has been the subject of debate. In 2001, a mouse enzyme was identified and characterized that cleaved β-carotene at the 9′,10′-double bond, producing β-apo-10′-carotenal and β-ionone.6 Recent research identified β-apo-8′-carotenal in human plasma after β-carotene administration.10 We have recently detected β-apo-8′-, 10′-, 12′-, and 14′-carotenals in human plasma at nanomolar concentrations (unpublished observations). These in vivo and in vitro experiments demonstrate the existence of apocarotenoids in the plasma and tissue and support the existence of the eccentric cleavage pathway of β-carotene metabolism.
There is evidence suggesting apocarotenoid compounds resulting from asymmetric cleavage of β-carotene might have effects on nuclear receptors. β-Apo-8′-carotenal induced DNA damage and the expression of cytochrome P450 (CYP) 1A2 in rats.11 Cleavage at the C9′, C10′ double bond of β-Carotene produces β-apo-10′-carotenal and β-ionone. β-Ionone was found to inhibit cell proliferation and induce apoptosis in human and murine gastric cancer cell lines, as well as in the mammary gland of rats.12,13 The exact pathway for β-ionone-mediated effects on cell proliferation and apoptosis has not been established and no study has related such cellular responses to the activation of RARs. BIA and BIAA, the short-chain products resulting from the cleavage of the C11′, C12′ double bond of β-carotene, reduced rhodopsin regeneration from their apo-proteins14 and inhibited the development of bean embryonic axes,15 respectively. Cleavage of β-carotene at the C13′, C14′ double bond produces β-apo-14′-carotenal and C13 ketone. β-apo-14′-carotenoids have been shown to inhibit RXRα, PPARα, γ and δ, LXRα and LXRβ activation.8
This research tested both short-chain apocarotenoids (BI, BCL, BIA, BIAA, C13 ketone and BCA) and long-chain apocarotenoids (β-apo-14-CA′ and β-apo-8-CA) that were readily available or that we were able to synthesize. β-Apo-10′- and β-apo-12′-carotenoids need to be tested for potential biological functions in future experiments.
Although several studies have examined apocarotenoid biological activity, information remains limited with respect to the basic mechanisms of their biological function. Whether they bind to nuclear receptors or are further metabolized to retinoic acid has not been clarified. We approached this research with the idea of investigating potential biological functions the apocarotenoids could serve within the cell. The possible regulation of RAR by apocarotenoids has not been previously described. Since apocarotenoids with certain chain lengths share some structural similarities with retinoic acid, we hypothesized that these eccentric cleavage products are able to activate RAR.
We investigated the effect of both long- and short-chain apocarotenals or apocarotenoic acids and all-trans RA on the activation of RARα and β transfected into monkey kidney fibroblast cells (CV-1 cells). The transactivation between RAR and the RARE promoter of firefly luciferase DNA led to the transcription of firefly luciferase DNA and eventually firefly luciferase protein production. The constant ratio among Renilla luciferase, firefly luciferase and RAR DNA cotransfected into cells at the same time made sure that the transfection efficiency and cell density were well controlled. Ethanol and all-trans retinoic acid acted as negative and positive controls, respectively, for the synthetic apocarotenals and apocarotenoic acids. The dual luciferase system represented a useful model for testing the transactivation activity of apocarotenoids on RARs. This kind of assay using CV-1 cells was first developed by Chambon and colleagues16 to assess the potency of retinoids in RAR-mediated transcriptional activation. CV-1 cells have low endogenous expression of RARS and have been widely used as a model transfection system to study transcriptional activation by RARs (see reference17 and references therein).
Our study showed that long-chain apocarotenoic acids did not transactivate RARα and β. In comparison, previous studies have demonstrated that they had weak biological activity on cell growth inhibition and cell differentiation. β-Apo-14′-CA was able to stimulate the differentiation of human acute promyelocytic leukemia cells and inhibit the growth of normal human bronchial epithelial cells and breast cancer cells. β-Apo-12′-CA inhibited the growth of HL-60 cells.18 In addition, previous studies indicated that long-chain apocarotenoic acids were not able to activate RARs. Tibaduiza et al.19 showed that long-chain β-apocarotenoic acids (8′, 10′, 12′ and 14′) were not able to effectively compete with all-trans RA binding with RARs. Therefore, long-chain apocarotenoic acids possibly carry out their biological activity independent of RAR transcriptional activation. In fact, long-chain apocarotenoids were demonstrated to activate other transcription systems, such as the AP-1 transcription system and PPAR/RXR nuclear receptors.8 Future studies investigating the transactivation on AP-1, PPAR/RXR or other nuclear receptors might help reveal the mechanism of the biological activities of long-chain apocarotenoic acids.
Short-chain apocarotenoids were also not able to transactivate RARα and β. Some short-chain apocarotenoids have biological activities, such as inhibiting cell growth and proliferation of cancer cells, inducing apoptosis in mammary gland of rat and reducing rhodopsin regeneration.14 Our experiments on the transactivation activity of β-carotene derivatives suggest that the biological effects of these apocarotenoids are through mechanisms other than transactivation of RARα and β. These might include the activation of other ligand-dependent transcription factors.
Acknowledgments
This work was supported by NIH grants R01HL049879 and R01DK044498.
Footnotes
Author contributions: RSM cloned and prepared the plasmid DNAs and wrote the draft of the manuscript using YY’s thesis as a template. YY conducted the transactivation experiments with the technical assistance of VMR. DH synthesized and purified the apocarotenoids under the direction of RWC. EHH designed the studies and prepared the final version of the paper.
References
- 1.Goodwin TW. Metabolism, nutrition and function of carotenoids. Annu Rev Nut. 1986;6:273–97. doi: 10.1146/annurev.nu.06.070186.001421. [DOI] [PubMed] [Google Scholar]
- 2.Packer L. The Antioxidant Miracle. The Controversial Carotenoids. New York: John Wiley & Sons, Inc; 2000. pp. 133–41. [Google Scholar]
- 3.Bieri JG, Brown ED, Smith JC. Determination of individual carotenoids in human plasma by high peformance liquid chromatography. J Liq Chromatogr. 1985;8:473–84. [Google Scholar]
- 4.Olson JA. The effect of bile and bile salts on the uptake and cleavage of β-carotene into retinol ester (vitamin A ester) by intestinal slices. J Lipid Res. 1964;5:402–8. [PubMed] [Google Scholar]
- 5.Sharma RV, Mathur SN, Ganguly J. Studies of the relative biopotenices and intestinal absorption of different apo-β-carotenoids in rats and chickens. Biochim Biophys Acta. 1976;158:377–83. doi: 10.1042/bj1580377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiefer C, Sumser E, Wernet MF, von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in drosophila. Proc Natl Acad Sci USA. 2002;99:10581–6. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prakash P, Liu C, Hu KQ, Krinsky IN, Russell RM, Wang XD. β-Carotene and β-apo-14′-carotenoic acid prevent the reduction of retinoic acid receptor β in benzol[a]pyrene-treated normal human bronchial epithelial cells. J Nutr. 2004;134:377–83. doi: 10.1093/jn/134.3.667. [DOI] [PubMed] [Google Scholar]
- 8.Ziouzenkova O, Orasanu G, Sukhova G, Lau E, Berger JP, Tang GW, Krinsky NI, Dolnikowki GG, Plutzky J. Asymmetric cleavage of β-carotene yields a transcriptional repressor of retinoid X and peroxisome proliferator-activated receptor responses. Mol Endocrin. 2007;21:77–88. doi: 10.1210/me.2006-0225. [DOI] [PubMed] [Google Scholar]
- 9.Nervi C, Grippo JF, Sherman MI, George MD, Jetten AM. Identification and characterization of nuclear retinoic acid-binding activity in human myeloblastic leukemia HL-60 cells. Proc Natl Acad Sci USA. 1989;86:5854–8. doi: 10.1073/pnas.86.15.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho CC, de Moura FF, Kim S-H, Clifford AJ. Excentral cleavage of β-carotene in vivo in a healthy man. Am J Clin Nutr. 2007;85:770–7. doi: 10.1093/ajcn/85.3.770. [DOI] [PubMed] [Google Scholar]
- 11.Yeh SL, Wu SH. Effects of quercetin on β-apo-8′-carotenal-induced DNA damage and cytochrome P1A2 expression in A549 cells. Chem Biol Interact. 2006;163:199–206. doi: 10.1016/j.cbi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Liu JR, Chen BQ, Yang BF, Dong HW, Sun CH, Wang Q, Song G, Song YQ. Apoptosis of human gastric adenocarcinoma cells induced by β-ionone. World J Gastroenterol. 2004;10:348–51. doi: 10.3748/wjg.v10.i3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JR, Yang BF, Chen BQ, Yang YM, Dong HW, Song YQ. Inhibition of β-ionone on SGC-7901 cell proliferation and upregulation of metalloproteinases-1 and 2 expression. World J Gastroenterol. 2004;10:167–71. doi: 10.3748/wjg.v10.i2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towner P, Gaertner W, Walckhoff B, Oesterhelt D, Hopf H. Regeneration of rhodopsin and bacteriorhodopsin: the role of retinal analogues as inhibitors. Eur J Biochem. 1981;117:353–9. doi: 10.1111/j.1432-1033.1981.tb06345.x. [DOI] [PubMed] [Google Scholar]
- 15.Sondheimer E, Walton DC. Structure–activity correlations with compounds related to abscissic acid. Plant Physiol. 1970;45:244–8. doi: 10.1104/pp.45.3.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astrom A, Pettersson U, Krust A, Chambon P, Voorhees JJ. Retinoic acid and synthetic analogs differentially activate retinoic acid receptor dependent transcription. Biochem Biophys Res Commun. 1990;173:339–45. doi: 10.1016/s0006-291x(05)81062-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z-P, Hutcheson JM, Poynton HC, Gabriel JL, Soprano KJ, Soprano DR. Arginine of retinoic acid receptor β which coordinates with the carboxyl group of retinoic acid functions independent of the amino acid residues responsible for retinoic acid receptor subtype specificity. Arch Biochem Biophys. 2003;409:375–84. doi: 10.1016/s0003-9861(02)00638-0. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, Matsui M, Murayama A. Biological activity of (all-E)-β-apo-12′-carotenoic acid and the geometrical isomers on human acute promyelocytic leukemia cell line HL-60. J Nutr Sci Vitaminol (Tokyo) 1995;41:575–85. doi: 10.3177/jnsv.41.575. [DOI] [PubMed] [Google Scholar]
- 19.Tibaduiza EC, Fleet JC, Russell RM, Krinsky IN. Excentric cleavage products of β-carotene inhibit estrogen receptor positive and negative breast tumor cell growth in vitro and inhibit activator protein-1-mediated transcriptional activation. J Nutr. 2002;132:1368–75. doi: 10.1093/jn/132.6.1368. [DOI] [PubMed] [Google Scholar]