Abstract
Despite its wide existence, the adaptive role of aneuploidy (the abnormal state of having unequal number of different chromosomes) has been a subject of debate. Cellular aneuploidy has been associated with enhanced resistance to stress, whereas on the organismal level it is detrimental to multi-cellular species. Certain aneuploid karyotypes are deleterious for specific environments, but karyotype diversity in a population potentiates adaptive evolution. To reconcile these paradoxical observations, this review distinguishes the role of aneuploidy in cellular versus organismal evolution. Further, it proposes a population genetics perspective to examine the behavior of aneuploidy on a populational versus individual level. By altering the copy number of a significant portion of the genome, aneuploidy introduces large phenotypic leap that enables small cell populations to explore a wide phenotypic landscape, from which adaptive traits can be selected. The production of chromosome number variation can be further increased by stress- or mutation-induced chromosomal instability, fueling rapid cellular adaptation.
Introduction
An E. coli cell contains a single DNA molecule of 5 million base pairs. S. cerevisiae (budding yeast), a unicellular eukaryote, has a DNA content of 24 million base pairs in its diploid form. This number increases to 6 billion base pairs in humans, providing the genetic coding to support the unsurpassed functional complexity. One of the important evolutionary advances in eukaryotes seems to be segmentation of the genome into multiple DNA molecules, which form highly compact, organized structures called chromosomes. With the increasing amount of DNA, the workload of DNA segregation during cell division rises accordingly. Eukaryotes utilize intricate machinery, consisting of the mitotic spindle, and the kinetochore associated with each segregating chromosome, to ensure accurate transmission of genome information[1,2]. Along with this machinery a sensitive monitoring system, the spindle assembly checkpoint (SAC), has evolved to deal with errors and perturbations of spindle mechanics[3,4]. Even so, chromosome segregation remains a weak link in genome transmission: in a normal yeast population, aneuploidy represents a large fraction of genetic variations available (See below).
Error in genome transmission is usually harmful to the fitness of an individual cell or organism, but in a population with a large number of individuals, imperfect genome transmission produces genetic variants, which are essential for adaptive evolution under selection. The most commonly considered genetic variants during evolutionary processes are point mutations. Chromosome segregation also produces genetic variants, not in single gene sequences but in the copy number of chromosomes, which contain hundreds of genes. This genetic variation is referred to as whole chromosome aneuploidy[5-8]. We note that “aneuploidy” in this review refers to whole chromosome number variation without the inclusion of segmental chromosome aneuploidy. By changing the dosage of many genes, aneuploidy leads to dramatic consequences. In human, aneuploidy is initially recognized in diseases, such as Down's syndrome[9] and cancer[10]. Aneuploidy's roles in these two cases are drastically different (see below). Moreover, there are many other cases, where aneuploidy is not a pathological but physiological state[11-13]. It is speculated that such physiological aneuploidy may have beneficial effects on cells or organism. In this essay, we propose a model to explain how aneuploidy can be an effective mode of adaptation during somatic evolution, where the population size is usually restricted. The rate of production of aneuploids is not always constant, and we discuss the findings that certain environmental stress could induce chromosome instability, leading to resistance to stress itself or increased evolvability to a broad spectrum of cytotoxic reagents.
Aneuploidy is a widespread genetic variation in nature
Early studies on aneuploidy related it solely to the disease state, which predicts that aneuploidy is unlikely to be persistent in the population as a result of the detrimental effect on fitness. In recent years, the advent of genome technology, especially comparative genome hybridization and DNA sequencing, reveals a different picture (Table 1). In lab strains of S. cerevisiae, it was estimated that 8% of the strains from the genome-wide ORF knockout library are aneuploid[14]. In wild yeasts isolated from the environment such as oak tree soil, aneuploidy was also identified[15]. In industrial strains, the deviation of DNA content from euploidy is a common feature documented decades ago[16]. High-resolution genomic analysis techniques such as array-based comparative genomic hybridization (aCGH) and next-generation sequencing has revealed the detailed genome structure and copy number variation, which include whole chromosome aneuploidy, in strains used for diverse industrial applications, such as sake and beer brewing[15,17,18], wine fermentation[18], and sherry-wine aging[19]. In pathogenic yeast/fungi, aneuploidy is associated with drug resistance[5,20]. For example, more than 50% fluconazole-resistant Candida albicans isolates from patients were found to harbor either whole chromosome or segmental duplication of Chr 5[21]. Whole chromosome aneuploidy was also found in fluconazole-resistant strains of another pathogenic yeast, Cryptococcus neoformans[22].
Table 1.
Examples of aneuploidy associated with environmental stress
| Species | Conditions | Associated stress | Ref | |
|---|---|---|---|---|
| Saccharomyces cerevisiae | Lab | Gene deletion | various growth defect | 14, 36 |
| Evironmental stress | proteotoxic stress | 43 | ||
| Industrial | sake production (1/9)* | ethanol, high sugar (osmotic stress) | 15, 18, | |
| wine production (4/26)* | ethanol, acetaldehyde, high sugar | 15, 18, 19 | ||
| beer brewing (3/4)* | ethanol, high sugar | 17, 18 | ||
| Candida albicans | fluconazole resistance(21/42)* | membrane defect | 21 | |
| Cryptococcus neoformans | fluconazole resistance | membrane defect | 22 | |
| Leishemania | different species | 24 | ||
| Human | most cancer | growth restriction, immune attack | 28-30 | |
| liver | metabolic stress | 13 | ||
Frequency of aneupoidy reported in the parentheses (Aneuploids/Total tested)
Beyond yeast and fungi, aneuploidy has been documented in many other contexts. It is long thought that due to their erroneous transmission during meiosis, aneuploid karyotypes are unlikely to be maintained during long-term adaptation and speciation in natural history. However, an outlier exists. Leishmaniasis is a form of clinical pathology ranging from disfiguring cutaneous lesions to fatal visceral infection, caused by different Leishmania protozoan parasites associated with varied pathology features[23]. Interestingly, it was found that four different Leishmanias have little variation in DNA sequence, yet exhibit dramatic difference in chromosome copy numbers[24]. The aneuploid Leishmania can perform sexual reproduction nonetheless[25], but the mechanistic details have yet to be elucidated.
In multicellular organisms such as mammals, aneuploidy is present in both the germline and somatic tissues. Germline aneuploidy is rare and when present it causes severe developmental abnormalities. In humans, chromosome number variation in fertilized oocytes causes rare birth defects such as Down's syndrome (trisomy 21, incidence at 1 in 2,000 births), Edwards syndrome (trisomy 18, 1 in 6,000 births), and Patau syndrome (trisomy 13, less than 1 in 10,000 births). However, it is intriguing that several studies reported that aneuploidy is highly prevalent in the early blastomeres of developing human or mouse embryos[26,27], raising a question as to at what stage aneuploidy impairs developmental programs and how aneuploid cells are cleared during later development. On the other hand, aneuploidy in somatic cells is not rare at all. Aneuploidy is a hallmark of cancer, one of the leading causes of death. It is present in more than 70% tumors[28-30]. Evidence indicates that aneuploidy may drive the tumorigenesis through its adaptive effect in a cell population (see following). But somatic aneuploid is not limited to cancer cells – work in recent years revealed that several normal human tissues bear a surprisingly high-level diversity of karyotypes. For example, normal human liver contains 25%-50% aneuploids[13]. In the fetal brain, it was estimated that 30-35% neurons are aneuploidy[31,32]. Compared with animal organisms, plant such as Arabidopsis thaliana, tolerates germline aneuploidy well and can cause substantial phenotypic variations[33,34]. In summary, aneuploidy is observed from yeasts to human. With the increasing application of quantitative DNA technology, it is likely that further evidence to emerge from diverse contexts illustrating the wide existence of aneuploidy.
Why is aneuploidy, which defines an “abnormal” genome, widespread in nature?
1) “Abnormal” karyotypes can be beneficial under abnormal environment
The effect of aneuploidy on fitness is context specific[6]. Aneuploidy is thought to bring abnormality due to an imbalance in gene dosage. It is assumed that the “normal” functionality of molecular complexes or pathways made of more loosely interacting molecules relies on the correct stoichiometry of their protein components. When the normal stoichiometry is skewed, the functionality, such as efficiency, timing or specificity, of the system would be reduced or altered in someway. However, normalcy is relative and in the context of physiology it refers to the preferred state or the state of highest fitness under a given condition. Thus, a cell with a genome imbalanced (i.e. with suboptimal stoichiometry) for one condition, say the “normal” environment, may indeed have the altered functionality that gives rise to optimal fitness under an altered, for instance, stressful condition[6,35]. In many cases, the prevalence of aneuploidy as discussed above was found indeed in association with stress (Table 1). For example, the wine brewing/aging process imposes potent proteotoxic stress due to high concentrations of ethanol and acetaldehyde[19]. Fluconazole impairs the synthesis of ergosterol, an essential component of Candida albicans’ membrane[36]. Even the normal tissue environment in an animal organism is usually repressive for cellular proliferation, which cancer cells must overcome (see following).
The mechanism by which aneuploidy can bring adaptive phenotypic change has been extensively studied in single cell organisms. In different manners, aneuploidy can cause expression change manifested on both mRNA[14,35,37-41] and protein[35,42] levels. Although altered chromosome stoichiometry leads to expression changes of many genes, in some cases the adaptive effect of aneuploidy can be attributed to dosage change of a single gene. For example, homozygous deletion of RPS24A gene on yeast Chr V causes a growth defect. However, large, fast-growing colonies occasionally appear among a group of small colonies. It was found that cells in these large colonies had gained a copy of Chr IX, carrying the RPS24B gene that is 97% identical in sequence to RPS24A[14]. An advantage of achieving adaptive functions through aneuploidy over that through mutations of specific genes is that genes contributing to the same physiological outcome may be present on the same aneuploid chromosome, and this allows combination of adaptive dosage changes of two or several genes through a single chromosome dosage change. In C. albicans, the fluconazole resistance associated with Chr V duplication can be mimicked by increasing the dosage of ERG11 (encoding the drug target) and TAC1 (encoding a regulator of the drug efflux system), which are both located on this chromosome[21,36]. Euploid budding yeast can adapt to lethal-level Hsp90 inhibitor, radicicol, through gain of Chr XV[43]. Much of the enhanced resistance is due to the synergistic effect of the increased dosage of two genes located on Chr XV: STI1, encoding an Hsp90 co-chaperone and PDR5, encoding a drug pump)[43].
The dosage change of genes located on an aneuploid chromosome can also bring adaptive traits by altering the expression of genes on other chromosome. Myo1 is a motor protein required for constriction of the bud neck during cytokinesis, whose gene deletion leads to cytokinesis failure and in most cases lethality[44]. In the rare Δmyo1 survivors, some are able to restore cytokinesis through gradual thickening of the cell wall at the bud neck. In these adapted strains, the expression of several genes involved in cell wall biogenesis was increased up to 16-fold compared to that in the isogenic wild-type haploid strain. Interestingly, these genes are located on multiple different chromosomes, but a commonly amplified chromosome in these strains is Chr XVI. It turned out that Chr XVI carries the genes encoding two upstream regulators of cell wall biogenesis, Rlm1 and its upstream activator Mkk2, a MAP kinase kinase[36]. Thus, through altering the dosage of regulatory factors aneuploidy can cause broad gene expression changes well beyond a direct DNA dosage effect.
Even though aneuploidy can bring adaptive traits into the population, it is also noticed that in any given environment, such as the presence of proteotoxic stress[37,43] or a DNA damaging agent[35,45], or low temperature[35], most aneuploid karyotypes tested are not adaptive, but only some aneuploid karyotypes show enhanced growth compared to the euploid. This reminds us of the fact that phenotypic changes generated by mutations are usually deleterious[46]; nonetheless mutations are a necessary ingredient of the force that drives adaptive evolution. In other words, chromosome number variation or any other type of mutations does not guarantee enhanced cell fitness, but rather the adaptive value of genetic variation is best appreciated on the population level, where the adaptive variant is selected through competition.
2) Aneuploidy impacts organismal versus cellular fitness differently in multicellular species
Despite evidence in unicellular organisms demonstrating how changes in chromosome stoichiometry bring about adaptation, it remains elusive whether similar mechanisms exist in metazoans. It has been shown that aneuploidy leads to gene expression variation in mammalian cell lines in a manner similar to that in yeast[40,41,47], but the counter argument has been that aneuploidy causes debilitating diseases such as Down's syndrome and cancer. One way to reconcile this paradox is to distinguish cellular fitness from organismal fitness.
In natural history, the appearance of multi-cellularity loosed the tie between organismal evolution and celluar evolution. The former considers relative fitness between individuals, whereas the later considers fitness between cells in the same individuals. In order to survive organismal competition, strict developmental programs tightly control cell proliferation, death and morphogenesis in order to form and maintain homeostasis of functional structures. Thus, organismal fitness occurs at the expense of the proliferative ability or even viability of individual component cells. Oncogenic mutations, on the other hand, promote the cellular proliferation and survivability of cancer cells at the expense of the fitness of the host organism (Fig. 1A). For example, the Ras protein, which controls cellular mitogenic signals, is mutated to hyperactive forms in 25% of cancers and renders abnormally high growth potential for the cancer cells harboring the mutations[48]. The extrinsic barrier to cell proliferation, such as limited vesicular accessibility, can also be lifted by enhanced expression of VEGF in tumors[48]. These examples highlight the apparent conflict between cellular versus organismal fitness.
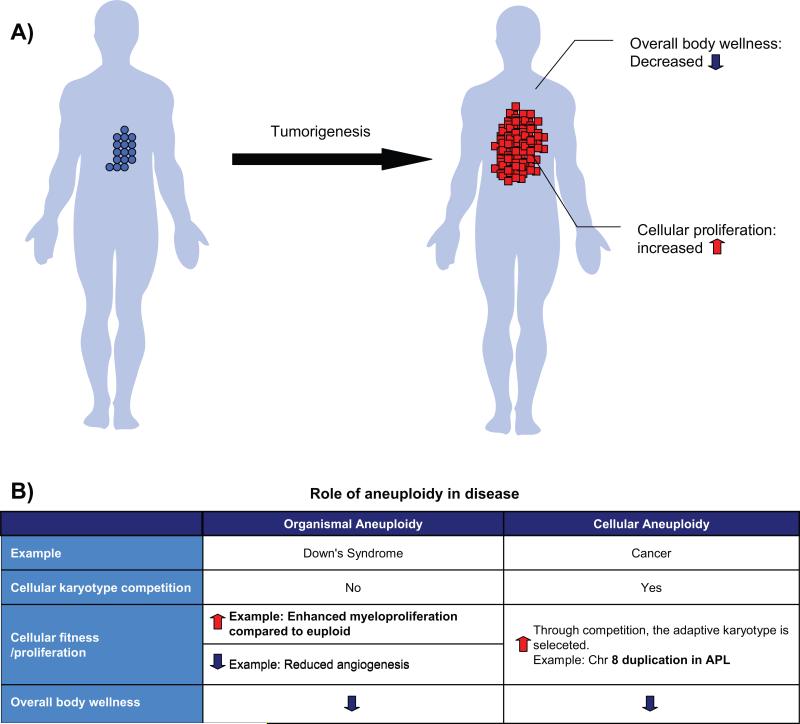
Figure 1. Aneuploidy can exert opposing effect on overall body wellness and cellular fitness in disease.
A: In tumorigenesis, the cellular fitness/proliferation of tumor tissue is enhanced at the expense of overall body wellness. B: Aneuploidy can have different roles on cellular versus organismal level. Organismal aneuploidy originates from karyotype alteration in parental germ line/gametes. Cellular aneuploidy results from errors in somatic cell mitosis.
Whole organism aneuploidy such as Down's syndrome originates from karyotype alteration in parental germ cells, which leads to drastic gene expression changes that disrupt the intricate developmental program evolved during long-term natural history, resulting in disease of the organism[49]. On the other hand, tumorigenesis involves fierce selection and competition between normal cells and cancer cells as well as between cancer cells of diverse karyotypes[50,51]. As the tissue environment for cancer cells is hostile, this presents the natural selective force for different types of genetic variants that could survive and improve the fitness of the cell population at the expense of organismal fitness. As the well known cancer hallmark, karyotype abnormality is a major source of genetic variation in cancer [10,29].
The direct causative relationship between specific karyotype and overproliferation phenotype of tumor has been captured in a handful cases (Fig. 1B). Trisomy 8 was observed in 12% of human acute promyelocytic leukemia (APL) [52,53]. It was long speculated that trisomy 8 brings the growth advantage through introduction of an additional copy of the oncogene, MYC. Interestingly, in an APL mouse model, 64% of the cases were trisomy for chromosome 15, which also contains the mouse MYC. MYC retrovirus transduction facilitates myeloid leukemogenesis and suppressed gain of chromosome 15. Meanwhile, the induction efficiency for APL in MYC heterozygous background was reduced. Remarkably, in MYC heterozygous mouse where APL was inducted, a preferential amplification of the chromosome 15 containing the wild-type MYC, but not the one missing the gene, was observed. These data strongly suggest that the elevated copy number of MYC through aneuploidy directly participates in the progression of APL[54]. Another case comes from the well-characterized Down's syndrome-associated predisposition to leukemia. Down's syndrome patients have a reduced incidence of most tumors compared with euploid population[55,56], but their incidence of pediatric acute megakaryoblastic leukemia (AMKL) is increased 500 times[57]. Accordingly, the mouse model of Down's syndrome, which contains trisomic chromosome region syntenic to human chromosome 21, also shows excessive cell proliferation in myeloid linage, which may progress into AMKL[58]. Later, it was found that by deleting the trisomic copy of Erg, a transcription factor necessary for platelet development and stem cell function, the myeloproliferation was restored to normal[59]. This case highlights that a karyotype (trisomy 21) that is detrimental in organismal level, can nontheless increased fitness and proliferation at cellular level under certain context (Fig. 1B). In spite of a few well-studied cases, the direct causative link between karyotype and tumor phenotype in many cases remains elusive due to the high level karyotype complexity associated with even a single cancer. This may reflect the existence of different ecological niche in a tumor [50]. In addition, different karyotypes can bring adaptation to the same stress, as shown recently in budding yeast[35,38]. The tools that monitor the karyotype in single cell level, such as spectral karyotyping (SKY) or single-cell sequencing[60], will provide insight into how karyotype heterogeneity evolves during the tumor progression or cancer treatment. An ability to dissect the contribution of specific karyotypes to tumor phenotypes in a karyotypically heterogeneous population will be crucial for understanding the role of aneuploidy in tumorigenesis.
Aneuploidy drives adaptation in small cell populations by phenotypic leap
We propose the following model to rationalize the effectiveness of aneuploidy in rapid cellular adaptation as observed in experimental studies in yeast. First, aneuploidy represents a readily occurring form of genetic variation in a population. The rate of chromosome missegregation in yeast is estimated to be 1 in 100,000 chromosome segregation events[61], which is 5 orders of magnitude higher than the point mutation rate per base pair per generation[62,63]. Considering a haploid yeast genome (16 chromosomes) with its coding region sized at107 base pairs, the rate of chromosome missgregation per cell division is likely to be ~10% of the rate of a random point mutation occurring in the genome. However, one chromosome missgregation event has the probability of 100% in causing the expression change of hundreds of genes in the resulting aneuploid progeny. Experimentally, the spectrum of mutations in yeast was analyzed in a study where the mutations were accumulated in 32 individual cultures growing for 4,800 generations in a selection-neutral process[63]. 200 population bottlenecks were introduced to allow unbiased accumulation of mutations. Whole genome re-sequencing revealed 33 point mutations, with 18 being non-silent and may alter protein function in the affected genes. This experiment also captured two aneuploidization events, each causing dosage change of over a hundred genes (Chromosome I and IX). In addition, other types of large changes in chromosome structure were also observed. Thus, aneuploidy and changes in chromosome structure represent a considerable portion of genetic variation in a non-stressed yeast population.
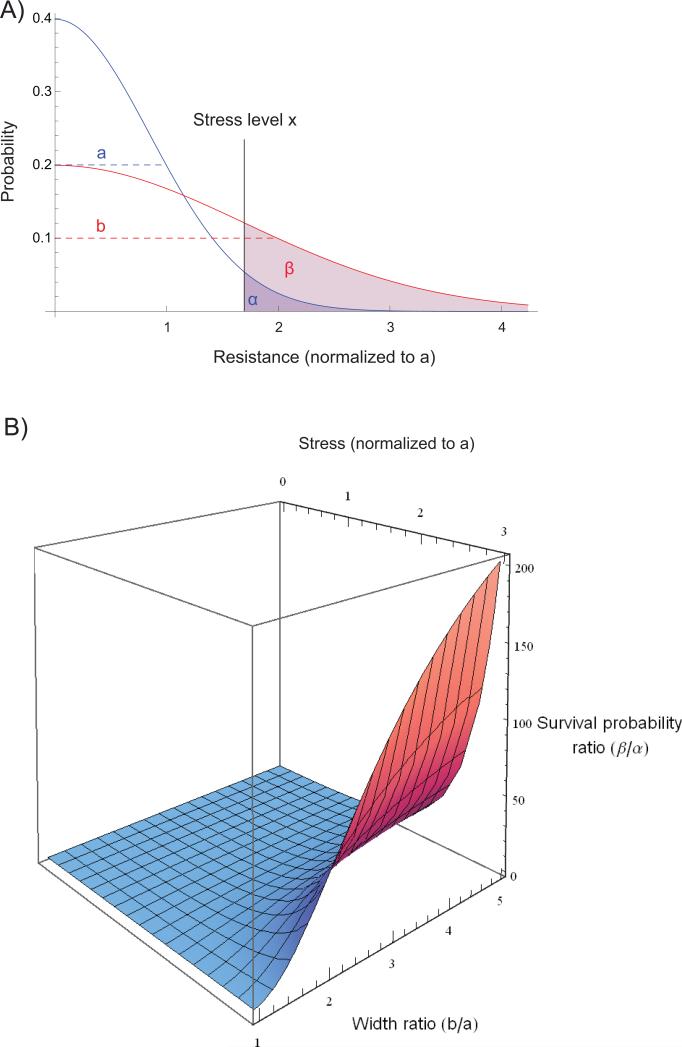
The model presented in Figure 2 compares the probability of adaptation caused by two classes of mutations, one with large and the other with relatively small phenotypic variation (Fig 2A). The relative adaptive probability between the two classes varies dramatically depending on the level of stress (i.e. selection) (Fig 2B). Aneuploidy modulates the expression of a large number of genes. One or multiple of these changes could interact with the stress to cause large phenotypic change, akin to phenotypic leap, which enables the cell to explore a wide region of phenotypic landscape[6]. Moreover, in a diploid genetic background, the common basal ploidy for many multicellular organisms, recessive mutations will be masked, further limiting the phenotypic impact of nucleotide substitutions.
Figure 2. Potent selection favors mutation with large phenotypic variation.
Class B mutation's probability of survival is calculated similarly.
Given these considerations, we speculate that adaptive evolution in relatively small populations under strong selective force, which limits the number of mutations with sufficient phenotypic effect to achieve adaptation, favors the selection of aneuploidy over point mutations. Certain somatic evolution processes, such as the clonal expansion in early tumor progression or relapse after drug treatment, may fall into this category. Gross chromosomal structure change represents another type of genomic change that can cause large phenotypic variation. Also like whole chromosome aneuploidy, gross chromosomal structure changes are frequently observed in cancer.
Chromosome instability can be induced by stress
A major cause of aneuploidy is “chromosome instability” (CIN), which result from errors in the chromosome segregation process during mitosis or meiosis. The rate of CIN is non-zero in well adapted euploid cell populations and can be further increased due to genetic aberrations or under certain stressful conditions as shown recently. Genes that cause CIN when mutated are called CIN genes, many of which encode components of kinetochore, centrosome or mitotic checkpoint, which directly participate in chromosome segregation process. In mammals, there is considerable evidence CIN gene mutations are tumorigenetic, even though the exact tumorigenetic karyotypes that arise in the presence of these mutations have not been identified. For example, Mad2 overexpression in mouse, which delays mitotic progression, promotes the occurrence of aneuploidy and leads to a wide spectrum of tumors[64]. Human genetics also discovered mutations in checkpoint component BUB1B[65], or centrosomal protein CEP57[66] cause mosaic variegated aneuploidy and hereditary cancer. However, CIN that is too high can inhibit tumorigenesis. In mouse, the haploinsufficiency of CENP-E, a kinechore component, modestly increase CIN in various tissues[67]. It drastically increases the incidence of spleen and lung tumors in aged animals. However, in liver, it inhibits the formation of spontaneous cancer. As liver's basal level of CIN is high[12,68], it is speculated that CIN level that is too high to even maintain the tumorigenetic karyotype can suppress the tumor formation[67].
Our recent study in budding yeast showed that other than genetic mutation, certain stress can escalate CIN and potentiate rapid cellular adaptation to this or other unrelated types of stress[43]. Assays using an artificial chromosome revealed that many stress conditions, including hydrogen peroxide (oxidative stress), cycloheximide (translational stress), tunicamycin (ER stress), etc., elevated the chromosome loss rate to a level similar to that caused by benomyl, a microtubule inhibitor that disrupts the mitotic spindle. Surprisingly, radicicol, an Hsp90 inhibitor, was an exceptionally effective CIN inducer: the chromosome loss rate was hundreds of times above the control and ~10 fold higher than that induced by benomyl, even at a radicicol concentration with only minor effect on growth. This CIN-inducing effect is likely to be due to a crucial role for Hsp90 in kinetochore assembly[69,70]. High concentration of Hsp90 inhibitor resulted in emergence of drug-resistant colonies with chromosome XV gain. It is noticed that even though most yeast aneuploids grow slower than the euploid counterpart under Hsp90 inhibition[37], rare adaptive aneuploidy yeasts (with Chr XV gain) can nonetheless emerge and be selected from the population with diverse karyotypes during the long-term adaptation process. More disturbingly, short-term exposure to moderate Hsp90 stress, which generates a karyotypically mosaic cell population, potentiated adaptation to unrelated cyto-toxic compounds through different aneuploid chromosome stoichiometries. In pathogenic yeast Candida albicans, exposure to oxidative stress, heat stress, and antifungal drugs elevates chromosome loss rate, which may also fuel the emergence of drug resistance in which aneuploidy is one of the major mechanisms[71].
The possibility of targeting Hsp90 in tumor therapy has been actively investigated in recent years[72]. Recent report showed that Hsp90 inhibitor can specifically antagonize the proliferation of certain trisomy cells as well as CIN cell lines with high level aneuploidy but spare the euploids, in short-term cell culture[73]. This acute effect may reflect that most aneuploids are sensitive to Hsp90 inhibition. However, as in yeast, Hsp90 was reported to be required for kinetochore function in mammalian cell lines[74,75], raising the possibility that in long term selection resistant cancer cells with rare adaptive karyoytpe may appear as a result of CIN induced by the drug itself.
Summary and perspective
Aneuploidy is a genetic alteration existing in somatic cell populations. The occurrence of aneuploidy can be further increased by either mutation in CIN genes or certain environmental stress. By altering the expression of hundreds of genes at the same time, aneuploidy imposes phenotypic consequences in general much larger than that by random single nucleotide mutations. This phenotypic leap makes aneuploidy an important mode of adaptation for somatic cell populations. Despite the observation of aneuploidy in cancer for over a hundred years, only in a handful cases the causative relation between specific karyotype and the tumorigenetic phenotype has been established. The karyotype-phenotype relationship in cancer is complicated by the complexity of karyotype in many cases and is clearly a challenge in future research. Further, whether the stress-induced chromosomal instability occurs in animal organisms and whether it could underlie rapid tumor cell evolution remains to be elucidated. It raises a question as to whether the stress caused by drugs is in fact a facilitator of the genetic instability promoting the evolution of drug resistance. The observation of aneuploidy in normal tissues has gained increasing attention in recent years. It remains unclear how some normal tissue can maintain high-level karyotype mosaicism. Whether this genetic diversity is required for the function of these tissues or helps the cells to cope with stressful tissue microenvironment are also interesting questions for future investigation.
Acknowledgement
We would like to thank Mushegian A. and Potapova T. for helpful discussion. This work is supported by NIH grant RO1-GM059964 to RL.
Footnotes
The authors declare no conflict of interest.
Reference
- 1.Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitagawa K, Hieter P. Nat Rev Mol Cell Biol. Nature Publishing Group; 2001. Evolutionary conservation between budding yeast and human kinetochores. pp. 678–87. [DOI] [PubMed] [Google Scholar]
- 3.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci. 2010;123:825–35. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lew DJ, Burke DJ. THE SPINDLE ASSEMBLY AND SPINDLE POSITION CHECKPOINTS. Annu Rev of Genet. 2003;37:251–82. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 5.Selmecki A, Forche A, Berman J. Genomic Plasticity of the Human Fungal Pathogen Candida albicans. Eukaryotic Cell. 2010;9:991–1008. doi: 10.1128/EC.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Current Opinion in Cell Biology. 2010;22:809–15. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 8.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends in Genet. 2011;27:446–53. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson D, Costa ACS. Down syndrome and genetics [mdash] a case of linked histories. Nat Rev Genet. 2005;6:137–47. doi: 10.1038/nrg1525. [DOI] [PubMed] [Google Scholar]
- 10.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingsbury MA, Yung YC, Peterson SE, et al. Aneuploidy in the normal and diseased brain. Cell Mol Life Sci. 2006;63:2626–41. doi: 10.1007/s00018-006-6169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan AW, Taylor MH, Hickey RD, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–10. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan AW, Hanlon Newell AE, et al. Frequent Aneuploidy Among Normal Human Hepatocytes. Gastroenterology. 2012;142:25–8. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes TR, Roberts CJ, Dai H, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–7. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 15.Kvitek DJ, Will JL, Gasch AP. Variations in Stress Sensitivity and Genomic Expression in Diverse S. cerevisiae Isolates. PLoS Genet. 2008;4:e1000223. doi: 10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codon AC, Benitez T, Korhola M. Chromosomal polymorphism and adaptation to specific industrial environments of Saccharomyces strains. Applied Microbiology and Biotechnology. 1998;49:154–63. doi: 10.1007/s002530051152. [DOI] [PubMed] [Google Scholar]
- 17.Bond U, Neal C, Donnelly D, James T. Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Current Genetics. 2004;45:360–70. doi: 10.1007/s00294-004-0504-x. [DOI] [PubMed] [Google Scholar]
- 18.Borneman AR, Desany BA, Riches D, et al. Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Infante JJ, Dombek KM, Rebordinos L, et al. Genome-Wide Amplifications Caused by Chromosomal Rearrangements Play a Major Role in the Adaptive Evolution of Natural Yeast. Genetics. 2003;165:1745–59. doi: 10.1093/genetics/165.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu G, Liu I, Sham A, Stajich J, et al. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans. Genome Biology. 2008;9:R41. doi: 10.1186/gb-2008-9-2-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selmecki A, Forche A, Berman J. Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science. 2006;313:367–70. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sionov E, Lee H, Chang YC, et al. Cryptococcus neoformans Overcomes Stress of Azole Drugs by Formation of Disomy in Specific Multiple Chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray HW, Berman JD, Davies CR, et al. Advances in leishmaniasis. The Lancet. 366:1561–77. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 24.Rogers MB, Hilley JD, Dickens NJ, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Research. 2011 doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akopyants NS, Kimblin N, Secundino N, et al. Demonstration of Genetic Exchange During Cyclical Development of Leishmania in the Sand Fly Vector. Science. 2009;324:265–8. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanneste E, Voet T, Le Caignec C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–83. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 27.van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Human Reproduction Update. 2011;17:620–7. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- 28.Mitelman F JBaMFE Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2012. 2012.
- 29.Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Current Opinion in Cell Biology. 2006:658–67. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Ozery-Flato M, Linhart C, Trakhtenbrot L, et al. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biology. 2011;12:R61. doi: 10.1186/gb-2011-12-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yurov YB, Iourov IY, Vorsanova SG, et al. Aneuploidy and Confined Chromosomal Mosaicism in the Developing Human Brain. PLoS ONE. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehen SK, Yung YC, McCreight MP, et al. Constitutional Aneuploidy in the Normal Human Brain. The Journal of Neuroscience. 2005;25:2176–80. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry IM, Dilkes BP, Miller ES, et al. Phenotypic Consequences of Aneuploidy in Arabidopsis thaliana. Genetics. 2010;186:1231–45. doi: 10.1534/genetics.110.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huettel B, Kreil DP, Matzke M, et al. Effects of Aneuploidy on Genome Structure, Expression, and Interphase Organization in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000226. doi: 10.1371/journal.pgen.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavelka N, Rancati G, Zhu J, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selmecki A, Gerami-Nejad M, Paulson C, et al. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Molecular Microbiology. 2008;68:624–41. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 37.Torres EM, Sokolsky T, Tucker CM, et al. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 38.Rancati G, Pavelka N, Fleharty B, et al. Aneuploidy Underlies Rapid Adaptive Evolution of Yeast Cells Deprived of a Conserved Cytokinesis Motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao C, Furge K, Koeman J, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proceedings of the National Academy of Sciences. 2007:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams BR, Prabhu VR, Hunter KE, et al. Aneuploidy Affects Proliferation and Spontaneous Immortalization in Mammalian Cells. Science. 2008;322:703–9. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Upender MB, Habermann JK, McShane LM, et al. Chromosome Transfer Induced Aneuploidy Results in Complex Dysregulation of the Cellular Transcriptome in Immortalized and Cancer Cells. Cancer Research. 2004;64:6941–9. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres EM, Dephoure N, Panneerselvam A, et al. Cell. Cell Press; 2010. Identification of Aneuploidy-Tolerating Mutations. pp. 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G, Bradford WD, Seidel CW, et al. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012 doi: 10.1038/nature10795. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolliday N, Pitcher M, Li R. Direct Evidence for a Critical Role of Myosin II in Budding Yeast Cytokinesis and the Evolvability of New Cytokinetic Mechanisms in the Absence of Myosin II. Mol. Biol. Cell. 2003:798–809. doi: 10.1091/mbc.E02-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheltzer JM, Blank HM, Pfau SJ, et al. Aneuploidy Drives Genomic Instability in Yeast. Science. 2011;333:1026–30. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–8. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 47.Geiger T, Cox J, Mann M. Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells. PLoS Genet. 2010;6:e1001090. doi: 10.1371/journal.pgen.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 49.Roper RJ, Reeves RH. Understanding the Basis for Down Syndrome Phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merlo LMF, Pepper JW, Reid BJ, et al. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 51.Heng HHQ, Bremer SW, Stevens JB, et al. Genetic and epigenetic heterogeneity in cancer: A genome-centric perspective. Journal of Cellular Physiology. 2009;220:538–47. doi: 10.1002/jcp.21799. [DOI] [PubMed] [Google Scholar]
- 52.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C. The Importance of Diagnostic Cytogenetics on Outcome in AML: Analysis of 1,612 Patients Entered Into the MRC AML 10 Trial. Blood. 1998;92:2322–33. others. [PubMed] [Google Scholar]
- 53.Le Beau MM, Bitts S, Davis EM, et al. Recurring chromosomal abnormalities in leukemia inPML-RARA transgenic mice parallel human acute promyelocytic leukemia. Blood. 2002;99:2985–91. doi: 10.1182/blood.v99.8.2985. [DOI] [PubMed] [Google Scholar]
- 54.Jones L, Wei G, Sevcikova S, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. The Journal of Experimental Medicine. 2010;207:2581–94. doi: 10.1084/jem.20091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. The Lancet. 2000;355:165–9. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 56.Yang Q, Rasmussen SA, Friedman JM. Mortality associated with Down's syndrome in the USA from 1983 to 1997: a population-based study. The Lancet. 2002;359:1019–25. doi: 10.1016/s0140-6736(02)08092-3. [DOI] [PubMed] [Google Scholar]
- 57.Zipursky A, Brown EJ, Christensen H, et al. Transient myeloproliferative disorder (transient leukemia) and hematologic manifestations of Down syndrome. Clin Lab Med. 1999;19:157–67. vii. [PubMed] [Google Scholar]
- 58.Kirsammer G, Jilani S, Liu H, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–75. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng AP, Hyland CD, Metcalf D, et al. Trisomy of Erg is required for myeloproliferation in a mouse model of Down syndrome. Blood. 2010;115:3966–9. doi: 10.1182/blood-2009-09-242107. [DOI] [PubMed] [Google Scholar]
- 60.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esposito MS, Maleas DT, Bjornstad KA, et al. Current Genetics. Springer Berlin / Heidelberg; 1982. Simultaneous detection of changes in chromosome number, gene conversion and intergenic recombination during mitosis of Saccharomyces cerevisiae: spontaneous and ultraviolet light induced events. pp. 5–11. [DOI] [PubMed] [Google Scholar]
- 62.Lang GI, Murray AW. Estimating the Per-Base-Pair Mutation Rate in the Yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lynch M, Sung W, Morris K, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proceedings of the National Academy of Sciences. 2008;105:9272–7. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
64.Sotillo R, Hernando E, D
 az-Rodr
az-Rodr  guez E et al. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
guez E et al. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar] - 65.Hanks S, Coleman K, Reid S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 66.Snape K, Hanks S, Ruark E, et al. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet. 2011;43:527–9. doi: 10.1038/ng.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weaver BAA, Silk AD, Montagna C, et al. Aneuploidy Acts Both Oncogenically and as a Tumor Suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 68.Putkey FR, Cramer T, Morphew MK, et al. Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Developmental Cell. 2002;3:351–65. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 69.Stemmann O, Neidig A, KÃcher T, et al. Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8585–90. doi: 10.1073/pnas.082223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodrigo-Brenni MC, Thomas S, Bouck DC, et al. Sgt1p and Skp1p Modulate the Assembly and Turnover of CBF3 Complexes Required for Proper Kinetochore Function. Molecular Biology of the Cell. 2004;15:3366–78. doi: 10.1091/mbc.E03-12-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forche A, Abbey D, Pisithkul T, et al. Stress Alters Rates and Types of Loss of Heterozygosity in Candida albicans. mBio. 2011;2 doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 73.Tang Y-C, Williams BR, Siegel JJ, et al. Identification of Aneuploidy-Selective Antiproliferation Compounds. Cell. 2011 doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davies AE, Kaplan KB. Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. The Journal of Cell Biology. 2010;189:261–74. doi: 10.1083/jcb.200910036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niikura Y, Ohta S, Vandenbeldt KJ, et al. 17-AAG, an Hsp90 inhibitor, causes kinetochore defects: a novel mechanism by which 17-AAG inhibits cell proliferation. Oncogene. 2006;25:4133–46. doi: 10.1038/sj.onc.1209461. [DOI] [PubMed] [Google Scholar]