Abstract
The variation in mutations in exons 3, 6, 7, 11 and 12 of the phenylalanine hydroxylase (PAH) gene was investigated in 59 children with phenylketonuria (PKU) and 100 normal children. Three single nucleotide polymorphisms were detected by sequence analysis. The mutational frequencies of cDNA 696, cDNA 735 and cDNA 1155 in patients were 96.2%, 76.1% and 7.6%, respectively, whereas in healthy children the corresponding frequencies were 97.0%, 77.3% and 8.3%. In addition, 81 mutations accounted for 61.0% of the mutant alleles. R111X, H64 > TfsX9 and S70 del accounted for 5.1%, 0.8% and 0.8% mutation of alleles in exon 3, whereas EX6-96A > G accounted for 10.2% mutation of alleles in exon 6. R243Q had the highest incidence in exon 7 (12.7%), followed by Ivs7 + 2 T > A (5.1%) and T278I (2.5%). G247V, R252Q, L255S, R261Q and E280K accounted for 0.8% while Y356X and V399V accounted for 5.9% and 5.1%, respectively, in exon 11. R413P and A434D accounted for 5.9% and 2.5%, respectively, in exon 12. Seventy-two variant alleles accounted for the 16 mutations observed here. The mutation characteristics and distributions demonstrated that EX6-96A > G and R243Q were the hot regions for mutations in the PAH gene in Shanxi patients with PKU.
Keywords: gene mutation, phenylalanine hydroxylase, phenylketonuria
Introduction
Phenylketonuria (PKU) is the most common inborn error of amino acid metabolism and shows autosomal recessive inheritance. Mutations in the gene encoding phenylalanine hydroxylase (PAH; EC1.14.16.1) cause phenylalanine accumulation in the body fluids and damage to the central nervous system that can result in severe mental retardation and neurobehavioral abnormalities (Xu and Yang, 2005). PKU, which was first described by Folling in 1934, is a worldwide disease with marked regional and ethnic variation in its incidence. The prevalence is high among whites and low amongst blacks and asians (Gu, 2003). A prevalence of 1 in 10,000 was reported in Caucasians (Scriver and Kaufman, 2001) while that in Indians was approximately 1:18,300 (Rama Devi and Naushad, 2004). PKU is more frequent in Tunisia (1:7,631) (Khemir et al., 2011). The incidence of PKU varies considerably across China, with an overall prevalence of 1:11,144 from 1985 to 2001 (Gu and Wang, 2004). The prevalence in the provinces of Shanxi, Jiangxi, Heilongjiang and Shandong was reported to be 1:7,849 (Li et al., 2009), 1:11,188 (Wang and Zhang, 2006), 1:5,346 (Li, 2005) and 1:7,408 (Chen and Tian, 2006), respectively. The incidence among neonates in Shanxi province from 2004 to 2009 was 1:3,425 (Shanxi Provincial Health Bureau, 2009), which suggests that the incidence in northern China is higher than in southern China.
The PAH gene was first cloned in 1983 (Woo et al., 1983). To date, 564 mutations of the PAH gene (12q22–q24) have been deposited in the PAH Mutation Analysis Consortium Database, of which 60.5% are missense mutations, 13.5% are deletion mutations, 11% are splicing mutations and 5.0% are nonsense mutations, inserts and silent mutations. The PAH gene mutations show considerable ethnic and regional variation (Zhang et al., 1995). In the present study, we examined the variation in mutations in exons 3, 6, 7, 11 and 12 of the PAH gene in 59 children with PKU from Shanxi province.
Subjects and Methods
Subjects
Fifty-nine unrelated children with PKU were recruited for this study after obtaining informed consent from the parents. The patients (30 males, 29 females) were identified during treatment at the Neonatal Screening Center of the Shanxi Province Women and Childrens Hospital in Taiyuan and came from various regions of Shanxi province. The subjects were ≤ 10 years old and were classified as < 2 years old (n = 38), 2–6 years old (n = 18) and 6–10 years old (n = 3). All 59 cases exhibited significant clinical manifestations of PKU and fulfilled the diagnostic criteria for PKU, with blood phenylalanine concentrations > 20 mg/dL (Qian and Wang, 2010). Urinary pterin analysis and blood neopterin dihydropteridine reductase assays were used to exclude tetrahydrobiopterin deficiency. Twenty-five patients were placed on a low Phe diet to decrease Phe levels following the initial diagnosis. The healthy children included 53 males and 47 females, with ages ranging from 1 month to 10 years old. This study was approved by Taiyuan Centre Hospital Ethical Comimittee.
DNA extraction
Genomic DNA was extracted from peripheral blood using a DNA extraction kit (Shanghai Ying Jun Biotechnology Co., Ltd., Shanghai, China). Purity was assessed by electrophoresis in 1% agarose gels and the DNA was stored at −20 °C.
PCR amplification
Exons 3, 6, 7, 11 and 12 of the PAH gene were amplified. The primers for exons 3, 6, 11 and 12 were based on a previous report (Gu, 2003) whereas those for exon 7 were designed specifically for this study. All primers were synthesized by Shanghai Jin Kang Biotechnology (Shanghai, China) (Table 1). The PCR mixture consisted of 10X buffer, 2.5 mM Mg2+, 0.2 mM dNTP, 0.5 μM primer, 1 U of Taq polymerase and 1 μL of template DNA in a total volume of 20 μL. The reaction conditions were: denaturation at 94 °C for 3 min followed by 35 cycles of amplification at 94 °C for 30 s and annealing (Table 1) for 30 s with a final extension at 72 °C for 5 min. The PCR products were detected by electrophoresis in 2% agarose gels.
Table 1.
Primer sequences for PKU exons 3, 6, 7, 11 and 12.
| Exon | Sequences | Annealing temperature (°C) | Product length (bp) |
|---|---|---|---|
| 3 | F: 5′-GTTAGGTTTTCCTGTTCTGG-3′ | 60 | 300 |
| R: 5′-CTTATGTTGCAAAATTCCTC-3′ | |||
| 6 | F: 5′-CACAGGTTCTGGTCCCCGAC-3′ | 61 | 354 |
| R: 5′-CTCTCCTCTCCTCAATCCTC-3′ | |||
| 7 | F: 5′-CTCCTAGTGCCTCTGACTCA-3′ | 60 | 273 |
| R: 5′-CAAACCTCATTCTTGCAGCAGG-3′ | |||
| 11 | F: 5′-AAGGAATCGGGGTGAGATGAGAGAAGGGGC-3′ | 64 | 357 |
| R: 5′-GGTACAAAGTTGCTGTAGACATTGGAGTCC-3′ | |||
| 12 | F: 5′-ATGCCACTGAGAACTCTCTT-3′ | 60 | 245 |
| R: 5′-AGTCTTCGATTACTGAGAAA-3′ |
Sequencing
The PCR products were sequenced by Shanghai Ying Jun Biotechnology (Shanghai, China) and compared with the human genomic DNA sequence in GenBank to identify the mutations.
Statistical analysis
All statistical analyses, including the χ2-test, were done using SPSS 13.0 statistical software. A p value ≤ 0.05 indicated significance.
Results
Exons 3, 6, 7, 11 and 12 of the PAH gene from 59 children with PKU and 100 healthy children were aligned with the GenBank cDNA sequence (GI: 209364518) of the PAH gene. Three single nucleotide polymorphisms (SNPs) [Q232 (CAA→CAG), V245V (GTG→GTA), and L385L (CTG→CTC)] were identified in the patients and healthy children. The incidence of the 696 locus SNP in children with PKU (96.2%) was not significantly different from that in healthy children (97%; χ2 = 0.79, p > 0.05). Similarly, there was no significant difference in the incidence of the 735 locus SNP in the patients compared to the healthy controls (76.1% vs. 77.3%; (χ2 = 0.327, p > 0.05) or in the incidence of the 1155 locus SNP (7.6% in patients vs. 8.3% in healthy controls; χ2 = 0.111, p > 0.05).
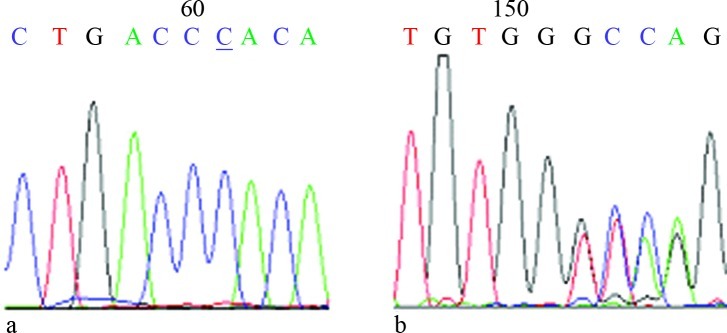
In children with PKU, 81 mutations belonging to 16 different types of mutations were observed and accounted for 61% of the PAH gene mutations. The mutations R111X, H64 > TfsX9 and S70 deletion were found in exon 3, with frequencies of 5.1%, 0.8% and 0.8%, respectively. Only the EX6-96A > G mutation was found in exon 6, with a frequency of 10.2%. The R234Q mutation in exon 7 had the highest frequency followed by Ivs7 + 2 T > A (5.1%) and T278I (2.5%). G247V, R252Q, L255S, R261Q and E280K accounted for 0.8% each. Y356X in exon 11 accounted for 5.9%, whereas V399V accounted for 1%. R413P and A434D in exon 12 accounted for 5.9% and 2.5%, respectively (Table 2). Among the 16 mutations, nine were missense, three involved splice sites, two were nonsense and two involved deletions, of which H64 > TfsX9 (Figure 1) was the first to be discovered. All of these mutations have been included in the PAH Mutation Analysis Consortium Database. As shown in Table 3, three alleles were identified in one patient, two alleles were identified in 16 patients and the remaining cases were identified in 33 samples. Six patients with homozygous mutations were identified. The intelligence of the 25 patients in the low Phe diet intervention group is developing well while the remaining children show some degree of mental retardation.
Table 2.
Shanxi PAH 3, 6, 7, 11 and 12 exon mutations.
| Exon site | Amino acid change | cDNA change | Type of mutation | Number of alleles | Mutation frequency (%) |
|---|---|---|---|---|---|
| 3 | R111X | c. 331C > T | nonsense | 6 | 5.1 |
| 3 | H64 > TfsX9 | c. 190delC | loss | 1 | 0.8 |
| 3 | S70 del | c. 208–210delTCT | loss | 1 | 0.8 |
| 6 | EX6-96A > G | c. 611A > G | splice site | 12 | 10.2 |
| 7 | R243Q | c. 728G > A | missense | 15 | 12.7 |
| 7 | G247V | c. 740G > T | missense | 1 | 0.8 |
| 7 | R252Q | c. 755G > A | missense | 1 | 0.8 |
| 7 | L255S | c. 764T > C | missense | 1 | 0.8 |
| 7 | R261Q | c. 782G > A | missense | 1 | 0.8 |
| 7 | T278I | c. 833C > T | missense | 3 | 2.5 |
| 7 | E280K | c. 838G > A | missense | 1 | 0.8 |
| IVS7 | IVS7+2T > A | c. 842 +2T > A | splice site | 6 | 5.1 |
| 11 | V399V | c. 1197A > T | splice site | 6 | 5.1 |
| 11 | Y356X | c. 1068C > A | nonsense | 7 | 5.9 |
| 12 | A434D | c. 1301C > A | missense | 3 | 2.5 |
| 12 | R413P | c. 1238G > C | missense | 7 | 5.9 |
| Number | 72 | 61.0 | |||
| Total | 118 | 100 |
Figure 1.
Sequencing of the H64 > TfsX9 mutation. A: Normal samples for the forward primer sequence. B: Absence of specimens sequenced with the reverse primer.
Table 3.
Mutations identified in 51 PKU patients.
| Patient | Sex | Age (years) |
PHA allele 1
|
PHA allele 2
|
||
|---|---|---|---|---|---|---|
| Codon | Het/Hom | Codon | Het/Hom | |||
| 17 | Male | 1.5 | Y356X | Het | ||
| 18 | Male | 3 | Y356X | Het | ||
| 21 | Male | 5 | R111X | Hom | ||
| 22 | Female | 2 | R252Q | Het | ||
| 25 | Male | 2.5 | H64 > TfsX9 | Het | ||
| 26 | Female | 7 | T278I | Het | G247V | Het |
| 27 | Female | 7 | IVS7+2T > A | Het | ||
| 28 | Male | 6 | R413P | Het | ||
| 33 | Male | 1.5 | EX6-96A > G | Het | ||
| 36 | Male | 5 | EX6-96A > G | Het | ||
| 37 | Male | 5 | R111X | Het | R243Q | Het |
| 38 | Female | 4 | EX6-96A > G | Het | Y356X | Het |
| 40 | Female | 10 | R413P | Het | ||
| 41 | Male | 2.5 | A434D | Het | ||
| 47 | Female | 4 | EX6-96A > G | Het | R243Q | Het |
| 50 | Male | 35 days | T278I | Het | R243Q | Het |
| 54 | Female | 35 days | T278I | Het | ||
| 60 | Male | 6 | R261Q | Het | ||
| 63 | Male | 2 | EX6-96A > G | Het | R413P | Het |
| 69 | Female | 1 | R243Q | Het | ||
| 72 | Female | 18 days | IVS7+2T > A | Het | ||
| 78 | Male | 7 months | R111X | Het | ||
| 81 | Female | 23 days | R243Q | Het | ||
| 84 | Female | 1 | IVS7+2T > A | Het | R243Q | Het |
| 87 | Female | 4 | Y356X | Hom | ||
| 90 | Male | 3 | EX6-96A > G | Hom | ||
| 99 | Male | 15 months | V399V | Het | ||
| 104 | Female | 1 | IVS7+2T > A | Het | R413P | Het |
| 107 | Female | 1 | R111X | Hom | ||
| 112 | Female | 18 days | EX6-96A > G | Het | ||
| 115 | Male | 15 days | V399V | Het | ||
| 118 | Male | 34 days | R243Q | Het | Y356X | Het |
| 121 | Female | 1 month | R413P | Het | ||
| 124 | Female | 8 months | EX6-96A > G | Het | R243Q | Het |
| 130 | Female | 12 months | A434D | Het | ||
| 132 | Female | 32 days | EX6-96A > G | Het | R243Q | Het |
| 134 | Male | 34 days | IVS7+2T > A | Het | R243Q | Het |
| 141 | Female | 4 | V399V | Het | R413P | Het |
| 144 | Male | 13 months | R243Q | Het | ||
| 147 | Male | 1 | IVS7+2T > A | Het | ||
| 153 | Male | 2 | R243Q | Het | ||
| 166 | Male | 6 months | Y356X | Het | ||
| 172 | Male | 1 | V399V | Het | ||
| 175 | Female | 23 days | R243Q | Het | E280K V399V | Het |
| 180 | Male | 8 months | EX6-96A > G | Het | R413P | Het |
| 186 | Female | 2.5 | R413P | Het | L255S | Het |
| 192 | Male | 36 days | EX6-96A > G | Het | ||
| 195 | Male | 43 days | S70 del | R243Q | Het | |
| 198 | Female | 11 months | V399V | Hom | ||
| 201 | Female | 4.5 | A434D | Het | ||
| 204 | Male | 3.5 | R243Q | Hom | ||
Discussion
Various PAH gene mutations have been identified in 13 exons, with those of exon 7 accounting for 20% of mutations (Zhang et al., 1995; Song et al., 2001; Fang et al., 1992; Güttler and Guldberg, 1996). Nearly 30 species of PAH gene mutations have been found in the Chinese population and involve mainly exons 7 and 12 (Zhang et al., 1995; Song et al., 2001). In the present study, we identified two relatively high-frequency mutations, EX6-96A > G and R243Q, with frequencies of 10.2% and 12.7%, respectively. Two hot spots for mutations in the PAH gene were identified in Shanxi province patients with PKU and generally agreed with the findings of previous studies (Zhang et al., 2004). Several studies have shown that there is considerable regional and ethnic variation in the distribution of PAH gene mutations in China (Fang et al., 1992; Güttler and Guldberg, 1996; Zhang et al., 1997). In agreement with this, the frequencies of the R243Q mutation in exon 7 of the PAH gene in northern and southern China were found to be 24% and 9.5%, respectively. The frequency of R243Q in exon 7 has been reported to be 18.2% in Chinese and 12% in Koreans; in contrast, R413P is a high-frequency mutation in Japanese (30.5%) (Zhang et al., 1997).
PAH mutations can be classified into disease-causing and silent mutations, depending on the extent to which enzymatic activity is affected by the mutation. In contrast to disease-causing mutations, silent mutations do not affect PAH activity. Disease-causing mutations often occur in the crosslink areas between exons or between an exon and intron and affect PAH gene transcription and translation; such interference affects protein folding and aggregation and accelerates degradation, thereby influencing the catalytic activity of PAH (Bai and Song, 2003). In the present study, 16 mutations were found, including nine missense mutations, three splice site mutations and two nonsense mutations, as well as two types of deletions. The nonsense, splice site and deletion mutations interfere with protein transcription or translation and are generally considered to be pathogenic mutations. The pathogenicity of missense mutations requires further investigation involving gene expression and protein structure analysis (Song et al., 2008). To our knowledge, this is the first report to identify the mutant site H64 > TfsX9, a single base deletion that resulted in a frameshift mutation in exon 3 of the PAH gene. The mutation at codon 64 (c. 190), which replaces histidine by threonine, may change the PAH structure and affect the corresponding enzymatic activity.
PKU should be detected and treated early since the earlier the treatment, the better the prognosis (Lu and Jiang, 2003). Early treatment is the key factor to changing the outcome of this disease (Zhang and Qi, 2001). The prenatal diagnosis of PAH gene mutations using the approach described here could be useful in reducing the incidence of PKU in Shanxi province. Screening for mutations could also be useful in the detection, genetic counseling and treatment of potential PKU carriers, thereby improving the quality of life of affected individuals.
Acknowledgments
The study was supported by the Scientific and Technological Project of Shanxi Province (grant no. 20080311065) and Shanxi Provincial Health Department Project (grant no. 200705).
Footnotes
Associate Editor: Mara H. Hutz
References
- Bai JL, Song F. Effect of gene mutation on the function and structure of phenylalanine hydroxylase. Int J Pediat. 2003;25:142–144. [Google Scholar]
- Chen GQ, Tian Y. Clinical analysis of 48 children with phenylketonuria. J Appl Clin Pediatr. 2006;21:483–484. [Google Scholar]
- Fang B, Yuan L, Wang M, Huang S, Wang T, Miao S, Ye J, Sun N, Lo H, Savio LC. Detection of point mutation of the phenylalanine hydroxylase gene and prenatal diagnosis of phenylketonuria. Clin Med Sci J. 1992;7:205–206. [PubMed] [Google Scholar]
- Gu XF. Newborn Disease Screening. Scientific and Technological Literature Publishing House; Shang Hai: 2003. p. 1215. [Google Scholar]
- Gu XF, Wang ZG. Screening for phenylketonuria and congenital hypothyroidism in 5.8 million neonates in China. Chin Prev Med. 2004;38:99–102. [PubMed] [Google Scholar]
- Güttler F, Guldberg P. The influence of mutations of enzyme activity and phenylalanine tolerance in phenylalanine hydroxylase deficiency. Eur J Pediatr. 1996;155:6–10. doi: 10.1007/pl00014253. [DOI] [PubMed] [Google Scholar]
- Khemir S, EI Asmi M, Sanhaji H, Feki M, Jemaa R, Tebib N, Dhondt JL, Ben Dridi MF, Mebazaa A, Kaabachi N. Phenylketonuria is still a major cause of mental retardation in Tunisia despite the possibility of treatment. Clin Neurol Neurosurg. 2011;113:727–730. doi: 10.1016/j.clineuro.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Lee DH, Koo SK, Lee KS, Yeon YJ, Oh HJ, Kim SW, Lee SJ, Kim SS, Lee JE, Jo I, et al. The molecular basis of phenylketonuria in Koreans. Hum Genet. 2004;49:617–621. doi: 10.1007/s10038-004-0197-5. [DOI] [PubMed] [Google Scholar]
- Li FX, Liu HL, Xiang L. Analyzing on screening and the effect of prevention from neonatal disorders. Chin J Child Health Care. 2009;17:464–465. [Google Scholar]
- Li ZY. Study on phenylketonuria in 138,988 neonatal patients. HeiLong Jiang Med J. 2005;29:554–559. [Google Scholar]
- Lu MW, Jiang HZ. Effect of early diagnosis and treatment of patients with phenylketonuria. Chin J Child Health Care. 2003;11:118–119. [Google Scholar]
- Qian ZL, Wang JF. Analysis of results of screening in 124,810 newborn with phenylketonuria. HaiNan Med. 2010;20:126–127. [Google Scholar]
- Rama Devi AR, Naushad SM. Newborn screening in India. Indian J Pediatr. 2004;71:157–160. doi: 10.1007/BF02723099. [DOI] [PubMed] [Google Scholar]
- Scriver CR, Kaufman S. Hyperphenylalaninemia: Phenylalanine hydroxylase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. 8th edition. McGraw-Hill; New York: 2001. pp. 1667–1724. [Google Scholar]
- Song F, Qu Y, Okano Y, Ye Z, Zhang Y, Jin Y, Wang H. In vitro expression and structural analysis of four missense mutations (G2147S, E280G, P362T, A434D) of phenylalanine hydroxylase gene. Chin J Med Genet. 2008;25:1–5. [PubMed] [Google Scholar]
- Wang Q, Zhang ZY. Screening and treatment of hypothyrosis phenylketonuria in Jiangxi Province. Chin J Birth Health Hered. 2006;14:111–112. [Google Scholar]
- Woo SL, Lidsky AS, Guttler FD, Chandra T, Robson KJ. Cloned human phenylalanine hydroxylase gene allows prenatal diagnosis and carrier detection of classical phenylketonuria. Nature. 1983;306:151–155. doi: 10.1038/306151a0. [DOI] [PubMed] [Google Scholar]
- Xu HP, Yang CH. Screening and follow-up of neonatal phenylketonuria. Foreign Med Sci (Section of Maternal and Child Health) 2005;16:343. [Google Scholar]
- Zhang J, Meng J, Zhai X, Fang G, Gao J, Shi M, Wang Y. Identification of novel mutations in the phenylalanine hydroxylase gene of classical phenylketonuria. Chin J Med Genet. 2005;22:134–137. [PubMed] [Google Scholar]
- Zhang M, Gu XF, Zhang MH. Mutations and their frequencies in exon 7 of phenylalanine hydroxylase gene of phenylketonuria in southern Chinese. Chin J Med Genet. 1995;12:324–326. [Google Scholar]
- Zhang M, Hsiao K, Su T, Chao H, Chen R, Gu X. Two novel mutations in phenylalanine hydroxylase gene and in vitro expression analysis of mutation Arg252Glu. Chin Med Sci J. 1997;12:22–25. [PubMed] [Google Scholar]
- Zhang YM, Qi JL. The monitor and analysis on 43 cases of PKU low phenylalanine diet. Chin J Child Health Care. 2001;2:5–6. [Google Scholar]
- Zhang YM, Qin JL, Qiu L. The PKU neonatal screening, diagnosis, treatment and genic mutational analysis in Beijing area. Chin J Child Health Care. 2004;11:366–388. [Google Scholar]
- Zhang Z, He YS. Advances in the studies of molecular heredity of phenylketonuria. Hereditas. 2004;26:729–734. [PubMed] [Google Scholar]
Internet Resources
- The PAH Mutation Analysis Consortium Database, http://www.mcgill.ca/pahdb (Aug 31, 2009).
- Shanxi Provincial Health Bureau http://www.sxws.cn/bureau/ShowLeaderActionBeta.asp?strNewsId=5252 (May 27, 2009).