Abstract
Carcharhinus limbatus has a cosmopolitan distribution and marked genetic structuring, mainly because of its philopatric behavior. However, analysis of this structuring has not previously included South American populations. In the present study, we analyzed a sample of adult individuals collected on the northern coast of Brazil and compared the sequences of the mitochondrial control region with those of populations already genotyped. Relatively high haplotype diversity (12 haplotypes, genetic diversity of 0.796) was observed, similar to that in other populations but with a much larger number of private alleles. In contrast to populations studied previously, which were represented by neonates, the pronounced allelic variability found in the South American individuals may have resulted from migrations from other populations in the region that have yet to be genotyped. This population was also genetically distinct from the other Atlantic populations (Fst > 0.8), probably because of female philopatry, and apparently separated from the northwestern Atlantic group 1.39 million years ago. These findings indicate that the C. limbatus population from northern Brazil is genetically distinct from all other populations and should be considered as a different management unit for the protection of stocks.
Keywords: Carcharhinus limbatus, control region, fishery stocks, mitochondrial DNA, population genetics
Introduction
Sharks are vulnerable to overexploitation by fisheries because of their low fecundity and slow maturation (Musick et al., 2000; Myers and Worm, 2005; Tsai et al., 2010; Tillett et al., 2011) and their limited potential for recruitment. This combination of traits has resulted in a sharp decline in populations in many parts of the world (Baum and Myers, 2004; Bradshaw et al., 2008; Ferretti et al., 2010). Given these considerations, the effective management of shark species should also consider good knowledge of the ecological characteristics and population structure of current stocks (Vaudo and Heithaus, 2009). Identification of the genetic structuring of populations is fundamental for determining whether and to what degree reproductive isolation exists, and can identify subpopulations that need to be treated as separate management units (Palsboll et al., 2007). The lack of information on the migration and dispersion patterns of sharks and the difficulties in obtaining an adequate sample of the distribution of a species hinder the evaluation of fishery stocks and potentially contribute to the application of management policies that are incompatible with the biological characteristics of each species, leading to possible overexploitation of stocks (Feldheim et al., 2001; Hueter et al., 2004; Gore et al., 2008).
The large populations, ample geographic distribution and dispersal capacity of many marine species, together with the general lack of physical barriers to dispersion in the world’s oceans, all contribute to long-distance gene flow, characterized by a reduced genetic structuring of populations (Palumbi, 1994; Ward et al., 1994; Rosenbaum et al., 2009). This is emphasized by the fact that only a few migrants are needed per generation to avoid the effects of genetic drift, which results in segregation of the genetic characteristics of populations at different localities (Waples, 1998). On the other hand, traits such as the lack of mobile larvae or philopatric behavior may limit gene flow and result in the genetic divergence of populations of marine organisms, even when they have a continuous distribution (Meylan et al., 1990).
The blacktip shark, Carcharhinus limbatus, has a cosmopolitan distribution, and is typically found in tropical and subtropical coastal waters (Compagno et al., 2005). In the western Atlantic, the species is found from Massachusetts to southern Brazil (Compagno, 1984). This shark is highly mobile, being able to travel distances of over 2,000 km, and migrates seasonally (Kohler et al., 1998; Carlson and Brusher, 1999).
The blacktip shark uses shallow coastal waters as nurseries where the juveniles spend the first few months of their lives (Hueter et al., 2005). These areas are normally relatively productive, with abundant dietary resources, and are also well protected from predators (Simpfendorfer and Milward, 1993; Heupel et al., 2004; Heupel et al., 2007; Faunce and Layman, 2009; Heupel and Simpfendorfer, 2011). An analysis of the genetic structure of different nurseries of C. limbatus in North America revealed that the females do not disperse randomly but are philopatric, returning to reproduce in the nurseries where they were born. This behavior can lead to marked genetic heterogeneity among nurseries (Keeney et al., 2003).
Genetic structuring has been recorded among nurseries of C. limbatus in North America, the Gulf of Mexico and the Caribbean (Keeney et al., 2005). These authors concluded that females copulate with males from different regions and then return to their natal nurseries to breed. In addition to this local structuring, a global analysis by Keeney and Heist (2006), which included new samples from Africa and the Indo-Pacific region, demonstrated pronounced structuring between these populations and those of the eastern Atlantic. However, this analysis did not include any South American populations.
Although C. limbatus is common along the coast of Brazil, little is known about its population structure or reproductive behavior in this region. The occurrence of pregnant females and juveniles off the coast of the Brazilian states of Rio Grande do Norte and Paraná indicates that nurseries exist in these areas (Yokota and Lessa, 2006; Bornatowski, 2008).
In the present study, sequences of the control region of mitochondrial DNA were analyzed to evaluate the genetic variability in C. limbatus on the northern coast of Brazil. The data were also used to assess the degree of divergence in relation to other populations (Keeney et al., 2003, 2005) and the possible existence of distinct genetic lineages within the geographic distribution of the species.
Materials and Methods
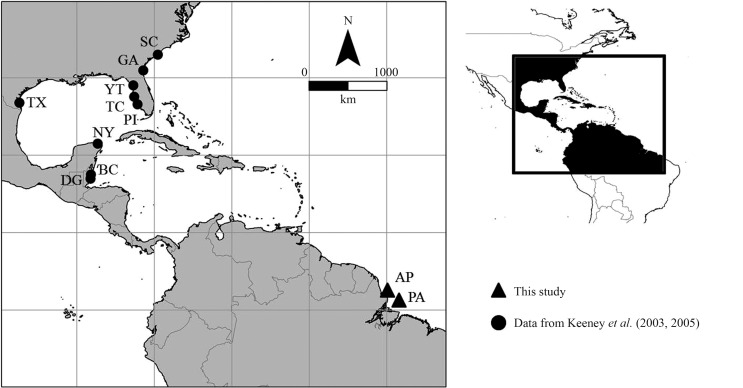
The specimens of C. limbatus analyzed were all adults collected during a boat expedition on the coast of the Brazilian states of Pará (0°28′38.82″ N, 48°18′55.38″ W) and Amapá (01°31′56.8″ N, 49°42′14.1″ W) and were identified based on Compagno (1984) (Figure 1). A sample of muscle tissue was extracted from each specimen and stored in 95% ethanol at −20 °C. Total DNA was extracted using a standard protocol (Sambrook et al., 1989) in which the tissue was digested with ribonucleases for 40 min at 37 °C and then dissolved in a solution of SDS and proteinase K at 55 °C. The material was subsequently washed in phenol-chloroform and precipitated with sodium acetate and isopropanol. The resulting total DNA was hydrated and frozen.
Figure 1.
Sampling localities for this phylogeographic study. Circles represent populations from Keeney et al. (2003, 2005) (BC – Belize City; DG – Dangriga; GA – Georgia; NY – Laguna Yalahau, northern Yucatan; PI – Pine Island Sound, Florida; SC – Bulls Bay, South Carolina; TC – Terra Ceia Bay, Florida; TX – Padre Island, Texas; YT – Yankeetown, Florida) and triangles represent locations sampled in this study (AP – Amapá and PA – Pará).
The control region of the mitochondrial DNA of 25 specimens of C. limbatus was amplified using the polymerase chain reaction (PCR). The primers and the amplification protocol used were described by Keeney et al. (2003). The PCR products were purified using an ExoSAP-IT kit (Amersham Pharmacia Biotech. Inc., UK) and then sequenced using the dideoxynucleotide method (Sanger et al., 1977) with a Big Dye reagent kit (ABI PrismTM Dye Terminator Cycle Sequencing Ready Reaction; Applied Biosystems, Foster City, CA, USA). The sequences were aligned automatically with CLUSTAL W (Larkin et al., 2007) run in the BIOEDIT sequence editor (Hall, 1999), for visual inspection and possible corrections. The nucleotide composition, number of transitions and transversions were calculated using MEGA 4.0 (Tamura et al., 2007).
Genetic (H) and nucleotide (π) diversity indices were determined with ARLEQUIN 3.1 (Excoffier et al., 2005). Pairwise genetic divergence between populations was estimated by Fst (Excoffier et al., 1992), with the significance being tested by 1000 permutations. Analysis of molecular variance (AMOVA), run in ARLEQUIN 3.1, was used to assess the genetic variability among populations. For this, the data provided by Keeney et al. (2003, 2005) and Keeney and Heist (2006) were added to those collected during this study. The data from previous studies were grouped into seven populations [BC – Belize City, Belize; DG – Dangriga, Belize; EA/IP – eastern Atlantic/Indo-Pacific (Africa, India, Australia, Philippines, Hawaii, USA); EG – eastern Gulf of Mexico (Pine Island, Terra Ceia and Yankeetown, USA); NA – northwestern Atlantic (South Carolina and Georgia; USA); NY – northern Yucatan (Laguna Yalahau northern Yucatan; Mexico); WG – western Gulf of Mexico (Padre Island, USA)], in addition to the northern Brazilian samples (Table 1). The haplotype network was constructed using the median joining (MJ) method in the NETWORK program, version 4.0 (Bandelt et al., 1999).
Table 1.
Position of the polymorphic sites and geographic distribution of the haplotypes. The haplotype numbers (Hap) are listed in the first column, and N indicates the number of individuals in which the haplotype was observed. Nucleotides identical to haplotype 1 are indicated by dots (BC – Belize City; BR – northern Brazil; DG – Dangriga; EA/IP – eastern Atlantic/Indo-Pacific; EG – eastern Gulf of Mexico (Pine Island, Terra Ceia and Yankeetown); NY – northern Yucatan (Laguna Yalahau northern Yucatan); WA – western Atlantic (Bulls Bay and Georgia); WG – western Gulf of Mexico (Padre Island)). Note that the different haplotype groups are shaded in different tones of gray.
| Hap | Nucleotide position
|
N | Population
|
|||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 5 | 8 | 1 | 1 | 1 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 6 | 7 | 7 | 7 | 7 | 8 | 8 | 9 | 1 | 1 | ||||||||||
| 8 | 5 | 7 | 3 | 8 | 3 | 4 | 5 | 0 | 8 | 0 | 2 | 4 | 6 | 7 | 7 | 7 | 7 | 4 | 5 | 7 | 9 | 2 | 8 | 5 | 5 | 6 | 7 | 2 | 4 | 1 | 0 | 0 | ||||||||||
| 9 | 4 | 0 | 8 | 0 | 6 | 4 | 5 | 8 | 8 | 9 | 2 | 3 | 9 | 0 | 3 | 3 | 7 | 6 | 6 | 7 | 0 | 7 | 6 | 5 | 1 | 5 | 5 | |||||||||||||||
| 4 | 5 | WA | EG | WG | NY | BC | DG | EA/IP | BR | |||||||||||||||||||||||||||||||||
| H1 | T | C | A | T | T | T | C | A | C | A | T | G | T | A | T | A | C | G | G | A | A | - | C | A | A | G | A | T | G | T | A | - | A | 125 | 35 | 53 | 14 | 23 | ||||
| H2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | C | . | - | . | 52 | 12 | 34 | 6 | |||||
| H3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | C | . | - | G | 21 | 15 | 6 | ||||||
| H4 | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | C | - | - | . | 29 | 11 | 13 | 5 | |||||
| H6 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | C | - | - | . | 17 | 4 | 7 | 6 | |||||
| H7 | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | . | . | - | . | 3 | 3 | |||||||
| H8 | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H9 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | G | . | - | . | . | . | . | . | . | . | . | . | - | . | 4 | 4 | |||||||
| H10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | G | . | - | . | . | . | . | . | . | . | C | . | - | G | 13 | 2 | 11 | ||||||
| H11 | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | . | . | - | . | 2 | 2 | |||||||
| H12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | C | . | C | . | - | G | 3 | 3 | |||||||
| H13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | - | . | . | . | . | . | . | . | . | . | - | G | 2 | 2 | |||||||
| H14 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | G | . | . | . | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | C | . | . | . | . | G | . | . | . | . | . | . | . | C | . | TA | G | 9 | 8 | 1 | ||||||
| H16 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | A | . | C | . | . | . | . | G | . | . | . | . | . | . | . | C | . | TA | G | 5 | 5 | |||||||
| H17 | . | . | . | . | . | . | T | . | . | G | . | . | . | . | . | . | C | . | . | . | . | G | . | . | . | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | G | . | G | . | . | . | . | . | . | . | . | . | - | G | 1 | 1 | |||||||
| H19 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | G | . | G | . | . | . | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H20 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | G | . | . | . | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | C | . | . | . | . | G | . | . | . | . | . | . | . | . | . | TA | G | 16 | 7 | 9 | ||||||
| H22 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | A | . | C | . | . | . | . | G | . | . | . | . | . | . | . | . | . | TA | G | 12 | 2 | 10 | ||||||
| H23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | C | . | . | G | . | G | . | . | . | . | . | . | . | . | . | TA | G | 3 | 3 | |||||||
| H24 | C | . | - | C | . | . | . | G | T | . | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | C | . | - | . | 20 | 20 | |||||||
| H25 | C | . | - | C | . | . | . | G | . | . | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H26 | C | . | - | C | . | . | . | G | T | . | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | . | . | - | . | 1 | 1 | |||||||
| H27 | C | . | - | C | . | . | . | G | T | . | C | . | . | G | A | . | C | . | . | T | . | G | T | C | G | . | . | . | . | C | . | - | . | 2 | 2 | |||||||
| H28 | C | . | . | C | . | . | . | G | T | . | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | C | . | - | . | 2 | 2 | |||||||
| H29 | C | . | . | C | C | . | . | G | T | . | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H30 | C | . | . | C | C | . | . | G | T | G | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H31 | C | . | - | C | . | . | . | G | T | . | C | A | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | . | . | - | . | 1 | 1 | |||||||
| H32 | C | . | - | C | . | . | . | G | T | G | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H33 | C | . | - | C | . | . | . | G | T | . | C | . | . | G | A | . | C | . | A | T | . | G | . | C | G | A | . | . | . | C | . | - | . | 5 | 5 | |||||||
| H34 | C | . | - | C | . | . | . | G | T | G | C | A | . | G | A | . | C | . | A | T | . | G | . | C | G | A | . | . | A | . | . | - | . | 2 | 2 | |||||||
| H35 | C | . | - | C | . | . | . | G | T | . | C | . | . | G | A | . | C | . | . | T | . | G | . | C | G | A | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H36 | C | . | - | C | . | . | . | G | . | . | C | . | . | G | A | . | C | . | A | T | . | G | . | C | G | A | C | . | A | C | . | - | . | 1 | 1 | |||||||
| H37 | C | . | . | C | . | . | . | G | T | . | C | A | . | G | A | . | C | . | A | T | . | G | . | C | G | A | . | . | A | C | . | - | . | 2 | 2 | |||||||
| H38 | . | . | . | . | . | A | T | . | . | . | C | . | . | . | . | . | T | A | . | . | . | G | . | . | G | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H39 | C | . | . | . | . | A | T | . | T | G | C | . | C | . | . | . | T | A | . | . | . | G | . | . | . | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H40 | C | . | . | . | . | A | T | . | T | G | C | . | . | . | . | . | T | A | . | . | . | G | . | . | . | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H41 | . | . | . | . | . | A | T | . | . | G | C | . | . | . | . | . | T | A | . | . | . | G | . | . | G | . | . | . | . | C | . | - | . | 11 | 11 | |||||||
| H42 | . | . | . | . | . | A | T | . | . | G | C | . | . | . | . | G | T | A | . | . | . | G | . | . | G | . | . | . | . | . | . | - | . | 1 | 1 | |||||||
| H43 | . | . | . | . | . | A | T | . | . | G | C | . | . | . | A | . | T | A | . | . | . | G | . | . | G | . | . | . | . | C | . | - | . | 1 | 1 | |||||||
| H44 | C | . | . | . | . | A | T | . | T | G | C | . | C | . | . | . | T | A | . | . | . | G | . | . | G | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H45 | . | . | . | . | . | A | T | . | . | G | C | . | . | . | . | . | T | A | . | . | G | G | . | . | G | . | . | . | . | C | . | - | . | 4 | 4 | |||||||
| H46 | . | . | . | . | . | A | T | . | . | G | C | . | . | . | . | . | T | A | . | . | G | G | . | . | G | . | . | . | . | C | . | TA | G | 1 | 1 | |||||||
| H47 | C | . | . | . | . | A | T | . | T | G | C | . | . | . | . | . | T | A | . | . | . | G | . | . | G | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H48 | . | . | . | . | . | A | T | . | T | G | C | . | . | . | . | G | T | A | . | . | . | G | . | . | G | . | . | . | . | C | - | - | . | 1 | 1 | |||||||
| H49 | . | . | . | . | . | A | T | . | . | G | C | . | . | . | . | . | T | A | . | . | . | G | . | . | G | . | . | . | . | . | . | - | . | 1 | 1 | |||||||
Fu’s Fs neutrality test (Fu, 1996) was applied in ARLEQUIN 3.1 to assess possible deviations from neutrality and provide inferences on the demographic history of the samples analyzed. The mismatch distribution (Rogers and Harpending, 1992) was constructed in DNAsp 5.0 (Librado and Rozas, 2009). The coalescence approach available in LAMARC 2.0 (Kuhner, 2006) was used to estimate demographic parameters such as the exponential growth rate (g) and the θ parameter. The estimates were based on runs of 2,000,000 steps, sampled at every 100 generations, with 10% of the samples discarded as burn-in. The results were evaluated using TRACER v. 1.4 (Rambaut and Drummond, 2007).
Divergence between populations was estimated using PAUP*, version 4.0b10 (Swofford, 2003), based on the evolutionary model chosen according to the Akaike criterion, as determined by jModelTest (Posada, 2008). To estimate the divergence time between samples, we followed the approach of Keeney and Heist (2006), who assumed that the eastern Atlantic/Indo-Pacific and northwestern Atlantic were isolated from one another by the rise of the Isthmus of Panama (3.2 million years ago – Mya) to arrive at a mutation rate of 0.43% per million years.
Results
Sequences of the control region 1067–1070 base pairs in length were obtained for 25 specimens of C. limbatus. Thirteen polymorphic sites were identified, nine of which were transitions, with one transversion and three deletions/insertions that resulted in 12 haplotypes (GenBank access number JX025760-71); the most common haplotype was found in 11 individuals (Table 1). Haplotype diversity was relatively high (H = 0.7967), whereas nucleotide diversity was low (π = 0.0021). These values were consistent with those reported for other populations and nurseries of C. limbatus and other shark species (Table 2).
Table 2.
Diversity indices for the mitochondrial control region in different populations of C. limbatus and other shark species: π = nucleotide diversity, H = haplotype diversity, N = sample size (individuals) and HAP = number of haplotypes recorded.Note that samples from the eastern Atlantic and Indo-Pacific are excluded here because of their small size. Localities are coded as in Figure 1 and Table 1.
| Species | π | H | N | HAP | Source |
|---|---|---|---|---|---|
| Carcharhinus limbatus (BR) | 0.0021 | 0.796 | 25 | 12 | Present study |
| Carcharhinus limbatus (SC) | 0.0035 | 0.371 | 34 | 2 | Keeney et al. (2003) |
| Carcharhinus limbatus (GA) | 0.0004 | 0.461 | 12 | 2 | Keeney et al. (2003) |
| Carcharhinus limbatus (PI) | 0.0120 | 0.785 | 45 | 10 | Keeney et al. (2003) |
| Carcharhinus limbatus (TC) | 0.0106 | 0.720 | 45 | 8 | Keeney et al. (2003) |
| Carcharhinus limbatus (YT) | 0.0134 | 0.796 | 45 | 9 | Keeney et al. (2003) |
| Carcharhinus limbatus (TX) | 0.0016 | 0.813 | 49 | 8 | Keeney et al. (2003) |
| Carcharhinus limbatus (NY) | 0.0029 | 0.790 | 49 | 7 | Keeney et al. (2003) |
| Carcharhinus limbatus (BC) | 0.00077 | 0.680 | 13 | 4 | Keeney et al. (2005) |
| Carcharhinus limbatus (DG) | 0.00049 | 0.526 | 19 | 2 | Keeney et al. (2005) |
| Cetorhinus maximus | 0.0013 | 0.720 | 62 | 6 | Hoelzel et al. (2006) |
| Carcharias taurus | 0.003 | 0.717 | 26 | 4 | Stow et al. (2006) |
| Sphyrna lewini | 0.013 | 0.800 | 271 | 24 | Duncan et al. (2006) |
| Rhincodon typus | 0.011 | 0.974 | 69 | 44 | Castro et al. (2007) |
| Triakis semifasciata | 0.0067 | - | 169 | 5 | Lewallen et al. (2007) |
| Galeorhinus galeus | 0.0071 | 0.92 | 116 | 38 | Chabot et al. (2009) |
With regard to population structuring, the Fst values (data not shown) between the western Atlantic populations analyzed by Keeney et al. (2005) and the Brazilian samples were high (> 0.80) and significant, effectively the largest observed. The arrangement that included the six western Atlantic groups analyzed by Keeney et al. (2005) (northern Atlantic, eastern Gulf of Mexico, western Gulf of Mexico, northern Yucatan, Belize City and Dangriga) and the Brazilian samples was tested using AMOVA (only samples from the western Atlantic were used here because of the small number of individuals in the samples from the other populations). This analysis indicated that around 60% of the total variation (Fct = 0.6064, p < 0.05) derived from that between groups, while the remaining 40% (Fst = 0.6008, p < 0.05) resulted from variation within populations. Variation between populations of the same group was not significant (Fsc = −0.0143, p > 0.05).
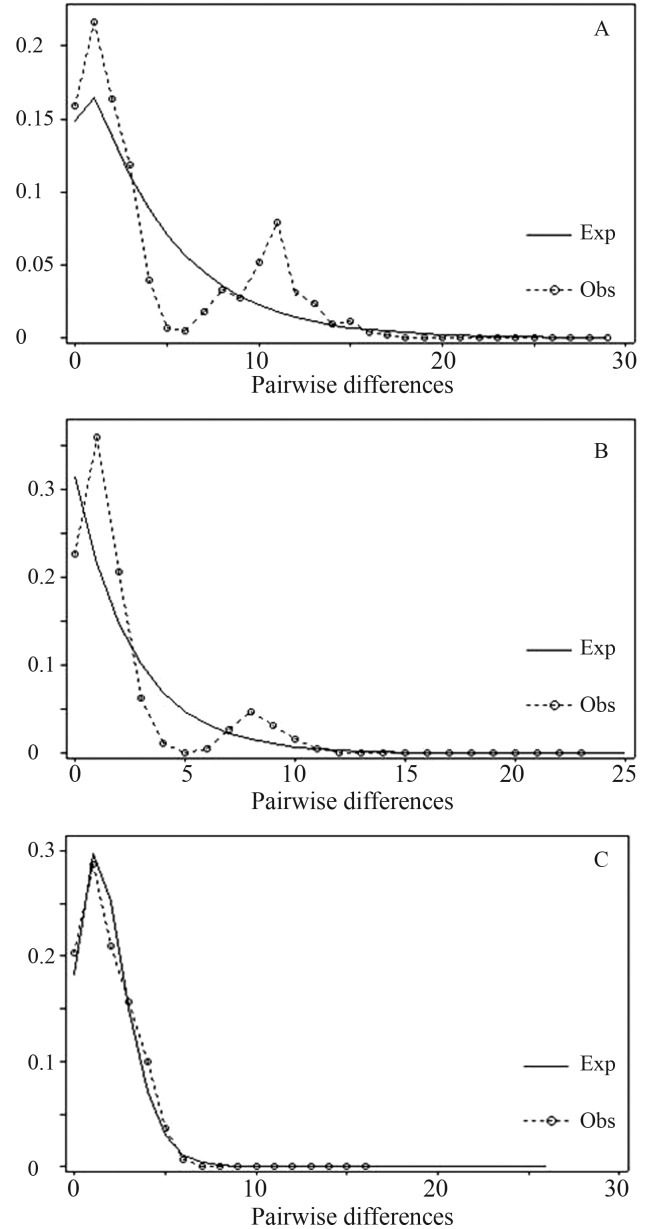
A plot of the mismatch distribution that included all of the samples genotyped worldwide (Keeney et al., 2003, 2005; Keeney and Heist, 2006) and the Brazilian samples resulted in a multimodal curve (Figure 2A) that could be accounted for by structuring of the different groups of haplotypes. However, when only the western Atlantic samples were included, the curve was bimodal (Figure 2B), indicating the existence of two groups of haplotypes that corresponded to the populations genotyped by Keeney et al. (2003, 2005) and the Brazilian samples, respectively. This finding further emphasized the differentiation of these two groups.
Figure 2.
Mismatch distribution of all populations (A), eastern Atlantic populations (B) and Brazilian samples (C). The solid line represents the curve expected (Exp) based on the expansion model. Obs – observed.
The unimodal curve obtained for the Brazilian samples (Figure 2C) suggested a process of expansion (Slatkin and Hudson, 1991; Rogers and Harpending, 1992). This conclusion was supported by the sum of the squared deviations, which did not reject this hypothesis (0.0117, p = 0.707), and by Fu’s Fs value, which indicated a significant deviation from neutrality (Fs = −5.7438, p = 0.001). Since these results supported a population expansion model, the coalescent model with constant exponential population growth provided by LAMARC 2.0 was chosen to estimate the demographic parameters of the Brazilian samples. The value of theta (θ) was 0.005859 and the exponential growth rate (g) was 606.02, indicating a relatively high rate of demographic growth.
The divergence among the haplotypes recorded in the northwestern Atlantic, northern Brazil and eastern Atlantic/Indo-Pacific was evaluated using the model (HKY+I+G: Lset Base = (0.3178 0.2024 0.1352) Nst = 2 TRatio = 8.1339 Rates = gamma, Shape = 0.5869 Pinvar = 0.9469) selected by jModelTest. The mean corrected divergence values for the three comparisons were 1.47% (0.6–2.7%) for northwestern Atlantic vs. northern Brazil, 2.62% (1.4–4.4%) for northwestern Atlantic vs. eastern Atlantic/Indo-Pacific and 3.68% (2.2–5.7%) for northern Brazil vs. eastern Atlantic/Indo-Pacific. The mean within-group divergence among the haplotypes was similar in the three groups, at 0.3% in the northwestern Atlantic and northern Brazil and 0.37% in the eastern Atlantic/Indo-Pacific.
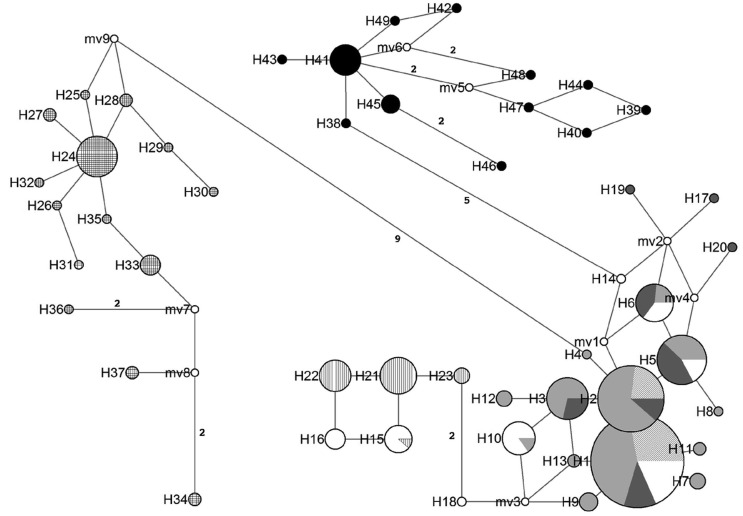
The haplotype network analysis revealed a clear separation of the Brazilian samples from the other populations of C. limbatus (Figure 3), given both the lack of shared haplotypes and the large number of mutations that typically separated the Brazilian haplotypes from those of other populations (five mutation steps from other western Atlantic populations). Based on the rate of 0.43% per million years, the most conservative estimate for the separation of the Brazilian and northwestern Atlantic populations would be 1.38 Mya.
Figure 3.
Haplotype network in which each circle represents a different haplotype (as labeled) and the circle size is proportional to its recorded frequency (based on all samples). The length of the branch joining different haplotypes is proportional to the number of mutations that separate them. The number of mutations is indicated in each branch, except where only a single mutation is involved. The relative frequencies of the haplotypes in each population are represented by different types of shading:diagonal stripes = northwestern Atlantic (SC and GA); light gray = eastern Gulf of Mexico (YT, TC and PI); dark gray = western Gulf of Mexico (TX); white = northern Yucatan (NY); vertical stripes = Belize City (BC); horizontal stripes = Dangriga (DG); crossed hatching = eastern Atlantic and Indo-Pacific; black = northern Brazil.
Discussion
Despite the intense exploitation of stocks throughout the world and the relative vulnerability of the sharks to such pressure (Baum and Myers, 2004; Myers et al., 2007), studies of many populations worldwide, including those of C. limbatus analyzed here, have found relatively high levels of haplotypic diversity. Furthermore, the number of haplotypes recorded in the northern Brazilian sample was relatively large when compared with the populations analyzed by Keeney et al. (2003, 2005), which could be accounted for by a process of expansion. However, as only one Brazilian population was genotyped, it is possible that the large number of unique haplotypes resulted from migrations among different South American populations not yet sampled but distinct from those sampled by Keeney et al. (2003, 2005). New data, including new sampling strategies and nuclear markers, would be necessary to test this hypothesis.
In contrast to the results of this study, recent analyses of shark populations from the coastal regions of Southern and Central America have not detected any differentiation between these areas (Mendonça et al., 2011; Karl et al., 2012). Mendonça et al. (2011) found no evidence of population structure in Rhizopriodon porosus from the Caribbean and northern Brazil. According to these authors, this lack of structuring may be related to the high migration rates across this region, particularly the high rates of primary production in the delta of the Amazon River, which attracts the sharks to this area.
This interpretation is supported by the fact that, in contrast to the studies of Keeney et al. (2003, 2005), the sample analyzed in the present study consisted of adults and is thus likely to have included potential migrants. Given this situation, a more reliable evaluation of this question will depend on the genotyping of other populations from South and Central America and the inclusion of juveniles from nursery grounds.
Obviously, it is difficult to sample the full geographic variation of cosmopolitan marine species such as the blacktip shark, so the inclusion of the present samples from Brazil is an important contribution to understanding the level of structuring that currently exists among populations of the western Atlantic. Keeney et al. (2003, 2005) investigated the genetic structuring of C. limbatus from the northwestern Atlantic, Gulf of Mexico and Caribbean by analyzing mitochondrial (control region) and nuclear (microsatellite) markers in neonates and found significant heterogeneity among nurseries. The differences observed in the frequency of mitochondrial haplotypes and microsatellites indicated the existence of philopatric behavior in the females, given the greater differentiation in maternally-mediated genetic traits in comparison with biparental traits.
An analysis of a wider spread of samples, including the Pacific, concluded that the subdivision found in the Atlantic was not repeated in the Pacific. In addition, shallow population structuring was found between the lineages of the eastern Atlantic and the Pacific. The addition of new Atlantic populations, such as the samples from northern Brazil, revealed additional structuring, as demonstrated by AMOVA, although these samples were genetically more divergent than those genotyped by Keeney et al. (2003, 2005). This was clearly shown by the mismatch plots in which the general multimodal curve was replaced by a bimodal arrangement when only the western Atlantic samples were included. Despite the relative differentiation of the Caribbean populations (Belize City and Dangriga) in relation to the other northwestern Atlantic samples (as emphasized by the haplotype network), these populations were still part of the northwestern Atlantic mismatch curve and shared common haplotypes. This observation confirmed that the samples from northern Brazil represent a diverse (separated by five mutations) and completely independent unit in relation to the remaining Atlantic populations, probably as a consequence of philopatric behavior, as previously noted in literature (Keeney et al., 2003, 2005; Hueter et al., 2004, 2005; Keeney and Heist, 2006).
The divergence time between the northeastern Brazilian and northwestern Atlantic populations was estimated at 1.38 Mya. However, as the mutation rate of the control region of C. limbatus is still unknown and since this value was based on the proposition of Keeney and Heist (2006), this divergence time may have been underestimated. The data nevertheless indicate that at least one separation event has occurred between northern and southern Atlantic populations since the formation of the Isthmus of Panama (3.3 Mya). As C. limbatus is adapted to warmer tropical and subtropical waters, shifts in ocean temperatures during the Pleistocene may have contributed to the isolation of populations (Keeney and Heist, 2006). The possibility of contact between the eastern and western Atlantic populations through a southern corridor, including the Brazilian coast, is not supported by the data presented here.
The ability of sharks to travel long distances and the fact that the marine environment has few physical barriers to dispersal (Palumbi, 1994; White et al., 2009) make it extremely difficult to define the limits of a species geographic distribution. As shown here, although C. limbatus can disperse over distances > 2,000 km (Carlson and Brusher, 1999), its populations can be genetically highly differentiated. The samples analyzed here were taken from a stock distinct from those examined in previous studies and the differentiation can be accounted for not only by the migratory behavior of the females but also by their fidelity to breeding sites, a pattern confirmed in other studies.
The results of this study emphasize the marked variability of the Brazilian samples, which is characteristic of a population in expansion, as well as considerable differences in comparison with other populations of the western Atlantic. This suggests that the Brazilian population should be treated as a distinct management unit, although such conclusions should be considered tentative given the fact that the study was based on a single molecular marker and only one population. Obviously, more representative sampling and the analysis of additional markers would provide more conclusive evidence on the genetic characteristics of the South American populations of C. limbatus.
Acknowledgments
This research was supported by Fundação de Amparo à Pesquisa do Estado do Pará (FAPESPA) through a scholarship to D.S. We also thank to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)- Edital Universal and the Research Fellowship to M.V.
Footnotes
Associate Editor: Fausto Foresti
References
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Baum J, Myers R. Shifting baselines and the decline of pelagic sharks in the Gulf of Mexico. Ecol Lett. 2004;7:135–145. [Google Scholar]
- Bornatowski H. A parturition and nursery area for Carcharhinus limbatus (Elasmobranchii, Carcharhinidae) off the coast of Paraná, Brazil. Braz J Oceanogr. 2008;56:317–319. [Google Scholar]
- Bradshaw C, Fitzpatrick B, Steinberg C, Brook B, Meekan M. Decline in whale shark size and abundance at Ningaloo Reef over the past decade: The world’s largest fish is getting smaller. Biol Conserv. 2008;141:1894–1905. [Google Scholar]
- Carlson J, Brusher J. An index of abundance for coastal species of juvenile sharks from the northeast Gulf of Mexico. Mar Fish Rev. 1999;61:37–45. [Google Scholar]
- Castro AL, Stewart S, Wilson G, Hueter RE, Meekan MG, Motta PJ, Bowen BW, Karl S. Population genetic structure of Earth’s largest fish, the whale shark (Rhincodon typus) Mol Ecol. 2007;16:5183–5192. doi: 10.1111/j.1365-294X.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- Chabot CL, Allen LG. Global population structure of the tope (Galeorhinus galeus) inferred by mitochondrial control region sequence data. Mol Ecol. 2009;18:545–552. doi: 10.1111/j.1365-294X.2008.04047.x. [DOI] [PubMed] [Google Scholar]
- Compagno LJV. Sharks of the World. An annotated and illustrated catalogue of shark species known to date. FAO Fish Synopsis. 1984;4:251–655. [Google Scholar]
- Compagno L, Dando M, Fowler S. A field Guide to the Sharks of the World. Harper-Collins; London: 2005. p. 368. [Google Scholar]
- Duncan K, Martin A, Bowen B, De Couet H. Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini) Mol Ecol. 2006;15:2239–2251. doi: 10.1111/j.1365-294X.2006.02933.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin, ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Faunce CH, Layman CA. Sources of variation that affect perceived nursery function of mangroves. In: Nagelkerken I, editor. Ecological Connectivity Among Tropical Coastal Ecosystems. Springer; the Hague: 2009. pp. 401–421. [Google Scholar]
- Feldheim K, Gruber S, Ashley M. Population genetic structure of the lemon shark (Negaprion brevirostris) in the western Atlantic: DNA microsatellite variation. Mol Ecol. 2001;10:295–303. doi: 10.1046/j.1365-294x.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- Ferretti F, Worm B, Britten GL, Heithaus MR, Lotze HK. Patterns and ecosystem consequences of shark declines in the ocean. Ecol Lett. 2010;13:1055–1071. doi: 10.1111/j.1461-0248.2010.01489.x. [DOI] [PubMed] [Google Scholar]
- Fu YX. New statistical tests of neutrality for DNA samples from a population. Genetics. 1996;143:557–570. doi: 10.1093/genetics/143.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M, Rowat D, Hall J, Gell F, Ormond R. Transatlantic migration and deep mid-ocean diving by basking shark. Biol Lett. 2008;4:395–398. doi: 10.1098/rsbl.2008.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Heupel MR, Simpfendorfer CA. Estuarine nursery areas provide a low-mortality environment for young bull sharks Carcharhinus leucas. Mar Ecol Progr Ser. 2011;433:237–244. [Google Scholar]
- Heupel MR, Simpfendorfer CA, Hueter RE. Estimation of shark home ranges using passive monitoring techniques. Environ Biol Fishes. 2004;71:135–142. [Google Scholar]
- Heupel MR, Carlson JK, Simpfendorfer CA. Shark nursery areas: Concepts, definition, characterization and assumptions. Mar Ecol Prog Ser. 2007;337:287–297. [Google Scholar]
- Hoelzel A, Shivji M, Magnussen J, Francis M. Low worldwide genetic diversity in the basking shark (Cetorhinus maximus) Biol Lett. 2006;2:639–642. doi: 10.1098/rsbl.2006.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueter R, Heupel M, Heist E, Keeney D. The implications of philopatry in sharks for the management of shark fisheries. J Northwest Atl Fish Sci. 2004;35:239–247. [Google Scholar]
- Hueter R, Heupel M, Heist E, Keeney D. Evidence of philopatry in sharks and implications for the management of shark fisheries. J Northwest Atl Fish Sci. 2005;35:239–247. [Google Scholar]
- Karl SA, Castro ALF, Garla RC. Population genetics of the nurse shark (Ginglymostoma cirratum) in the western Atlantic. Mar Biol. 2012;159:489–498. [Google Scholar]
- Keeney D, Heist E. Worldwide phylogeography of the blacktip shark (Carcharhinus limbatus) inferred from mitochondrial DNA reveals isolation of western Atlantic populations coupled with recent Pacific dispersal. Mol Ecol. 2006;15:3669–3679. doi: 10.1111/j.1365-294X.2006.03036.x. [DOI] [PubMed] [Google Scholar]
- Keeney D, Heupel M, Hueter R, Heist E. Genetic heterogeneity among blacktip shark, Carcharhinus limbatus, continental nurseries along the US Atlantic and Gulf of Mexico. Mar Biol. 2003;143:1039–1046. [Google Scholar]
- Keeney D, Heupel M, Hueter R, Heist E. Microsatellite and mitochondrial DNA analyses of the genetic structure of blacktip shark (Carcharhinus limbatus) nurseries in the northwestern Atlantic, Gulf of Mexico, and Caribbean Sea. Mol Ecol. 2005;14:1911–1923. doi: 10.1111/j.1365-294X.2005.02549.x. [DOI] [PubMed] [Google Scholar]
- Kohler N, Casey J, Turner P. NMFS Cooperative Shark Tagging Program, 1962–93: An atlas of shark tag and recapture data. Mar Fish Rev. 1998;60:1–87. [Google Scholar]
- Kuhner MK. LAMARC 2.0: Maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R. Clustal W and Clustal X ver. 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lewallen EA, Anderson TW, Bohonak AJ. Genetic structure of leopard shark (Triakis semifasciata) populations in California waters. Mar Biol. 2007;152:599–609. [Google Scholar]
- Librado P, Rozas J. DnaSP, ver. 5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Mendonça FF, Oliveira C, Gadig OBF, Foresti F. Phylogeography and genetic population structure of Caribbean sharpnose shark Rhizoprionodon porosus. Rev Fish Biol Fish. 2011;21:799–814. [Google Scholar]
- Meylan A, Bowen B, Avise J. A genetic test of the natal homing versus social facilitation models for green turtle migration. Science. 1990;248:724–727. doi: 10.1126/science.2333522. [DOI] [PubMed] [Google Scholar]
- Musick J, Burgess G, Cailliet G, Camhi M, Fordham S. Management of sharks and their relatives (Elasmobranchii) Fisheries. 2000;25:9–13. [Google Scholar]
- Myers R, Worm B. Extinction, survival or recovery of large predatory fishes. Philos Trans R Soc Lond B Biol Sci. 2005;360:13–20. doi: 10.1098/rstb.2004.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RA, Baum JK, Shepherd TD, Powers SP, Peterson CH. Cascading effects of the loss of apex predatory sharks from a coastal ocean. Science. 2007;315:1846–1850. doi: 10.1126/science.1138657. [DOI] [PubMed] [Google Scholar]
- Palsboll PJ, Berube M, Allendorf FW. Identification of management units using population genetic data. Trends Ecol Evol. 2007;22:11–16. doi: 10.1016/j.tree.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Palumbi S. Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst. 1994;25:547–572. [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. Tracer ver. 1. 4. University of Oxford; Oxford: 2007. [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rosenbaum H, Pomilla C, Mendez M, Leslie M, Best P, Findlay K, Minton G, Ersts P, Collins T, Engel M. Population structure of humpback whales from their breeding grounds in the South Atlantic and Indian oceans. PLoS One. 2009;4:e7318. doi: 10.1371/journal.pone.0007318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpfendorfer C, Milward N. Utilisation of a tropical bay as a nursery area by sharks of the families Carcharhinidae and Sphyrnidae. Environ Biol Fishes. 1993;37:337–345. [Google Scholar]
- Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow A, Zenger K, Briscoe D, Gillins M, Peddemors V, Otaway N, Harcourt R. Isolation and genetic diversity of endangered grey nurse shark (Carcharias taurus) populations. Biol Lett. 2006;2:308–311. doi: 10.1098/rsbl.2006.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), ver. 4.0 b10. Sinauer Associates Inc., Publishers; Sunderland: 2003. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software ver. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tillett BJ, Meekan MG, Field IC, Hua Q, Bradshaw CJA. Similar life history traits in bull (Carcharhinus leucas) and pig-eye (C. amboinensis) sharks. Mar Freshw Res. 2011;62:850–860. [Google Scholar]
- Tsai WP, Liu KM, Joung SJ. Demographic analysis of the pelagic thresher shark, Alopias pelagicus, in the northwestern Pacific using a stochastic stage-based model. Mar Freshw Res. 2010;61:1056–1066. [Google Scholar]
- Vaudo J, Heithaus M. Spatiotemporal variability in a sandflat elasmobranch fauna in Shark Bay, Australia. Mar Biol. 2009;156:2579–2590. [Google Scholar]
- Waples R. Separating the wheat from the chaff: Patterns of genetic differentiation in high gene flow species. J Hered. 1998;89:438–450. [Google Scholar]
- Ward R, Woodwark M, Skibinski D. A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. J Fish Biol. 1994;44:213–232. [Google Scholar]
- White TA, Stefanni S, Stamford J, Hoelzel A. Unexpected panmixia in a long-lived, deep-sea fish with well-defined spawning habitat and relatively low fecundity. Mol Ecol. 2009;18:2563–2573. doi: 10.1111/j.1365-294X.2009.04218.x. [DOI] [PubMed] [Google Scholar]
- Yokota L, Lessa RP. A nursery area for sharks and rays in northeastern Brazil. Environ Biol Fishes. 2006;75:349–360. [Google Scholar]