Abstract
Parodontidae fish show few morphological characteristics for the identification of their representatives and chromosomal analyses have provided reliable features for determining the interrelationships in this family. In this study, the chromosomes of Apareiodon hasemani from the São Francisco River basin, Brazil, were analyzed and showed a karyotype with 2n = 54 meta/submetacentric chromosomes, and a ZZ/ZW sex chromosome system. The study revealed active NORs located on pair 11 and additional 18S rDNA sites on pairs 7 and 22. The 5S rDNA locus was found in pair 14. It showed a pericentric inversion regarding the ancestral condition. The satellite DNA pPh2004 was absent in the chromosomes of A. hasemani, a shared condition with most members of Apareiodon. The WAp probe was able to detect the amplification region of the W chromosome, corroborating the common origin of the system within Parodontidae. These chromosomal data corroborate an origin for the ZW system of Parodontidae and aid in the understanding of the differentiation of sex chromosome systems in Neotropical fishes.
Keywords: cytogenetics, FISH, 18S rDNA, 5S rDNA, Neotropical fishes
Introduction
Fishes are the largest group of vertebrates, with a wide variety of adaptive responses to diversified aquatic habitats and ecological limitations. Also with respect to sex determination, fishes exhibit much variety. Unlike the situation in most vertebrates, where the male and female sexes are represented by two different individuals (gonochorism), hundreds of fish species are known to be hermaphroditic (Devlin and Nagahama, 2002; Schartl, 2004).
Sex chromosomes are fundamental in many vertebrate species for the development of either a male or a female (Livernois et al., 2012). Few species of fish are carriers of heteromorphic chromosomal systems when compared to the enormous taxa diversity (Oliveira et al., 2007). However, fishes have a remarkable variety of sex determination chromosome systems (XY, X0, X1X2Y, XY1Y2, ZW, Z0, ZW1W2, Z1Z1Z2Z2/Z1Z2, W1W2, WXZ) (Moreira-Filho et al., 1993; Devlin and Nagahama, 2002; Oliveira et al., 2008), in which the ZZ/ZW simple system with female heterogamety is the most frequent one among Neotropical fishes (Artoni and Bertollo, 2002; Bellafronte et al., 2011; Machado et al. 2011).
The family Parodontidae occurs throughout South America and its species are classified in three genera: Parodon, Apareiodon and Saccodon (Pavanelli, 2003). Generally they are robust fish, with strong pectoral, ventral and caudal fins, a fusiform body, and a dorsal profile which is more arched than the ventral one (Travassos, 1957). Parodontidae taxonomy is controversial because the family members lack diagnostic morphological traits that are sufficiently reliable for accurate phylogenetic analysis (Pavanelli and Britski, 2003; Ingenito LFS, 2008, Ph.D Thesis, Universidade Federal do Rio de Janeiro, Brazil). Ingenito argued that existing morphological phylogenetic evidence for the genus Apareiodon is insufficient to support its maintenance, and that Apareiodon should be regarded as a junior synonym of Parodon.
Cytogenetic analyses of both Parodon and Apareiodon revealed a conserved diploid number of 54 chromosomes, but with remarkable heterogeneity in the distribution of heterochromatin, NOR activity, number and location of 18S rDNA, 5S rDNA and satellite DNA familiy sites, and morphologically differentiated sex chromosome systems (for a review see Bellafronte et al., 2011). From the Parodontidae species that have been cytogenetically analyzed, six have sex chromosomes: ZZ/ZW1W2 for Apareiodon affinis Steindachner, 1879, and ZZ/ZW for Parodon hilarii Reinhardt, 1867, Parodon moreirai Ingenito & Buckup, 2005, Apareiodon vladii Pavanelli, 2006, Apareiodon sp. and Apareiodon ibitiensis Campos, 1944 (Moreira-Filho et al., 1980, 1993; Centofante et al., 2002; Vicente et al., 2003; Rosa et al., 2006; Vicari et al., 2006; Bellafronte et al., 2009).
In Parodontidae, satellite DNA has great importance for the understanding of karyotype differentiation. So far, two satellite DNAs have been described for this fish group: pPh2004 (Vicente et al., 2003) and WAp (Schemberger et al., 2011). The physical mapping of the pPh2004 sequence showed that this satellite DNA is present on chromosomes Z and W (also on autosomal chromosomes); however, it is not a part of the heterochromatic amplified region of the W chromosome heteromorphism (Bellafronte et al., 2011). The WAp satellite DNA probe is capable of detecting the amplification region and the chromosome W heteromorphism, corroborating the common origin of this system in Parodontidae, apart from homologies to other chromosome sites (Schemberger et al., 2011).
The integrated analysis of chromosomal markers has also allowed to deduce the chromosomal differentiation of the family, which is organized in: (i) species without morphologically differentiated sex chromosomes (Apareiodon piracicabae Eigenmann, 1907 and Apareiodon vittatus Garavello, 1977), (ii) species with differentiated sex chromosomes and without satellite DNA pPh2004 (A. ibitiensis, Apareiodon sp., A. vladii), and (iii) species with proto sex chromosomes and/or heteromorphic sex chromosome systems and presence of satellite DNA pPh2004 (A. affinis, P. hilarii, P. moreirai, Parodon nasus Kner, 1859 and Parodon pongoensis Allen, 1942) (Schemberger et al., 2011).
Based on these data, the present study aimed at characterizing the species Apareiodon hasemani Eigenmann, 1916 (from the São Francisco River basin, Brazil) with respect to chromosome number, morphology, banding, sex system, presence or absence of certain molecular cytogenetic markers, as well as to establish karyoevolutionary relationships based in other cytogenetic studies performed in the family Parodontidae.
Materials and Methods
Chromosome studies were carried out in 18 specimens (7 males and 11 females) of A. hasemani, collected in the main channel of the São Francisco River, in Pirapora city, state of Minas Gerais, Brazil (17°21′17.47″ S and 44°57′18.45″ W). The analyzed samples were deposited in the Núcleo de Pesquisas em Limnologia, Ictiologia e Aquicultura (Nupelia), Universidade Estadual de Maringá, Brazil, with voucher number NUP11512. Samples were collected in compliance with the Ethics Committee on Animal Experimentation (Process CEUA 07/2011) of the Universidade Estadual de Ponta Grossa, Brazil) and with authorization to collect the biological material (Brazilian Federal license: Ministério do Meio Ambiente/Instituto Brasileiro do Meio Ambiente e dos Recursos Renováveis MMA/IBAMA/SISBIO number 10538-1) and the Instituto Estadual de Florestas de Minas Gerais.
Mitotic chromosomes were obtained from the anterior kidney, according to the methodology described by Bertollo et al. (1978). The heterochromatin was detected using the methodology of C-banding (Sumner, 1972). C-banded chromosomes were stained with propidium iodide (50 μg mL−1) according to Lui et al. (2009). The nucleolar organizer regions were detected using the silver nitrate method (Ag-NOR) described by Howell and Black (1980).
Fluorescence in situ hybridization (FISH) experiments were performed in the Parodontidae specimens using probes for 18S rDNA (Hatanaka and Galetti Jr, 2004), 5S rDNA (Martins and Galetti Jr, 1999), pPh2004 satellite DNA (Vicente et al., 2003) and WAp satellite DNA (Schemberger et al., 2011). The 18S rDNA, 5S rDNA and pPh2004 cloned probes were labeled with biotin or digoxigenin via PCR, using plasmid vector primers (T7 promoter and M13 reverse). The PCR amplification was done with: 20 ng of template DNA, 1X reaction buffer, 2 mM MgCl2, 40 μM dATP, dGTP and dCTP, 28 μM dTTP, 12 μM biotin 16-dUTP, or digoxigenin 11 dUTP (Roche Applied Science), 0.3 μM of each primer and 1 U Taq DNA Polymerase (Invitrogen). The WAp probe was labeled via degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR), using 11-dUTP-digoxigenin (Roche Applied Science). This PCR amplification was done with: 100 ng of template DNA, 1X reaction buffer, 2 mM MgCl2, 40 μM dATP, dGTP and dCTP, 28 μM dTTP, 12 μM 11-dUTP-digoxigenin, 2 μM DOP primer, and 1 U Taq DNA polymerase (Invitrogen). The FISH procedure was performed under high stringency conditions (2.5 ng/μL probe, 50% formamide, 2X SSC, 10% dextran sulfate) following the methodology described by Pinkel et al. (1986). The signal was detected using the anti-streptavidin antibody conjugated to Alexa Fluor 488 (Invitrogen) and an anti-digoxigenin antibody conjugated to rhodamine (Roche Applied Science). The chromosomes were counterstained with DAPI (0.2 μg/mL), covered with Vectashield mounting medium (Vector) and analyzed with the aid of an Olympus BX41 epifluorescence microscope equipped with a DP71 digital image capture system (Olympus).
Chromosomes were identified based on the system proposed by Levan et al. (1964) and classified as metacentric (m), submetacentric (sm), or subtelocentric (st).
Results
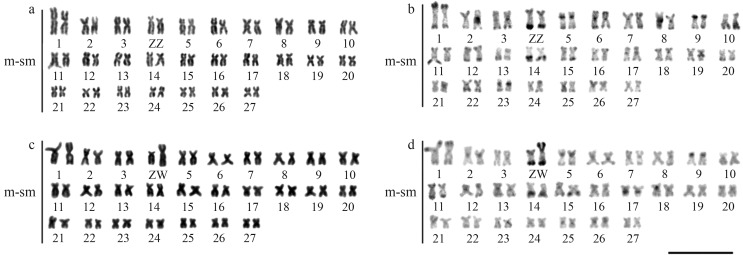
The species A. hasemani showed 2n = 54 meta-submetacentric chromosomes, with a fundamental number equal to 108 for both sexes (Figure 1). However, a heteromorphism in size and morphology was observed for chromosome pair 4, restricted to females, which had a medium-sized submetacentric chromosome (Z) and a large metacentric chromosome (W) (Figure 1). The C-banding technique revealed centromeric heterochromatin regions in some chromosomes, as well as interstitial and terminal bands (Figure 1b and 1d). The ZZ pair showed centromeric and terminal C-bands on both chromosome arms. In all metaphases of females, the W chromosome had similar centromeric and terminal heterochromatin bands on the long arm, when compared to the Z chromosome. The heterochromatin segments, however, occupied almost the entire short arm of the W chromosome, being twice as large as in the Z chromosome (Figure 1b, d). The nucleolar organizer regions stained with silver (Ag-NORs) showed terminal signals on the long arm of one chromosome pair, pair 11, which was subtelocentric (Figure 2, box), while FISH with 18S rDNA probes showed up to five terminal signals on the long arm of pairs 11 (larger and more evident marks), 7 and 22 (smaller in size), and all these signals generally coinciding with GC-rich regions (Figure 2a). FISH with 5S rDNA probes showed only one marked pair, located in the interstitial region of the long arm of chromosome pair 14 (Figure 2a). Apareiodon hansemani did not show pPh2004 satellite DNA sites (data not shown), but WAp satellite DNA sites were found in the terminal regions of some chromosomes of the karyotype (Figure 2b). The Z chromosome showed the satellite DNA WAp in the terminal region of the short and long arms and in the proximal region of the short arm (Figure 2bB). In turn, the W chromosome also showed a WAp amplification from the proximal region of the short arm (Figure 2b).
Figure 1.
Karyotypes of males and female Apareiodon hasemani arranged from Giemsa-staining (a, c) and C-banding (b, d). Bar = 10 μm.
Figure 2.
Chromosomal markers in Apareiodon hasemani (a) representative double-FISH karyotype, showing five 18S rDNA sites and chromosome pair 14 bearing 5S rDNA sites. (b) representative the WAp signals. The nucleolar organizer regions revealed by silver nitrate (Ag-NOR) staining are shown in the insert box. Bar = 10 μm.
Discussion
Karyotype analyses of Apareiodon hasemani (Figure 1) show a diploid number of 54 chromosomes, similarly to other species of Parodontidae, except for the population of A. affinis from the Upper Paraná River basin (Moreira-Filho et al., 1980), which has 2n = 54/55 chromosomes. The chromosome morphology of A. hasemani consists mainly of meta-submetacentric chromosomes, a characteristic shared with A. affinis (Moreira-Filho et al., 1980), P. hilarii (Moreira-Filho et al., 1993) and P. moreirai (Centofante et al., 2002). In addition to meta-submetacentric chromosomes, other species also have subtelocentric chromosomes (Moreira-Filho et al., 1985; Jesus and Moreira-Filho 2000a, b; Bellafronte et al., 2005; Vicari et al., 2006; Rosa et al., 2006). Populations of A. affinis from the Lower Paraná and Paraguay River basins showed a structural polymorphism with the presence of acrocentric chromosomes, probably caused by pericentric inversions (Jesus et al., 1999; Jorge and Moreira-Filho, 2000).
The location of the major NORs sites of A. hasemani in the terminal region of the long arm of a subtelocentric chromosome pair, revealed by silver nitrate staining and the 18S rDNA probes (Figure 2a, box), is a character shared by other species of the genus Apareiodon and by P. nasus (Bellafronte et al., 2005; Vicari et al., 2006; Rosa et al., 2006; Bellafronte et al., 2009, 2011). In populations of A. affinis in the Lower Paraná and Paraguay basins, acrocentric chromosomes originated from pericentric inversions are NOR carriers (Jesus et al., 1999; Jorge and Moreira-Filho, 2004). Additional sites were also found in A. hasemani, in a characteristic shared with A. ibitiensis and A. vittatus.. Dispersal events may have originated the additional 18S rDNA sites in these species, since they are found in more than one chromosome pair (Jesus and Moreira-Filho, 2000a; Bellafronte et al., 2009, 2011, this study). Considering that NORs are generally associated with terminal heterochromatin, chromosome rearrangements and transpositions become more likely, which may result in dispersion in the genome (Moreira Filho et al., 1984).
FISH with 5S rDNA probes in Parodontidae revealed that these cistrons are often located on the short arm of one of the submetacentric chromosome pairs (Centofante et al., 2002; Vicari et al., 2006; Bellafronte et al., 2009, 2011) (Figure 2a). Additional sites may, however, occur in different locations in A. vladii, P. hilarii, P. pongoensis and P. nasus, which show a case of synteny with the 18S rDNA gene (Vicente et al., 2001; Bellafronte et al., 2005; Rosa et al., 2006). Apareiodon hasemani and most species of Parodontidae show only one submetacentric pair carrying 5S rDNA sites; however, the cistron is located in the subterminal region of the long arm. It is likely that this pattern originated from a pericentric inversion, since in other Parodontidae species and in the outgroup Anostomidae the 5S rDNA sites are located on the short arm (Bellafronte et al., 2011). A similar case was observed in the genus Astyanax (Kavalco et al. 2004), where A. intermedius shows all 5S rDNA sites located on acrocentric chromosomes, while A. guiton has a marked subtelocentric pair besides the aforementioned acrocentric chromosomes, probably originated by a pericentric inversion.
Apareiodon hasemani shares a ZZ/ZW heteromorphic sex chromosome system with P. hilarii, P. moreirai, A. vladii, A. ibitiensis and Apareiodon sp. (Bellafronte et al., 2011). The distribution of constitutive heterochromatin in A. hasemani (Figure 1b,d) is characterized by centromeric bands in some chromosome pairs, large interstitial and terminal blocks, and blocks adjacent to the NORs, which have also been found in several other species of Parodontidae (Moreira-Filho et al., 1984; Jesus and Moreira-Filho 2000a,b; Vicente et al., 2001, 2003; Centofante et al., 2002; Bellafronte et al., 2005; Rosa et al., 2006; Vicari et al., 2006). Moreover, the Z sex chromosome exhibited a correspondence of proximal and terminal bands on short arms with those of other parodontids bearing sex chromosomes (Bellafronte et al., 2011; Schemberger et al., 2011). Schemberger et al. (2011) inferred that the Z and W chromosomes of Parodontidae were initially a homologous pair, and that heterocromatinization through the accumulation of WAp sequences was a decisive step in the differentiation of the W chromosome.
On the other hand, the terminal heterochromatic block in the long arm of the Z and W chromosomes of A. hasemani (Figure 2b) led to a size increase of the long arm of these chromosomes, in relation to other species in the group. Thus, with the increase in the long arm of the Z chromosome due to this terminal heterochromatin, the heterochromatic differentiation of the W chromosome in the short arm region did not lead to a sufficient size increase to exceed the total length of the long arm (Figure 3), a common state observed among the other parodontid carriers of heteromorphic sex chromosomes. In this proposed scenario, not only would the W chromosome change in shape and size, but the Z chromosome would also have increased in size in relation to a possible ancestral condition. This mechanism is in agreement with the model for the evolution of heteromorphic sex chromosomes (Muller, 1964; Charlesworth, 1978; Green, 1990).
Figure 3.
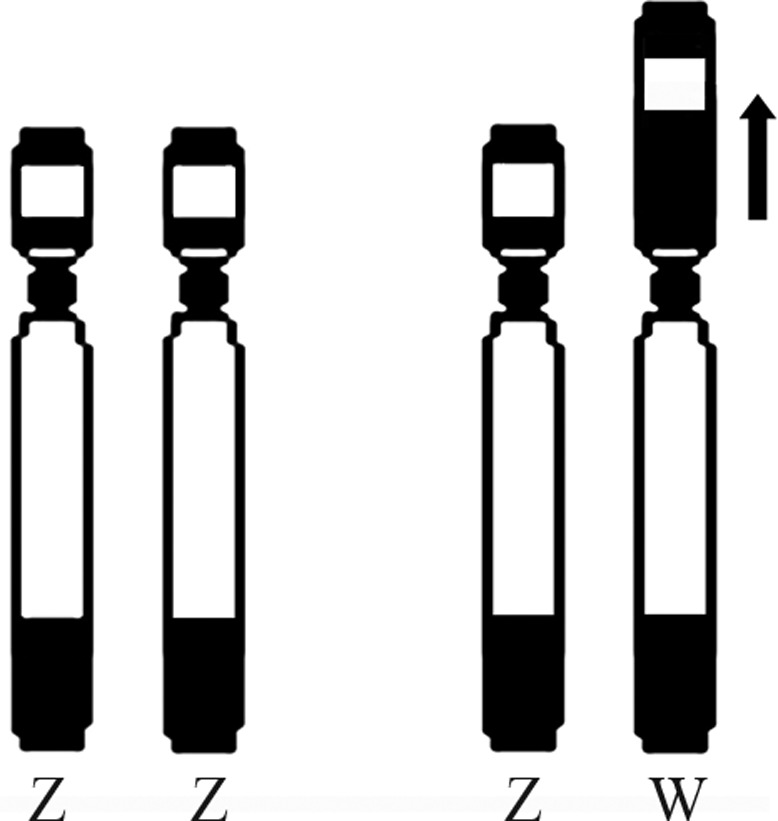
Hypothetical derivation of the W chromosome from an ancestral chromosome similar to the Z chromosome in Apareiodon hasemani.
During the evolution of the heteromorphic sex chromosomes XY or ZW, there is a suppression of meiotic recombination between a pair of homomorphic chromosomes (homologues), which progressively accumulate sex determining genes, followed by structural chromosomal rearrangements such as deletions, insertions, accumulation of transposable elements, and expansion of repetitive sequences (Schartl, 2004; Kasahara, 2009; Livernois et al., 2012). In recent years, repetitive DNA sequences have been widely studied in fishes, providing important information on the differentiation and evolution of sex chromosomes. These studies have shown that the differentiation of sex chromosomes is frequently associated with the accumulation of such DNA sequences on chromosomes (Vicente et al., 2003; Parise-Maltempi et al., 2007; Machado et al., 2011). FISH with the sequence pPh2004 in A. hasemani showed that this satellite DNA is not present in the karyotype or in species with heteromorphic ZW sex chromosomes : A. ibitiensis, Apareiodon sp. and A. vladii (Bellafronte et al., 2011).
Furthermore, the WAp satellite DNA probe (Schemberger et al., 2011) detected the W chromosome amplification region of A. hasemani, suggesting that this system is common to other parodontid carriers of sex chromosomes. In the family Parodontidae, all species studied so far have positive sites for this marker, suggesting a common origin of this satellite DNA and therefore of the sex chromosome systems (Schemberger et al., 2011). The simple ZZ/ZW sex chromosome systems may have originated from a paracentric inversion of a WAp terminal site to the proximal region of the short arm of a metacentric chromosome pair, with the subsequent amplification of this sequence leading to differentiation of the W sex chromosome in most species. Through the use of chromosome markers, Schemberger et al. (2011) demonstrated the existence of closely related species groups: (i) those without heteromorphic sex chromosomes (A. piracicabae and A. vittatus); (ii) those with proto sex chromosomes and the presence of satellite DNA pPh2004 (P. nasus and P. pongoensis); (iii) those with a heteromorphic sex chromosome system (A. ibitiensis and A. vladii); (iv) those with a heteromorphic sex chromosome system and the presence of satellite DNA pPh2004; and (v) a species with a multiple sex chromosome system and satellite DNA pPh2004 (A. affinis). Apareiodon hasemani shows chromosomal markers similar to those found in A. ibitiensis, Apareiodon sp. and A. vladii (Bellafronte et al., 2011; Schemberger et al., 2011); however, the location of the 5S rDNA can be considered derived in A. hasemani.
The main Brazilian rivers are separated by barriers that prevent the dispersal of species and populations, favoring the occurrence of events that cause the isolation of groups (Weitzman et al., 1988). By such conditions, unique karyotypes and distinct sex chromosomes systems in fish may have become fixed independently in different species of a genus, promoting diversification and contributing to the speciation process (Centofante et al., 2001). Although allopatry may have contributed to the origin of sex chromosome systems, Moreira-Filho et al. (1985) reported the occurrence of four species of Parodontidae in sympatry and syntopy, so the different sex chromosomes systems in some species would play an important role in their isolation. This study found that the repetitive DNA in Parodontidae seemed to play an important role in the chromosome diversification of the studied species, as in A. hasemani. Furthermore, A. hasemani presented derived chromosomal regions with respect to 5S rDNA, contributing to the genetic isolation of the population and differentiation of strains.
Acknowledgments
The authors thank the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA-License number: 10538-1) and the Instituto Estadual de Florestas (IEF/MG) for authorization to collect the material. This study was financed by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Fundação Araucária (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná), and Instituto Estadual de Florestas de Minas Gerais.
Footnotes
Associate Editor: Yatiyo Yonenaga-Yassuda
References
- Artoni RF, Bertollo LAC. Evolutionary aspects of the ZZ/ZW sex chromosome system in the Characidae fish, genus Triportheus. A monophyletic state and NOR location on the W chromosome. Heredity. 2002;89:15–19. doi: 10.1038/sj.hdy.6800081. [DOI] [PubMed] [Google Scholar]
- Bellafronte E, Margarido VP, Moreira-Filho O. Cytotaxonomy of Parodon nasus and Parodon tortuosus (Pisces, Characiformes). A case of synonymy confirmed by cytogenetic analyses. Genet Mol Biol. 2005;28:710–716. [Google Scholar]
- Bellafronte E, Vicari MR, Artoni RF, Margarido VP, Moreira-Filho O. Differentiated ZZ/ZW sex chromosomes in Apareiodon ibitiensis (Teleostei, Parodontidae): Cytotaxonomy and biogeography. J Fish Biol. 2009;75:2313–2325. doi: 10.1111/j.1095-8649.2009.02488.x. [DOI] [PubMed] [Google Scholar]
- Bellafronte E, Schemberger MO, Moreira-Filho O, Almeida MC, Artoni RF, Margarido VP, Vicari MR. Chromosomal markers in Parodontidae: An analysis of new and reviewed data with phylogenetic inferences. Rev Fish Biol Fisheries. 2011;21:559–570. [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae) Braz J Genet. 1978:2103–120. [Google Scholar]
- Centofante L, Bertollo LAC, Moreira-Filho O. Comparative cytogenetics among sympatric species of Characidium (Pisces, Characiformes). Diversity analysis with the description of a ZW sex chromosome system and natural triploidy. Caryologia. 2001;54:253–260. [Google Scholar]
- Centofante L, Bertollo LAC, Moreira-Filho O. A ZZ/ZW sex chromosome system in a new species of the genus Parodon (Pisces, Parodontidae) Caryologia. 2002;55:139–150. [Google Scholar]
- Centofante L, Bertollo LAC, Buckup PA, Moreira Filho O. Chromosomal divergence and maintenance of sympatric Characidium fish species (Crenuchidae, Characidiinae) Hereditas. 2003;138:213–218. doi: 10.1034/j.1601-5223.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci USA. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Green DM. Muller’s Ratchet and the evolution of supernumerary chromosomes. Genome. 1990;33:818–824. [Google Scholar]
- Hatanaka T, Galetti PM., Jr Mapping 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1929 (Characiformes, Prochilodontidae) Genetica. 2004;122:239–244. doi: 10.1007/s10709-004-2039-y. [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia. 1980;36:1014–1015. doi: 10.1007/BF01953855. [DOI] [PubMed] [Google Scholar]
- Ingenito LFS, Buckup PA. A new species of Parodon from the Serra da Mantiqueira, Brazil (Teleostei, Characiformes, Parodontidae) Copeia. 2005;4:765–771. [Google Scholar]
- Jesus CM, Bertollo LAC, Moreira-Filho O. Comparative cytogenetics in Apareiodon affinis (Pisces, Characiformes) and considerations regarding diversification of the group. Genetica. 1999;105:63–67. [Google Scholar]
- Jesus CM, Moreira-Filho O. Cytogenetic studies in some Apareiodon species (Pisces, Parodontidae) Cytologia. 2000a;65:397–402. [Google Scholar]
- Jesus CM, Moreira-Filho O. Karyotypes of three species of Parodon (Teleostei, Parodontidae) Ichthyol Explor Freshwaters. 2000b;11:75–80. [Google Scholar]
- Jorge LC, Moreira-Filho O. Cytogenetic studies on Apareiodon affinis (Pisces, Characiformes) from Paraná river basin: Sex chromosomes and polymorphism. Genetica. 2000;109:267–273. doi: 10.1023/a:1017522914023. [DOI] [PubMed] [Google Scholar]
- Jorge LC, Moreira-Filho O. Nucleolar organizer regions as markers of chromosomal polymorphism in Apareiodon affinis (Pisces, Parodontidae) Caryologia. 2004;57:203–207. [Google Scholar]
- Kasahara S. Introdução à Pesquisa em Citogenética de Vertebrados. Sociedade Brasileira de Genética; Ribeirão Preto: 2009. p. 160. [Google Scholar]
- Kavalco KF, Pazza R, Bertollo LAC, Moreira-Filho O. Gene mapping of 5S rDNA sites in eight fish species from the Paraíba do Sul river basin, Brazil. Cytogenet Genome Res. 2004;106:107–110. doi: 10.1159/000078567. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]
- Livernois AM, Graves JAM, Waters PD. The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity. 2012;108:50–58. doi: 10.1038/hdy.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui RL, Blanco DR, Margarido VP, Moreira-Filho O. First description of B chromosomes in the family Auchenipteridae, Parauchenipterus galeatus (Siluriformes) of the São Francisco River basin (MG, Brazil) Micron. 2009;40:552–559. doi: 10.1016/j.micron.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Machado TC, Pansonato-Alves JC, Pucci MB, Nogaroto V, Almeida MC, Oliveira C, Foresti F, Bertollo LAC, Moreira-Filho O, Artoni RF, et al. Chromosomal painting and ZW sex chromosomes differentiation in Characidium (Characiformes, Crenuchidae) BMC Genetics. 2011;12:e65. doi: 10.1186/1471-2156-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C, Galetti PM., Jr Chromosomal localization of 5S DNAr genes in Leporinus fish (Anostomidae, Characiformes) Chromosome Res. 1999;7:363–367. doi: 10.1023/a:1009216030316. [DOI] [PubMed] [Google Scholar]
- Moreira-Filho O, Bertollo LAC, Galetti PM., Jr Evidences for a multiple sex chromosome system with female heterogamety in Apareiodon affinis (Pisces, Parodontidae) Caryologia. 1980;33:83–91. [Google Scholar]
- Moreira-Filho O, Bertollo LAC, Galetti PM., Jr Structure and variability of nucleolar organizer regions in Parodontidae fish. Can J Genet Cytol. 1984;26:564–568. [Google Scholar]
- Moreira-Filho O, Bertollo LAC, Galetti PM., Jr Karyotypic study of some species of family Parodontidae (Pisces-Cypriniformes) Caryologia. 1985;38:47–55. [Google Scholar]
- Moreira-Filho O, Bertollo LAC, Galetti PM., Jr Distribution of sex chromosome mechanisms in Neotropical fish and description of a ZZ/ZW system in Parodon hilarii (Parodontidae) Caryologia. 1993;46:115–125. [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Oliveira RR, Feldberg E, Anjos MB, Zuanon J. Karyotype characterization and ZZ/ZW sex chromosome heteromorphism in two species of the catfish genus Ancistrus Kner, 1854 (Siluriformes, Loricariidae) from the Amazon basin. Neotrop Ichthyol. 2007;5:301–306. [Google Scholar]
- Oliveira RR, Feldberg E, Anjos MB, Zuanon J. Occurrence of multiple sexual chromosomes (XX/XY1Y2 and Z1Z1Z2Z2/Z1Z2W) in catfishes of the genus Ancistrus (Siluriformes, Loricariidae) from the Amazon basin. Genetica. 2008;13:243–249. doi: 10.1007/s10709-007-9231-9. [DOI] [PubMed] [Google Scholar]
- Parise-Maltempi PP, Martins C, Oliveira C, Foresti F. Identification of a new repetitive element in the sex chromosomes of Leporinus elongatus (Teleostei, Characiformes, Anostomidae): New insights into the sex chromosomes of Leporinus. Cytogenet Genome Res. 2007;116:218–223. doi: 10.1159/000098190. [DOI] [PubMed] [Google Scholar]
- Pavanelli CS. Family Parodontidae (Parodonts) In: Reis RE, Kullander SO, Ferraris CJ Jr, editors. Check List of the Freshwaters Fishes of South and Central America. EDIPUCRS; Porto Alegre: 2003. pp. 46–50. [Google Scholar]
- Pavanelli CS, Britski HA. Apareiodon Eigenmann, 1916 (Teleostei, Characiformes), from the Tocantins-Araguaia Basin, with description of three new species. Copeia. 2003;2:337–348. [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa R, Bellafronte E, Moreira-Filho O, Margarido VP. Description of the ZZ/ZW sex chromosome system and localization of 5S and 18S DNAr genes in Apareiodon sp. (Pisces, Characiformes, Parodontidae) Genetica. 2006;128:159–166. doi: 10.1007/s10709-005-5700-1. [DOI] [PubMed] [Google Scholar]
- Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genet Dev. 2004;14:634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Schemberger MO, Bellafronte E, Nogaroto V, Almeida MC, Schuhli GS, Artoni RF, Moreira-Filho O, Vicari MR. Differentiation of repetitive DNA sites and sex chromosome systems reveal closely related group in Parodontidae (Actinopterygii, Characiformes) Genetica. 2011;139:1499–1508. doi: 10.1007/s10709-012-9649-6. [DOI] [PubMed] [Google Scholar]
- Sumner AT. A simple techinique for demonstrating centromeric heterocromatin. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Travassos H. Sobre um novo “Parodontidae” do Estado de Goiás, Brasil (Cypriniformes, Characoidei) Rev Bras Biol. 1957;17:147–151. [Google Scholar]
- Vicari MR, Moreira-Filho O, Artoni RF, Bertollo LAC. ZZ/ZW sex chromosome system in an undescribed species of the genus Apareiodon (Characiformes, Parodontidae) Cytogenet Genome Res. 2006;114:163–168. doi: 10.1159/000093333. [DOI] [PubMed] [Google Scholar]
- Vicari MR, Artoni RF, Moreira-Filho O, Bertollo LAC. Colocalization of repetitive DNAs and silencing of major rRNA genes. A case report of the fish Astyanax janeiroensis. Cytogenet. Genome Res. 2008;122:67–72. doi: 10.1159/000151318. [DOI] [PubMed] [Google Scholar]
- Vicente VE, Jesus CM, Moreira-Filho O. Chromosomal localization of 5S and 18S rRNA genes in three Parodon species (Pisces, Parodontidae) Caryologia. 2001;54:365–369. [Google Scholar]
- Vicente VE, Bertollo LAC, Valentini SR, Moreira-Filho O. Origin and differentiation of sex chromosome system in Parodon hilarii (Pisces, Parodontidae). Satellite DNA, G and C-banding. Genetica. 2003;119:115–120. doi: 10.1023/a:1026082904672. [DOI] [PubMed] [Google Scholar]
- Weitzman SH, Menezes NA, Weitzman MJ. Phylogenetic biogeography of the Glandulocaudini (Teleostei, Characiformes, Characidae) with comments on the distributions of other freshwater fishes in Eastern and Southeastern Brazil. In: Vanzolini PE, Heyer WR, editors. Proceedings of a Workshop on Neotropical Distribution Patterns. Academia Brasileira de Ciências; Rio de Janeiro: 1988. pp. 379–427. [Google Scholar]