Abstract
Intramuscular fat (IMF) content plays an important role in meat quality. Many genes involved in lipid and energy metabolism were identified as candidate genes for IMF deposition, since genetic polymorphisms within these genes were associated with IMF content. However, there is less information on the expression levels of these genes in the muscle tissue. This study aimed at investigating the expression levels of sterol regulating element binding protein-1c (SREBP-1c), diacylglycerol acyltransferase (DGAT-1), heart-fatty acids binding protein (H-FABP), leptin receptor (LEPR) and melanocortin 4 receptor (MC4R) genes and proteins in two divergent Banna mini-pig inbred lines (BMIL). A similar growth performance was found in both the fat and the lean BMIL. The fat meat and IMF content in the fat BMIL were significantly higher than in the lean BMIL, but the lean meat content was lower. The serum triacylglycerol (TAG) and free fatty acid (FFA) contents were significantly higher in the fat than in the lean BMIL. The expression levels of SREBP-1c, DGAT-1 and H-FABP genes and proteins in fat BMIL were increased compared to the lean BMIL. However, the expression levels of LEPR and MC4R genes and proteins were lower.
Keywords: intramuscular fat (IMF), expression levels, Banna mini-pig inbred line (BMIL)
Introduction
In pig production, intramuscular fat (IMF) content is an important determinant of meat quality characteristics such as tenderness, juiciness and flavor (Fernandez et al., 1999). Because of the importance of IMF on the economics of pig production, it is important to elucidate the molecular mechanisms underlying intramuscular fat deposition (Wood et al., 2008). Particularly, in the Chinese local pigs, the IMF content is higher than in other commercial pig breeds (Zhao et al., 2009), which makes them an ideal model for elucidating the mechanism of IMF deposition.
During the past decades, many efforts have been made to appoint possible candidate genes for IMF deposition (Schwab et al., 2009). Sterol regulating element binding protein-1c (SREBP-1c), acyl CoA:diacylglycerol acyltransferase (DGAT-1) (Mourot and Kouba, 1998; Ding et al., 2000; Chen et al., 2008; Switonski et al., 2010) and the intracellular heart fatty acid binding protein (H-FABP) affect the IMF content in pigs (Gerbens et al., 1997, 1998, 1999, 2000; Glatz et al., 2003; Gardan et al., 2007; Li et al., 2007; Li et al., 2010; Switonski et al., 2010). Genetic variants of the leptin receptor (LEPR) and the melanocortin 4 receptor (MC4R) genes were reported to be associated with IMF deposition (Mackowski et al., 2005; Ovilo et al., 2005; Bruun et al., 2006; Jokubka et al., 2006; Kim et al., 2006; Piórkowska et al., 2010; Fan et al., 2009, 2010; Li et al., 2010; Switonski et al., 2010). Although LEPR and MC4R are mainly studied in hypothalamus, some reports indicated that they could be expressed in muscle tissue as well (Stinckens et al., 2009; Li et al., 2010; Larkina et al., 2011; Tyra et al., 2011). Global genomics analyses also led to the hypothesis that IMF deposition could be affected by both lipid metabolism and energy metabolism (Liu et al., 2009; Cánovas et al., 2010). However, most of the studies only focused on lipogenic genes, ignoring energy metabolism genes (Mourot and Kouba, 1998; Zhao et al., 2009). Moreover, these reports usually used different pig breeds as fat and lean models, which were fed diets according to their individual nutritional standard. These studies could not rule out the possibility that the different expressions of genes might be due to breed characteristics rather than to actual variations in IMF deposition.
The Banna Mini-pig Inbred Line (BMIL) was established in China in 1980 (Huo et al., 2012). The original ancestors were a sow and her son. The further reproduction occurred by full-sibling or parent-offspring mating. Progressive inbreeding generated the fat and the lean BMIL. The body weight of mature pigs reaches about 45.00 ± 1.50 kg. They are ideal experimental models for a better understanding of the genetic mechanisms leading to divergent fat deposition phenotypes, because they have the same or a very similar background genotype and were fed the same diets in the same environment (Guo et al., 2008). The aim of this study was to compare the expression levels of the SREBP-1c, DGAT-1, H-FABP, LEPR and MC4R genes and proteins in fat and lean BMIL and to explain the phenotypic variations of IMF deposition in these two divergent BMILs.
Materials and Methods
The animal experiments and protocols used in this study were approved by the Yunnan Agricultural University Institutional Animal Care and Use Committee. The fat and lean BMIL were fed in the Yunnan Key Laboratory of BMIL, Yunnan Agricultural University, China.
Fat and lean BMIL animals of the 25th generation were selected, with an inbreeding coefficient of 99.56%. A total of twelve healthy pigs were fed ad libitum the same corn-soybean diets (Table 1) in the same environmental conditions for 1.5–1.6 ± 0.45 years. Their body weights were determined every week, and their growth performances were calculated. Then the pigs were slaughtered by exsanguination after electrical stunning, and their hot carcass weight was measured. The fat-free lean and fat contents were calculated from the left side of the carcass, after a 20 h chill at 2 °C. The average backfat thickness of the first, tenth, last-rib and last-lumbar sites were calculated. Longissimus muscle samples from the last ribs were collected from each one of the animals, snap-frozen in liquid nitrogen, and stored at −80 °C prior to analysis.
Table 1.
Composition of diets.
| Percentage (%) | |
|---|---|
| Corn | 68.98 |
| Wheat bran | 22.53 |
| Soybean meal | 5.62 |
| Fish meal | 0.50 |
| Limestone | 0.97 |
| Monocalcium phosphate | 0.10 |
| Salt | 0.30 |
| Premix1 | 1.00 |
| Calculated nutritional composition | |
| Crude protein | 10.42 |
| Total Lysine | 0.69 |
| Digestive energy (MJ/kg) | 13.11 |
| Calcium | 0.46 |
| Total phosphorus | 0.37 |
| Available phosphorus | 0.14 |
Vitamin premix contents per kilogram of diet: vitamin A - 8,267 IU; vitamin D2 - 2,480 IU; vitamin E - 66 IU; menadionine (as menadionine pyrimidinol bisulfite complex) - 6.2 mg; riboflavin - 10 mg; Ca d-pantothenic acid - 37 mg; niacin - 66 mg; vitamin B12 - 45 μg; d-biotin - 331 μg; folic acid - 2.5 mg; pyridoxine - 3.31 mg; thiamine - 3.31 mg; vitamin C - 83 μg; Trace mineral premix contents per kilogram of diet: Zn - 127 mg; Fe - 127 mg; Mn - 20 mg; Cu - 12.7 mg; I - 0.80 mg, in the form of zinc sulfate, ferrous sulfate, manganese sulfate, copper sulfate, ethylenediamine dihydriodide, respectively, with calcium carbonate as carrier, besides 0.3 mg Se per kilogram of diet.
IMF content
The IMF content, expressed as weight percentage of dry muscle tissue, was evaluated 24 h after slaughter, using the Soxhlet petroleum-ether extraction method.
Serum parameters
Blood samples were centrifuged at 600 × g for 15 min. Serum was obtained and stored at −20 °C for posterior analyses. Serum FFA and TG concentrations were determined using the respective commercial kit assays (Nanjing Jiancheng Biochemical Reagent Co., Nanjing, China) according to the protocols provided by the manufacturer.
RNA extraction and Reverse transcription (RT)
Total RNA was extracted from the muscle tissues using Trizol reagent according to the manufacturer’s protocol (Takala, Japan). Total RNA concentration was then quantified by measuring the absorbance at 260 nm in an Eppendorf Biospectrophotometer (Eppendorf AG, Hamburg, Germany). The absorption ratios (260/280 nm) ranged from 1.8 to 2.0. The integrity of the RNA was verified by 1.4% agarose-formaldehyde gel electrophoresis. Total RNA (2 μg) was then reverse-transcribed by incubation at 42 °C for 1 h in a 25 μL mixture consisting of MMLV (Promega Co., Shanghai, China), RNase inhibitor (Promega), and oligo dT. The reaction was terminated by heating to 95 °C for 5 min and quick cooling on i.e. cDNA was stored at −20 °C for PCR amplification.
Real-Time Quantitative PCR (RT-qPCR)
RT-qPCR was performed to quantify the mRNA expression abundance of genes SREBP1c, H-FABP, DGAT1, LEPR and MC4R. ß-actin was used as internal control. The 25 μL PCR mixture contained 12.5 μL of 2iQ SYBR Green Supermix, 0.5 μL (10 mM) of each primer (Table 2), and 1 μL of cDNA. Mixtures were incubated in an iCyler iQ Real-time Detection system (Bio-Rad Laboratories, Hercules, CA, USA). A melting curve analysis was performed to ensure that only a single PCR product was amplified. Control reactions were set for each sample. PCR amplification efficiencies were between 91% and 103%. The amount of specific target was calculated according to the following formula: ratio = 2−ΔCTtarget (sample - calibrator)/2−ΔCT ß-actin (sample - calibrator). All primers used were designed by Primer Premier 5 and synthesized by Shanghai Shenggong Biological Company (Shanghai, China).
Table 2.
Specific primers used for real-time quantitative PCR (RT-qPCR)
| Gene name | Sequence | Product size | Position | Accession number |
|---|---|---|---|---|
| β-actin | F: 5′-GCCGCACCACTGGCATTGTC-3′ | 399 bp | Exon 2 | DQ845171 |
| R: 5′-AGGTAGTTTCGTGGATGCCGCAG-3′ | Exon 3 | |||
|
| ||||
| SREBP-1c | F: 5′-GCGACGGTGCCTCTGGTAGT-3′ | 218 bp | Exon 1 | AY496867 |
| R: 5′-CGCAAGACGGCGGATTTA-3′ | Exon 2 | |||
|
| ||||
| H-FABP | F: 5′-CCTGGAAGCTAGTGGACAGC-3′ | 227 bp | Exon 1 | AJ416019 |
| R: 5′-TGCCTCTTTCTCGTAAGTGC-3′ | Exon 2 | |||
|
| ||||
| DGAT-1 | F: 5′-AAGGACGGACACGACGATG-3′ | 289 bp | Exon 1 | AY093657 |
| R: 5′-GGAACGCAGTCACAGCAAAG-3′ | Exon 5 | |||
|
| ||||
| LEPR | F: 5′-GTGATAACTGCATTTGACTTGGC-3′ | 285 bp | Exon 1 | NM001024587 |
| R: 5′-CTGCAATGTTGTCTGCATGTACAG-3′ | Exon 1 | |||
|
| ||||
| MC4R | F: 5′-TGGAGAAAATCGCTGAGGCTACC-3′ | 632 bp | Exon1 | NM214173 |
| R: 5′-ATGATGAACAAAACACCCGACACC-3′ | Exon2 | |||
Western blotting
Muscle samples were collected and homogenized on ice in 700 μL buffer A [50 mM Tris-HCl (pH 7.5), 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM EDTA, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 10% glycerol] containing 1% Triton X-100, 5 μM aprotinin, leupeptin and pepstatin. The lysates were centrifuged at 6000 g for 20 min at 4 °C to remove insoluble material. Thereafter, supernatant extracts were collected and protein concentration determined, using the method described by Bradford (1976). Then the extracts were frozen at −80 °C until the western blot analyses were performed.
To measure SREBP-1c, DGAT-1, H-FABP, LEPR and MC4R protein expression, 50 μg of total whole-cell protein extract were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% resolving gel), transferred to a nitrocellulose membrane and probed overnight, with rabbit polyclonal anti- SREBP-1c, anti-DGAT-1, anti-H-FABP, anti-LEPR, and anti-MC4R antibodies (Sigma, USA) at 1:500, 1:500, 1:1000, 1:1000 and 1:2000 dilutions, respectively. The membranes were then probed with a goat anti-rabbit IgG-horseradish peroxidase conjugate (1:20,000 dilution) (Sigma, USA) for 1 h at room temperature. Blots were developed using the SuperSignal West Pico Chemiluminescent Substrate system (Bio-Rad, Hercules, CA, USA) and imaged on X-film, for image analysis and densitometry. Signal intensity was quantified using the Quantity One 1-D analysis software (Bio-Rad, Hercules, CA, USA).
Statistical analysis
Data regarding growth performance, sera parameters and mRNA expression were analyzed using paired t-tests with the Statistical Packages for Social Science (SPSS) 12.0 software. All data were presented as mean ± S.E.M. Differences were considered statistically significant if p < 0.05.
Results
Growth performance, carcass traits and serum parameters
Growth performance, carcass traits and serum parameters are presented in Table 3. There was no significant difference in growth performance between the fat and lean BMIL (p > 0.05). A significant reduction in fat percentage was found in the fat BMIL compared to the lean BMIL (p < 0.05). A significant increase of lean meat was observed in the fat BMIL (p < 0.05). The serum concentrations of TG and FFA as well as the IMF content were significantly higher in the fat BMIL than in the lean BMIL (p < 0.05).
Table 3.
Growth performance, carcass traits and serum parameters
| Indexes | Lean BMIL | Fat BMIL | |
|---|---|---|---|
| Growth performance | Daily feed intake (g/d) | 22.57 ± 0.06 | 28.67 ± 0.01 |
| Daily gain (g/d) | 122.00 ± 24.98 | 121.00 ± 25.02 | |
| Gain: feed ratio | 4.11 ± 0.01 | 4.20 ± 0.01 | |
|
| |||
| Carcass traits | Lean meat percentage (%) | 41.75 ± 1.01* | 31.00 ± 0.77 |
| Fat meat percentage (%) | 34.33 ± 1.68 | 53.10 ± 0.62* | |
| Average backfat thickness (cm) | 1.31 ± 0.20 | 4.61 ± 0.13* | |
| IMF(%) | 4.51 ± 0.10 | 7.82 ± 0.08* | |
|
| |||
| Serum parameters | Triacylglycerol (mg/100 mL) | 52.63 ± 1.77 | 61.97 ± 2.01* |
| EFFA (μEq/L) | 211.34 ± 19.99 | 266.99 ± 20.17* | |
Values are expressed as means ± SE;
values in the same row with asterisk are significantly different at p < 0.05.
Expression levels of genes and proteins
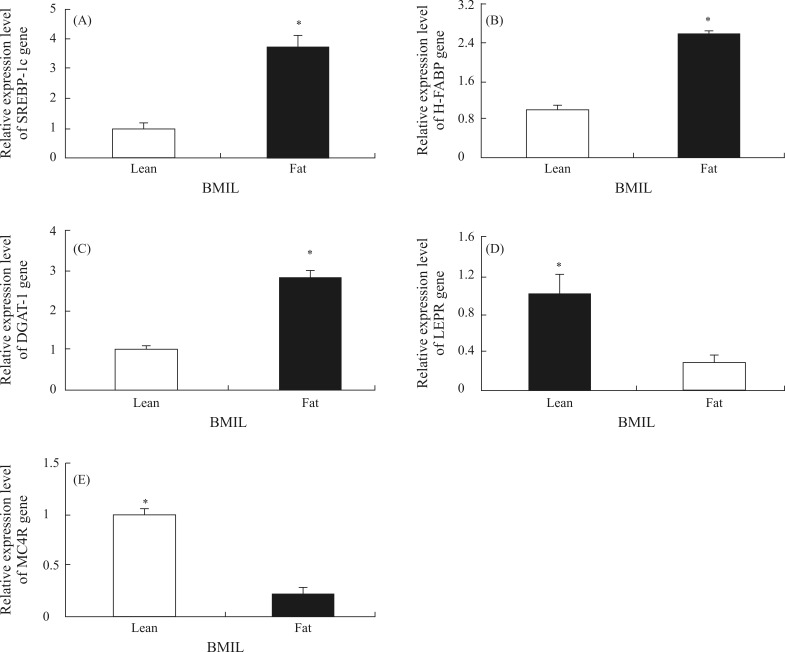
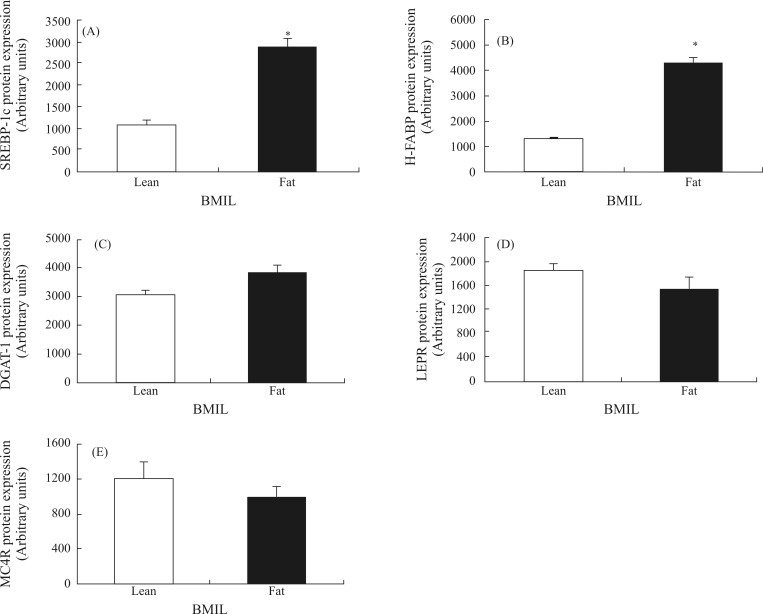
Figures 1 and 2 show the expression levels of the SREBP1c, H-FABP, DGAT1, LEPR and MC4R genes and proteins in muscle tissue of fat and lean BMIL animals. The expression levels of the H-FABP, SREBP-1c and DGAT1 genes and proteins were significantly higher in the fat BMIL than in the lean BMIL (p < 0.05). The mRNA and protein abundance of MC4R and LEPR was lower in the fat BMIL compared to the lean BMIL. The difference in LEPR gene expression between the fat and lean BMIL was significant (p < 0.05).
Figure 1.
mRNA expression levels of SREBP-1c, H-FABP, DGAT1, LEPR and MC4R (A, B, C, D and E, respectively) in the fat and lean BMIL. Means ± SE with asterisk indicate a significant difference between the two groups (p < 0.05).
Figure 2.
Expression levels of proteins SREBP-1c, H-FABP, DGAT1, LEPR and MC4R (A, B, C, D and E, respectively) in the fat and lean BMIL. Means ± SE with asterisk indicate a significant difference between the two groups (p < 0.05).
Discussion
In this study, the growth performance of fat and lean BMIL animals showed no significant difference. However, an increase in fat meat as well as in IMF content and reduced lean meat content in the fat BMIL compared to the lean BMIL was found. As a rule, animal obesity models weigh more than their lean counterparts. However, a similar feed intake was also reported in the lean Yorkshire and fat Ossabaw pigs (Wangsness et al., 1980). This may suggest that the different fat deposition in the fat and lean BMIL pigs does not result from their feed intake, but rather from their genotypes for fatness traits.
TAG, the major component of IMF in muscles, is stored within intramuscular adipocytes (Gao and Zhao, 2009). It could be synthesized using fatty acids which were de novo synthesized or originated from feed nutrients. Hauser et al. (1997) reported that the plasma of fat pigs had a higher percentage of triglycerides than the plasma of lean pigs. Our results also showed that the fat BMIL had higher serum TAG and FFA concentrations as well as IMF content compared with the lean BMIL. These data indicate that more TAG or FFA from feed nutrients may be allotted to the muscle tissue of fat BMIL, leading to a higher IMF deposition than in the lean BMIL.
In general, an increase in IMF content is mainly due to an increase in TG contents (Fernandez et al., 1999). Consequently, the TG metabolism in muscle tissue should be a target for identification of genes involved in IMF deposition, especially the TAG synthesis metabolism. SREBP-1c can stimulate the transcription of genes encoding acetyl-CoA carboxylase (ACC) (Zhuang et al., 2003) and fatty acid synthase (FAS) enzymes (Magana et al., 2000). H-FABP plays a role in transporting the fatty acids, and has been regarded as a candidate gene for IMF deposition in pigs (Gerbens et al., 1998; Glatz et al., 2003; Chmurzynska, 2006; Damon et al., 2006; Gardan et al., 2007). DGAT are microsomal enzymes which catalyze the final step of the triglyceride synthesis pathway (Yen et al., 2008). Overexpression of DGAT enzymes in transgenic mice is sufficient to increase cellular TAG storage in skeletal muscle (Koliwad et al., 2010). Our results show that the expression levels of the SREBP-1c, H-FABP and DGAT-1 genes and proteins in the fat BMIL pigs were higher than in the lean BMIL pigs. However, the difference in DGAT-1 protein expression was not significant. The expression of the DGAT-1 gene and protein was positively correlated (Coleman and Lee, 2004). The results suggest that, in the fat BMIL, more fatty acids would be synthesized de novo and transported to store TAG.
Intramuscular TG is a major form of energy storage and represents a significant fuel for cellular metabolism (Gao and Zhao, 2009). MC4R and LEPR play an important role in the control of energy homeostasis. Many reports have shown that the genetic variants of MC4R and LEPR are associated with porcine fat deposition (Mackowski et al., 2005; Ovilo et al., 2005; Bruun et al., 2006; Jokubka et al., 2006; Kim et al., 2006; Fan et al., 2009, 2010; Li et al., 2010; Piórkowska et al., 2010; Switonski et al., 2010). The LEPR gene mediates the regulation of leptin effects (Halaas and Friedman, 1997; Houseknecht and Portocarrero, 1998;Li et al., 2010). The MC4R gene has been most closely linked to controlling the energy balance in rodents. The interaction between melanocortins and their receptors in the hypothalamus is one of the main neuroendocrine pathways controlling energy balance (Yeo et al., 2000; Wardlaw, 2001). However, some studies have indicated that these genes could also be expressed in the muscle tissue (Stinckens et al., 2009; Li et al., 2010; Larkina et al., 2011; Tyra et al., 2011). Our data show that the mRNA and protein abundance of LEPR and MC4R in the fat BMIL was lower than in the lean BMIL. However, the difference in MC4R protein expression was not significant. This may be due to the fact that the main function of MC4R is to regulate the food intake (Kim et al., 2006). Since in the present study the feed intake by the fat and the lean BMIL pigs was similar,this may suggestthat the IMF deposition could be affected by energy metabolism-related genes.
In conclusion, fat and lean BMIL with divergent phenotypes for IMF content did not result from feeding differences, but rather were influenced by feed nutrients TAG and FFA and the expression levels of genes and proteins which participate in the TAG synthesis process and the energy metabolism.
Acknowledgments
This work was supported by the National Key Program of Transgenic Project of China (No. 2009ZX08009-140B), the National Key Foundation Research Development Project of China (973 Project, number 2007CB116201), the National Natural Science Foundation of China (No.30660132 and 31060331) and the Yunnan Natural Science Foundation of China (No. 2009CD056). The authors would like to thank Dr. Marinus F.W. te Pas for editing the manuscript.
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruun CS, Jørgensen CB, Nielsen VH, Andersson L, Fredholm M. Evaluation of the porcine melanocortin 4 receptor (MC4R) gene as a positional candidate for a fatness QTL in a cross between Landrace and Hampshire. Anim Genet. 2006;37:359–362. doi: 10.1111/j.1365-2052.2006.01488.x. [DOI] [PubMed] [Google Scholar]
- Cánovas A, Quintanilla R, Amills M, Pena RN. Muscle transcriptomic profiles in pigs with divergent phenotypes for fatness traits. BMC Genomics. 2010;11:372–387. doi: 10.1186/1471-2164-11-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang XJ, Xia D, Chen J, Wegner J, Jiang Z, Zhao RQ. Sterol regulatory element binding transcription factor 1 expression and genetic polymorphism significantly affect intramuscular fat deposition in the longissimus muscle of Erhualian and Sutai pigs. J Anim Sci. 2008;86:57–63. doi: 10.2527/jas.2007-0066. [DOI] [PubMed] [Google Scholar]
- Chmurzynska A. The multigene family of fatty acidbinding proteins (FABPs): Function, structure and polymorphism. J Appl Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Damon M, Louveau I, Lefaucheur L, Lebret B, Vincent A, Leroy P, Sanchez MP, Herpin P, Gondret F. Number of intramuscular adipocytes and fatty acid binding protein-4 content are significant indicators of intramuscular fat level in crossbred Large White x Duroc pigs. J Anim Sci. 2006;84:1083–1092. doi: 10.2527/2006.8451083x. [DOI] [PubMed] [Google Scholar]
- Ding ST, Schinckel AP, Weber TE, Mersmann HJ. Expression of porcine transcription factors and genes related to fatty acid metabolism in different tissues and genetic populations. J Anim Sci. 2000;78:2127–2134. doi: 10.2527/2000.7882127x. [DOI] [PubMed] [Google Scholar]
- Fan B, Lkhagvadorj S, Cai W, Young J, Smith RM, Dekkers JC, Huff-Lonergan E, Lonergan SM, Rothschild MF. Identification of genetic markers associated with residual feed intake and meat quality traits in the pig. Meat Sci. 2010;84:645–650. doi: 10.1016/j.meatsci.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Fan B, Onteru SK, Plastow GS, Rothschild MF. Detailed characterization of the porcine MC4R gene in relation to fatness and growth. Anim Genet. 2009;40:401–409. doi: 10.1111/j.1365-2052.2009.01853.x. [DOI] [PubMed] [Google Scholar]
- Fernandez X, Monin G, Talmant A, Mourot J, Lebret B. Influence of intramuscular fat content on the quality of pig meat-1. Composition of the lipid fraction and sensory characteristics of m. longissimus lumborum. Meat Sci. 1999;53:59–65. doi: 10.1016/s0309-1740(99)00037-6. [DOI] [PubMed] [Google Scholar]
- Gao SZ, Zhao SM. Physiology, affecting factors and strategies for control of pig meat intramuscular fat. Recent Pat Food Nutr Agric. 2009;1:59–74. [PubMed] [Google Scholar]
- Gardan D, Louveau I, Gondret F. Adipocyte- and heart-type fatty acid binding proteins are both expressed in subcutaneous and intramuscular porcine (Sus scrofa) adipocytes. Comp Biochem Physiol B. 2007;148:14–19. doi: 10.1016/j.cbpb.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Gerbens F, Rettenberger G, Lenstra JA, Veerkamp JH, te Pas MFW. Characterization, chromosomal localization, and genetic variation of the porcine heart fatty acid-binding protein. Mamm Genome. 1997;8:328–331. doi: 10.1007/s003359900433. [DOI] [PubMed] [Google Scholar]
- Gerbens F, Jansen A, van Erp AJM, Harders F, Meuwissen THE, Rettenberger G, Veerkamp JH, te Pas MF. The adipocyte fatty acid-binding protein locus: Characterization and association with intramuscular fat content in pigs. Mamm Genome. 1998;9:1022–1026. doi: 10.1007/s003359900918. [DOI] [PubMed] [Google Scholar]
- Gerbens F, van Erp AJM, Harders FL, Verburg FJ, Meuwissen THE, Veerkamp JH, te Pas MFW. Effect of genetic variants of the heart fatty acid-binding protein gene on intramuscular fat and performance traits in pigs. J Anim Sci. 1999;77:846–852. doi: 10.2527/1999.774846x. [DOI] [PubMed] [Google Scholar]
- Gerbens F, de Koning DJ, Harders FL, Meuwissen THE, Janss LLG, Groenen MA, Veerkamp JH, Van Arendonk JA, te Pas MF. The effect of adipocyte and heart fatty acid-binding protein genes on intramuscular fat and backfat content in Meishan crossbred pigs. J Anim Sci. 2000;78:552–559. doi: 10.2527/2000.783552x. [DOI] [PubMed] [Google Scholar]
- Glatz JF, Schaap FG, Binas B. Cytoplasmic fatty acid-binding protein facilitates fatty acid utilization by skeletal muscle. Acta Physiol Scand. 2003;178:367–371. doi: 10.1046/j.1365-201X.2003.01166.x. [DOI] [PubMed] [Google Scholar]
- Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. 2008;47:367–373. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- Halaas JL, Friedman JM. Leptin and its receptor. J Endocrinol. 1997;155:215–216. doi: 10.1677/joe.0.1550215. [DOI] [PubMed] [Google Scholar]
- Hauser N, Mourot J, De Clercq L, Genart C, Remacle C. The cellularity of developing adipose tissues in Pietrain and Meishan pigs. Reprod Nutr Dev. 1997;37:617–625. doi: 10.1051/rnd:19970601. [DOI] [PubMed] [Google Scholar]
- Houseknecht KL, Portocarrero CP. Leptin and its receptors: Regulators of whole body energy homeostasis. Domest Anim Endocrinol. 1998;15:457–475. doi: 10.1016/s0739-7240(98)00035-6. [DOI] [PubMed] [Google Scholar]
- Huo J, Huo H, Wang P, Zeng Y, Xiao H. The Association of MC1R gene with coat color of Banna Mini-Pig Inbred Line (BMIL) J Anim Vet Adv. 2012;1:503–508. [Google Scholar]
- Jokubka R, Maak S, Kerziene S, Swalve HH. Association of a melanocortin 4 receptor (MC4R) polymorphism with performance traits in Lithuanian White pigs. J Anim Breed Genet. 2006;123:17–22. doi: 10.1111/j.1439-0388.2006.00559.x. [DOI] [PubMed] [Google Scholar]
- Kim KS, Lee JJ, Shin HY, Choi BH, Lee CK, Kim JJ, Cho BW, Kim TH. Association of melanocortin 4 receptor (MC4R) and high mobility group AT-hook 1 (HMGA1) polymorphisms with pig growth and fat deposition traits. Anim Genet. 2006;37:419–421. doi: 10.1111/j.1365-2052.2006.01482.x. [DOI] [PubMed] [Google Scholar]
- Koliwad SK, Streeper RS, Monetti M, Cornelissen I, Liana C, Terayama K, Naylor S, Rao M, Hubbard B, Farese RV. DGAT1-dependent triacylglycerol storage by macrophages protects mice from diet-induced insulin resistance and inflammation. J Clin Invest. 2010;120:756–767. doi: 10.1172/JCI36066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkina TA, Sazanova AL, Fomichev KA, Olu Barkova, Sazanova AA, Malwski T, Jaszczak K. Expression profiling of candidate genes for abdominal fat mass in domestic chicken Gallus gallus. Genetika. 2011;47:1140–1144. [PubMed] [Google Scholar]
- Li B, Zerby HN, Lee K. Heart fatty acid binding protein is upregulated during porcine adipocyte development. J Anim Sci. 2007;85:1651–1659. doi: 10.2527/jas.2006-755. [DOI] [PubMed] [Google Scholar]
- Li X, Kim SW, Choi JS, Lee YM, Lee CK, Choi BH, Kim TH, Choi YI, Kim JJ, Kim KS. Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content. Mol Biol Rep. 2010;37:3931–3939. doi: 10.1007/s11033-010-0050-1. [DOI] [PubMed] [Google Scholar]
- Liu J, Damon M, Gutton N, Guisle I, Ecolan P, Vincent A, Cherel P, Gondret F. Differentially-expressed genes in pig longissimus muscles with contrasting levels of fat, as identified by combined transcriptomic, reverse transcription PCR, and proteomic analyses. J Agric Food Chem. 2009;57:3808–3817. doi: 10.1021/jf8033144. [DOI] [PubMed] [Google Scholar]
- Mackowski M, Szymoniak K, Szydlowski M, Kamyczek M, Eckert R, Rozycki M, Switonski M. Missense mutations in exon 4 of the porcine LEPR gene encoding extracellular domain and their association with fatness traits. Anim Genet. 2005;36:135–137. doi: 10.1111/j.1365-2052.2005.01247.x. [DOI] [PubMed] [Google Scholar]
- Magana MM, Koo SH, Towle HC, Osborne TF. Different sterol regulatory element-binding protein-1 isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J Biol Chem. 2000;275:4726–4733. doi: 10.1074/jbc.275.7.4726. [DOI] [PubMed] [Google Scholar]
- Mourot J, Kouba M. Lipogenic enzyme activities in muscles of growing Large White and Meishan pigs. Livest Prod Sci. 1998;55:127–133. [Google Scholar]
- Ovilo C, Fernández A, Noguera JL, Barragán C, Letón R, Rodríguez C, Mercadé A, Alves E, Folch JM, Varona L. Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet Res. 2005;85:57–67. doi: 10.1017/s0016672305007330. [DOI] [PubMed] [Google Scholar]
- Piórkowska K, Tyra M, Rogoz M, Ropka-Molik K, Oczkowicz M, Rózycki M. Association of the melanocortin-4 receptor (MC4R) with feed intake, growth, fatness and carcass composition in pigs raised in Poland. Meat Sci. 2010;85:297–301. doi: 10.1016/j.meatsci.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Schwab CR, Mote BE, Du ZQ, Amoako R, Baas TJ, Rothschild MF. An evaluation of four candidate genes for use in selection programmes aimed at increased intramuscular fat in Duroc swine. J Anim Breed Genet. 2009;126:228–236. doi: 10.1111/j.1439-0388.2008.00770.x. [DOI] [PubMed] [Google Scholar]
- Stinckens A, Luyten T, Van den Maagdenberg K, Janssens S, De Smet S, Georges M, Buys N. Interactions between genes involved in growth and muscularity in pigs: IGF-2, myostatin, ryanodine receptor 1, and melanocortin-4 receptor. Domest Anim Endocrinol. 2009;37:227–235. doi: 10.1016/j.domaniend.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Switonski M, Stachowiak M, Cieslak J, Bartz M, Grzes M. Genetics of fat tissue accumulation in pigs: A comparative approach. J Appl Genet. 2010;51:153–168. doi: 10.1007/BF03195724. [DOI] [PubMed] [Google Scholar]
- Tyra M, Ropka-Molik K, Eckert R, Piorkowska K, Oczkowicz M. H-FABP and LEPR gene expression profile in skeletal muscles and liver during ontogenesis in various breeds of pigs. Domest Anim Endocrinol. 2011;40:147–154. doi: 10.1016/j.domaniend.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Wangsness PJ, James LG, Sherritt GW. Feeding behavior of lean and obese pigs. Physiol Behav. 1980;24:407–410. doi: 10.1016/0031-9384(80)90107-9. [DOI] [PubMed] [Google Scholar]
- Wardlaw SL. Obesity as a neuroendocrine disease: Lessons to be learned from proopiomelanocortin and melanocortin receptor mutations in mice and men. J Clin Endocrinol Metabol. 2001;86:1442–1446. doi: 10.1210/jcem.86.4.7388. [DOI] [PubMed] [Google Scholar]
- Wood JD, Enser M, Fisher AV, Nute GR, Sheard PR, Richardson RI, Hughes SI, Whittington FM. Fat depositon, fatty acid composition and meat quality: A review. Meat Sci. 2008;78:343–358. doi: 10.1016/j.meatsci.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV. Thematic review series: Glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GS, Farooqi IS, Challis BG, Jackson RS, O’Rahilly S. The role of melanocortin signalling in the control of body weight: Evidence from human and murine genetic models. QJM 2000. 2000;93:7–14. doi: 10.1093/qjmed/93.1.7. [DOI] [PubMed] [Google Scholar]
- Zhao SM, Liu LY, Zhang X, Ge CR, Liu YG, Gao SZ. Effects of monoclonal antibody on fat tissue development, carcass composition, growth performance and fat metabolism of pigs by subcutaneous injection. Livest Sci. 2009;122:8–15. [Google Scholar]
- Zhao SM, Ren LJ, Chen L, Zhang X, Cheng ML, Li WZ, Zhang YY, Gao SZ. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids. 2009;44:1029–1037. doi: 10.1007/s11745-009-3356-9. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Yin L, Hillgartner FB. SREBP-1 integrates the actions of thyroid hormone, insulin, cAMP, and medium chain fatty acids on ACC alpha transcription in hepatocytes. J Lipid Res. 2003;44:356–368. doi: 10.1194/jlr.M200283-JLR200. [DOI] [PubMed] [Google Scholar]