Abstract
Purpose
The phosphatidylinositol 3-kinase (PI3K)/PTEN/AKT/mTOR and Ras/Raf/MEK/ERK pathways have been implicated in endometrial tumorigenesis. In this candidate pathway analysis, we investigated associations between genetic variations in these two pathways and both risk and clinical outcomes of endometrial cancer.
Methods
We genotyped a total of 48 potentially functional SNPs in 11 key genes (AKT1, AKT2, AKT3, BRAF, FRAP1, KRAS, PDPK1, PIK3CA, PIK3CB, PIK3R1, and PTEN) with the Sequenom genotyping platform in 115 endometrial cancer patients and 230 cancer-free women to evaluate their associations with risk, survival, and recurrence of endometrial cancer.
Results
We found the following: (1) PIK3CA rs6443624 and rs9838411 variants either borderline or significantly decreased risk of endometrial cancer in a dominant model (adjusted odds ratio [OR], 0.62; 95% CI, 0.39–1.00 and 0.59; 95% CI, 0.36–0.95, respectively). Furthermore, there was a statistically significant multiplicative interaction (P int = 0.036) between these two loci in risk of endometrial cancer. In contrast, the AKT1 rs2498801 genotype significantly increased risk of endometrial cancer (adjusted OR, 1.94; 95% CI, 1.02–3.67 in a recessive model). (2) In Cox regression analyses, three SNPs (PIK3R1 rs1862162, AKT2 rs892119, and PIK3CA rs2699887) showed significant associations with survival of endometrial cancer patients. (3) KRAS rs7312175 and PIK3CA rs6443624 had significant effects on recurrence of endometrial cancer individually and combined in a locus–dosage manner (adjusted P trend = 0.003).
Conclusion
These results suggest that common genetic variations in these pathways may modulate risk and clinical outcomes of endometrial cancer. Further replication and functional studies are needed to confirm these findings.
Electronic supplementary material
The online version of this article (doi:10.1007/s00432-011-1103-0) contains supplementary material, which is available to authorized users.
Keywords: PI3K/PTEN/AKT/mTOR and RAS/RAF/MEK/ERK pathways, Polymorphisms, Endometrial cancer risk, Survival, Recurrence
Introduction
Endometrial cancer is the most common invasive gynecologic malignancy and the fourth most common cancer among women in developed countries. In the United States, it is estimated that there will be 46,470 new diagnoses of endometrial cancer and 8,120 disease-related deaths in 2011 (Siegel et al. 2011). The accumulated evidence indicates that exposure to high-level estrogen is an important risk factor for endometrial cancer (Akhmedkhanov et al. 2001). However, individuals with a family history of endometrial cancer have a 1.3–1.8-fold increased risk of endometrial cancer, suggesting genetic susceptibility to the development of endometrial cancer (Paynter et al. 2005).
In the past few decades, important biological pathways have been investigated for their involvement in the development of endometrial carcinoma, including the phosphatidylinositol 3-kinase (PI3K)/PTEN/AKT/mTOR and RAS/RAF/MEK signaling pathways (Ninomiya et al. 2004; Steelman et al. 2008). These two pathways are activated by many growth factors and cytokines and subsequently play critical roles in driving cell proliferation and preventing apoptosis (Nicholson and Anderson 2002; McCubrey et al. 2006). The estrogen receptor further interacts with the PI3K/PTEN/AKT/mTOR pathway at multiple levels, supporting potential crosstalk between estrogens and the PI3K pathway (Kirkegaard et al. 2005; Antico-Arciuch et al. 2010). Abnormal regulation of the PI3K and RAS pathways induced by mutations in Ras and B-Raf as well as other genes (e.g., PI3K, PTEN, and AKT) occurs in a wide range of tumor types, and there is extensive evidence validating various components of these pathways as molecular targets for cancer therapy (Li et al. 1997; Lynch et al. 2004; Samuels et al. 2004; Carpten et al. 2007; Steelman et al. 2010). For endometrial cancer, inactivating mutations in PTEN and activating mutations in KRAS and PIK3CA have been reported to occur in 30–50, 10–30, and 30–40% of endometrial cancers, respectively (Yuan and Cantley 2008). Furthermore, mutations or overexpressions of genes involved in these pathways have been associated with invasion, metastasis, and prognosis of a variety of cancers, including endometrial cancer (Minaguchi et al. 2001; Catasus et al. 2008; Chen et al. 2009; Rudd et al. 2011; Urick et al. 2011). The role of somatic mutations in these pathways in determining patient outcomes is both complex and dependent on interactions with other events (Mori et al. 2007; Catasus et al. 2008, 2009; Konstantinova et al. 2010; Murayama-Hosokawa et al. 2010; Urick et al. 2011) but suggests that evaluation of the role of germline mutations in initiation and progression of endometrial cancer is clearly warranted.
Common genetic variations, such as single-nucleotide polymorphisms (SNPs), may modulate the function of genes in PI3K and RAS/RAF signaling pathways, resulting in both predisposition to and altered clinical outcomes of endometrial cancer. Some SNPs in genes involved in these two pathways have been implicated in risk of ovarian and colorectal cancer (Li et al. 2008; Quaye et al. 2009), and other SNPs with prognosis of esophageal and lung cancer (Hildebrandt et al. 2009; Pu et al. 2010). However, few studies have investigated the association between SNPs of genes in these pathways and endometrial cancer development (Treloar et al. 2007; Wang et al. 2009). Therefore, we conducted this pilot case–control study to examine the associations between 48 potentially functional SNPs in 11 core genes (AKT1, AKT2, AKT3, BRAF, FRAP1, KRAS, PDPK1, PIK3CA, PIK3CB, PIK3R1, and PTEN) in these two pathways and risk and clinical outcomes of endometrial cancer.
Materials and methods
Study subjects
MD Anderson Cancer Center is a tertiary referral center, and the patient population may be skewed toward cases of advanced or recurrent cancers and therefore not representative of cases from the general population. The 115 incident patients with newly diagnosed and histologically confirmed endometrial cancer included in this study therefore represented only approximately 3% of all uterine cancer patients seen at MD Anderson during the period of 2000–2008 with both blood samples collected and clinical follow-up data available for analysis. Peripheral blood samples were collected during surgery by the Gynecologic Cancer Tumor Bank at The University of Texas M.D. Anderson Cancer Center. The 230 healthy cancer-free women were randomly selected as controls from a large repository of controls enrolled in an ongoing lung cancer case–control study, recruited from a multispecialty physician group in Houston. Controls were selected from the same time period as the cases, frequency-matched to the patients on age, ethnicity, and smoking status (never, former, or current). One milliliter of whole blood was used for genomic DNA extraction with a DNA blood Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s instructions. Demographic data and environmental exposure history were obtained from structured questionnaires administered to the controls and from a core self-administered questionnaire and chart review for the cases. Tumors were staged according to the TNM classification and grouped as early clinical stage (I–II) or advanced clinical stage (III–IV). All cases were followed up after surgical treatment until death, disease recurrence, or date of last follow-up. The average follow-up time was 29.24 months, and the last follow-up date was October 8, 2009.
SNP selection and genotyping
Polymorphisms were selected by an approach combining both tagging SNPs and potentially functional SNPs for 11 genes (AKT1, AKT2, AKT3, BRAF, FRAP1, KRAS, PDPK1, PIK3CA, PIK3CB, PIK3R1, and PTEN). SNPs selected met one of the following criteria: (1) tagging SNPs chosen from genotyped SNPs of European populations in the HapMap database (MAF [Minor Allele Frequency] ≥ 0.05 and r 2 = 0.8) and further identified as potentially functional SNPs (influencing protein function, mRNA splicing, miRNA binding, or promoter activity) through the online software Pupasuite 2 (http://pupasview.bioinfo.ochoa.fib.es/) and FuncPred (http://manticore.niehs.nih.gov/snpfunc.htm); (2) common SNPs (MAF ≥ 0.05) located in the 5′ and 3′ UTRs (untranslated regions) reported by the NCBI dbSNP database, which may be important in regulating gene expression; (3) SNPs that had been investigated for association with cancer risk/prognosis in previously published studies. Therefore, a total of 48 SNPs were selected and genotyped with the Sequenom genotyping platform (Supplemental Table 1). The information about assay conditions, primers, and probes is available upon request. All samples were run in duplicates with call rates of >95%, and all output spectra were visually inspected.
Statistical analysis
The chi-square (χ2) test was used to determine whether genotype distributions were in Hardy–Weinberg equilibrium (HWE) in the controls. The associations between genotypes and endometrial cancer risk were estimated by computing odds ratios (ORs) and 95% confidence intervals (CIs) from both univariate and multivariate logistic regression analyses with adjustment for age, ethnicity, and smoking. The Kaplan–Meier method and the log-rank test were used to analyze associations between survival and demographic characteristics, clinical features, and SNPs. Univariate or multivariate Cox regression analyses were performed to determine predictive factors of endometrial cancer prognosis by estimating crude and adjusted hazard ratios (HRs) and their 95% confidence intervals (CIs). All statistical analyses were two-sided, and P < 0.05 was considered statistically significant. Statistical Analysis System software (version 9.2; SAS Institute, Cary, NC) and PLINK whole-genome association analysis toolset were used for the statistical analyses.
Results
Patient characteristics
The demographic features of the 115 endometrial cancer patients and 230 cancer-free women controls are summarized in Table 1. The majority (76.2%) of the participants were non-Hispanic whites, the others included 10 (8.7%) African-Americans and 17 (14.8%) Mexican-Americans. The distributions of age and ethnicity between the cases and controls were almost identical (P = 0.94 and 0.93, respectively). About 48.2% (54/112) and 32.4% (36/111) of the cases received radiotherapy and chemotherapy after surgery, respectively. Of the total 115 endometrial cancer cases, 60.9% had early stages of I–II, 39.1% had advanced stages (III–IV), 67 (58.3%) were pure endometrioid, 37 (32.1%) were mixed (endometrioid with clear cell or papillary or serous, etc.), and 11 (9.6%) were other types including MMMT (malignant mixed Müllerian tumor) and other sarcomas.
Table 1.
Characteristics and clinical features for cases and controls
| Variables | Cases (n = 115) | Controls (n = 230) | P* | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age (year) | |||||
| ≤60 | 48 | 41.7 | 97 | 42.2 | 0.939 |
| >60 | 67 | 58.3 | 133 | 57.8 | |
| Ethnicity | |||||
| Non-Hispanic white | 88 | 76.5 | 175 | 76.1 | 0.929 |
| Other | 27 | 23.5 | 55 | 23.9 | 0.331 |
| Smoking status | |||||
| Never | 81 | 70.4 | 150 | 65.2 | |
| Ever | 34 | 29.6 | 80 | 34.8 | |
| Histology | |||||
| Pure endometrioid | 67 | 58.3 | |||
| Mixed | 37 | 32.1 | |||
| Other | 11 | 9.6 | |||
| Clinical stage | |||||
| I–II | 70 | 60.9 | |||
| III–IV | 45 | 39.1 | |||
| Radiotherapy | |||||
| Yes | 54 | 48.2 | |||
| No | 58 | 51.8 | |||
| Chemotherapy | |||||
| Yes | 36 | 32.4 | |||
| No | 75 | 67.6 | |||
* Two-sided χ2 test
Association between SNPs and susceptibility to endometrial cancer
Analysis for single locus
Among the 48 SNPs listed in Supplemental Table 1, three SNPs (rs7636454 in PIK3CA, rs61764374 in KRAS, and rs10866 in PDPK1) had a MAF <0.05 in controls, seven SNPs were not in HWE (P < 0.05; rs8100018 in AKT2, rs1770345 in FRAP1, rs7960917, rs8720 and rs7312175 in KRAS, and rs12573787 and rs12357281 in PTEN), and two SNPs (rs12124983 in FRAP1 and rs4854955 in PIK3CA) had a high LD (linkage disequilibrium) with rs2295080 in FRAP1 and rs2865084 in PIK3CA, respectively. As a result, 12 SNPs were excluded from the final analysis for susceptibility to endometrial cancer.
We used three genetic models (additive, dominant and recessive) to investigate the associations between genotypes of the remaining 36 SNPs and endometrial cancer risk (Supplemental Table 1). Three SNPs (AKT1 rs2498801, PIK3CA rs6443624, and PIK3CA rs9838411) had a P value <0.05 in at least one model and were selected for further analysis. After adjustment for age, ethnicity, and smoking status, logistic regression analysis (Table 2) demonstrated that both PIK3CA rs6443624 and rs9838411 remained borderline or significantly associated with decreased risk of endometrial cancer in a dominant model (adjusted OR, 0.62; 95% CI, 0.39–1.00 for rs6443624 and adjusted OR, 0.59; 95% CI, 0.36–0.95 for rs9838411); in contrast, AKT1 rs2498801 was significantly associated with increased risk of endometrial cancer (adjusted OR, 1.94; 95% CI, 1.02–3.67 in a recessive model) (Table 2). No significant associations were found between the other 33 SNPs and endometrial cancer risk (detailed data not shown).
Table 2.
The associations between genotypes of genes in PI3K and RAS/RAF pathways and risk of endometrial cancer
| Genotypes | Cases | Controls | Crude OR (95% CI) | Adjusted OR (95% CI)* | P* |
|---|---|---|---|---|---|
| AKT1 rs2498801 | n = 114 | n = 229 | |||
| AA | 48 | 99 | 1.00 | 1.00 | |
| AG | 45 | 106 | 0.88 (0.54–1.43) | 0.90 (0.55–1.48) | 0.669 |
| GG | 21 | 24 | 1.81 (0.92–3.56) | 1.83 (0.92–3.65) | 0.086 |
| AA/AG | 93 | 205 | 1.00 | 1.00 | |
| GG | 21 | 24 | 1.93 (1.02–3.64) | 1.94 (1.02–3.67) | 0.043 |
| PIK3CA rs6443624 | n = 114 | n = 227 | |||
| CC | 73 | 120 | 1.00 | ||
| CA | 34 | 87 | 0.64 (0.39–1.05) | 0.63 (0.39–1.04) | 0.071 |
| AA | 7 | 20 | 0.58 (0.23–1.43) | 0.58 (0.23–1.47) | 0.252 |
| CC | 73 | 120 | 1.00 | 1.00 | |
| CA/AA | 41 | 107 | 0.63 (0.40–1.00) | 0.62 (0.39–1.00) | 0.049 |
| PIK3CA rs9838411 | n = 115 | n = 229 | |||
| GG | 81 | 134 | 1.00 | 1.00 | |
| GA | 29 | 82 | 0.59 (0.35–0.97) | 0.58 (0.35–0.96) | 0.035 |
| AA | 5 | 13 | 0.64 (0.22–1.85) | 0.65 (0.21–1.97) | 0.443 |
| GG | 81 | 134 | 1.00 | 1.00 | |
| GA/AA | 34 | 95 | 0.59 (0.37–0.96) | 0.59 (0.36–0.95) | 0.032 |
| Combined effect (favorable alleles) † | n = 113 | n = 227 | |||
| 0–1 | 46 | 65 | 1.00 | 1.00 | |
| 2 | 35 | 67 | 0.74 (0.43–1.29) | 0.77 (0.43–1.35) | 0.354 |
| >2 | 32 | 95 | 0.48 (0.28–0.82) | 0.48 (0.27–0.83) | 0.009 |
| P trend | 0.008 | 0.009 | |||
* Logistic regression models adjusted for age, ethnicity, and smoking status
† The combined effects were grouped according to numbers of favorable alleles (rs2498801 A, rs6443624 A, and rs9838411 A were considered as favorable alleles)
Analysis for combined effects and interactions of SNPs
To evaluate any combined effects of these three variants, we summed the number of protective alleles (i.e., rs2498801 A, rs6443624 A, and rs9838411 A) and observed a significant locus–dosage effect between favorable alleles and endometrial cancer risk (adjusted P trend = 0.009). As shown in Table 2, those subjects with 2 and >2 protective alleles had 23 and 52% reduced risk (OR, 0.77; 95% CI, 0.43–1.35 and OR, 0.48; 95% CI, 0.27–0.83, respectively) of endometrial cancer, compared to those with 0–1 protective alleles. Furthermore, a statistically significant multiplicative interaction (P int = 0.036) between two loci of PIK3CA (rs6443624 and rs9838411) was found in risk of endometrial cancer.
Association between SNPs and clinical outcomes of endometrial cancer
Analysis for associations between clinical features and overall survival
Complete follow-up data were available for 115 patients with 20 deaths. The follow-up time ranged from 0.2 to 118 months with the mean of 29.4 months. Clinical stage, radiotherapy, and histology were significantly associated with survival time (log-rank P = 0.004, 0.025, and <0.001, respectively); ethnicity and chemotherapy had no association with survival (log-rank P = 0.156 and 0.059, respectively); age was borderline significant (P = 0.051). Univariate Cox regression analysis showed that the hazard of death increased in patients with advanced stage (compared with patients with early stage, HR, 3.73; 95% CI, 1.43–9.73), patients not receiving radiotherapy (compared with patients with radiotherapy, HR, 2.54; 95% CI, 0.97–6.60), and patients with other types of histology (compared with patients with pure endometrioid carcinoma, HR, 6.70; 95% CI, 2.50–18.00).
Analysis for effects of genetic variants on survival of endometrial cancer patients
Of the 45 common SNPs, Cox regression analyses showed that three SNPs (rs1862162 in PIK3R1, rs892119 in AKT2, and rs2699887 in PIK3CA) exhibited significant associations with the hazard of death. As shown in Table 3, after adjustment for age, ethnicity, smoking status, stage, radiotherapy, chemotherapy, and histology, rs1862162 and rs892119 variant genotypes remained significantly associated with increased hazard ratios (adjusted HR, 2.75; 95% CI, 1.03–7.37 for rs1862162 in a dominant model; and adjusted HR, 11.58; 95% CI, 1.07–125.04 for rs892119 in a recessive model); in contrast, rs2699887 variant genotypes was significantly associated with a lower hazard (AG vs. GG: adjusted HR, 0.24; 95% CI, 0.07–0.87). No statistically significant associations were found between other SNPs and survival (detailed data not shown).
Table 3.
Association between genotypes of genes in PI3K and RAS/RAF pathways and survival of endometrial cancer
| Genotypes | Patients | Deaths | Crude HR (95% CI) | Adjusted HR* (95% CI) | P* |
|---|---|---|---|---|---|
| PIK3R1 rs1862162 | 113 | 20 | |||
| TT | 65 | 8 | 1.00 | 1.00 | |
| TC | 38 | 11 | 2.67 (1.07–6.65) | 3.59 (1.28–10.10) | 0.015 |
| CC | 10 | 1 | 0.64 (0.08–5.12) | 0.94 (0.11–8.28) | 0.954 |
| TT | 65 | 8 | 1.00 | 1.00 | |
| TC/CC | 48 | 12 | 2.05 (0.84–5.02) | 2.75 (1.03–7.37) | 0.044 |
| AKT2 rs892119 | 112 | 20 | |||
| GG | 77 | 13 | 1.00 | 1.00 | |
| GA | 31 | 5 | 1.05 (0.37–2.99) | 1.23 (0.37–4.06) | 0.735 |
| AA | 4 | 2 | 6.21 (1.37–28.15) | 13.79 (1.13–168.89) | 0.040 |
| GG/GA | 108 | 18 | 1.00 | 1.00 | |
| AA | 4 | 2 | 5.94 (1.35–26.06) | 11.58 (1.07–125.04) | 0.044 |
| PIK3CA rs2699887 | 115 | 20 | |||
| GG | 66 | 12 | 1.00 | 1.00 | |
| AG | 35 | 4 | 0.48 (0.15–1.55) | 0.24 (0.07–0.87) | 0.029 |
| AA | 14 | 4 | 1.48 (0.46–4.73) | 0.73 (0.20–2.66) | 0.633 |
* Cox regression models with adjustment for age, ethnicity, smoking status, histology, stage, and treatment status
To identify independent prognostic factors for endometrial cancer, we next performed a multivariate stepwise Cox regression analysis including demographic characteristics (ethnicity and age), clinical features (stage, histology, radiotherapy, and chemotherapy), and genotypes of above-mentioned three significant SNPs on survival of endometrial cancer patients. At last, five variables (histology, radiotherapy, age, PIK3R1 rs1862162 heterozygosity, and PIK3CA rs2699887 heterozygosity) remained as significant prognosticators in the regression model with a significance level of P < 0.05 (Table 4).
Table 4.
Stepwise Cox regression model on survival of endometrial cancer patients
| Variables* | Βeta | SE | HR | 95% CI | P |
|---|---|---|---|---|---|
| Histology† | 1.083 | 0.289 | 2.95 | 1.67–5.21 | 0.0004 |
| Radiotherapy (no vs. yes) | 2.056 | 0.607 | 7.82 | 2.38–25.68 | 0.0088 |
| Age (≥64 vs. <64)‡ | 1.64 | 0.591 | 5.13 | 1.61–16.34 | 0.0250 |
| PIK3R1 rs1862162 (TC vs. TT) | 1.841 | 0.591 | 6.31 | 2.07–19.23 | 0.0096 |
| PIK3CA rs2699887 (AG vs. GG) | −1.614 | 0.669 | 0.20 | 0.05–0.74 | 0.0111 |
* Variables including age, ethnicity, smoking status, histology, stage, radiotherapy, chemotherapy, and significant genotypes of rs1862162, rs892119, and rs2699887 were included in the stepwise multivariate analysis
† Ranked data with pure endometrioid, mixed, and others as ordinal
‡ Dichotomized by the median age in patients
Analysis for effects of genetic variants on endometrial cancer recurrence
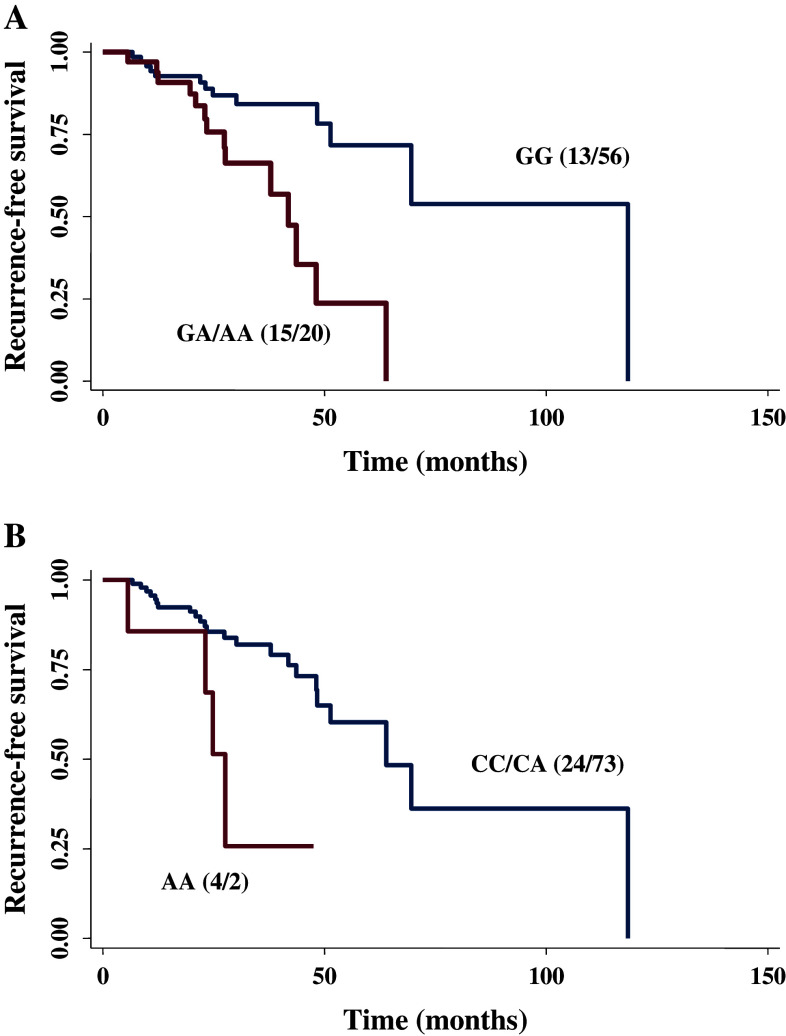
There were data on recurrence for 104 patients, and 28 of them had such a recurrence. On Cox regression analyses, variant genotypes of these two SNPs exhibited increased risk of recurrence (adjusted HR, 3.82; 95% CI, 1.58–9.23 for rs7312175 in a dominant model; and adjusted HR, 5.22; 95% CI, 1.43–19.03 for rs6443624 in a recessive model). Furthermore, when we combined these two variants, we found a significant locus–dosage effect on recurrence risk (adjusted P trend = 0.003). As shown in Table 5, compared to patients with “0” risk alleles, patients with “1” or “2–4” risk alleles of these 2 SNPs had a 2.92- and 7.36-fold increased recurrence risk (95% CI, 0.99–9.61 and 1.99–27.15, respectively) after adjustment for age, ethnicity, smoking status, histology, stage, and treatment status. Figure 1 showed the Kaplan–Meier curves of recurrence-free survival in 104 endometrial cancer patients by KRAS rs7312175 genotypes (GA/AA vs. GG: log-rank P = 0.001) and by PIK3CA rs6443624 genotypes (AA vs. CC/CA: log-rank P = 0.005).
Table 5.
Association between genotypes of genes in PI3K and RAS/RAF pathways and recurrence of endometrial cancer
| Genotypes | Recurrence | No recurrence | HR(95% CI) | Adjusted HR* (95% CI) | P* |
|---|---|---|---|---|---|
| KRAS rs7312175 | 28 | 76 | |||
| GG | 13 | 56 | 1.00 | 1.00 | |
| GA | 13 | 18 | 3.33 (1.45–7.63) | 3.59 (1.44–8.92) | 0.006 |
| AA | 2 | 2 | 5.47 (1.17–25.69) | 6.80 (1.27–36.36) | 0.025 |
| GG | 13 | 56 | 1.00 | 1.00 | |
| GA/AA | 15 | 20 | 3.51 (1.57–7.83) | 3.82 (1.58–9.23) | 0.003 |
| PIK3CA rs6443624 | 28 | 75 | |||
| CC | 17 | 47 | 1.00 | 1.00 | |
| CA | 7 | 26 | 0.87 (0.35–2.13) | 1.15 (0.46–2.92) | 0.766 |
| AA | 4 | 2 | 4.07 (1.32–12.59) | 5.86 (1.52–22.63) | 0.010 |
| CC/CA | 24 | 73 | 1.00 | 1.00 | |
| AA | 4 | 2 | 4.2 (1.40–12.58) | 5.22 (1.43–19.03) | 0.012 |
| Combined effect of risk alleles † | |||||
| 0 | 7 | 34 | 1.00 | 1.00 | |
| 1 | 13 | 30 | 2.06 (0.77–5.51) | 2.92 (0.99–9.61) | 0.053 |
| 2–4 | 8 | 11 | 4.57 (1.55–13.48) | 7.36 (1.99–27.15) | 0.003 |
| P trend | 0.006 | 0.003 |
* Cox regression models with adjustment for age, ethnicity, smoking status, histology, stage, and treatment status
† The combined effects were grouped according to numbers of risk alleles (rs7312175 A and rs6443624 A were considered as risk alleles)
Fig. 1.
Kaplan–Meier curves of recurrence-free survival in endometrial cancer patients by genotypes. a By KRAS rs7312175 genotypes (GA/AA vs. GG: log-rank P = 0.001). b By PIK3CA rs6443624 genotypes (AA vs. CC/CA: log-rank P = 0.005). The recurrence information is available for 104 patients, and the numbers in parentheses are the numbers of recurrence/no recurrence patients with different genotypes
Discussion
The PI3K/PTEN/AKT/mTOR and Ras/Raf/MEK/ERK signaling pathways play an important role in cell cycle progression, gene expression, apoptosis, drug resistance, and sensitivity to targeted therapy (McCubrey et al. 2006; Abrams et al. 2010; Steelman et al. 2010). In these pathways, PI3K is first activated by growth factors and cytokine receptor ligation and subsequently induces a kinase cascade through AKT and mTOR (Manning and Cantley 2007). PI3Ks are members of an intracellular lipid kinase family, composed of catalytic and regulatory subunits encoded by separate genes and alternative splicing (Philp et al. 2001). Among those, PIK3CA encodes the p110α catalytic subunit of class IA PI3Ks, and the genetic alterations in the PIK3CA gene have been reported in a wide variety of human malignancies, including endometrial cancer (Hayes et al. 2006; Catasus et al. 2008).
Recently, epidemiological studies showed that a SNP in PIK3CA (rs2865084) was associated with risk of endometrioid ovarian cancer (P trend = 0.0344) (Quaye et al. 2009), PIK3CA rs2677764 tagSNP was statistically significantly associated with endometrial cancer (Lacey et al. 2011) and another SNP in PIK3CA (rs2699887) was associated with toxicity for lung cancer patients receiving platinum-based chemotherapy (Pu et al. 2010).
In our study, we found that three SNPs (rs6443624, rs9838411, and rs2699887) located in the intron of PIK3CA were significantly associated with susceptibility, survival, or recurrence of endometrial cancer, supporting the importance of this gene in cancer development and occurrence. Several reasons may explain the observed associations in our study: (1) these potentially functional tagging SNPs are the proxies of other functional SNPs within that region in the genome; and (2) bioinformatics tools have showed that rs9838411 and rs2699887 may affect the binding of transcription factors (http://pupasuite2.bioinfo.cipf.es/ and (http://manticore.niehs.nih.gov/snpfunc.htm). Therefore, these potentially functional variants may alter normal splicing patterns or transcription of the PIK3CA gene. Intriguingly, our results showed that rs6443624 had an inconsistent effect on the disease risk and recurrence of endometrial cancer, possibly due to different biological roles of this gene in the initiation and progression of endometrial cancer. Furthermore, the presence of a locus–locus interaction between rs6443624 and rs9838411 in PIK3CA suggested the complexity of effects of genetic variants on the development of endometrial cancer and warrants additional mechanistic investigations.
PIK3R1 is another member of the PI3Ks family and encodes the p85α regulatory subunit (Arcaro and Guerreiro 2007). Functional studies showed that a SNP in codon326 of PIK3R1 (rs3730089, Met > Ile) results in reduced p85α protein expression and increased binding to IRS-1, which negatively regulates the PI3K signaling and super-activates this pathway (Almind et al. 2002; Luo and Cantley 2005). Moreover, a case–control study including 421 colon cancer cases and 483 controls reported that this SNP significantly increased the risk of colon cancer in American population (Li et al. 2008). Nevertheless, our results did not find an association between rs3730089 and risk of endometrial cancer. However, we found the heterozygosity of another SNP of PIK3R1, rs1862162, was an unfavorable prognostic factor for endometrial cancer. The SNP rs1862162 is located in the 5′ region near the PIK3R1 gene, and this variant may play a role in survival by affecting the transcription and expression of PIK3R1. In addition, we also found that rs2498801 in the 3′ UTR of AKT1 was associated with an increased risk of endometrial cancer. AKT1 is one of three isoforms of AKTs (AKT1, AKT2, and AKT3), which are major downstream targets of growth factor receptor tyrosine kinases that signal through PI3K (Testa and Bellacosa 2001). Mounting evidence indicates that alterations in AKT proteins play an important role in regulating cell survival and growth, thus contributing to the pathogenesis of cancer (Bellacosa et al. 2005).
KRAS is a small G protein member of RAS family (N-, H-, and KRAS). Mutations of the KRAS gene have been implicated in the development and/or progression of numerous endometrial malignancies (Mizuuchi et al. 1992; Oehler et al. 2003; Abal et al. 2006; Lax 2007). However, we only found that rs7312175 in KRAS was associated with the recurrence of endometrial cancer. Bioinformatics tools show that rs7312175 in KRAS is located in the transcription factor binding site. Therefore, this SNP may result in the variation of transcription activity and expression of KRAS, which can affect the recurrence of endometrial cancer.
In this pilot study, we investigated potentially functional SNPs in 11 key genes involved in the PI3K/PTEN/AKT/mTOR and RAS/RAF/MEK/ERK pathways and provided the evidence that variations in these pathways play a role in modulating susceptibility, survival, or recurrence of endometrial cancer. However, several limitations exist in our present study. Firstly, the small sample size limits the statistical power of our study, especially for subgroups. Therefore, we did not present the preliminary findings from the stratification analysis. For example, we found that there were significant differences in PIK3R1 rs3756668 and PTEN rs701848 genotype frequencies between endometrioid and non-endometrioid carcinomas (P = 0.007 and 0.003, respectively). Both SNPs were located in 3′-UTR of the genes, which may have effects on the gene and protein expression. This might be a clue for different carcinogenic pathways in histology types. However, the functions of these regulatory SNPs need to be investigated in cell lines or xenografts in additional mechanistic studies. We have also performed the analysis on all subjects and in Whites only separately. The trends in the analysis of both risk assessment and clinical outcomes were similar. Therefore, we did not exclude the 23% minorities from the analysis to avoid diminishing of the power. Secondly, we do not have the complete information such as age at menarche/menopause, overweight/obese status, use of estrogen therapy, nulliparity, family history, and the presence of HNPCC (hereditary non-polyposis colorectal cancer), diabetes and PCOS (polycystic ovary syndrome) for most of controls and some of the cases. Therefore, we only included the available data on demographic and exposure in the analysis. Thirdly, multiple SNPs were included in the analyses, which might result in false-positive findings due to multiple comparisons. The significant SNPs PIK3CA rs6443624 and rs9838411 (in LD with rs7641983, r 2 = 0.966) associated with susceptibility to endometrial cancer in the present analysis were not validated in a population-based case–control study in Poland (PECS) (Lacey et al. 2011). Finally, the functional significance of these identified SNPs was not clear. For example, how do the three SNPs rs6443624, rs9838411, and rs2699887 play different roles in susceptibility, survival, or recurrence of endometrial cancer in the same PIK3CA gene? In our correlation analysis of SNPs and available corresponding gene expression data from 9 tumor tissues, we found that PIK3CA rs2699887 (in LD with rs2699905, r 2 = 1.0) was significantly associated with the PIK3CA gene expression levels measured with three probes in Affymatrix gene expression platform (data not shown). Therefore, additional profound studies with large sample sizes, genome-wide association, and mechanistic studies are warranted to unravel the biological relevance of these SNPs and the underlying molecular mechanisms for the observed associations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported in part by National Institute of Health (NIH) grants ES11740 and CA131274 (Q. Wei), Stand Up to Cancer/American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR—DT0209, the Uterine SPORE CA098258 and the Kleberg Center For Molecular Markers (G. B. Mills), CA127219 (M. R. Spitz), and a Cancer Center Core grant from the National Cancer Institute to M. D. Anderson (CA016672). We would like to thank Joseph Celestino, Yang Li, Min Zhao, and Kejing Xu for preparing the DNA samples.
References
- Abal M, Planaguma J, Gil-Moreno A, Monge M, Gonzalez M, Baro T, Garcia A, Castellvi J, Ramon YCS, Xercavins J, Alameda F, Reventos J (2006) Molecular pathology of endometrial carcinoma: transcriptional signature in endometrioid tumors. Histol Histopathol 21(2):197–204 [DOI] [PubMed] [Google Scholar]
- Abrams SL, Steelman LS, Shelton JG, Wong EW, Chappell WH, Basecke J, Stivala F, Donia M, Nicoletti F, Libra M, Martelli AM, McCubrey JA (2010) The Raf/MEK/ERK pathway can govern drug resistance, apoptosis and sensitivity to targeted therapy. Cell Cycle 9(9):1781–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmedkhanov A, Zeleniuch-Jacquotte A, Toniolo P (2001) Role of exogenous and endogenous hormones in endometrial cancer: review of the evidence and research perspectives. Ann N Y Acad Sci 943:296–315 [DOI] [PubMed] [Google Scholar]
- Almind K, Delahaye L, Hansen T, Van Obberghen E, Pedersen O, Kahn CR (2002) Characterization of the Met326Ile variant of phosphatidylinositol 3-kinase p85alpha. Proc Natl Acad Sci USA 99(4):2124–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antico-Arciuch VG, Dima M, Liao XH, Refetoff S, Di Cristofano A (2010) Cross-talk between PI3K and estrogen in the mouse thyroid predisposes to the development of follicular carcinomas with a higher incidence in females. Oncogene [DOI] [PMC free article] [PubMed]
- Arcaro A, Guerreiro AS (2007) The phosphoinositide 3-kinase pathway in human cancer: genetic alterations and therapeutic implications. Curr Genomics 8(5):271–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Kumar CC, Di Cristofano A, Testa JR (2005) Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv Cancer Res 94:29–86 [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE (2007) A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448(7152):439–444 [DOI] [PubMed] [Google Scholar]
- Catasus L, Gallardo A, Cuatrecasas M, Prat J (2008) PIK3CA mutations in the kinase domain (exon 20) of uterine endometrial adenocarcinomas are associated with adverse prognostic parameters. Mod Pathol 21(2):131–139 [DOI] [PubMed] [Google Scholar]
- Catasus L, Gallardo A, Cuatrecasas M, Prat J (2009) Concomitant PI3K-AKT and p53 alterations in endometrial carcinomas are associated with poor prognosis. Mod Pathol 22(4):522–529 [DOI] [PubMed] [Google Scholar]
- Chen JS, Wang Q, Fu XH, Huang XH, Chen XL, Cao LQ, Chen LZ, Tan HX, Li W, Bi J, Zhang LJ (2009) Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: association with MMP-9. Hepatol Res 39(2):177–186 [DOI] [PubMed] [Google Scholar]
- Hayes MP, Wang H, Espinal-Witter R, Douglas W, Solomon GJ, Baker SJ, Ellenson LH (2006) PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res 12(20 Pt 1):5932–5935 [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Yang H, Hung MC, Izzo JG, Huang M, Lin J, Ajani JA, Wu X (2009) Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol 27(6):857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard T, Witton CJ, McGlynn LM, Tovey SM, Dunne B, Lyon A, Bartlett JM (2005) AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol 207(2):139–146 [DOI] [PubMed] [Google Scholar]
- Konstantinova D, Kaneva R, Dimitrov R, Savov A, Ivanov S, Dyankova T, Kremensky I, Mitev V (2010) Rare mutations in the PIK3CA gene contribute to aggressive endometrial cancer. DNA Cell Biol 29(2):65–70 [DOI] [PubMed] [Google Scholar]
- Lacey JV Jr, Yang H, Gaudet MM, Dunning A, Lissowska J, Sherman ME, Peplonska B, Brinton LA, Healey CS, Ahmed S, Pharoah P, Easton D, Chanock S, Garcia-Closas M (2011) Endometrial cancer and genetic variation in PTEN, PIK3CA, AKT1, MLH1, and MSH2 within a population-based case-control study. Gynecol Oncol 120(2):167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax SF (2007) Molecular genetic changes in epithelial, stromal and mixed neoplasms of the endometrium. Pathology 39(1):46–54 [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275(5308):1943–1947 [DOI] [PubMed] [Google Scholar]
- Li L, Plummer SJ, Thompson CL, Tucker TC, Casey G (2008) Association between phosphatidylinositol 3-kinase regulatory subunit p85alpha Met326Ile genetic polymorphism and colon cancer risk. Clin Cancer Res 14(3):633–637 [DOI] [PubMed] [Google Scholar]
- Luo J, Cantley LC (2005) The negative regulation of phosphoinositide 3-kinase signaling by p85 and it’s implication in cancer. Cell Cycle 4(10):1309–1312 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350(21):2129–2139 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129(7):1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D’Assoro AB, Salisbury JL, Mazzarino MC, Stivala F, Libra M (2006) Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul 46:249–279 [DOI] [PubMed] [Google Scholar]
- Minaguchi T, Yoshikawa H, Oda K, Ishino T, Yasugi T, Onda T, Nakagawa S, Matsumoto K, Kawana K, Taketani Y (2001) PTEN mutation located only outside exons 5, 6, and 7 is an independent predictor of favorable survival in endometrial carcinomas. Clin Cancer Res 7(9):2636–2642 [PubMed] [Google Scholar]
- Mizuuchi H, Nasim S, Kudo R, Silverberg SG, Greenhouse S, Garrett CT (1992) Clinical implications of K-ras mutations in malignant epithelial tumors of the endometrium. Cancer Res 52(10):2777–2781 [PubMed] [Google Scholar]
- Mori N, Kyo S, Sakaguchi J, Mizumoto Y, Ohno S, Maida Y, Hashimoto M, Takakura M, Inoue M (2007) Concomitant activation of AKT with extracellular-regulated kinase 1/2 occurs independently of PTEN or PIK3CA mutations in endometrial cancer and may be associated with favorable prognosiss. Cancer Sci 98(12):1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama-Hosokawa S, Oda K, Nakagawa S, Ishikawa S, Yamamoto S, Shoji K, Ikeda Y, Uehara Y, Fukayama M, McCormick F, Yano T, Taketani Y, Aburatani H (2010) Genome-wide single-nucleotide polymorphism arrays in endometrial carcinomas associate extensive chromosomal instability with poor prognosis and unveil frequent chromosomal imbalances involved in the PI3-kinase pathway. Oncogene 29(13):1897–1908 [DOI] [PubMed] [Google Scholar]
- Nicholson KM, Anderson NG (2002) The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal 14(5):381–395 [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Kato K, Takahashi A, Ueoka Y, Kamikihara T, Arima T, Matsuda T, Kato H, Nishida J, Wake N (2004) K-Ras and H-Ras activation promote distinct consequences on endometrial cell survival. Cancer Res 64(8):2759–2765 [DOI] [PubMed] [Google Scholar]
- Oehler MK, Brand A, Wain GV (2003) Molecular genetics and endometrial cancer. J Br Menopause Soc 9(1):27–31 [DOI] [PubMed] [Google Scholar]
- Paynter RA, Hankinson SE, Colditz GA, Kraft P, Hunter DJ, De Vivo I (2005) CYP19 (aromatase) haplotypes and endometrial cancer risk. Int J Cancer 116(2):267–274 [DOI] [PubMed] [Google Scholar]
- Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, Thomas RJ, Phillips WA (2001) The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res 61(20):7426–7429 [PubMed] [Google Scholar]
- Pu X, Hildebrandt MA, Lu C, Lin J, Stewart DJ, Ye Y, Gu J, Spitz MR, Wu X (2010) PI3K/PTEN/AKT/mTOR pathway genetic variation predicts toxicity and distant progression in lung cancer patients receiving platinum-based chemotherapy. Lung Cancer [DOI] [PMC free article] [PubMed]
- Quaye L, Song H, Ramus SJ, Gentry-Maharaj A, Hogdall E, DiCioccio RA, McGuire V, Wu AH, Van Den Berg DJ, Pike MC, Wozniak E, Doherty JA, Rossing MA, Ness RB, Moysich KB, Hogdall C, Blaakaer J, Easton DF, Ponder BA, Jacobs IJ, Menon U, Whittemore AS, Kruger-Kjaer S, Pearce CL, Pharoah PD, Gayther SA (2009) Tagging single-nucleotide polymorphisms in candidate oncogenes and susceptibility to ovarian cancer. Br J Cancer 100(6):993–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino MJ, Bell DW (2011) A unique spectrum of somatic PIK3CA (p110{alpha}) mutations within primary endometrial carcinomas. Clin Cancer Res 17(6):1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304(5670):554 [DOI] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, Jemal A (2011) Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 61(4):212–236 [DOI] [PubMed] [Google Scholar]
- Steelman LS, Stadelman KM, Chappell WH, Horn S, Basecke J, Cervello M, Nicoletti F, Libra M, Stivala F, Martelli AM, McCubrey JA (2008) Akt as a therapeutic target in cancer. Expert Opin Ther Targets 12(9):1139–1165 [DOI] [PubMed] [Google Scholar]
- Steelman LS, Abrams SL, Shelton JG, Chappell WH, Basecke J, Stivala F, Donia M, Nicoletti F, Libra M, Martelli AM, McCubrey JA (2010) Dominant roles of the Raf/MEK/ERK pathway in cell cycle progression, prevention of apoptosis and sensitivity to chemotherapeutic drugs. Cell Cycle 9:8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa JR, Bellacosa A (2001) AKT plays a central role in tumorigenesis. Proc Natl Acad Sci USA 98(20):10983–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treloar SA, Zhao ZZ, Le L, Zondervan KT, Martin NG, Kennedy S, Nyholt DR, Montgomery GW (2007) Variants in EMX2 and PTEN do not contribute to risk of endometriosis. Mol Hum Reprod 13(8):587–594 [DOI] [PubMed] [Google Scholar]
- Urick ME, Rudd ML, Godwin AK, Sgroi DC, Merino M, Bell DW (2011) PIK3R1 (p85-alpha/p85{alpha}) is somatically mutated at high frequency in primary endometrial cancer. Cancer Res [DOI] [PMC free article] [PubMed]
- Wang T, Ho G, Ye K, Strickler H, Elston RC (2009) A partial least-square approach for modeling gene–gene and gene-environment interactions when multiple markers are genotyped. Genet Epidemiol 33(1):6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC (2008) PI3K pathway alterations in cancer: variations on a theme. Oncogene 27(41):5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.