Abstract
Saliva is a readily available biofluid that may contain metabolites of interest for diagnosis and prognosis of diseases. In this work, a differential 13C-/12C-isotope dansylation labeling method, combined with liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry (LC-FTICR-MS), is described for quantitative profiling of the human salivary metabolome. New strategies are presented to optimize the sample preparation and LC-MS detection processes. The strategies allow the use of as little of 5 μL of saliva sample as a starting material to determine the concentration changes of an average of 1058 ion pairs or putative metabolites in comparative saliva samples. The overall workflow consists of several steps including acetone-induced protein precipitation, 12C-dansylation labeling of the metabolites, and LC-UV measurement of the total concentration of the labeled metabolites in individual saliva samples. A pooled sample was prepared from all the individual samples and labeled with 13C-dansylation to serve as a reference. Using this metabolome profiling method, it was found that compatible metabolome results could be obtained after saliva samples were stored in tubes normally used for genetic material collection at room temperature, -20°C freezer, and -80°C freezer over a period of one month, suggesting that many saliva samples already collected in genomic studies could become a valuable resource for metabolomics studies, although the effect of much longer term of storage remains to be determined. Finally, the developed method was applied for analyzing the metabolome changes of two different groups: normal healthy older adults and comparable older adults with mild cognitive impairment (MCI). Top-ranked 18 metabolites successfully distinguished the two groups, among which seven metabolites were putatively identified while one metabolite, taurine, was definitively identified.
Introduction
Saliva is an oral fluid secreted mainly from the salivary glands and plays a pivotal role in several physiologic functions related to the oral cavity system, such as food swallowing and antibacterial and antiviral protection.1, 2 It is composed of many secretory products including proteins and metabolites. It is believed that many compounds present in the blood are also present in saliva due to chemical diffusion and transport processes involved in salivary glands or through the gingival sulcus.1 Thus the chemical constitution of saliva, like blood, can be influenced by the physiological state of an individual, and hence used for disease diagnosis and prognosis. With the development of very sensitive analytical tools for detection of trace amounts of chemical components in saliva, the field of saliva diagnostics is being advanced rapidly.1-7 Many biomarkers of cancer, cardiovascular, and other diseases can potentially be detected in saliva.7 Compared to other biofluids, collection of saliva is easy and not invasive. It can be done in private or a remote site or in clinically challenging situations where blood sampling is not possible.
Saliva has also been widely used as a source of DNA and RNA for genetic studies8-10 and, as a result, large repositories of human saliva samples, often accompanied with important clinical information of the donors, are available. These are great resources not only for genomics, but also for proteomic and metabolomic studies, particularly for the discovery of new biomarkers for disease diagnosis and prognosis. Indeed, salivary proteomics is an active research field where a variety of analytical techniques are being developed and applied to profile the proteomes of saliva samples.5, 11-14 In contrast, there are only a few reports focusing on metabolome analyses of saliva using NMR15-19, gas chromatography mass spectrometry (GC-MS)3, 20, 21, liquid chromatography MS (LC-MS)3, 22-25, and capillary electrophoresis MS (CE-MS).26 The major challenge in saliva metabolome profiling lies in the relatively low abundance of metabolites present in saliva. Even with metabolite extraction and concentration, a much smaller number of metabolites have been detected in saliva, compared to other biofluids such as blood or urine. For example, recently reported metabolome profiles generated by LC-MS or CE-MS were generally composed of less than 100 metabolite peaks.3, 22-26 To increase the probability of discovering specific biomarkers of diseases using salivary metabolomics, there is a clear need to develop more sensitive analytical tools to profile a large number of metabolites, i.e., to cast a bigger net to find the biomarkers.
In this work, we report a high performance isotope-labeling LC-MS approach, based on dansylation labeling27, for quantitative and more comprehensive profiling of the salivary metabolome. An analytical workflow was developed to process saliva samples with as little as 5 μL of starting material. On average, more than 1000 putative metabolites could be detected and the relative concentrations of these metabolites could be determined using differential isotope labeling of individual samples (12C2-labeling) and a pooled saliva reference sample (13C2-labeling). This method was applied to the study of the sample storage effect on metabolome profiling. The application of this method for salivary metabolomics was demonstrated in the study of the effect of mild cognitive impairment (MCI) on metabolome changes of the individuals with MCI, compared to age- and gender-matched controls.
Experimental Section
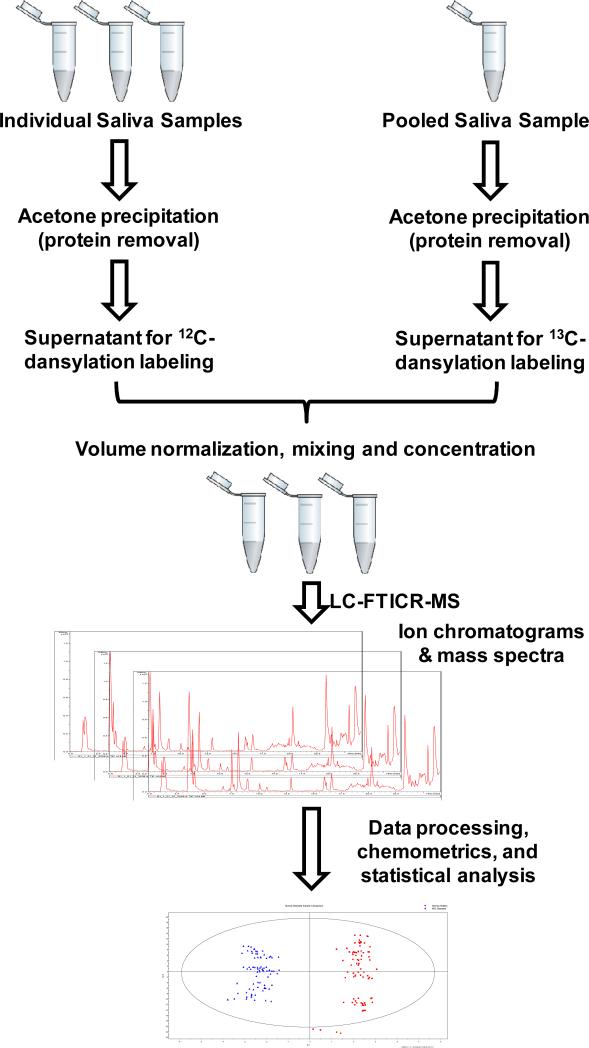
Figure 1 depicts the workflow used for saliva metabolome analysis. The key experimental processes are highlighted below and some details of the experimental steps are given in Supplemental Notes N1-N3.
Figure 1.
Workflow of the high performance isotope labeling LC-MS method developed for salivary metabolome profiling.
Saliva Sample Collection
All biosamples were collected in active and certified compliance with prevailing human research ethics guidelines. All human participants contributed signed informed consent forms. The participants were enrolled in the Victoria Longitudinal Study (VLS), a large-scale epidemiological study of biological, neurocognitive, and biomedical human aging.28 The present groups were classified on the basis of objective and standardized procedures for clinically differentiating a normal aging from a probable mild cognitive impairment (MCI) group.29-32 The present classifications included 4-year follow-up independent assessments confirming stability of initial clinical status. The saliva samples were collected initially for genomic research, and all samples have been successfully processed for DNA extraction and initial genotyping.33 Specifically, the saliva samples were collected by Oragene®•DNA collection kits (DNS Genotek Inc., Ottawa, ON, Canada), as administered by trained VLS staff. The collection tube of each kit contains 2 mL of stabilizing reagents mainly composed of ethanol (10-30%) and Tris-buffer (1-5%), according to the manufacturer. Up to 2 mL of saliva can be collected. The majority of the samples were collected in the afternoon, following the protocol given in the collection kit (e.g., rinsing mouth with water to clear particles and collecting saliva to the 2-mL mark which was done usually within 5 to 15 min). The samples were stored at room temperature in a locked laboratory archive room.
Sample Processing
For processing the saliva sample to remove proteins, acetone was cooled to -20°C in advance. An aliquot of saliva sample was placed in 600-μL Eppendorf tube, and four times the sample volume of cold acetone was added to the tube. The solution was vortexed, and then incubated at -20°C overnight. After incubation, the solution was centrifuged at 20000 g for 30 min. The resulting supernatant was aliquoted out for dansylation labeling.
Dansylation Labeling
The synthesis of 13C-dansyl chloride as the isotope labeling reagent has been described by K. Guo et al.27 The dansylation labeling reaction has also been described27, but with some minor changes tailored to the saliva sample labeling (see Supplemental Note N1).
LC-FTICR-MS
An Agilent 1100 series capillary HPLC system (Agilent, Palo Alto, CA) and an Agilent reversed-phase Eclipse C18 column (2.1 mm × 100 mm, 1.8 μm particle size, 95 Å pore size) were used for online LC-MS using a Bruker 9.4 Tesla Apex-Qe Fourier-transform ion-cyclotron resonance (FT-ICR) mass spectrometer (Bruker, Billerica, MA, USA). All MS spectra were obtained in the positive ion mode. For the LC-MS work, LC solvent A was 0.1% (v/v) formic acid in 5% (v/v) ACN, and solvent B was 0.1% (v/v) formic acid in ACN. The gradient elution profile was as follows: t = 0 min, 20% B; t = 3.50 min, 35% B; t = 18.00 min, 65% B; t = 21.00 min, 95% B; t = 21.50 min, 95% B; t = 23.00 min, 98% B; t = 24.00 min, 98% B; t = 26.50 min, 99% B. The flow rate was 180 μL/min, and the sample injection volume was 2 μL. The flow from HPLC was split 1:3 and a 60 μL/min flow was loaded to the ESI source of the FT-ICR mass spectrometer, while the rest of the flow was delivered to waste.
LC-UV
An ACQUITY UPLC® system (Waters Corporation, Milford, MA) including binary solvent manager, sampler manager, and photo diode array (PDA) detector, and a Waters ACQUITY UPLC™ BEH (Ethylene Bridged Hybrid) C18 column (2.1 mm × 50 mm, 1.7 μm particle size) were used for online LC-UV.34 The detection wavelength was set at 338 nm.
Data Processing and Statistical Analysis
The data files obtained from LC-FTICR-MS analysis were first converted to NetCDF format by Bruker Compass DataAnalysis software (Bruker Daltonics). XCMS,35 a public-domain software, was used to analyze the NetCDF files to pick up the 12C-/13C-ion pairs of the same metabolites in two comparative samples. All the processed files by XCMS were aligned together in one Excel file. SIMCA-P+ 12.0 software (Umetrics, Umeå, Sweden) was used for multivariate statistical analysis.
Metabolite Identification
Several metabolites were selected according to their ranks of VIP value (see Results) which may serve as potential biomarkers for differentiating two different groups of individuals. Accurate mass of each underivatized metabolite was calculated by subtracting the mass of dansyl group from that of dansylation labeled metabolite. An in-house developed web-based software MyCompoundID was used to search the accurate mass within the human matabolome database (HMDB),36 using a mass accuracy tolerance of 5 ppm. The potential biomarker was definitively identified if both the retention time and the accurate mass could be matched with those of the authentic standard.
Results and Discussion
Dansylation Labeling
In the workflow shown in Figure 1, dansylation labeling is a key step. Saliva, like other biofluids, contains many highly polar and poorly ESI-ionizable metabolites that are also present in low abundance. The use of dansylation labeling to form dansyl derivatives of metabolites overcomes some of technical issues related to LC-MS analysis of these metabolites.27, 37 Dansylation mainly targets the amine. and phenol-containing metabolites.38-40 Upon labeling, the hydrophobicity of these metabolites is altered to an extent that they can be separated efficiently by reversed-phase liquid chromatography (RPLC). In addition, the ESI responses of these metabolites are enhanced by 10- to 1000-fold, depending on the metabolite type. An isotope tag (12C2 or 13C2) can be built into the dansylation reagent such that the light- (i.e., 12C2-) and heavy- (i.e., 13C2-) labeling reagents can be used to differentially react with two comparative samples for relative quantification of the metabolomes. Using differential isotope labeling, peaks belonging to the labeled amine- or phenol-containing metabolites can be picked up according to the accurate mass difference of the ion pair. For example, for a metabolite pair labeled with one dansyl group, the mass difference should be 2.0067 Da, with a measurement accuracy of <2 ppm in mass difference. The mass difference for n-tag singly charged ion pair is n×2.0067 Da. Using the in-house developed program, redundant peaks such as adduct ions or multiply charged ions of the same metabolite and the noise and background peaks are eliminated to retain only one ion pair from a putative metabolite at a given retention time.27, 37
In applying this isotope labeling technique for profiling the metabolome of saliva samples, our goal was to use a small volume of starting material to generate a maximum number of peak pairs. Thus, we examined several variables that could influence the analytical performance and subsequently developed the strategies to optimize or control these variables. In order to compare the effect of individual variables on the total number of peak pairs detected, we needed to ensure that the same amount of labeled metabolites from differently processed samples was analyzed by LC-MS for fair comparison. Thus, the first step in our process of developing an optimal saliva metabolome analysis method was to develop a quantification method to measure the total amount or concentration of labeled amine- or phenol-containing metabolites in a sample. However, because a saliva standard with known total concentration of metabolites is not available, it was not possible to directly determine the total concentration of salivary metabolites in an individual sample.
Total Concentration of Salivary Metabolites
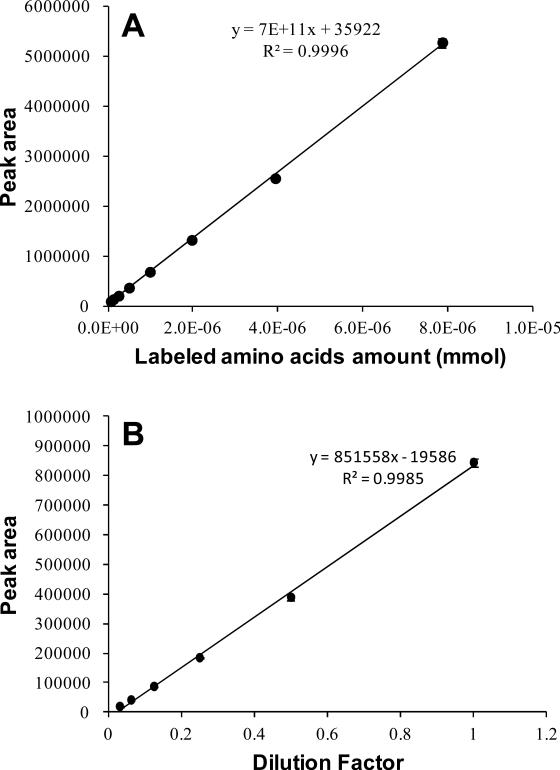
Because human saliva contains amino acids that are present in relatively higher concentrations, compared to other metabolites,15 we expected that a mixture of amino acid standards might serve well as a reference standard for estimating the total concentration of salivary metabolites. To test this expectation, an amino acid standard mixture (aa-mixture) was labeled with 12C-dansyl chloride and the UV absorbance of the labeled aa-mixture, with various dilutions, was analyzed using the fast step-gradient LC-UV method described in the Experimental Section and Supplemental Note N1. The calibration curve obtained is shown in Figure 2A. Meanwhile, another curve was generated from dilution of a labeled saliva sample and is shown in Figure 2B. It is clear that the two calibration curves have different slopes, indicating that the labeled aa-mixture and the labeled saliva sample have different absorption coefficients. Thus, the calibration curve of the labeled aa-mixture cannot be used to determine the absolute concentration of the labeled salivary metabolites. It is apparent that there are many other metabolites than amino acids influencing the overall absorptivity of the labeled saliva sample. Because identification of all the major metabolites contributing to the UV absorbance is very difficult, it is not possible to determine the absolute concentration of the total metabolites present in saliva.
Figure 2.
(A) Calibration curve built with 12C-dansyl labeled amino acid standard mixture solution. (B) Calibration curve built with 12C-dansyl labeled saliva used.
However, for sample comparison and controlling the amount of labeled metabolites being injected into LC-MS, only the relative concentrations among different samples are needed to be determined. Therefore, we resorted to the use of the dilution curve of a labeled saliva sample (Figure 2B) as the calibration curve for relative quantification. In the course of developing an optimal workflow for salivary metabolome profiling, several experimental variables were carefully examined.
Sample Size and Sample Processing
One of the important variables in determining the outcome of saliva analysis is the initial sample size used for profiling. The use of lower volumes of saliva sample is desirable for multiple measurements of the same sample collected from an individual. Aside from genomic and proteomic measurements, even for metabolome analysis alone, multiple aliquots are often required to analyze a wide range of metabolites to achieve high metabolome coverage. For example, the dansylation chemistry described in this work targets the amine- and phenol-containing metabolites. For analyzing other classes of metabolites, different isotope labeling chemistries41 are used with each experiment consuming an aliquot of a valuable sample. In addition, to generate better statistics to reveal subtle concentration differences of the metabolome among different individual, replicate measurements of the same sample are needed.
To determine the minimal size of the initial saliva sample that can be handled conveniently, we tested three different volumes of the saliva sample collected from an healthy individual using the Oragene®•DNA collection kit. They were 5, 10, and 20 μL samples, corresponding to the original saliva volumes of 2.5, 5, and 10 μL, respectively. First, the extraction efficiency of the metabolites from different volumes of saliva was examined. In this case, two aliquots of 20 μL, two aliquots of 10 μL and four aliquots of 5 μL saliva underwent protein precipitation using 4 times of volumes of cold acetone. The supernatants from the 20-μL samples were then aliquoted into 8 vials with each containing 25 μL. For the 10-μL samples or the 5-μL samples, the supernatants were aliquoted into 4 vials with each containing 25 μL. To each 25 μL aliquot, 25 μL of water was added to a total volume of 50 μL for dansylation labeling. Two μL of the labeled sample was injected into LC-UV for UV measurement. Table 1 shows the total concentration of the labeled metabolites in each aliquot in terms of the dilution factor that was determined from the calibration curve shown in Figure 2B. These aliquots gave similar concentrations, indicating that the extraction efficiency of metabolites from saliva through the acetone protein precipitation process was almost the same, even when different volumes of saliva were used.
Table 1.
Comparison of metabolite extraction efficiency with different initial saliva volumes examined by UV absorbance.
| Aliquot* | UV peak area | Dilution factor |
|---|---|---|
| 20-1 | 462632 | 0.529 |
| 20-2 | 434492 | 0.501 |
| 20-3 | 442564 | 0.509 |
| 20-4 | 418833 | 0.485 |
| 20-5 | 444456 | 0.511 |
| 20-6 | 461126 | 0.527 |
| 20-7 | 433296 | 0.500 |
| 20-8 | 431449 | 0.498 |
| 10-1 | 445360 | 0.511 |
| 10-2 | 459850 | 0.526 |
| 10-3 | 449206 | 0.515 |
| 10-4 | 473283 | 0.539 |
| 5-1 | 416713 | 0.483 |
| 5-2 | 431406 | 0.498 |
| 5-3 | 439281 | 0.506 |
| 5-4 | 416750 | 0.483 |
| 0.508±0.017 | ||
x-y refers to experiment replicate #y with the use of x μL of saliva sample as the starting material..
Second, the possibility of sample loss during the process of drying down the supernatant after acetone precipitation was studied. According to the volume requirement of an optimized dansylation labeling reaction condition,27 50 μL of sample solution was needed. While dealing with different volumes of saliva, especially 20 μL or even higher volumes of samples, final volumes of the supernatants were always more than 50 μL, leading to the necessity to first dry them down, and then re-dissolve it to 50 μL for the reaction. It was found that the average dilution factor of the labeled metabolites from the 5-μL saliva sample was 0.202±0.013 after the sample was dried down completely and then re-dissolved before dansylation, compared to 0.492±0.011 for the ones without drying down. This indicates that there was sample loss in the drying step. To avoid this problem, in all subsequent experiments, supernatants were used directly for labeling after acetone precipitation.
Finally, we optimized the labeling reaction for the 5-μL saliva. After acetone precipitation, the volume of the supernatant was about 25 μL. In our experiment, 25 μL of the supernatant was used directly for dansylation labeling without dilution with water to 50 μL. In other words, the scale of the labeling reaction was halved. LC-UV measurements of three 5-μL saliva samples labeled using this reduced scale showed that the average dilution factor was 1.010±0.014, compared to 0.508±0.017 in Table 1. As expected, the total concentration of the labeled metabolites was doubled. The influence of the dansyl chloride concentration was also investigated. Instead of 18 mg/mL of dansyl chloride for labeling the saliva sample, concentration at 36 mg/mL was tested. LC-UV measurement results showed that the average peak area from three replicates was 849639±42407 and 825491±20683, respectively, for the low and high concentration dansyl labeling. From the comparison, it was clear that, the concentration of dansyl chloride used, i.e., 18 mg/mL, was sufficiently high for labeling saliva samples and any increase in dansyl chloride concentration did not improve the product yield.
Sample Injection in LC-MS
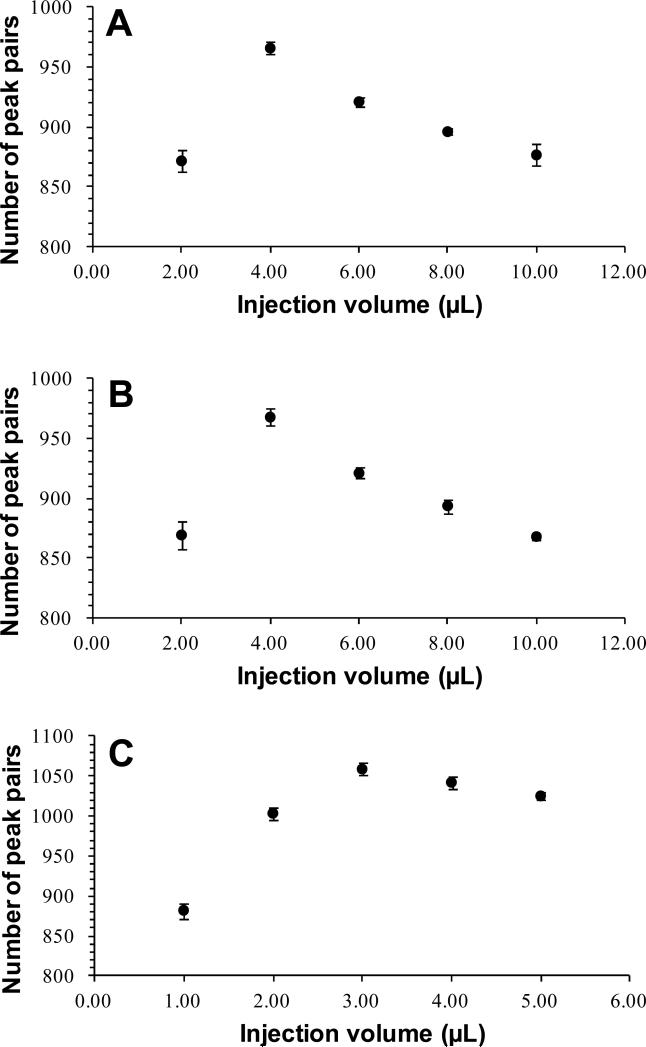
The amount and volume of sample injected into LC-MS can affect the detectability of the metabolites, which can be gauged by examining the number of peak pairs detected from a mixture of the same sample labeled differentially with 12C- and 13C-dansyl chloride. In our work, the individual 12C-labeled sample from 5-μL saliva was mixed with the 13C-labeled control prepared from a 25-μL saliva sample in a 1:1 ratio. The mixture was injected into LC-MS with different volumes: 2 μL, 4 μL, 6 μL, 8 μL and 10 μL. The results are shown in Figure 3A. The maximum number of peak pairs (966±5; n=3) was detected with an injection volume of 4 μL. When the volume of sample injection increased, a decrease in peak pair number was observed, likely due to the peak broadening in chromatographic separation that decreased the detectability of some of low abundance ions.
Figure 3.
Number of peak pairs detected as a function of injection volume from a sample prepared from (A) 5 μL saliva labeled by a scaled down dansylation reaction (i.e., 1/2 volume as normal), (B) 10 μL saliva labeled by normal volume dansylation reaction, and (C) 5 μL saliva labeled by a scaled down dansylation reaction, followed by reducing the labeled saliva sample volume by half via SpeedVac.
For comparison, three aliquots of 10 μL saliva were individually subjected to acetone precipitation, and 50 μL of supernatant from each aliquot was taken out for 12C-dansyl chloride labeling. The average dilution factor of these three solutions was found to be 0.954±0.017, which was close to those prepared from 5-μL saliva and done using the half-scale labeling reaction. After spiking in the 13C-labeled control solution, the mixture was directly injected into LC-FTICR-MS for analysis, with an injection volume of 2 μL, 4 μL, 6 μL, 8 μL or 10 μL. The results are shown in Figure 3B. The two datasets shown in Figure 3A and 3B are very similar, indicating that similar results could be obtained using a 5 μL of saliva starting material, compared to the use of larger volumes of samples. Note that the volume of the original saliva used was actually only 2.5 μL, taking into account the twice dilution of the original saliva during collection using the collection kit that contains 2 mL of stabilizing reagents including ethanol.
As indicated earlier, injection volume can affect the detectability to some extent (for example, in Figure 3A, there was about 10% reduction when the volume was increased from 4 μL to 8 μL). We recognized that the injected sample contained 50% of aqueous solution and 50% organic solvent (ACN); the organic solvent could broaden the chromatographic peaks, resulting in the reduction of the total number of peak pairs detected in LC-MS. We thus tried to concentrate the sample by evaporating solvent to reduce the volume by half. The resultant sample was injected in 1, 2, 3, 4, or 5 μL separately for LC-FTICR-MS analysis. Figure 3C shows the results obtained from the injections of this concentrated sample. Interestingly, more peak pairs were detected from 2, 3, 4, or 5 μL of injection of the concentrated sample, compared to 4, 6, 8, or 10 μL of injection of the non-concentrated sample, respectively. For the 1 μL injection of the concentrated sample, the number of peak pairs detected was similar to that from 2 μL injection of the non-concentrated sample. This suggests that there was no significant loss of samples during the concentration step. The results also indicate that injecting a more concentrated sample in a smaller volume has a benefit of increasing the overall number of peak pairs detected. This is likely related to the chromatographic peak shapes in LC-MS. Injecting a more concentrated solution in a smaller volume, the chromatographic peaks that appeared during LC separation were not as broad as those obtained while injecting a larger volume of a lower concentration solution. Especially, the non-concentrated solution directly after dansylation labeling, which contained 50% of aqueous solvent and 50% of organic solvent, had stronger eluting strength than the initial condition of mobile phase in the LC separation containing 20% of organic solvent. In contrast, the concentrated solution was mostly aqueous with little organic solvent remaining. From these results, it can be concluded that concentrating the sample solutions after labeling to remove the organic solvent before injecting into LC-MS is a preferred approach to increase the number of peak pairs detected.
Method Reproducibility
All the experiments discussed above were performed in triplicate for sample preparation, labeling reaction, or LC-FTICR-MS analysis. Good reproducibility was obtained, with coefficients of variation (CVs) of (a) less than 6% for the sample preparation and labeling reaction by judging the UV measurement of the labeled metabolites in each individual sample prepared under the same condition, and (b) less than 7% for the LC-MS results in terms of the total number of detected peak pairs.
Overall Workflow
The above results and discussion indicate that 5 μL of saliva sample could be processed using the protocol described to generate a similar number of peak pairs as observed from larger volumes of starting materials (i.e., over 1000 peak pairs or putative metabolites). Further reduction of the volume of the starting material is possible. However, 5 μL saliva was already found to be convenient to work with and, even with multiple measurements, the total sample amount required for metabolome profiling work should be manageable to collect. A summary of the overall workflow is given in Supplemental Note N2, along with the procedure of using LC-UV measurement of the labeled samples to normalize sample concentrations prior to mixing the 12C-labeled individual sample with the 13C-labeled pooled reference sample (see Supplemental Note N3).
Pilot Test of Metabolome Profiling
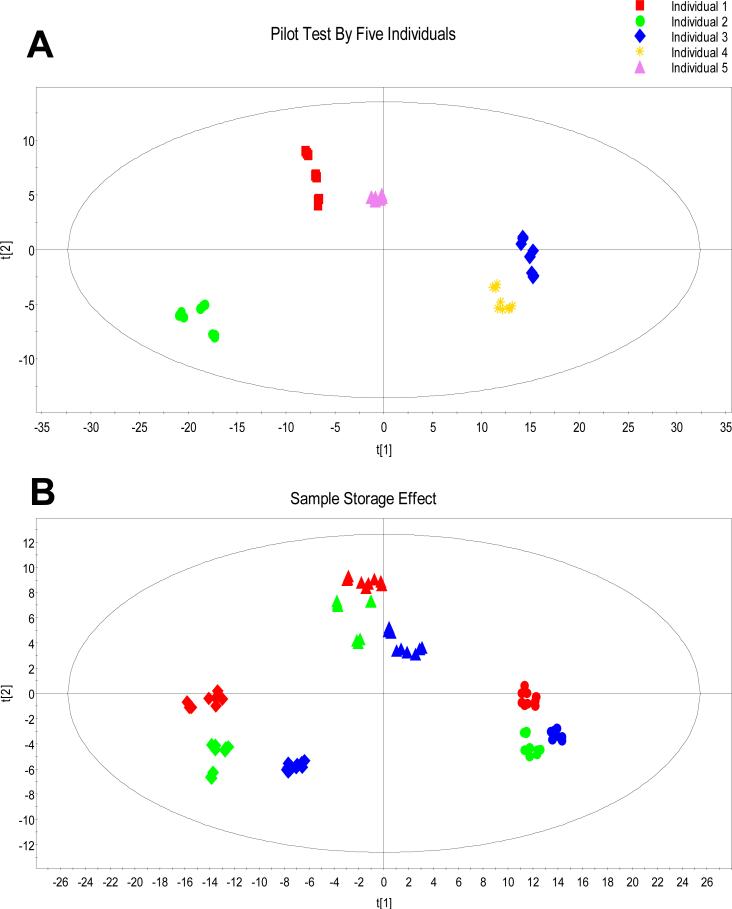
To examine the performance of the developed method for saliva metabolome profiling, 5 different saliva samples obtained from normal individuals were analyzed in a pilot test. A pooled sample was generated by taking an aliquot from each individual sample and labeled with 13C-dansyl chloride. Each individual saliva sample was prepared in triplicate and labeled with 12C-dansyl chloride, and injected three times for LC-FTICR-MS analysis. Quantitative data processing and statistical analysis were applied to the obtained LC-MS datasets. The PCA result (R2X[cum]: 0.840, Q2[cum]: 0.811) is shown in Figure 4A which demonstrates that the data points from 9 injections representing the same individual saliva sample cluster closely, while the 5 different individual saliva samples tested can be distinguished and separated into 5 clusters. This example illustrates that the developed protocol can be used to handle 5 μL of saliva sample and generate reproducible results from individual saliva samples.
Figure 4.
(A) PCA analysis result for the pilot test with 5 individual saliva samples. Each colour represents 9 replicates of the same individual sample. (B) PCA analysis result for the storage condition comparison test with 3 individual saliva samples. Each color indicates one specific storage condition, and each shape represents twenty-seven LC-MS injections of one individual sample: nine replicates of freshly collected analysis (red), nine replicates of analysis after four weeks room temperature storage (green), and nine replicates of analysis after four weeks of -80°C freezer storage (blue).
Sample Storage Effect
As the first application of the developed method for saliva metabolome analysis, we examined the effect of saliva sample storage on the metabolome profile. Specifically, saliva samples used for genetic testing or other applications are often stored at room temperature prior to analysis. Because of a lower cost for storage and shipping, saliva samples stored at room temperature is preferred over that in a -80°C freezer. However, during the storage, properties of metabolites may change. To address this issue, three freshly collected saliva samples from three healthy individuals were individually divided into three fractions. One fraction was analyzed immediately after being collected, and the other two fractions were stored at room temperature and in a -80°C freezer, separately, for 4 weeks.
Figure 4B shows the metabolome comparison among all three individual samples, each with three storage conditions described above. The PCA plot (R2X[cum]: 0.837, Q2[cum]: 0.808) shows clear separation between individual samples (the threshold scores commonly used to define a good separation are 0.500 for both R2X[cum] and Q2[cum]), while the data obtained from all three different storage conditions from the same individual are clustered together. This result indicates that the variations among different individuals are larger than those caused by the sample storage conditions within individuals. Supplemental Tables S1 and S2 show the results of T-test and calculations of Pearson's correlation coefficient (r), respectively. The T-test results in Table S1 show that the p-values obtained from different-individual comparisons are much smaller than those obtained from comparisons of different storage conditions within the same individual. In terms of Pearson's correlation coefficient, the closer the r-value to 1, the more similar the two samples to each other. Calculation results shown in Table S2 demonstrate that the r-values of individual-to-individual range from 0.41 to 0.59, while those of different storage condition comparisons of the same individual are from 0.70 to 0.93. Both tests reach the same conclusion as the visual appearance shown in the PCA plot in Figure 4B that room temperature saliva storage does not change the metabolome profile of an individual to a significant extent that affects the comparison of the metabolomes among different individuals. This finding is very significant, as this work suggests that many saliva samples currently collected for other purposes and stored at room temperature could potentially be used for metabolomics studies. Of course, this work only examined the effect of storage conditions on metabolome changes over a 4-weeks period. The longer term storage effect remains to be determined.
Metabolome Comparison of Healthy and MCI Saliva Samples
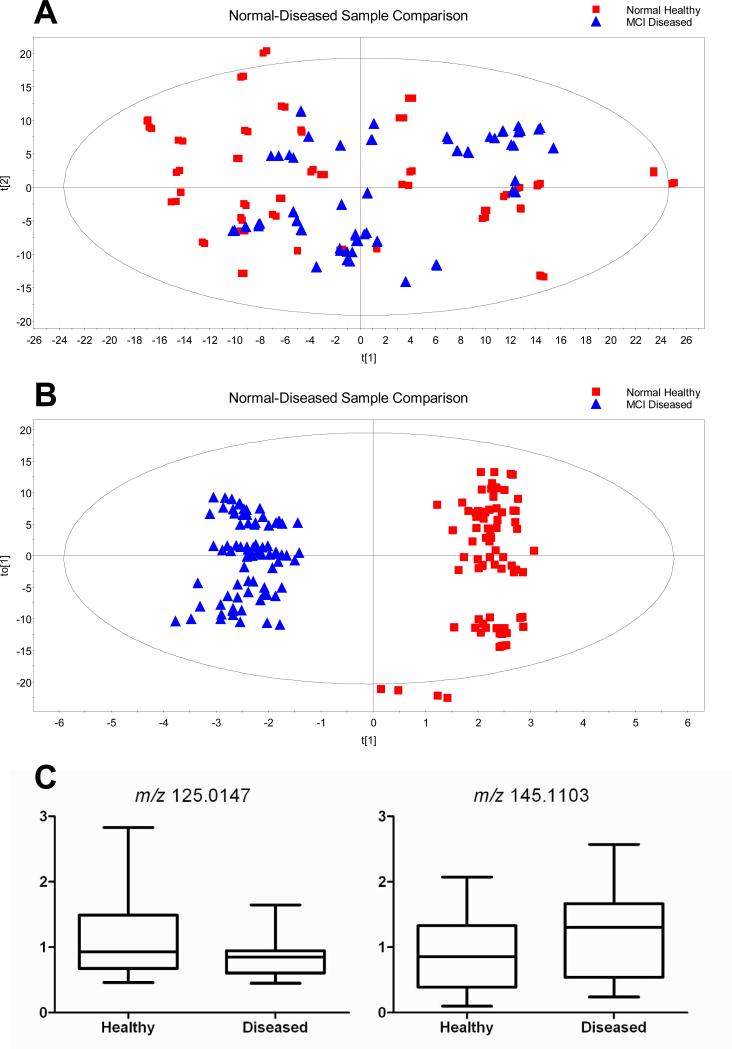
As an example of the potential applications of the developed method for saliva metabolomics, we investigated the metabolome difference between two groups of human subjects, namely 20 older adults classified with mild cognitive impairment (MCI) and 20 age- and gender-matched not-impaired controls. As noted earlier, strict clinical classification procedures were followed, and both groups demonstrated 4-year in-status stability.32, 33 The healthy adults were aged 64-75 with 10 males and 10 females and the diseased ones were aged 65-75 also with 10 males and 10 females. In this metabolomics work, individual saliva samples were processed in duplicate and the resultant samples were separately analyzed by LC-MS replicate runs. Thus, there are four peak-intensity ratios determined for each putative metabolite. Each ratio reflects the relative concentration of the metabolite present in an individual sample to that in the pooled reference sample. Because the same reference was used for all 40 samples, the peak-intensity ratio differences among different samples represent the relative concentration variations of the metabolite in these samples. The PCA plot of all the data obtained from the 40 individuals (each individual has 4 replicate points) is shown in Figure 5A. The separation of the two groups is not clear in the PCA plot. A supervised statistical analysis model, orthogonal partial least squares-discriminant analysis (OPLS-DA), was then applied to the metabolome data and the resulting scores plot is shown in Figure 5B. This plot displays two clusters separating clearly from each other. Four outliers of data points originated from the same saliva sample were observed and, upon further inspection of the data, it was found that these four outliers missed a large portion of peak intensity ratios. The reasons of missing ratios are unknown, but likely related to the nature of the single saliva sample.
Figure 5.
(A) PCA plot of all the data obtained from the 40 individuals. (B) Scores plot of the orthogonal partial least squares-discriminant analysis (OPLS-DA) model demonstrating the separation between the MCI diseased group and the normal healthy group. (C) Box plots of two discriminant metabolites in differentiating MCI from normal healthy control: a representative of down-regulated discriminant metabolite, taurine (left panel), and an up-regulated discriminant metabolite with m/z 145.1104 (right panel).
To examine the validity of the OPLS-DA model, some model fit criteria were examined. In the model, R2X and R2Y represent the fraction of the variance of X matrix and Y matrix, respectively, while Q2Y indicates the predictive accuracy of the built model upon a seven-fold cross validation conducted by leaving 1/7th samples out in each round. When the cumulative values of R2X, R2Y and R2Q (R2X[cum], R2Y[cum], and Q2Y[cum]) are close to 1, it implies an excellent model, while the values above 0.5 were considered to be a validated model. The above three values in our OPLS-DA model were found to be 0.851, 0.958, and 0.920, respectively, and thus the model was valid. An S-plot model was then built to select the significant metabolites that were expressed differently in the diseased group compared to the healthy group.
Table 2 lists the top 18 important discriminant metabolites with their VIP scores, a measure of their relative influence calculated by T-test, along with their fold change between the diseased group and the healthy group. Both the VIP scores and the p-values obtained from T-tests showed significant differences for each identified discriminant metabolite between the two groups. For the fold change data, 17 out of the 18 metabolites picked were down-regulated, while one of them was up-regulated. The relative concentration differences of an individual metabolite present in the two groups of samples can be examined by using a box-plot. As examples, the box-plots of one down-regulated (left panel) and one up-regulated (right panel) discriminant metabolites are shown in Figure 5C. As Table 2 shows, the fold change for the individual metabolites is relatively small. It is likely that the use of multiple metabolites, instead of one metabolite, would be needed to provide the sufficiently high discrimination power to distinguish the two groups.
Table 2.
Summary of the discriminant metabolites determined from VIP scores of the OPLS-DA model for variations between MCI diseased group and normal healthy group.
| Accurate mass | VIP | p-value | Fold change* | Putative Metabolite Match |
|---|---|---|---|---|
| 226.1687 | 2.52 | 1.91E-06 | -1.15 | 1,8-Diazacyclotetradecane-2,9-dione |
| 215.1637 | 2.33 | 4.45E-07 | -1.30 | |
| 250.0954 | 2.33 | 2.80E-04 | -1.20 | |
| 231.1225 | 2.26 | 2.22E-04 | -1.15 | Ala-Ala-Ala, Gly-Gly-Val, Val-Asn, Val-Gly-Gly, Gly-Val-Gly, Asn-Val |
| 262.1323 | 2.25 | 2.39E-05 | -1.24 | Phe-Pro, Pro-Phe |
| 156.0901 | 2.2 | 1.70E-03 | -1.10 | |
| 169.0982 | 2.19 | 1.46E-04 | -1.13 | |
| 213.1118 | 2.17 | 3.76E-06 | -1.18 | |
| 161.1052 | 2.08 | 3.24E-04 | -1.12 | |
| 125.0148 | 2.06 | 2.37E-04 | -1.31 | Taurine |
| 214.0962 | 2.05 | 4.98E-05 | -1.18 | |
| 158.1056 | 2.04 | 5.82E-04 | -1.16 | |
| 192.0750 | 1.99 | 4.90E-03 | -1.10 | |
| 234.1377 | 1.96 | 8.04E-04 | -1.20 | |
| 158.0691 | 1.94 | 1.15E-03 | -1.13 | Ser-Ser |
| 287.1961 | 1.93 | 1.26E-03 | -1.20 | Arg-Leu, Ile-Arg, Leu-Arg, Arg-Ile |
| 145.1104 | 1.87 | 2.27E-02 | 1.26 | 2-amino-heptanoic acid, L-Alanine-n-butylester, N-methyl-isoleucine |
| 152.0950 | 1.86 | 1.70E-03 | -1.18 | 4-(Hydroxylamino)-N,N-dimethylaniline |
A positive fold change indicates an up-regulation that has higher metabolite concentration in the MCI group, and a negative fold change represents a down-regulation that has higher metabolite concentration in the healthy group.
Metabolite Identification
Among the eighteen selected discriminant metabolites that contributed most to the separation of the MCI diseased group and the normal healthy group based on their VIP values, we were able to match 7 metabolites (see Table 2), based on their accurate masses, to the Human Metabolome Database (HMDB).36 Moreover, one metabolite, taurine, was definitively identified by matching the retention time and accurate mass with its authentic standard under the same experimental condition. Taurine is essential for cardiovascular function, skeletal muscle function, the retina and the central nervous system.42 It is involved in several physiological actions in the brain, such as osmoregulation,43 neurotransmission,44 and membrane stabilization.45 It has also been shown to have the function of protection against glutamate excitotoxicity46 and prevention of epileptic seizures.47 It has been reported that over-activation of glutamatergic transmission could mediate in several neurological diseases, relating the glutamate excitotoxicity to a series of chronic and acute neuronal diseases, e.g., Parkinson's, Huntington's and Alzheimer's diseases.48, 49 Given that taurine may prevent the neurotoxicity of β-amyloid and the excitotoxicity of glutamate, it may be neuroprotective for some neurodegenerative diseases.50 Notably, MCI is viewed as a probable precursor and high-risk condition of Alzheimer's disease;29 thus, its differential presence in normal vs. MCI groups is theoretically and clinically promising.
More identification work will be performed in the future upon the availability of standards. The use of preparative separation techniques for isolating and enriching the putative metabolites followed by tandem MS and/or NMR analysis will be needed for unknown metabolite identification. In addition, the number of subjects in both groups should be increased to verify and validate potential biomarkers found in this metabolomics work. Nevertheless, this example illustrates that the high performance isotope labeling LC-MS method can be used for salivary metabolome profiling of two different clinical groups of individuals for discovery of potential biomarkers of neurodegenerative disease.
Conclusions
An isotope labeling LC-MS method has been developed for human salivary metabolome analysis. In this method, 5 μL of saliva sample could be processed with acetone protein precipitation, dansyl chloride labeling, and then UV measurement of the total concentration of the labeled metabolites. Although the absolute concentration of the total metabolites in a saliva sample could not be determined due to the lack of a proper standard for calibration, relative quantification could be performed using a dilution curve of a labeled saliva sample, such as a labeled pooled sample. This way of relative quantification provided a means of normalizing the individual sample concentration by taking varying volumes of samples for labeling and mixing to ensure that the same amount of sample from each individual was used for metabolome comparison. In addition, the UV measurement values could be used to optimize the sample injection amount for LC-FTICR-MS analysis to maximize the number of metabolites detected. In a differential isotope labeling LC-MS approach, in which the concentrations of individual metabolites present in 12C-labeled individual samples were compared to those in a 13C-labeled pooled sample, very good reproducibility of both sample processing and LC-MS measurement could be obtained with CVs of less than 7% in terms of total concentration of metabolites and the number of peak pairs detected. In a mixture of 12C-labeled individual sample and 13C-labeled control, the number of peak pairs detected ranged from 1052 to 1067, with an average of 1058. This number is much higher than <100 metabolites detectable by the reported LC-MS3, 22-25 or CE-MS methods.26 Using this method, the effect of saliva sample storage on metabolome profile changes was investigated and it was found that room temperature sample storage did not cause a significant alteration to the metabolome profile, compared to the use of a -80°C freezer for sample storage. Finally, this method was applied for metabolome comparison of two different groups of individuals: normal healthy older adults vs. older adults with MCI disease. Using OPLS-DA, separation between the two groups was clearly observed, leading to the discovery of several discriminant metabolites that contributed most to the separation. Of particular interest, taurine was positively identified as one of the metabolites with lower concentrations in individuals with MCI, compared to the normal old adults.
Because of the ease and simplicity of obtaining saliva samples in a non-invasive manner and possibility of sample storage at room temperature, we envisage a wide use of this important biofluid for metabolomics studies, particularly in the field of disease biomarker discovery. The salivary metabolome profiling method described in this work opens the possibility of performing relative quantification of a large number of putative metabolites (~1000) using a small volume of starting materials. While this work focused on the use of dansylation chemistry to analyze the amine. and phenol-containing metabolites in saliva, other labeling chemistries targeted at carboxylic acids,41 adehydes and ketones have been recently developed and application of these labeling chemistries to saliva samples should significantly increase the metabolome coverage.
Supplementary Material
Acknowledgements
This work was funded by grants from Genome Canada (to LL), the Natural Sciences and Engineering Research Council of Canada (NSERC) (to LL), the Collaborative Health Research Projects of NSERC and Canadian Institute of Health Research (CIHR) (to LL and RAD), the Canada Research Chairs program (to RAD and LL), and the National Institutes of Health (R37 AG008235, to RAD). We acknowledge the contribution of Dr. Stuart MacDonald to clinical biosample selection.
Footnotes
Supporting Information Available
Supplemental Tables S1-S2 and Notes N1-N3 indicated in the text are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Spielmann N, Wong DT. Oral Dis. 2011;17:345–354. doi: 10.1111/j.1601-0825.2010.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima DP, Diniz DG, Moimaz SAS, Sumida DH, Okamoto AC. Int. J. Infect. Dis. 2009;14:E184–E188. doi: 10.1016/j.ijid.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 3.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu C, Rodriguez T, Thiravirojana-Thetkathuek A, Pearson M. Toxicological and Environmental Chemistry. 2008;90:315–325. [Google Scholar]
- 5.Castagnola M, Picciotti PM, Messana I, Fanali C, Fiorita A, Cabras T, Calo L, Pisano E, Passali GC, Iavarone F, Paludetti G, Scarano E. Acta Otorhinolaryngol. Ital. 2011;31:347–357. [PMC free article] [PubMed] [Google Scholar]
- 6.Hart RW, Mauk MG, Liu C, Qiu X, Thompson JA, Chen D, Malamud D, Abrams WR, Bau HH. Oral Dis. 2011;17:745–752. doi: 10.1111/j.1601-0825.2011.01808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Clin. Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- 8.Lee YH, Zhou H, Reiss JK, Yan XM, Zhang L, Chia D, Wong DTW. Clin. Chem. 2011;57:1295–1302. doi: 10.1373/clinchem.2010.159210. [DOI] [PubMed] [Google Scholar]
- 9.Hubel A, Aksan A, Skubitz APN, Wendt C, Zhong X. Biopreserv. Biobank. 2011;9:237–244. doi: 10.1089/bio.2010.0034. [DOI] [PubMed] [Google Scholar]
- 10.de Jong EP, van Riper SK, Koopmeiners JS, Carlis JV, Griffin TJ. Clin. Chim. Acta. 2011;412:2284–2288. doi: 10.1016/j.cca.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu S, Loo JA, Wong DT. In: Oral-Based Diagnostics. Malamud D, Niedbala RS, editors. Vol. 1098. Blackwell Publishing; Oxford: 2007. pp. 323–329. [Google Scholar]
- 12.Border MB, Schwartz S, Carlson J, Dibble CF, Kohltfarber H, Offenbacher S, Buse JB, Bencharit S. Mol. Biosyst. 2012;8:1304–1310. doi: 10.1039/c2mb05079j. [DOI] [PubMed] [Google Scholar]
- 13.Castagnola M, Cabras T, Iavarone F, Fanali C, Nemolato S, Peluso G, Bosello SL, Faa G, Ferraccioli G, Messana I. Expert Rev. Proteomics. 2012;9:33–46. doi: 10.1586/epr.11.77. [DOI] [PubMed] [Google Scholar]
- 14.Hu S, Loo JA, Wong DT. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda I, Stretch C, Barnaby P, Bhatnager K, Rankin K, Fu H, Weljie A, Jha N, Slupsky C. NMR Biomed. 2009;22:577–584. doi: 10.1002/nbm.1369. [DOI] [PubMed] [Google Scholar]
- 16.Bertram HC, Eggers N, Eller N. Anal. Chem. 2009;81:9188–9193. doi: 10.1021/ac9020598. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd GR, Wongravee K, Silwood CJL, Grootveld M, Brereton RG. Chemometrics Intell. Lab. Syst. 2009;98:149–161. [Google Scholar]
- 18.Peeters K, Lamers R, Penninks AH, Knol EF, Bruijnzeel-Koomen C, van Nesselrooij JHJ, Knulst AC. Int. Arch. Allergy Immunol. 2011;155:23–30. doi: 10.1159/000318654. [DOI] [PubMed] [Google Scholar]
- 19.Aimetti M, Cacciatore S, Graziano A, Tenori L. Metabolomics. 2012;8:465–474. [Google Scholar]
- 20.Sanchez M. d. N., Garcia EH, Pavon JLP, Cordero BM. Anal. Chem. 2012;84:379–385. doi: 10.1021/ac2026892. [DOI] [PubMed] [Google Scholar]
- 21.Penn DJ, Oberzaucher E, Grammer K, Fischer G, Soini HA, Wiesler D, Novotny MV, Dixon SJ, Xu Y, Brereton RG. J. R. Soc. Interface. 2007;4:331–340. doi: 10.1098/rsif.2006.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pesek JJ, Matyska MT, Loo JA, Fischer SM, Sana TR. J. Sep. Sci. 2009;32:2200–2208. doi: 10.1002/jssc.200900270. [DOI] [PubMed] [Google Scholar]
- 23.Yan SK, Wei BJ, Lin ZY, Yang Y, Zhou ZT, Zhang WD. Oral Oncol. 2008;44:477–483. doi: 10.1016/j.oraloncology.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez-Sanchez B, Priego-Capote F, de Castro MDL. J. Chromatogr. A. 2012;1248:178–181. doi: 10.1016/j.chroma.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 25.Barnes VM, Ciancio SG, Shibly O, Xu T, Devizio W, Trivedi HM, Guo L, Jonsson TJ. J. Dent. Res. 2011;90:1293–1297. doi: 10.1177/0022034511416240. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo K, Li L. Anal. Chem. 2009;81:3919–3932. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 28.Dixon RA, de Frias CM. Aging Neuropsychol. Cogn. 2004;11:346–376. [Google Scholar]
- 29.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC, Snyder PJ, Carrillo MC, Thies B, Phelps CH. Alzheimers. Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Frias CM, Dixon RA, Strauss E. Neuropsychology. 2009;23:778–791. doi: 10.1037/a0016743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon RA, Lentz TL, Garrett DD, MacDonald SWS, Strauss E, Hultsch DF. Neuropsychology. 2007;21:381–399. doi: 10.1037/0894-4105.21.3.381. [DOI] [PubMed] [Google Scholar]
- 32.Dolcos S, MacDonald SWS, Braslavsky A, Camicioli R, Dixon RA. Neuropsychology. 2012;26:209–223. doi: 10.1037/a0026760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon RA, DeCarlo CA, MacDonald SWS, Vergote D, Jhamandas J, Westaway D. 2012 submitted. [Google Scholar]
- 34.Wu Y, Li L. Anal. Chem. 2012 submitted (in revision) [Google Scholar]
- 35.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 36.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia JG, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong YP, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo K, Bamforth F, Li L. J Am Soc Mass Spectrom. 2011;22:339–347. doi: 10.1007/s13361-010-0033-4. [DOI] [PubMed] [Google Scholar]
- 38.Weber G. Biochem. J. 1952;51:155–168. doi: 10.1042/bj0510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartley BS, Massey V. Biochimica Et Biophysica Acta. 1956;21:58–70. doi: 10.1016/0006-3002(56)90093-2. [DOI] [PubMed] [Google Scholar]
- 40.Gros C. Labouess.B Eur. J. Biochem. 1969;7:463–&. doi: 10.1111/j.1432-1033.1969.tb19632.x. [DOI] [PubMed] [Google Scholar]
- 41.Guo K, Li L. Anal. Chem. 2010;82:8789–8793. doi: 10.1021/ac102146g. [DOI] [PubMed] [Google Scholar]
- 42.Huxtable RJ. Physiol. Rev. 1992;72:101–163. doi: 10.1152/physrev.1992.72.1.101. [DOI] [PubMed] [Google Scholar]
- 43.Nagelhus EA, Lehmann A, Ottersen OP. Neuroscience. 1993;54:615–631. doi: 10.1016/0306-4522(93)90233-6. [DOI] [PubMed] [Google Scholar]
- 44.McBride WJ, Frederickson RCA. Federation Proceedings. 1980;39:2701–2705. [PubMed] [Google Scholar]
- 45.Birdsall TC. Altern Med Rev. 1998;3:128–136. [PubMed] [Google Scholar]
- 46.Leon R, Wu H, Jin Y, Wei JN, Buddhala C, Prentice H, Wu JY. J. Neurosci. Res. 2009;87:1185–1194. doi: 10.1002/jnr.21926. [DOI] [PubMed] [Google Scholar]
- 47.El Idrissi A, Messing J, Scalia J, Trenkner E. In: Taurine 5: Beginning the 21st Century. Lombardini JB, Schaffer SW, Azuma J, editors. Vol. 526. Kluwer Academic/Plenum Publ; New York: 2003. pp. 515–525. [Google Scholar]
- 48.Lipton SA, Rosenberg PA. N. Engl. J. Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- 49.Mattson MP. Neuromol. Med. 2003;3:65–94. doi: 10.1385/NMM:3:2:65. [DOI] [PubMed] [Google Scholar]
- 50.Louzada PR, Lima ACP, Mendonca-Silva DL, Noel F, De Mello FG, Ferreira ST. Faseb J. 2004;18:511–518. doi: 10.1096/fj.03-0739com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.