SUMMARY
Many types of human tumor cells have overexpressed pyruvate kinase M2 (PKM2). However, the mechanism underlying this increased PKM2 expression remains to be defined. We demonstrate here that EGFR activation induces PLCγ1-dependent PKCε monoubiquitylation at Lys321 mediated by RINCK1 ubiquitin ligase. Monoubiquitylated PKCε interacts with a ubiquitin-binding domain in NEMO zinc finger and recruits the cytosolic IKK complex to the plasma membrane, where PKCε phosphorylates IKKβ at Ser177 and activates IKKβ. Activated RelA interacts with HIF1α, which is required for RelA to bind the PKM2 promoter. PKCε- and NF-κB-dependent PKM2 upregulation is required for EGFR-promoted glycolysis and tumorigenesis. In addition, PKM2 expression correlates with EGFR and IKKβ activity in human glioblastoma specimens and with grade of glioma malignancy. These findings highlight the distinct regulation of NF-κB by EGF, in contrast to TNFα, and the importance of the metabolic cooperation between the EGFR and NF-κB pathways in PKM2 upregulation and tumorigenesis.
Keywords: EGFR, PKM2, PKCε, NF-κB, RelA, NEMO, IKKβ, HIF1α, monoubiquitylation, phosphorylation, glycolysis, tumorigenesis
INTRODUCTION
Tumor cells have elevated rates of glucose uptake and higher lactate production in the presence of oxygen. This phenomenon, known as aerobic glycolysis, or the Warburg effect, supports tumor cell growth (Vander Heiden et al., 2009). Pyruvate kinase regulates the rate-limiting final step of glycolysis, which catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP, yielding pyruvate and ATP. Four pyruvate kinase isoforms exist in mammals and are derived from two distinct genes, PKLR and PKM (formerly PKM2). The R and L isozymes are expressed in erythrocytes and the liver, respectively, and are encoded by the PKLR gene, arising through the use of different tissue-specific promoters (Mazurek et al., 2005). The M1 and M2 isoforms result from mutually exclusive alternative splicing of the PKM pre-mRNA, reflecting inclusion of either exon 9 (PKM1) or exon 10 (PKM2). The splicing factors polypyrimidine tract binding protein (PTB, also known as PTBP1 or hnRNP I) and hnRNP A1/2 bind repressively to sequences flanking exon 9 to ensure exon 10 inclusion (Clower et al., 2010; David et al., 2010), but the specific molecular mechanisms through which PKM2 is transcriptionally regulated upon extracellular stimulation remain to be defined.
PKM2 is overexpressed in human cancers (Mazurek et al., 2005). Replacement of PKM2 with PKM1 in human lung cancer cells inhibits tumor formation in nude mouse xenografts (Christofk et al., 2008). Under hypoxic conditions, prolyl-hydroxylated PKM2 interacts with HIF1α to induce glycolytic gene expression that enhances glucose metabolism in cancer cells (Luo et al., 2011). We recently reported that PKM2 binds to phosphorylated β-catenin Y333 and is required for epidermal growth factor receptor (EGFR) activation-induced β-catenin transactivation, (Lu, 2012; Yang et al., 2011). In addition, we demonstrated that PKM2 phosphorylates histone H3-T11, leading to H3-K9 acetylation and expression of CCND1 (encoding for cyclin D1) and MYC, cell cycle progression, and tumorigenesis (Yang et al., 2012). These findings point to an essential role for PKM2 expression in aerobic glycolysis and cell proliferation and to the need to further understand the mechanisms regulating PKM2 expression during tumor development.
Activation of nuclear factor kappa enhancer binding protein (NF-κB), induced by pathogens, carcinogens, or inflammation, may play a direct or indirect role in tumor progression (Brown et al., 2008; Karin, 2006). The NF-κB family consists of p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), c-Rel, and RelB, which can associate with one another to form various heterodimeric and homodimeric combinations (Ghosh and Hayden, 2008). Stimulation of cells with tumor necrosis factor (TNF)α, interleukin-1β (IL-1β), or Toll-like receptor (TLR) ligands activates the canonical pathway of NF-κB activation (Skaug et al., 2009), leading to activation of tumor growth factor (TGF)β-activated kinase 1 (TAK1) complex through TNF receptor-associated factor (TRAF) proteins. TAK1 then activates the IκB kinase (IKK) complex, which consists of two catalytic subunits, IKKα and IKKβ, and regulatory subunit NF-κB essential modulator (NEMO, also known as IKKγ). The N-terminal CC1 domain of NEMO interacts with IKK, whereas the regulatory C-terminal half is involved in signal recognition (Ghosh and Hayden, 2008). The latter comprises the CC2-LZ domain, including a coiled-coil (CC2) and a leucine zipper (LZ) motif, and a CCHC-type zinc finger (ZF) domain. The CC2-LZ domain contains a ubiquitin-binding domain (UBD) (termed NOA/UBAN/NUB) that preferentially interacts with Lys-63-polyubiquitin chains (Ea et al., 2006; Wu et al., 2006). The ZF domain of NEMO also contains a UBD, which belongs to the ubiquitin-binding ZF (UBZ) type (Cordier et al., 2009). The NEMO-dependent IKK activation phosphorylates IκBα at serines 32 and 36, leading to polyubiquitylation of lysines 21 and 22 by the SCF-βTrCP E3 ligase complex and subsequent degradation by the 26S proteasome. Cytoplasmic RelA/p50 dimers are subsequently released from binding of IκBα and translocate to the nucleus, where they bind κB sites in the promoters or enhancers of target genes, leading to their transcription (Ghosh and Hayden, 2008).
NF-κB activation can be induced by the EGFR signaling pathway (Brown et al., 2008). Activation or overexpression of EGFR is observed in up to 30% of solid tumors and generally correlates with a poor clinical prognosis (Wykosky et al., 2011; Yang et al., 2011). In contrast to the intensively studied NF-κB activation during inflammatory response, the mechanism underlying EGFR-induced NF-κB activation is largely unknown (Brown et al., 2008). In addition, although the importance of activation of EGFR and NF-κB in tumor development has been separately revealed (Brown et al., 2008; Voldborg et al., 1997), it is still unclear whether NF-κB plays a role in EGFR-related cancer cell metabolism.
EGFR activates many downstream signaling molecules, including protein kinase C (PKC) (Hornia et al., 1999). The PKC family consists of at least ten members, divided into three subgroups: classical PKCs (α, βI, βII, γ), novel PKCs (δ, ε, η, θ), and atypical PKCs (ζ, ι/λ) (Breitkreutz et al., 2007). Novel PKCs require diacylglycerol (DG) or phorbol esters for activation (Breitkreutz et al., 2007). DG, in turn, is generated by phosphoinositide-specific phospholipase C (PLC) isozymes, which catalyze a membrane phospholipid, phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2). There are six PLC isotypes, β, γ, δ, ε, ζ, and η, which are composed of 14 mammalian PLC isozymes. PLCγ1 and PLCγ2 are the only isozymes that contain SH2 domains (Choi et al., 2007). In response to EGF stimulation, the SH2 domains of PLCγ1 bind to the autophosphorylated EGFR Y992, which leads to phosphorylation and activation of PLCγ1 by EGFR (Nogami et al., 2003).
In this report, we show that activation of EGFR in human cancer cells results in increased glucose uptake and lactate production in a PKM2-dependent manner. Intriguingly, EGFR activation leads to NF-κB-dependent upregulation of PKM2 expression; NF-κB activation, in turn, is mediated by PLCγ1 and PKCε monoubiquitylation–dependent IKKβ activation. This EGFR-initiated signaling cascade promotes tumor development.
RESULTS
EGFR Activation Results in Upregulation of PKM2 Expression
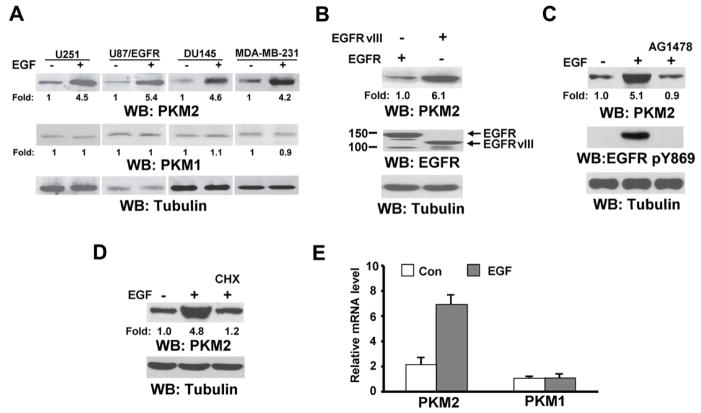
EGFR activation and PKM2 upregulation have been detected separately in many cancer types; however, the connection between these two tumorigenesis-related alterations remains unknown. To examine whether EGFR activation regulates PKM2 expression, we used EGF to stimulate DU145 human prostate cancer cells, MDA-MB-231 human breast carcinoma cells, and U251 and EGFR-overexpressed U87 (U87/EGFR) human glioblastoma (GBM) cells. EGF treatment increased expression of PKM2, but not PKM1 (Fig. 1A). In addition, U87 cells expressing constitutively active EGFRvIII mutant, which lacks 267 amino acids from its extracellular domain and is commonly found in GBM as well as in breast, ovarian, prostate, and lung carcinomas (Kuan et al., 2001), had significantly higher levels of PKM2 expression compared to U87/EGFR cells without EGF treatment (Fig. 1B). EGF-induced PKM2 upregulation was blocked by pretreatment with AG1478, an EGFR inhibitor (Fig. 1C), which indicates that EGFR activation is required for PKM2 upregulation. Pretreatment with cycloheximide, which blocks protein translation, inhibited EGF-induced PKM2 upregulation (Fig. 1D), suggesting that PKM2 expression is not primarily regulated by altering PKM2 stability. Real-time quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis using primers specific for mRNA of PKM2 or PKM1 showed an increase in the mRNA levels of PKM2, but not of PKM1, upon EGF treatment (Fig. 1E). These results suggest that EGFR activation enhances PKM2 protein expression by increasing its mRNA level.
Fig. 1. EGFR Activation Results in Upregulation of PKM2 Expression.
A–D, Immunoblotting analyses were performed with or without the indicated antibodies.
(A) The indicated cell lines were treated with EGF (100 ng/ml) for 12 h.
(B) U87 cells were stably transfected with plasmids expressing EGFR or EGFRvIII.
(C) U87/EGFR cells were pretreated with or without AG1478 (1 μM) for 30 min before EGF (100 ng/ml) treatment for 12 h.
(D) U87/EGFR cells were pretreated with or without cycloheximide (CHX) (200 μg/ml) for 30 min before EGF (100 ng/ml) treatment for 12 h.
(E) mRNA expression levels of PKM2 and PKM1 in U87/EGFR cells treated with or without EGF (100 ng/ml) for 12 h were measured by real-time quantitative RT-PCR analysis. β-actin mRNA from the same cDNA library was amplified as a control. The relative mRNA levels of PKM2 and PKM1 were normalized to the levels of untreated cells and β-actin mRNA. Data represent the means ± SD of three independent experiments.
EGF Increases PKM2 Expression in a PKC- and NF-κB-dependent Manner
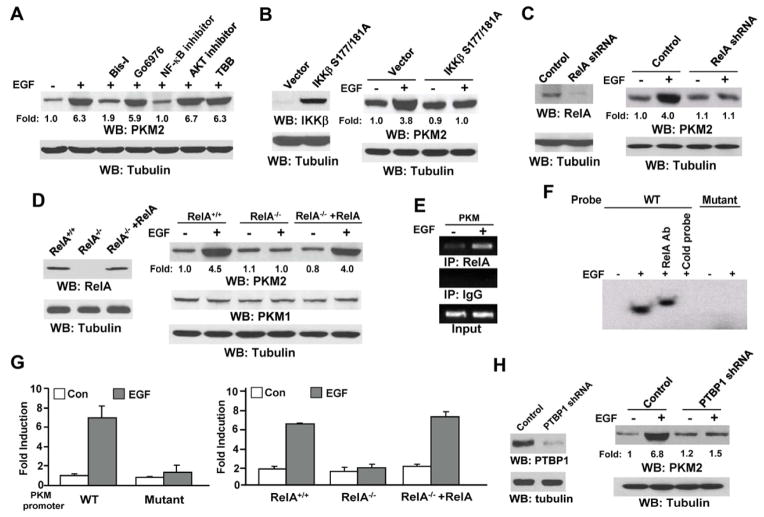
To determine how PKM2 expression is regulated by EGFR activation, we pretreated U87/EGFR cells with the following inhibitors: general PKC inhibitor Bis-I, PKCα/β inhibitor Go6976, NF-κB activation inhibitor II, an AKT inhibitor, and CK2 inhibitor TBB, which successfully blocked 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced and PKC kinase activity-dependent degradation of PKCε (Fig. S1A) and PKCα (Fig. S1B) (Lu et al., 1998), TNFα-induced and NF-κB-dependent IκBα promoter activation (Fig. S1C), EGF-induced AKT phosphorylation (Fig. S1D), and EGF-induced and CK2-dependent α-catenin S641 phosphorylation (Ji et al., 2009) (Fig. S1E), respectively. Inhibition of general PKC and NF-κB, but not of AKT or CK2 (Fig. 2A), largely abrogated EGF-induced PKM2 upregulation. These results implicate a role for PKC and NF-κB activation in the regulation of PKM2 expression. This finding was further validated by the observation that the stable expression of the dominant-negative kinase-dead IKKβ S177/181A mutant blocked EGF-induced PKM2 upregulation, which indicates that IKKβ activation is essential for this upregulation (Fig. 2B). Consistent with these findings, EGF treatment resulted in increased IKKβ activity (Fig. S2A) and enhanced cellular IkBα S32 phosphorylation and degradation of IkBα (Fig. S2B). In addition, RelA depletion by expressing RelA shRNA in U87/EGFR cells (Fig. 2C) or RelA deficiency blocked EGF-enhanced PKM2 expression without affecting PKM1 expression (Fig. 2D), whereas reconstituted expression of RelA in RelA−/− mouse embryonic fibroblasts (MEFs) restored the ability of EGF to induce PKM2 expression (Fig. 2D).
Fig. 2. EGF Increases PKM2 Expression in a PKC- and NF-κB-dependent Manner.
A–D, H, Immunoblotting analyses were performed with the indicated antibodies.
(A) U87/EGFR cells were pretreated with Bis-I (2 μM), Go6976 (2 μM), NF-κB activation inhibitor II (7 μM), an AKT inhibitor (10 μM), or TBB (50 μM), followed by EGF (100 ng/ml) stimulation for 12 h.
(B) 293T cells transiently transfected with pCep4 EGFR and vectors expressing IKKβ S177/181A were treated with or without EGF (100 ng/ml) for 12 h.
(C) U87/EGFR cells stably transfected with pGIPZ expressing a control or a RelA shRNA were treated with or without EGF (100 ng/ml) for 12 h.
(D) RelA+/+, RelA−/−, or RelA−/− fibroblasts with reconstituted RelA expression were treated with or without EGF (100 ng/ml) for 12 h.
(E) U87/EGFR cells treated with or without EGF (100 ng/ml) for 12 h. ChIP assay was performed with an anti-RelA antibody for immunoprecipitation, followed by PCR with PKM promoter–specific primers.
(F) An oligonucleotide containing the putative WT or mutated NF-κB binding sequence was labeled using [γ-32P] ATP and T4 polynucleotide kinase. Nuclear extracts of U87/EGFR cells treated with or without EGF (100 ng/ml) for 12 h were incubated with the 32P-labeled probe in the presence or absence of an anti-RelA antibody or unlabeled oligonucleotide. Samples were subjected to 5% polyacrylamide gel electrophoresis, and the dried gel was exposed to x-ray film.
(G) The luciferase reporter vector pGL3-promoter containing either the WT or a mutated PKM promoter fragment was transfected into U87/EGFR cells (left panel), or RelA+/+, RelA−/−, or RelA−/− fibroblasts with reconstituted RelA expression (right panel), which were treated with or without EGF (100 ng/ml) for 12 h. The relative levels of luciferase activity were normalized to the levels of untreated cells and to the levels of luciferase activity of the Renilla control plasmid. Data represent the mean ± standard deviation of three independent experiments.
(H) U251 cells infected with lentiviruses expressing a control or a PTBP1 shRNA were treated with or without EGF (100 ng/ml) for 12 h.
See also Figs. S1 and S2.
Analysis of the PKM promoter using TFSEARCH software (http://www.cbrc.jp/research/db/TFSEARCH.html) identified a single putative NF-κB binding sequence, -291 GCGACTTTCC -300, which is similar to the NF-κB binding consensus sequence GGGRNNYYCC (N, any base; R, purine; and Y, pyrimidine) (Hayden and Ghosh, 2004). Chromatin immunoprecipitation (ChIP) with an anti-RelA antibody showed that EGFR activation results in the binding of RelA to the PKM promoter (Fig. 2E). To more directly assess an EGF-dependent NF-κB regulation of PKM, we performed electrophoretic mobility shift assays (EMSA) with an oligonucleotide containing the putative wild-type (WT) or mutated NF-κB binding sequence (GCTACTTGTTT, highlighting the mutated nucleotides). The use of lysate derived from EGF-treated cells resulted in a marked increase in the NF-κB binding activity of the WT oligonucleotide, and the inclusion of an anti-RelA antibody resulted in a supershifted NF-κB-binding band (Fig. 2F). The inclusion of excess unlabelled oligonucleotide blocked the NF-κB binding activity, and the mutated oligonucleotide failed to bind to NF-κB. To examine the effect of NF-κB binding on the PKM promoter activity, we transiently expressed a luciferase reporter vector containing the PKM promoter (from -1959 to -11 nucleotide) with either the WT or mutated NF-κB binding sequence into U87/EGFR cells, RelA+/+ MEFs, or RelA−/− MEFs. As demonstrated in Fig. 2G, the activity of the WT, but not mutated, PKM promoter was significantly enhanced in EGF-treated U87/EGFR cells (left panel). Deficiency of RelA blocked EGF-induced PKM promoter activity, which was rescued by reconstituted expression of RelA in RelA−/− MEFs (right panel). Real-time quantitative RT-PCR analysis showed that RelA deficiency inhibited an EGF-induced increase in mRNA levels of PKM2, but not of PKM1 (Fig. S2C). These results support a mechanism whereby EGFR activation results in NF-κB binding to GCGACTTTCC in the PKM promoter and activation of transcription.
EGF treatment increased the mRNA levels of PKM2 but not of PKM1 (Fig. 1E), suggesting that predominantly isoform-specific splicing of PKM pre-mRNA may occur co-transcriptionally. PTBP1, which is associated with gliomagenesis (Cheung et al., 2006), binds repressively to PKM sequences flanking exon 9, resulting in exon 10 inclusion (Clower et al., 2010; David et al., 2010). EGF treatment significantly increased PTBP1 expression (Fig. S2D, left panel), and RNAi-mediated PTBP1 depletion (Fig. 2H, left panel) blocked EGF-enhanced mRNA (Fig. S2D, middle panel) and protein expression of PKM2 (Fig. 2H, right panel), which was accompanied by upregulated PKM1 mRNA levels (Fig. S2D, right panel). PTBP1 protein expression was not affected by treatment with NF-κB inhibitor (Fig. S2D), indicating that NF-κB is not involved in the regulation of PTBP1 expression in response to EGF stimulation. These results suggest that EGF-induced upregulation of PKM2, but not PKM1, is regulated by both EGF-induced NF-kB activation and upregulated PTBP1 expression, which subsequently increase PKM transcription and generation of PKM2 mRNA by splicing, respectively.
PKCε Downstream from PLCγ1, Rather Than TAK1, Activates IKKβ and Subsequently Increases PKM2 Expression
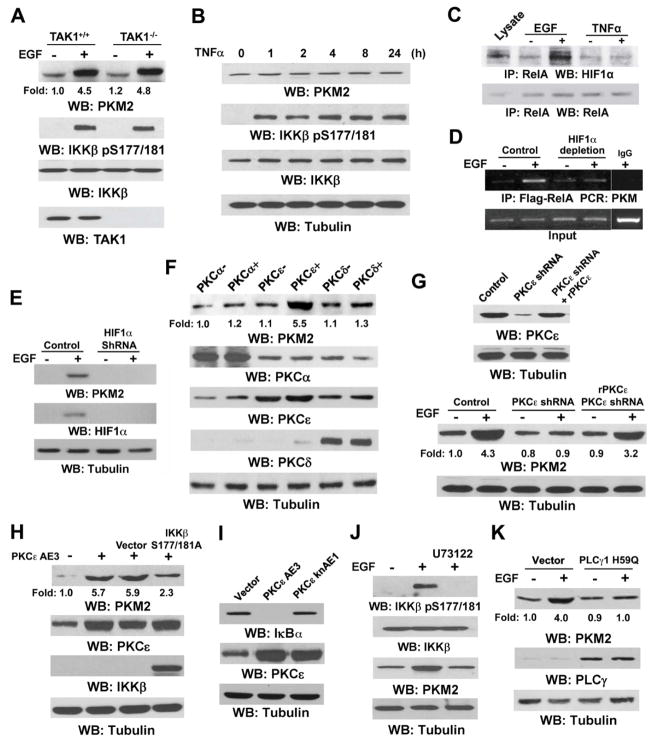
TAK1, which phosphorylates and activates IKKβ, is essential for canonical activation of RelA/p50 in response to inflammatory stimuli (Skaug et al., 2009). Nevertheless, the deficiency of TAK1 did not affect EGF-induced PKM2 expression or IKKβ activation, as reflected by its phosphorylation levels (Fig. 3A). These results indicate that EGF activates IKKβ/RelA via a mechanism that differs from the inflammatory stimuli-induced activation of IKKβ. In line with these findings, although both EGF and TNFα induced IKKβ activation, TNFα had no effect on PKM2 expression (Fig. 3B), whereas EGF induced significant PKM2 upregulation. In addition, a luciferase reporter assay showed that the promoter activity of IκBα, which is TNFα-induced and NF-κB-regulated, was enhanced by treatment with TNFα, but not EGF (Fig. S2E). These results suggest that EGFR activates IKKβ, thereby enhancing PKM2 expression, through a distinct signaling transmission, and NF-κB activation induced by EGF and TNFα regulates the expressions of different sets of downstream genes.
Fig. 3. PKCε Downstream from PLCγ1, Rather Than TAK1, Activates IKKβ and Subsequently Increases PKM2 Expression.
Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies.
(A) The indicated fibroblasts were treated with or without EGF (100 ng/ml) for 12 h.
(B) U87/EGFR cells were treated with TNFα (10 ng/ml) for the indicated time.
(C) U87/EGFR cells were treated with or without EGF (100 ng/ml) or TNFα (10 ng/ml) for 4 h.
(D) U87/EGFR cells were treated with or without EGF (100 ng/ml) for 12 h. HIF1α was immunodepleted from the cell lysates by incubation with an anti-HIF1α antibody, which was followed by a ChIP assay with an anti-Flag antibody for immunoprecipitation of Flag-RelA and PCR analysis with PKM promoter–specific primers.
(E) U87/EGFR cells transiently transfected with a control or a HIF1α siRNA were treated with or without EGF (100 ng/ml) for 12 h.
(F) 293T cells were transiently transfected with constitutively active (+) or kinase-dead (−) PKC mutants.
(G) U87/EGFR cells stably transfected with pGIPZ expressing a control or a PKCε shRNA were reconstituted with or without expression of rPKCε and were treated with or without EGF (100 ng/ml) for 12 h.
(H and I) 293T cells were transiently transfected with the indicated plasmids.
(J and K) U87/EGFR cells pretreated with or without U73122 (2 μM) for 30 min (J) or stably expressed PLCγ1 H59Q (K) were treated with or without EGF (100 ng/ml) for 12 h.
See also Figs. S2E, S3, S4, and S5.
NF-κB activation in response to different extracellular stimuli likely enables NF-κB to be in complex with different transcriptional coregulators and to induce different sets of gene expression (Ghosh and Hayden, 2008; Hoffmann et al., 2006). Given that HIF1α is implicated as a transcriptional factor that regulates PKM2 transcription (Luo et al., 2011; Sun et al., 2011), we have tested whether HIF1α is a coactivator with RelA in the regulation of PKM2. Fig. 3C shows that EGF, but not TNFα, induced an interaction between endogenous HIF1α and endogenous RelA. This interaction is required for HIF1α and RelA to bind to the PKM promoter, as demonstrated by ChIP and real-time quantitative PCR analyse, which showed that HIF1α increased its binding to the PKM promoter, which was blocked by RelA deficiency (Fig. S3A). Immunodepletion of HIF1α blocked EGF-induced binding of RelA to the PKM promoter (Figs. 3D and S3B) and histone H3 acetylation at this promoter (Fig. S3C). In addition, depletion of HIF1α inhibited EGF-induced PKM2 expression (Fig. 3E). These results indicate that HIF1α is a distinct coactivator for RelA in response to EGF, but not TNFα, to induce PKM2 expression.
EGF-induced PKM2 expression in normoxic conditions could be further enhanced by creating a hypoxic condition that increased HIF1α expression (Fig. S3D). Intriguingly, upregulation of HIF1α expression was also induced upon EGF stimulation, which could be largely blocked by NF-κB inhibition (Fig. S3E). These results are in line with a previous report of IKKβ-dependent HIF1α transcription (Rius et al., 2008). Immunodepletion of HIF1α does not affect the binding of RelA to the HIF1α promoter upon EGF treatment (Fig. S3F), suggesting that RelA regulates HIF1α expression in an HIF1α-independent manner and that HIF1α is involved in some, but not all, NF-kB-dependent gene expression.
The general PKC inhibitor Bis-I, but not the PKCα/β inhibitor Go6976, blocked EGF-induced PKM2 upregulation (Fig. 2A), suggesting that a non-α/β isoform of PKC is involved in PKM2 regulation. Transient expression of constitutively active or kinase-dead mutants of PKCα, PKCε, PKCδ (Fig. 3F), or PKCζ (Fig. S4A) in U87/EGFR cells showed that only expression of a constitutively active mutant of PKCε (PKCε AE3) enhanced PKM2 expression. In addition, expression of the dominant-negative kinase-dead PKCε knAE1 (Fig. S4B) or shRNA depletion of PKCε (Fig. 3G) blocked EGF-induced PKM2 upregulation in U87/EGFR or U251 (data not shown) cells, and the effect of PKCε depletion was rescued by the expression of RNAi-resistant (r) PKCε (rPKCε). Furthermore, expression of a dominant-negative kinase-dead mutant of IKKβ (IKKβ S177/181A) largely blocked active PKCε-induced PKM2 expression (Fig. 3H). That PKCε activates IKKβ was further evidenced by enhanced IκBα degradation by an active, but not inactive, mutant of PKCε (Fig. 3I). These results indicate that PKCε, which is upstream from IKKβ activation, is responsible for EGF-induced PKM2 upregulation. In addition, inhibition of PLCγ with the PLCγ inhibitor U73122 or through stable expression of a dominant-negative PLCγ1 H59Q mutant in U87/EGFR cells abrogated EGF-induced IKKβ activation and PKM2 expression (Figs. 3J and 3K). These results indicate that PLCγ1, which is downstream from EGFR and upstream from PKC activation, plays a key role in PKM2 upregulation.
To test whether other growth factors regulate PKM2 expression, we treated U87/EGFR cells with the platelet-derived growth factor (PDGF). Fig. S5 shows that PDGF induced PKM2 expression, which was blocked by inhibition of PLCγ, PKC, and NF-κB and depletion of RelA. These results indicate that both EGF and PDGF induce PKM2 expression in a PLCγ/PKC/NF-κB signal pathway-dependent manner.
PKCε Phosphorylates IKKβ at Ser177 and Activates IKKβ
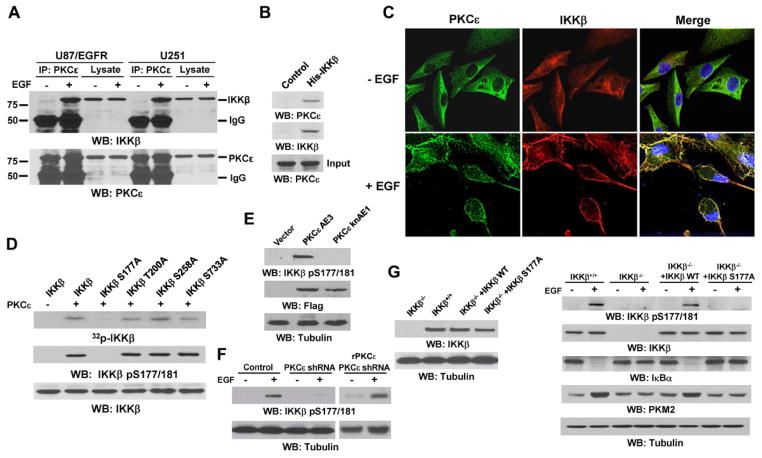
To determine whether PKCε directly activates IKKβ, we immunoblotted the immunoprecipitated PKCε, PKCα, or PKCζ from U87/EGFR or U251 cells with an IKKβ antibody. This experiment showed that EGF induces an increased binding of endogenous IKKβ to PKCε (Fig. 4A), but not to PKCα or PKCζ (Fig. S6A). Furthermore, bacterially purified His-IKKβ pulled down purified active GST-PKCε, indicating that these two protein kinases directly bind each other (Fig. 4B). Immunofluorescent studies showed that both PKCε and IKKβ translocated from the cytosol to the membrane and co-localized with each other upon EGF treatment (Fig. 4C), and co-immunoprecipitation analyses of membrane fractions showed that the proteins interact with each other on the membrane (data not shown).
Fig. 4. PKCε Phosphorylates IKKβ at Ser177 and Activates IKKβ.
(A, B, D–G) Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies.
(A) The indicated cells were treated with or without EGF (100 ng/ml) for 30 min.
(B) Bacterially purified His-IKKβ on nickel agarose beads was mixed with purified active GST-PKCε. A His-protein pull-down assay with nickel agarose beads incubated with GST-PKCε as a control was performed.
(C) U87/EGFR cells were treated with EGF (100 ng/ml) for 30 min and immunostained with the indicated antibodies. Nuclei were stained with Hoechst 33342 (blue).
(D) In vitro kinase assays were performed with purified active PKCε and bacterially purified WT His-IKKβ or different His-IKKβ mutants.
(E) 293T cells were transiently transfected with Flag-PKCε AE3 or Flag-PKCε knAE1.
(F) U87/EGFR cells stably transfected with pGIPZ expressing a control or a PKCε shRNA were reconstituted with or without expression of rPKCε and treated with or without EGF (100 ng/ml) for 30 min.
(G) IKKβ +/+ and IKKβ−/− fibroblasts reconstitutively expressing WT or S177A mutant of IKKβ were treated with or without EGF (100 ng/ml) for 30 min (top three panels) or 12 h (bottom two panels).
See also Fig. S6.
To examine whether PKCε phosphorylates IKKβ, we conducted an in vitro kinase assay, which showed that purified active PKCε phosphorylates bacterially expressed His-IKKβ (Fig. 4D). Analysis of IKKβ amino acid sequences by the motif-based profile-scanning ScanSite program revealed that IKKβ has several potential PKC phosphorylation motifs, at S177, T200, S258, and S733. Mutation of S177, but not T200, S258, or S733, into Ala largely reduced IKKβ phosphorylation by PKCε in vitro (Fig. 4D, upper panel). Immunoblotting analysis with an anti-phospho-IKKβ S177/181 antibody detected PKCε-phosphorylated WT IKKβ, but not IKKβ S177A mutant, indicating that S177, but not S181, is phosphorylated by PKCε (Fig. 4D, middle panel). In addition, expression of constitutively active PKCε AE3, but not the kinase-dead PKCε knAE1 mutant, resulted in phosphorylation of IKKβ at S177 (Fig. 4E). Furthermore, PKCε depletion (Fig. 3G) blocked EGF-induced IKKβ phosphorylation at S177 in U87/EGFR cells, which was rescued by reconstituted expression of rPKCε (Fig. 4F). Given that TAK1 activates IKKβ via phosphorylation of S177 in the activation loop of IKKβ (Wang et al., 2001), phosphorylation of IKKβ at S177 by PKCε might activate IKKβ, thereby inducing PKM2 expression. To test this hypothesis, we reconstituted the expression of WT or S177A mutant IKKβ in IKKβ−/− fibroblasts (Fig. 4G, left panel). IKKβ deficiency abrogated EGF-induced IKKβ phosphorylation at S177, IκBα degradation, and PKM2 upregulation, which were rescued by re-expression of WT IKKβ, but not IKKβ S177A mutant (Fig. 4G, right panel). These results indicate that PKCε phosphorylates IKKβ at S177 and activates IKKβ, which in turn induces PKM2 upregulation.
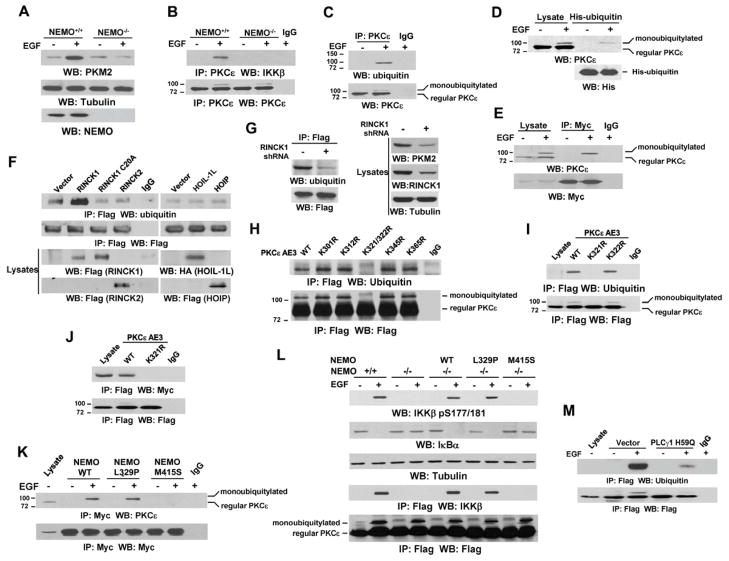
Binding of NEMO Zinc Finger to Monoubiquitylated PKCε at Lys321 Regulates the Interaction between PKCε and IKKβ
NEMO, functioning as an adaptor protein via binding of ubiquitylated proteins, is essential for TNFα-induced IKKβ phosphorylation and activation mediated by TAK1 (Skaug et al., 2009). NEMO deficiency completely blocked EGF-induced PKM2 expression (Fig. 5A), indicating its indispensable role in EGF-regulated PKM2 upregulation. Although IKKβ binds to PKCε directly in vitro (Fig. 4B), the membrane translocation of cytosolic IKKβ (Fig. 4C) and interaction with PKCε on the plasma membrane might need NEMO acting as a recruiting protein. To examine this hypothesis, we immunoprecipitated PKCε from both NEMO+/+ and NEMO−/− fibroblasts and immunoblotted it with an IKKβ antibody. As shown in Fig. 5B, NEMO deficiency abolished the EGF-induced association between endogenous PKCε and IKKβ. These results indicate that NEMO is required for PKCε binding to IKKβ in vivo. To determine whether NEMO or PKCε is ubiquitylated, we used an anti-ubiquitin antibody to immunoblot immunoprecipitated NEMO or PKCε from EGF-treated or -untreated U87/EGFR cells. EGF stimulation induced monoubiquitylation of PKCε (Fig. 5C, top panel), whereas no ubiquitylation of NEMO was detected (data not shown). Consistently, immunoblotting analyses showed that EGF treatment induced a slower migrating PKCε that was about 7 kD (a size of monoubiquitin) bigger than WT PKCε (Fig. 5C, bottom panel). Furthermore, His-protein pull-down analysis of 293T cells transiently expressing His-ubiquitin, which was followed by immmunoblotting with a PKCε antibody, showed that EGF treatment induced monoubiquitylated PKCε (Fig. 5D). In addition, the immunoblotting of immunoprecipitated Myc-tagged NEMO with a PKCε antibody showed that EGF treatment resulted in NEMO binding to monoubiquitylated PKCε but not to non-modified PKCε (Fig. 5E).
Fig. 5. Binding of NEMO Zinc Finger to Monoubiquitylated PKCε at Lys321 Regulates the Interaction Between PKCε and IKKβ.
Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies.
(A and B) The indicated cells were treated with or without EGF (100 ng/ml) for 12 h (A) or 30 min (B).
(C) U87/EGFR cells were treated with or without EGF (100 ng/ml) for 30 min.
(D) Nickel agarose beads were mixed with lysates of 293T cells transiently transfected with plasmids expressing EGFR and His-ubiquitin and treated with or without EGF (100 ng/ml) for 30 min.
(E) 293T cells were transiently transfected with plasmids expressing EGFR and Myc-NEMO and treated with or without EGF (100 ng/ml) for 30 min.
(F) A vector expressing Flag-PKCε AE3 was cotransfected with or without vectors expressing WT Flag-RINCK1, Flag-RINCK1 C20A, WT Flag-RINCK2, HA-HOIL-1L, or Flag-HOIP into 293T cells.
(G) U87/EGFR cells expressing Flag-PKCε AE3 were stably transfected with a vector expressing a control shRNA or RINCK1 shRNA.
(H and I) 293T cells were transiently transfected with plasmids expressing Flag-PKCε AE3 or the indicated PKCε AE3 mutants.
(J) 293T cells were transiently transfected with plasmids expressing Myc-NEMO and Flag-PKCε AE3 or the PKCε AE3 K321R mutant.
(K) U87/EGFR cells stably transfected with WT or the indicated NEMO mutants were treated with or without EGF (100 ng/ml) for 30 min.
(L) Flag-PKCε WT was transiently transfected into NEMO+/+ and NEMO−/− fibroblasts reconstitutively expressing WT or the indicated NEMO mutants, and the cells were treated with or without EGF (100 ng/ml) for 30 min.
(M) U87/EGFR cells with or without expression of PLCγ1 H59Q were treated with or without EGF (100 ng/ml) for 30 min.
See also Fig. S6.
RING-finger protein that interacts with C kinase (RINCK)1 and linear ubiquitin assembly complex (LUBAC) composed of HOIL-1L and HOIP are known E3 ubiquitin ligases for PKC (Chen et al., 2007; Nakamura et al., 2006). Immunoblotting anlaysis of immunoprecipitated Flag-PKCε AE3 with an anti-ubiquitin antibody showed that expressing WT RINCK1, but not inactive RINCK1 C20A, RINCK2, HOIL-1L, or HOIP, resulted in enhanced monoubiquitination of PKCε (Fig. 5F). In addition, RINCK1 depletion blocked EGF-induced Flag-PKCε mono-ubiquitination and PKM2 expression (Fig. 5G). These results indicate that RINCK1 mediates PKCε monoubiquitination upon EGFR activation. To identify the ubiquitylated residue, we treated the immunoprecipitated Flag-PKCε from EGF-stimulated U87/EGFR cells with cyanogen bromide, which hydrolyzes peptide bonds at the C-terminus of methionine residues (Schroeder et al., 1969). Immmunoblotting analysis with an anti-ubiquitin antibody suggested that the –278 to –387 fragment contains the ubiquitylated residue (data not shown). Mutation of K321/322, but not K301, K312, K345, or K365, into Arg abolished EGF-induced monoubiquitylation of Flag-tagged PKCε (Fig. 5H). A single mutation at K321 or K322 showed that mutation at K321, but not K322, abrogated monoubiquitylation of PKCε upon EGF stimulation (Fig. 5I). In addition, co-immunoprecipitation analyses showed that a K321R mutant of Flag-PKCε AE3 lost its binding to Myc-tagged NEMO (Fig. 5J), indicating that EGF-induced PKCε monoubiquitylation at K321 provides a binding motif for NEMO.
To determine whether the UBD domains of NEMO bind to monoubiquitylated PKCε, we stably expressed Myc-tagged NEMO WT, L329P mutant abrogated ubiquitin-binding ability of LZ motif (NOA/UBAN/NUB domain) of NEMO (Wu et al., 2006), and M415S mutant interrupted the UBD domain in ZF domain of NEMO (Cordier et al., 2009) in U87/EGFR cells. Immunoblotting of immunoprecipitated Myc-NEMO with a PKCε antibody showed that the mutation at M415, but not at L329, abolished the EGF-induced interaction between NEMO and PKCε (Fig. 5K). Furthermore, NEMO−/− fibroblasts with reconstituted expression of NEMO M415S, but not WT or L329P mutant (Fig. S6B), failed to rescue NEMO-deficiency-induced inhibition of EGF-enhanced IKKβ S177 phosphorylation, IκBα degradation, and binding of IKKβ to Flag-PKCε (Fig. 5L). These results indicate that NEMO zinc finger binding to monoubiquitylated PKCε plays a pivotal role in IKKβ activation by PKCε.
Inhibition of PLCγ1 abrogated EGF-induced IKKβ activation and PKM2 expression (Figs. 3J and 3K). To further investigate whether PLCγ1 regulates PKM2 expression via PKCε, we treated U87/EGFR cells stably expressing PLCγ1 H59Q mutant (Fig. 3K) with EGF, showing that expression of the dominant-negative PLCγ1 mutant significantly blocked EGF-induced PKCε monoubiquitylation (Fig. 5M). These results further support that PLCγ1 downstream from EGFR upregulates PKM2 expression via regulation of PKCε.
EGF Promotes Glycolysis and Tumorigenesis by PKCε- and NF-κB-dependent PKM2 Upregulation
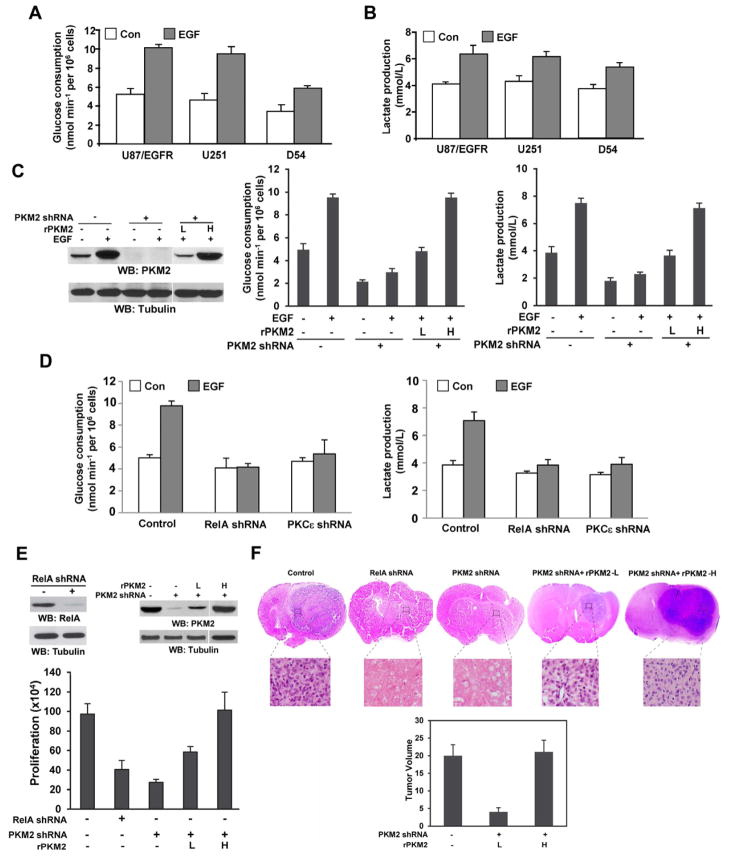
EGF treatment of U87/EGFR, U251, and D54 human GBM cells enhanced glucose consumption (Fig. 6A) and lactate production (Fig. 6B), which was also observed by overexpression of PKM2, but not PKM1, in U87/EGFR cells (Fig. S7A–C). In addition, overexpression of PKM2, but not PKM1, enhanced cyclin D1 expression (Fig. S7A), which is consistent with that PKM2 regulates β-catenin transactivation and expression of downstream CCND1 gene (encoding cyclin D1) (Yang et al., 2011). In contrast to PKM2 overexpression, PKM2 depletion (Fig. 6C, left panel) largely reduced both basal and EGF-enhanced glucose consumption and lactate production (Fig. 6C, middle and right panels). Reconstituted expression of RNAi-resistant PKM2 (rPKM2) restored EGF-promoted glycolysis in an rPKM2 expression level-dependent manner (Fig. 6C), indicating that increased PKM2 expression upon EGF stimulation plays an instrumental role in EGFR-promoted glycolysis. Consistently, depletion of RelA or PKCε, which largely reduced EGF-induced PKM2 upregulation (Figs. 2C and 3G), significantly reduced EGF-enhanced glucose consumption and lactate production (Fig. 6D).
Fig. 6. EGF Promotes Glycolysis and Tumorigenesis by PKCε- and NF-κB-dependent PKM2 Upregulation.
(A and B) The indicated cells in no-serum DMEM were treated with or without EGF (100 ng/ml) for 20 h. The media were collected for analysis of glucose consumption (A) or lactate production (B), which was normalized by cell numbers (per 106). Data represent the means ± SD of three independent experiments.
(C) Immunoblotting analyses of lysates of U87/EGFR cells stably transfected with pGIPZ expressing a control or a PKM2 shRNA with or without reconstituted expression of rPKM2 at different levels were performed with the indicated antibodies (left panel). These cells were treated with or without EGF (100 ng/ml) for 20 h. The media were collected for analysis of glucose consumption (middle panel) or lactate production (right panel), which was normalized by cell numbers (per 106). Data represent the means ± SD of three independent experiments. L, low expression. H, high expression.
(D) U87/EGFR cells with or without PKCε or RelA depletion were treated with or without EGF (100 ng/ml) for 20 h. The media were collected for analysis of glucose consumption (left panel) or lactate production (right panel), which was normalized by cell numbers (per 106). Data represent the means ± SD of three independent experiments.
(E) Immunoblotting analyses of lysates of U87/EGFRvIII cells with or without RNAi-depleted RelA or PKM2 or combined expression of rPKM2 were performed with the indicated antibodies (top panel). A total number of 2 × 104 cells from each cell line were plated and counted 7 days after seeding in DMEM with 2% bovine calf serum (bottom panel). Data represent the means ± SD of three independent experiments. L, low expression. H, high expression.
(F) U87/EGFRvIII cells (5 × 105), with or without RNAi-depleted RelA or PKM2 or combined expression of rPKM2, were intracranially injected into athymic nude mice. After 2 weeks, the mice were sacrificed and tumor growth was examined. H&E-stained coronal brain sections show representative tumor xenografts (top panel). Tumor volumes were calculated (bottom panel). L, low expression. H, high expression. Data represent the means ± SD of seven mice.
See also Fig. S7.
Depletion of RelA or PKM2 from U87/EGFRvIII cells inhibited proliferation of cells, which were in culture for 7 days (Fig. 6E). Reconstituted expression of rPKM2 in U87/EGFRvIII-PKM2 shRNA cells restored EGFRvIII-promoted cell proliferation in an rPKM2 expression level-dependent manner (Fig. 6E). These results indicate that EGFR-increased PKM2 expression is required for EGFR-promoted cell proliferation.
To determine the roles of RelA and PKM2 in brain tumor development, we intracranially injected U87/EGFRvIII cells, with or without depleted RelA or PKM2, or U87/EGFRvIII cells with depleted endogenous PKM2 and reconstituted expression of rPKM2 into athymic nude mice. Dissection of the mice 2 weeks after injection revealed that all of the animals injected with U87/EGFRvIII cells had rapid tumor growth (Fig. 6F). In contrast, no tumors were detected in the mice injected with U87/EGFRvIII cells with depleted RelA or PKM2. Reconstituted expression of rPKM2 in U87/EGFRvIII-PKM2 shRNA cells restored EGFRvIII-promoted tumor growth in an rPKM2 expression level-dependent manner (Fig. 6F). U87/EGFR cells injected into mice for 2 weeks did not lead to tumor formation. However, overexpression of PKM2, but not PKM1, in U87/EGFR cells elicited significant tumor growth (Fig. S7D). These results elucidate the significance of RelA-dependent PKM2 upregulation in EGFR-promoted brain tumor development.
Levels of PKM2 Correlate with Levels of EGFR Activity in Human GBM and with Grades of Glioma Malignancy and Prognosis
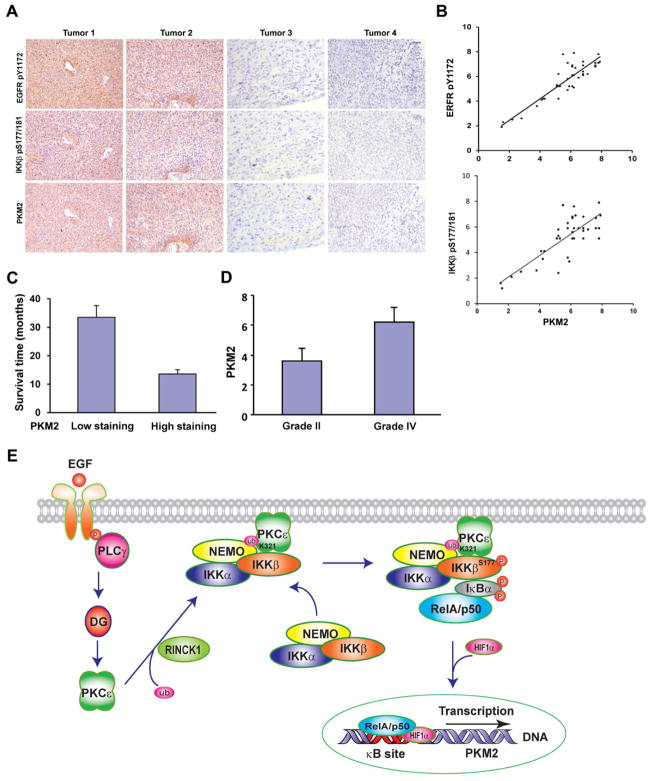
We demonstrated that EGFR activation results in IKKβ-dependent PKM2 upregulation. To further determine whether our findings have clinical relevance, we examined the activity of EGFR and IKKβ and PKM2 expression levels in serial sections of 55 human primary GBM specimens by immunohistochemical (IHC) analyses. As shown in Fig. 7A, the activity levels of EGFR and IKKβ reflected by their phosphorylation levels correlated with the levels of PKM2 expression. Quantification of the staining on a scale of 0–8 showed that the correlation between PKM2 expression levels and EGFR (Fig. 7B, upper panel: r = 0.80, p < 0.001) or IKKβ activity (Fig. 7B, bottom panel: r = 0.88, p < 0.001) was significant in different specimens. The survival durations for the 55 patients, all of whom received standard adjuvant radiotherapy after surgery, followed by treatment with an alkylating agent (temozolomide in the majority of cases), were analyzed with respect to low (2–5 staining) versus high (5.1–8 staining) expression of PKM2. Patients with low PKM2-expressing tumors (20 cases) had a median survival of 34.5 months, compared with 13.6 months for patients with high PKM2-expressing tumors (35 cases) (p < 0.001) (Fig. 7C). These results strongly support a role for EGFR activation in the IKKβ-dependent upregulation of PKM2 in human GBM and reveal a strong relationship between PKM2 expression and patient prognosis.
Figure 7. Levels of PKM2 Correlate with Activity Levels of EGFR and IKKβ in Human GBM and with Grades of Glioma Malignancy and Prognosis.
(A and B) IHC staining with anti–phospho-EGFR Y1172, anti–phospho-IKKβ S177/181, and anti-PKM2 antibodies was performed on 55 GBM specimens. Representative photos of four tumors are shown (A). Semi-quantitative scoring was performed (Pearson product moment correlation test, r = 0.88, p < 0.001, top panel; r = 0.89, p < 0.001, bottom panel). Note that some of the dots on the graphs represent more than one specimen (some scores overlapped) (B).
(C) Overall survival time of 55 patients with GBM by low (20 patients) and high (35 patients) PKM2 expression (P < 0.001). Data represent the mean ± SD of survival time (months) of 20 patients with low PKM2 expression and 35 patients with high PKM2 expression.
(D) Immunohistochemical staining of 27 diffuse astrocytoma specimens with a PKM2 antibody was performed and analyzed by comparing it with the staining of 38 GBM specimens (Student’s t test, two-tailed, P < 0.001). Data represent the mean ± SD of the staining scores for PKM2 from 27 astrocytoma specimens and 38 GBM specimens.
(E) A mechanism for EGFR-induced PKM2 upregulation. EGFR activation results in the binding of the SH2 domain of PLCγ1 to autophosphorylated EGFR and activation of PLCγ1. Diacylglycerol generated by PLCγ1 will activate PKCε, which results in RINCK1-dependent monoubiquitylation of PKCε at K321 and subsequent recruitment of the NEMO/IKKβ complex. PKCε phosphorylates IKKβ at S177 and activates IKKβ, leading to RelA/HIF1α-dependent transcriptional upregulation of PKM2.
To examine whether the level of PKM2 expression correlated with the grade of glioma malignancy, we compared PKM2 expression levels in low-grade diffuse astrocytoma (WHO grade II) and high-grade GBM (WHO grade IV) (Furnari et al., 2007). IHC analyses of 27 human low-grade diffuse astrocytoma specimens showed significantly lower levels of PKM2 in these low-grade gliomas than in the GBM specimens (Fig. 7D). Thus, the level of PKM2 correlated with the grade of glioma malignancy.
DISCUSSION
PKM2 plays an essential role in aerobic glycolysis and tumor growth (Christofk et al., 2008). Nevertheless, how PKM2 expression is regulated during tumor development remains largely unclear. Although the mechanisms and the role of NF-κB activation during inflammatory response have been extensively reported (Skaug et al., 2009), answers to the questions of how NF-κB is regulated in response to growth factor stimulation and whether this regulation contributes to cancer cell metabolism remain elusive (Brown et al., 2008). In this report, we reveal an important mechanism underlying the upregulation of PKM2 and the activation of NF-κB by EGFR activation in tumor cells.
NF-κB activation induced by inflammatory response has been intensively studied. For TNFα signaling, RIP1 is polyubiquitylated via the K63-linked ubiquitin chain, which is mediated by TRAF2/5 as well as the E2-conjugating enzyme Ubc13-Uev1A. Polyubiquitylated RIP1 provides docking sites to recruit proteins containing specific ubiquitin-binding domains, including the NZF domain of TAB2 in complex with TAK1 and the CC2-LZ domain of NEMO. The formation of this complex promotes TAK1-regulated phosphorylation and activation of IKKβ (Skaug et al., 2009). TNFα, through a yet-to-be-identified signaling mechanism, also induces HOIP-HOIL-1L ubiquitin ligase-dependent linear polyubiquitylation of NEMO, which leads to recruitment of TAB2–TAK1 for IKKβ phosphorylation (Rahighi et al., 2009; Tokunaga et al., 2009). Therefore, TAK1-dependent phosphorylation and activation of IKKβ through NEMO-involved protein interactions, which require K63-linked polyubiquitylation of RIP1 or linear polyubiquitylation of NEMO, are crucial regulatory events in TNFα-induced canonical NF-κB activation. In stark contrast, EGF-induced NF-κB activation is TAK1 independent, and IKKβ S177, which is phosphorylated by TAK1 in inflammatory response, is phosphorylated by PKCε instead. In addition, monoubiquitylated PKCε provides a docking site for binding of the NEMO zinc finger. The binding of NEMO to PKCε creates a direct interaction between PKCε and IKKβ, which results in IKKβ phosphorylation by PKCε and subsequent NF-κB activation.
In contrast to the unclearly defined location of TAK1 phosphorylation of IKKβ, EGFR activation results in the plasma membrane translocation of cytosolic PKCε and IKKβ and in the interaction of these proteins on the membrane, strongly suggesting that PKCε phosphorylates and activates IKKβ on the plasma membrane. In addition, EGF, but not TNFα, induced HIF1α expression and an interaction between RelA and HIF1α, which is required for the binding of RelA to PKM promoter and PKM2 expression. Although RelA by itself is sufficient to bind a nucleosome-unassociated oligonucleotide containing the NF-κB binding sequence, it needs HIF1α to act as a co-activator to induce PKM2 transcription, and HIF1α may facilitate and stabilize the transcription factor complexes at PKM2 promoter regions. These results indicate that EGF and TNFα activate NF-κB via distinct mechanisms and subsequently induce different sets of gene expression. NF-κB-dependent PKM transcription acts coordinately with splicing of the pre-mRNA, which is mediated by EGF-induced upregulation of PTBP1, leading to increased expression of PKM2, but not PKM1.
Aberrantly higher activity of EGFR due to gene amplification or mutation of EGFR has been detected in approximately 40% of GBM tumors, which are the most common and biologically aggressive types of brain tumors (Voldborg et al., 1997; Wykosky et al., 2011). We found that the activity levels of EGFR and IKKβ in human GBM cell lines correlate with the levels of PKM2 expression. In addition, the level of PKM2 in human glioma tissue correlates with the level of EGFR activity, grade of glioma malignancy, and patient prognosis, suggesting that PKM2 expression levels could be a biomarker for brain tumor malignancy and prognosis. Depletion of PKM2 or blocking PKM2 upregulation by expression of RelA shRNA largely inhibited EGFR-enhanced aerobic glycolysis in GBM cells and brain tumor growth, indicating that NF-κB activation-dependent PKM2 plays a crucial role in EGFR-promoted GBM cell metabolism and brain tumor growth.
Alterations in cell metabolism caused by PKM2, EGFR overexpression, or NF-κB activation have all been observed in human cancers. We propose a mechanistic model of tumor metabolism that integrates these different components. Our findings demonstrate that activation of EGFR in human cancer cells results in increased glucose uptake and lactate production in a PKM2 expression–dependent manner. Furthermore, EGF-induced PKM2 upregulation is dependent on activation of a PLCγ1-PKCε-IKKβ-RelA signaling cascade (Fig. 7E) in which NEMO zinc finger binds to monoubiquitylated PKCε at K321, mediated by RINCK1, leading to the direct interaction between IKKβ and PKCε and phosphorylation and activation of IKKβ by PKCε. Activated RelA in complex with its co-activator HIF1α is required for PKM2 expression. Increased PKM2 expression enhanced cyclin D1 expression, glycolysis, cell proliferation, and tumorigenesis, highlighting the essential role of PKM2 expression levels in tumor development. Our studies unearthed important mechanisms underlying EGFR-induced NF-κB activation and upregulated PKM2 expression during tumor development. The demonstration of a mechanistic interplay between the EGFR and NF-κB pathways in cancer metabolism provides an important insight for further understanding tumor development and may provide a molecular basis for treating activated EGFR- and upregulated PKM2-related tumors by interfering with this EGFR-induced signaling transmission at multiple levels.
EXPERIMENTAL PROCEDURES
Materials
Rabbit polyantibodies recognizing PKM1, EGFR, phospho-α-catenin S641, phospho-EGFR-Y1172, and RelA were obtained from Signalway Biotechnology (Pearland, TX), and rabbit polyantibodies recognizing PKM2, IKKβ, and phospho-IKKβ-S177/181 were obtained from Cell Signaling Technology (Danvers, MA). Polyclonal antibodies for PKCα, PKCε, PKCδ, PTBP1, and IkBα were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
In Vitro Kinase Assays
The kinase reactions were done by mixing purified active PKCε and bacterially purified WT His-IKKβ or different His-IKKβ mutants in 20 μl kinase assay buffer containing 10 μCi of [gamma-32P] ATP, 25 mM MOPS (pH 7.2), 12.5 mM β-glycerol-phosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT, and 2.5 μl PKC lipid activator (SignalChem, Richmond, BC, Canada) for 20 min at 30°C. Reactions were stopped by adding an equal volume of 2X SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer and boiling for 5 min. Samples were then separated by 6% SDS-PAGE and transferred onto nitrocellulose membranes for exposing to X-ray film. Biotinylated IκBα (Ser32) peptide was used for measuring IKKβ activity (HTScan IKKβ kinase assay kit, Cell Signaling Technology, Danvers, MA).
Luciferase Reporter Gene Assay
The luciferase reporter vector pGL3-promoter containing either the WT or a mutated PKM promoter fragment or a luciferase reporter vector containing the IκBα promoter was transfected into U87/EGFR cells, RelA+/+, or RelA−/− fibroblasts seeded in 24-well plates at 1.5×104 cells/well. 12 h after transfection, the medium was replaced with 0.1% serum for another 12–24 h, and EGF (100 ng/ml) was added 12 h before harvesting. 10 ml out of the 100 ml cell extract were used for measuring luciferase activity. The relative levels of luciferase activity were normalized to the levels of untreated cells and to the levels of luciferase activity of the Renilla control plasmid. Data represent the mean ± standard deviation of three independent experiments.
Supplementary Material
HIGHLIGHTS.
NF-κB is required for EGFR-induced transcriptional upregulation of PKM2.
Monoubiquitylated PKCε recruits IKK complex.
PKCε phosphorylates IKKβ at Ser177 and activates IKKβ.
PKM2 upregulation is required for EGFR-promoted glycolysis and tumorigenesis.
Acknowledgments
We thank Fabrice Agou (Unité de Biochimie Structurale et Cellulaire) for NEMO plasmids, Alexandra Newton (University of California at San Diego) for RINCK plasmids, Kazuhiro Iwai (Osaka University) for HOIP and HOIL-1L plasmids, and Keqiang Ye (Emory University) for PLCγ1 H59Q plasmid. We thank Xuede Lee for his technical assistance and Dawn Chalaire for her critical reading of this manuscript.
This work was supported by National Cancer Institute grants 2R01CA109035 (Z.L.) and CA16672 (Cancer Center Support Grant); a research grant (RP110252; Z.L.) from the Cancer Prevention and Research Institute of Texas (CPRIT), an American Cancer Society Research Scholar Award RSG-09-277-01-CSM (Z.L.), and the James S. McDonnell Foundation 21st Century Science Initiative in Brain Cancer Research Award (22002038; Z.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Breitkreutz D, Braiman-Wiksman L, Daum N, Denning MF, Tennenbaum T. Protein kinase C family: on the crossroads of cell signaling in skin and tumor epithelium. J Cancer Res Clin Oncol. 2007;133:793–808. doi: 10.1007/s00432-007-0280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Cohen J, Arun P, Chen Z, Van Waes C. NF-kappaB in carcinoma therapy and prevention. Expert Opin Ther Targets. 2008;12:1109–1122. doi: 10.1517/14728222.12.9.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Gould C, Garza R, Gao T, Hampton RY, Newton AC. Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J Biol Chem. 2007;282:33776–33787. doi: 10.1074/jbc.M703320200. [DOI] [PubMed] [Google Scholar]
- Cheung HC, Corley LJ, Fuller GN, McCutcheon IE, Cote GJ. Polypyrimidine tract binding protein and Notch1 are independently re-expressed in glioma. Mod Pathol. 2006;19:1034–1041. doi: 10.1038/modpathol.3800635. [DOI] [PubMed] [Google Scholar]
- Choi JH, Ryu SH, Suh PG. On/off-regulation of phospholipase C-gamma 1-mediated signal transduction. Adv Enzyme Regul. 2007;47:104–116. doi: 10.1016/j.advenzreg.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier F, Grubisha O, Traincard F, Veron M, Delepierre M, Agou F. The zinc finger of NEMO is a functional ubiquitin-binding domain. J Biol Chem. 2009;284:2902–2907. doi: 10.1074/jbc.M806655200. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Hornia A, Lu Z, Sukezane T, Zhong M, Joseph T, Frankel P, Foster DA. Antagonistic effects of protein kinase C alpha and delta on both transformation and phospholipase D activity mediated by the epidermal growth factor receptor. Mol Cell Biol. 1999;19:7672–7680. doi: 10.1128/mcb.19.11.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wang J, Nika H, Hawke D, Keezer S, Ge Q, Fang B, Fang X, Fang D, Litchfield DW, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36:547–559. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer. 2001;8:83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- Lu Z. Nonmetabolic functions of pyruvate kinase isoform M2 in controlling cell cycle progression and tumorigenesis. Chin J Cancer. 2012;31:5–7. doi: 10.5732/cjc.011.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Liu D, Hornia A, Devonish W, Pagano M, Foster DA. Activation of protein kinase C triggers its ubiquitination and degradation. Mol Cell Biol. 1998;18:839–845. doi: 10.1128/mcb.18.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Tokunaga F, Sakata S, Iwai K. Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem Biophys Res Commun. 2006;351:340–347. doi: 10.1016/j.bbrc.2006.09.163. [DOI] [PubMed] [Google Scholar]
- Nogami M, Yamazaki M, Watanabe H, Okabayashi Y, Kido Y, Kasuga M, Sasaki T, Maehama T, Kanaho Y. Requirement of autophosphorylated tyrosine 992 of EGF receptor and its docking protein phospholipase C gamma 1 for membrane ruffle formation. FEBS Lett. 2003;536:71–76. doi: 10.1016/s0014-5793(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder WA, Shelton JB, Shelton JR. An examination of conditions for the cleavage of polypeptide chains with cyanogen bromide: application to catalase. Arch Biochem Biophys. 1969;130:551–556. doi: 10.1016/0003-9861(69)90069-1. [DOI] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voldborg BR, Damstrup L, Spang-Thomsen M, Poulsen HS. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann Oncol. 1997;8:1197–1206. doi: 10.1023/a:1008209720526. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Wykosky J, Fenton T, Furnari F, Cavenee WK. Therapeutic targeting of epidermal growth factor receptor in human cancer: successes and limitations. Chin J Cancer. 2011;30:5–12. doi: 10.5732/cjc.010.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.