Abstract
Background:
Pseudomonas aeruginosa is a common cause of nosocomial infections. It exhibits innate resistance to a wide range of antibiotics. This study was performed to determine clonal characteristic of P. aeruginosa isolated from clinical specimens, hospital means, and hospital personnel by PCR- ribotyping patterns.
Methods:
A total of 104 P. aeruginosa were isolated from clinical and environmental samples (59 clinical, 45 hospital means and hospital personnel). P. aeruginosa was identified by standard bacteriological methods, mucoid colony morphotypes, and antibiotic resistance rate. The genomes of isolates were extracted and all considered species were confirmed by 16S rDNA- based PCR assay. Then all isolates were genotyped by the 16S - 23SrDNA and Hinf1 restriction enzyme technique.
Results:
Antibacterial sensitivity pattern of isolates showed clinical and environmental specimens were approximately identical (high antibiotic resistance to Ceftazidime and low antibiotic resistance to Amikacin). Colony morphotypes of specimens revealed that mucoid type of clinical isolates were more than that of environmental isolates. Among clinical and environmental strains P1; (570 bp) was the most prevalence pattern.
Conclusions:
Antibiotic resistance, phenotypic characterization, and PCR- ribotyping pattern showed there is clonal relatedness between clinical and environmental isolates and environment could be a main reservoir for P. aeruginosa infections in hospital.
Keywords: Hospital means, nosocomial infections, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa infrequently found as part of the human microflora in healthy individuals is a gram-negative, nonglucose fermenter rod. P. aeruginosa is widespread in natural environments and it is an opportunistic pathogen for humans lead to a broad spectrum of disease such as urinary, burn, respiratory infections, and septicemia.[1] It is the primary cause of ventilated, associated pneumonia in the intensive care unit.[2] In recent years nosocomial infections caused by P. aeruginosa have been recognized as an acute problem in hospitals due to its intrinsic resistance to many antibiotic classes and its capacity to acquire practical resistance to all effective antibiotics.[3] All these features in P. aeruginosa characterize it as a major microorganism to monitor antibiotic resistance in the clinical specimens. On the other hand, the spread of these bacteria in hospital personnel, wet places, could be a reservoir of resistance genes. Therefore, it is necessary to evaluate the contribution of hospital equipment and personnel in the dissemination route of resistance genes. Presently, genetic techniques supported by phenotypic tests enable us to be inform a detailed characteristic of strains isolated from clinical and environmental wards.[4] In order to determine the genetic relationship between clinical and environmental isolates of P. aeruginosa, there are some techniques such as Restriction endonucleases analysis, Multilocus enzyme electrophoresis, Biotyping, Pulsefield gel electrophoresis, and Ribotyping. Among them, the PCR-ribotyping is an efficient technique used during last 10 years and based on the amplification of spacer regions or intervening sequences between 16S and 23S rDNA genes.[5] There are some previous studies in Iran about the role of P. aeruginosa in nosocomial infection such as Khorvash and co-workers that indicate there are good tools to detect P. aeruginosa nosocomial infection by PCT and CRP.[6] Japoni et al. showed that there are some difficulty in treatment of MDR P. aeruginosa.[7] But these data do not show information about the role of P. aeruginosa isolated from the equipment in nosocomial infections and treatment of MDR isolates and our work is one of the first studies in this case in Iran.
Therefore, the aim of this study was to investigate phenotypic traits of P. aeruginosa isolated from clinical, hospital means, and personnel including colony morphotypes, antibiotic resistance, and genetic relatedness of it by the PCR-ribotyping technique.
METHODS
This study is a cross-sectional, descriptive kind that performed in Alzahra Medical Center, Esfahan, Iran 2009-2010 part of research project number 388093. Clinical specimens were collected from the environment of hospital, personnel, and patients, and samples were sent to the microbiology lab of bacterial and virology group, medical school, Isfahan University of medical science
Isolated bacteria
A total of 59 clinical and 45 hospital environmental isolates of P. aeruginosa were studied. Clinical specimens including urine (25), tracheal (16), blood (3), wound (6), CSF (2), peritoneum (4), stool, joint, and tissue (1) were obtained in Alzzahra hospital, Esfahan, Iran. Environmental specimens including 41 strains gained from 113 samples of the hospital means, showers, tap water, patient's rooms and so on as well as four specimens picked up from 123 throat, hands, coverlet samples of 91 personnel were also obtained in the hospital cited above. Sampling of environmental cases was performed by sterile moisture swab. All swab specimens were carried to microbiology laboratory by using TSB medium.
Isolation of strains
All of specimens were cultured on MacConkey-agar, Blood agar, selective agars such as Cetrimide agar and Pseudomonas agar in order to isolate P. aeruginosa strains. Then plates were incubated at 37°C for 24 h. P. aeruginosa colonies were identified by standard bacteriological methods[8,9,10,11,12] and confirmed by 16S rDNA- based PCR assay.[13]
Antibiotic susceptibility
Antibiotic susceptibility of all P. aeruginosa isolates was determined to following antibiotics; Gentamicin (GM). Ciprofloxacin (CP), Piperacillin (pip), Amikacin (AN), and Ceftazidime (CAZ) (Hi media, Padtan Teb), by using the Kirby Bauer method as recommended by CLSI.[14,15] P. aeruginosa ATCC 27853 served as the control strain. Each antibiogram test was performed 3 times and their mean was considered.
Molecular typing
DNA was extracted by boiling method. After 24 h, single colony was picked up from cultured on Mueller-Hinton agar and suspended in the microtube contained 100 μL of distilled water (Cinnagen Co.); then the tube was boiled for 15 min in boiling water. Equal volume of 24:1 chloroform: isoamyl alcohol (Qualigens) was added and centrifuged at 12 000 rpm for 15 min, the supernatant was used as crude DNA for 16S rDNA- based PCR assay and PCR- ribotyping.[16]
16S- rDNA based PCR assay
16S rDNA sequence has been used as a taxonomic “gold standard” in determining the phylogenies of bacterial species.[13] In order to confirm the isolates as P. aeruginosa, we used a PCR assay that based on 16S rDNA sequence with specific primers as described by Theodore et al.[13] PCR was carried out in 25 μL reaction volumes,(2 mM MgCl2, 250 μM (each) dNTP, 0.4, μM (each) primer, 1 U of Taq polymerase (Cinnagen Co.), and 2 μL of extracted DNA and adjusted to 25 μL by the addition of high-performance liquid chromatography-grade H2O in a Thermocycler (Eppendrof master cycler personal) as initial denaturization for 2 min at 95°C, with 25 cycles of 20s at 94°C, 20s at 58° C, and 40s at 72°C and a final extension of 1 min at 72°C. Amplicon measuring 5 μL was loaded on 1% agarose gel (Cinnagen Co.) prepared in 0.5X TBE and the results were read on the GEL-DOC system.
PCR- ribotyping and digestion with Hinf1 restriction enzyme
PCR-ribotyping of P. aeruginosa isolates and digesting with Hinf1 (fermentas) restriction enzyme was carried out as described by Agarwal et al.[16] PCR-ribotyping amplification reaction was performed in a total volume of 50 μL containing 200 μM dNTPs mix, 2.5 mM MgCl2, 2.5U Taq DNA polymerase, 20 pm of each primer, and 6 μL of the DNA template. Amplification programmed for 25 cycles consisting of 94°C for 1 min, 41°C for 1 min and 72°C for 1 min. A final extension at 72°C for 7 min was applied. Amplicon product was loaded on agarose gel. PCR-ribotyping amplification product measuring 10 μL was restricted with 5U Hinf1 restriction enzyme in 37°C for 24 h and restricted fragments were loaded on 2% agarose gel or on 10% polyacrilamide gel for better resolution.[16]
Statitical analysis
The chi square test was used to compare the antibiograms in different groups. The P value below 0.05 was considered significant. Sampling of the specimen was accidental.
RESULTS
Data from all 103 clinical and environmental specimens were included in the analysis. Table 1 showed the comparison of colony morphotype, antibiotic resistance and PCR-ribotyping pattern between P. aeruginosa isolated from clinical and environmental specimens. By chi-square test there were no any statistical differences between clinical and environmental specimens for any of colony morphotype, antibiotic resistance, and PCR-ribotyping pattern (P-values> 0.05).
Table 1.
Comparison of Colony morphotype, antibiotic resistance, and PCR-ribotyping pattern between P. aeruginosa isolated from clinical and environmental specimens
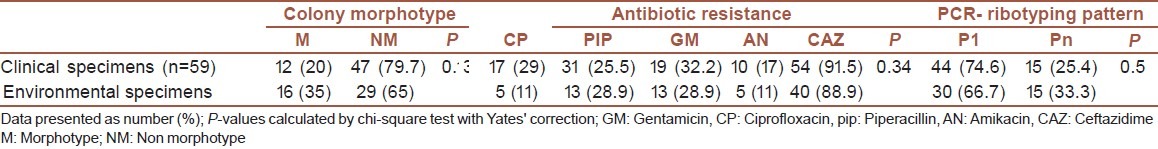
Among clinical isolates of P. aeruginosa colony morphotype: four strains (16%) out of 25 urine isolates, five (31.2%) strain out of 16 tracheal isonelates, o (33.3%) strain out of three blood isolates, two (33.3%) strain out of four wound isolates showed mucoidmorphotype [Table 2]. There was no statistical association between colony morphotype and origin of clinical isolates (P-value = 0.79). Also association between origin of clinical isolates with antibiotic resistance (P-value = 0.99) and PCR- ribotyping pattern (P-value = 0.17) was not statistically significant.
Table 2.
Colony morphotype, antibiotic resistance, PCR-ribotyping pattern of P. aeruginosa isolated from 103 clinical and environmental specimens
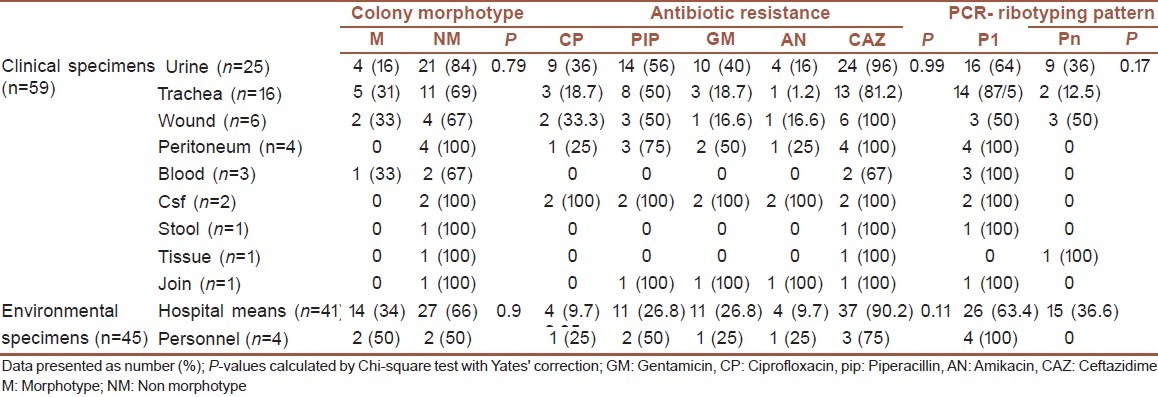
As shown in Table 2 among environmental isolates of P. aeruginosa 14 (34%) strains out of 41 hospital means and two (50%) strains out of four hospital personnel showed mucoidmorphotypes. There no statistical associations between hospital means and hospital personnel samples for colony morphotype, antibiotic resistance, and PCR- ribotyping pattern (P-values > 0.05).
16s rDNA-based PCR assay
All P. aeruginosa isolated from clinical and environmental specimens was identified by standard bacteriological methods, confirmed by 16s rDNA-based PCR assay. All isolates showed band corresponding to special band of P. aeruginosa (956 bp). P. aeruginosa ATCC (27853) was used as positive control.
PCR- ribotyping and digestion with Hinf1 restriction enzyme
Different PCR- ribotyping patterns were detected in strains of clinical and environmental isolates. Among clinical strains, P1 (570 bp on gel electrophoresis) was the most prevalence pattern so that 14 strains (87.5%) of tracheal isolates, 16 (64%) of urinary isolates, 1 (100%) of stool isolate, 3 (100%) of blood isolates, 3 (50%) of wound isolates, 4 (100%) of pleura, 1 (100%) of joint isolates, and 2 (100%) of CSF isolates showed p1 pattern while 9 (36%) of urinary isolates, 2 (12.5%) of tracheal isolates, 3 (50%) of wound isolates, 1 strain of tissue isolate showed other patterns than p1 that are presented in [Table 1] and [Figure 1].
Figure 1.
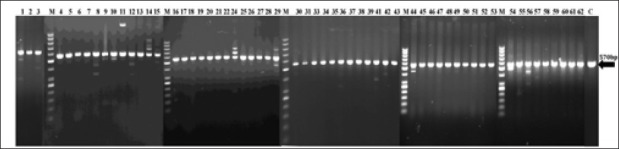
PCR-Ribotyping of P.aeruginosa isolated from Clinical specimens.9 strains of urinary isolates, 2 strains of tracheal isolates,3 strains of wound isolates, 1 strain of tissue isolate showed other pattern than P1 pattern; (Line;1,8,9,10,11,12,14,24,29,36,41,42,44,54,56). The magirity of strains Showed P1 pattern (570bp) C: Control M: Size marker (50bp)
Among environmental isolates p1 was the most prevalence pattern. 63.7% of hospital means and 100% of hospital personnel isolates showed p1 pattern. 36.3% of hospital means also showed other patterns than p1.
Digestion with Hinf1 restriction enzyme
Digestion profile of p1 pattern (570 bp) with Hinf1restriction enzyme yielded bands as (380 bp), (110 bp), (80 bp) that were detected in both clinical and environmental strains [Figures 2–4], and digestion profile of other PCR-ribotyping patterns (not nomenclature) with Hinf1 RE also showed in [Figures 2 and 4], as mentioned before these strains were detected in clinical and hospital means isolates.
Figure 2.
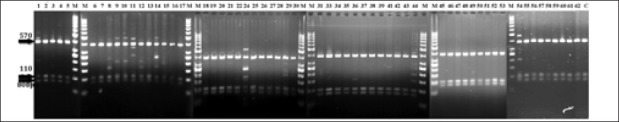
Hinf1 restriction enzyme profile of P. aeruginosa isolated from Clinical specements Clinical strains of P.aeruginosa that detected P1 pattern(570bp),yielded fragments as 80bp,110bp and 380bp in digestion with Hinf1 restriction enzyme. Clinical strains of P.aeruginosa that detected Pn pattern) lines;1,89,10,11,12,14,24,29,36,41,42,44,54,56),yielded other extra fragments in addition to P1 fragments with Hinf1 restriction enzyme; C: Control M: marker Pn; notnomenclature
Figure 4.
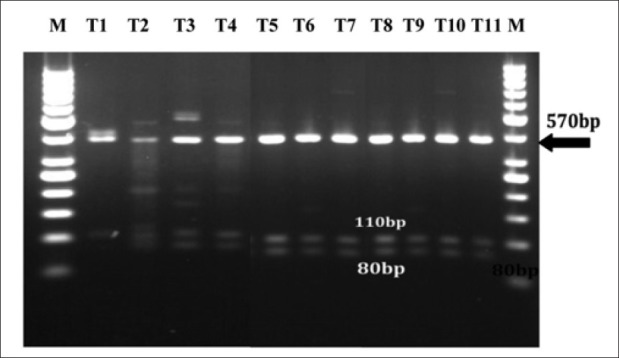
Hinf1 restriction enzyme profile of P. aeruginosa isolated from hospital means strains of P.aeruginosa tht detecte Pn pattern (lin; T1, T3, T7, T10), yield other extra fragments in addition to P1 fragments with Hinf1 restriction enzy C: Control M:marker Pn: notnomenclature
Figure 3.
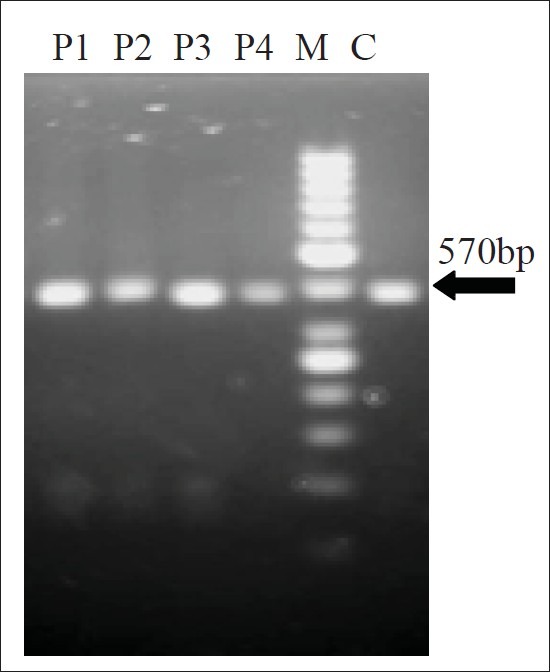
Hinf1 restriction enzyme profile of P. aeruginosa isolated from hospital persounnel All hospital personnel strains of P. aerugunosa that detected P1 pattern(570bp), yielded fragments as 80bp, 110bp and 380bp in digestion with Hinf1 restriction enzyme. C: Control M:Marker
DISCUSSION
P. aeruginosa is a ubiquitous microorganism that it could affect individual with immunocompromised situation and responsible for nosocomial infections.[17] It has not only metabolic versatility and remarkable ability to adaptation and colonization in wide variety of ecologic environments (water, soil, animals), but also notability for its intrinsic ability to resistance to wide variety of antimicrobial agents. Mucoid form is adaptation mechanisms of P. aeruginosa for survive in environments that are concerned to polysaccharide net as called Alginate. Mucoid strains of P. aeruginosa are found in CF patients in a high frequency while they are isolated from urinary tract infections in less frequency (10%) although these strains are rarely found in other patients.[18,19]
In this study, P. aeruginosa strains isolated from clinical specimens including urine (16%), tracheal (31.2%), wound (33.3%), and blood (33.3%) specimens have mucoid morphotype, although, such type is not detected in isolates of peritoneum, CSF, stool, tissue and joint specimens. In the other hand most of the isolates of hospital means (equipments, baths, showers, ward halls, patient's rooms) and personnel also were mucoid morphotypes. Because of low frequency of P. aeruginosa in CSF, tissue, join, and peritoneum specimens it is not reliable to compare of mucoid morphotype cases among different sample groups. It seems that CF patients are important resource for mucoid morphotypes of P. aeruginosa. The important issue is that the mucoid morphotypes are detected in both clinical and hospital environment isolates. There is controversial between researchers about origin of mucoid strains in clinical specimens and some studies such as Corona Nakamura suggest a common clone plays essential role in these cases.[18]
Our data showed the highest antibiotic resistance to Ceftazidime and the lowest resistance to Amikacin in both clinical and hospital environment isolates were significant. Carrol and Leone reported high antibiotic resistance rate to CAZ, PIP, CP, GM, and AN disks in both clinical and environment isolates.[20,21] According to the study performed by Mohammad Taheri in Iran antibiotic resistance rate of P. aeruginosa isolated from clinical and environment specimens to CAZ and AN were (77%) and (49.2%), respectively.[22] Ruiz et al. have been reported that clinical isolates were more resistance to antimicrobial agents than environmental isolates; this may be due to the fact that clinical strains had been submitted to the selective action of antibiotics.[23]
Genotype characterization of P. aeruginosa isolated from clinical and environment specimens represented that pattern1 (P1: 570 bp) is the predominant pattern of PCR- ribotyping in whole strains isolated from both clinical and environment specimens. P1 pattern of PCR-ribotyping in clinical specimens such as blood, peritoneum, joint and CSF was 100%. Using digestion with Hinf1 restriction enzyme, these bands were detected in the whole of these isolates. Their weight of fragment producing bands were (80 bp), (110 bp), and (380 bp). Bacteria isolated from tracheal, wound, urinary, environmental, and personnel had the P1 and non P1 pattern of PCR-ribotyping; therefore, it could be revealed that there is clone relatedness. Agarwal et al. have been reported that P1 of PCR-ribotyping in more than (90%) isolate of CF patients and other clinical specimens including urinary, tracheal, wound, and environment specimens had P1 and non P1 pattern of PCR-ribotyping.[8] More investigations need to survey patterns of PCR-ribotyping and it is essential to appoint the relation of these patterns for disease caused by P. aeruginosa.
Katarzyna et al. in their study on 62 strains of P. aeruginosa isolated from clinical specimens (especially urinary and throat swab specimens) have been reported that nine PCR- ribotyping detected patterns had been possess band as (220-900 bp) and the (560 bp) band has been the most prevalence (58.1%) case.[5] The important issue is that P1 pattern is the predominant pattern in both clinical and hospital environment specimens and this especially is important in hospital equipments and personnel that likely act as carrier. In clinic, hospital mediations and hosts are the fundamental agent in genotypic variety and outbreak new pattern of PCR- ribotyping in P. aeruginosa.
CONCLUSION
Mucoid forms of P. aeruginosa may be isolated in any clinical disease and environmental source in addition P. aeruginosa always subjects to antibiotic resistance and drug administration management influence on its rate. Similarity of antibiotic resistance pattern between clinical and environment isolates refer to distribution of P. aeruginosa and carrier role of hospital equipments and personnel. Also the kind of PCR- ribotyping pattern of strains isolated from clinical and hospital environment represent which one of this group act as contamination resource. Therefore, it can help to prevent Pseudomonas infection among immunocompromised individuals.
ACKNOWLEDGMENTS
The authors are grateful to Vice-chancellor for Research, Isfahan University of medical sciences for financial support of the present study (research project number 388093).
Footnotes
Source of Support: Isfahan University of medical sciences (research project number 388093)
Conflict of Interest: None declared.
REFERENCES
- 1.Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, et al. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A. 2011;108:7481–6. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nseir S, Ader F, Lubret R, Marquette CH. Pathophysiology of airway colonization in critically ill COPD patient. Curr Drug Targets. 2011;12:514–20. doi: 10.2174/138945011794751537. [DOI] [PubMed] [Google Scholar]
- 3.Fuentefria DB, Ferreira AE, Corção G. Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water.Are they genetically related? J Environ Manage. 2011;92:250–5. doi: 10.1016/j.jenvman.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Czekajło-Kołodziej U, Giedrys-Kalemba S, Medrala D. Phenotypic and genotypic characteristics of Pseudomonas aeruginosa strains isolated from hospitals in the north–west region of Poland. Pol J Microbiol. 2006;55:103–12. [PubMed] [Google Scholar]
- 5.Wolska K, Szweda P. Comparative evaluation of PCR ribotyping and ERIC PCR for determining the diversity of clinical Pseudomonas aeruginosa isolates. Pol J Microbiol. 2008;57:157–63. [PubMed] [Google Scholar]
- 6.Khorvash F, Abdi F, Dialami K, Mehrabi A. Can serum procalcitonin and C-reactive protein as nosocomial infection markers in hospitalized patients without localizing signs? J Res Med Sci. 2011;16:1280–5. [PMC free article] [PubMed] [Google Scholar]
- 7.Japoni A, Farshad S, Albozi A. Pseudomonas aeruginosa: Burn infection, Treatment and Antibacterial Resistance. IRCMJ. 2009;11:244–53. [Google Scholar]
- 8.Agarwal G, Kapil A, Kabra SK, Das BK, Dwivedi SN. Characterization of Pseudomonas aeruginosa isolated from chronically infected children with cystic fibrosis in India. BMC Microbial. 2005;5:43. doi: 10.1186/1471-2180-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pseudomonas Agars Pseudomonas Agar F • Flo Agar Pseudomonas Agar P • Tech Agar. [Last accessed on 2011 Dec 31]. Available from: http://www.bd.com/ds/technicalCenter/inserts/Pseudomonas_Agars.pdf .
- 10.Campbell ME, Farmer SW, Speert DP. New Selective Medium for Pseudomonas aeruginosa with Phenanthroline and 9-Chloro-9-[4-(Diethylamino) Phenyl]-9, 10- Dihydro-10-Phenylacridine Hydrochloride (C-390) J Clin Microbiol. 1988;26:1910–2. doi: 10.1128/jcm.26.9.1910-1912.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.70887 Cetrimide Agar (Pseudomonas Selectice Agar Base) [Last accessed on 2011 Dec 31]. Available from: https://www.sigmaaldrich.com/etc/medialib/docs/Fluka/Datasheet/70887dat.Par.0001.File.tmp/70887dat.pdf .
- 12.Govan JR, Collee JG, Fraser AG, Marmion BP, Simmons A. Pseudomonas, Stenotrophomonas, Burkholderia. 14th ed. New York: Churchill Livingstone; 1996. Practical Medical Microbiology; pp. 413–24. [Google Scholar]
- 13.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–9. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 15.Wayene, PA, USA: CLSI; 2005. Clinical and Laboratory Standards Institute Performance standards for antimicrobial testing: Document M10-S15. [Google Scholar]
- 16.Agarwal G, Kapil A, Kabra SK, Chandra R, Das B, Diwedi SN. Phenotypic and genotypic variants of Pseudomonas aeruginosa isolated from children with cystic fibrosis in India. Indian J Med Res. 2002;116:73–81. [PubMed] [Google Scholar]
- 17.Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, et al. Genetic features of pseudomonas aeruginosa isolates from Cystic Fibrosis patients compared with those isolates from other origins. J Med Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- 18.Corona-Nakamura AL, Miranda-Novales MG, Leaños-Miranda B, Portillo-Gómez L, Hernández-Chávez A, Anthor-Rendón J, et al. Epidemiologic Study of Pseudomonas aeruginosa in Critical Patients and Reservoirs. Arch Med Res. 2001;32:238–42. doi: 10.1016/s0188-4409(01)00267-3. [DOI] [PubMed] [Google Scholar]
- 19.Pugashetti BK, Metzger HM, Jr, Vadas L, Feingold DS. Phenotypic differences among clinically isolated mucoid Pseudomonas aeruginosa strains. J Clin Microbiol. 1982;16:686–91. doi: 10.1128/jcm.16.4.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone I, Chirillo MG, Raso T, Zucca M, Savoia D. Phenotypic and genotypic characterization of Pseudomonas aeruginosa from cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 2008;27:1093–9. doi: 10.1007/s10096-008-0551-1. [DOI] [PubMed] [Google Scholar]
- 21.O’ Carroll MR, Syrmis MW, Wainwright CE, Greer RM, Mitchell P, Coulter C, et al. Clonal strains of Pseudomonas aeruginusa in paediatric and adult cystic fibrosis units. Eur Respir J. 2004;24:101–6. doi: 10.1183/09031936.04.00122903. [DOI] [PubMed] [Google Scholar]
- 22.Mohammad Taheri Z, Shahbazi N, Khoddami M. Genetic Diversity of pseudomonas aeruginosa Strains isolated from Hospitalized patients. Tanaffos. 2008;7:32–9. [Google Scholar]
- 23.Ruiz L, Dominguez MA, Ruiz N, Vinas M. Relationship between clinical and environmental isolates of Pseudomonas aeruginosa in a hospital setting. Arch Med Res. 2004;35:251–7. doi: 10.1016/j.arcmed.2004.02.005. [DOI] [PubMed] [Google Scholar]


