Abstract
TDP-43 is an evolutionarily conserved RNA-binding protein currently under intense investigation for its involvement in the molecular pathogenesis of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). TDP-43 is normally localized in the nucleus, but translocated to the cytoplasm in diseased neurons. The endogenous functions of TDP-43 in the nervous system remain poorly understood. Here, we show that the loss of Drosophila TDP-43 (dTDP-43) results in an increased production of sensory bristles and sensory organ precursor (SOP) cells on the notum of some but not all flies. The location of ectopic SOPs varies among mutant flies. The penetrance of this novel phenotype is dependent on the gender and sensitive to environmental influences. A similar SOP phenotype was also observed on the wing and in the embryos. Overexpression of dTDP-43 causes both loss and ectopic production of SOPs. Ectopic expression of ALS-associated mutant human TDP-43 (hTDP-43M337V and hTDP-43Q331K) produces a less severe SOP phenotype than hTDP-43WT, indicating a partial loss of function of mutant hTDP-43. In dTDP-43 mutants, miR-9a expression is significantly reduced. Genetic interaction studies further support the notion that dTDP-43 acts through miR-9a to control the precision of SOP specification. These findings reveal a novel role for endogenous TDP-43 in neuronal specification and suggest that the FTD/ALS-associated RNA-binding protein TDP-43 functions to ensure the robustness of genetic control programs.
INTRODUCTION
TDP-43, an evolutionarily conserved RNA-binding protein, is currently under intense investigation for its contribution to the molecular pathogenesis of frontotemporal dementia and amyotrophic lateral sclerosis (ALS) (1–5). In both diseases, TDP-43 is depleted from the nucleus of affected neurons, implicating the loss of TDP-43’s normal nuclear function as a major pathogenic mechanism. However, the significance of TDP-43 in neural development and function is largely unknown.
During development, complex molecular interactions ensure the precision and reproducibility of patterning events, despite stochastic genetic and environmental fluctuations, a process called canalization (6). Sensory organ precursor (SOP) cells in Drosophila's peripheral nervous system are one of the best-studied models for understanding the molecular pathways that control early neurogenesis and the robustness of developmental processes (7–9). A key process in SOP specification is lateral inhibition mediated by the Notch-Delta signaling pathway and complex feedback regulatory loops in which dynamic expression of the transcription factor Senseless (Sens) plays a central role (10,11).
Proper levels of gene expression during development are controlled through multiple mechanisms, including microRNAs (miRNAs). These small, non-coding RNAs regulate mRNA translation or stability, mostly by binding to 3′ untranslated regions (12,13). miR-9a ensures the precise specification of SOPs, which is in part through suppressing Sens expression and thus functions as a component of the regulatory circuit that confers robustness in lateral inhibition (14–16). Another miRNA involved in this developmental process is miR-7 that targets enhancer of split, a key transcription factor upstream of Sens (17). These findings support the hypothesis that miRNAs play a key role in canalization (18,19). Here, we show that Drosophila TDP-43 (dTDP-43) acts through miR-9a to regulate the precision of SOP specification in Drosophila. Ectopic expression of ALS-associated mutant human TDP-43, hTDP-43M337V and hTDP-43Q331K (20) produces a less severe SOP phenotype than hTDP-43WT, indicating a partial loss of function of mutant hTDP-43. These results provide novel insights into the molecular functions of TDP-43 and suggest that the loss of TDP-43's normal function may disturb the robustness of genetic control programs.
RESULTS
dTDP-43 mutant flies produce ectopic SOPs on the notum
Previously, we obtained mutant flies with a nonsense mutation introduced into the codon encoding Glu-367 of the dTDP-43 gene and showed that dTDP-43 regulates dendritic branching (20). When examining viable adult dTDP-43Q367X mutant flies, we unexpectedly found that some of them had ectopic large sensory bristles called macrochaetes on the notum (Fig. 1B and C). The number (total 22) and position of these macrochaetes were precisely patterned in wild-type flies, an excellent example of canalization (9). For simplicity, our analysis focuses on scutellar (SC), dorsocentral (DC) and anterior postalar (aPA) bristles (Fig. 1A).
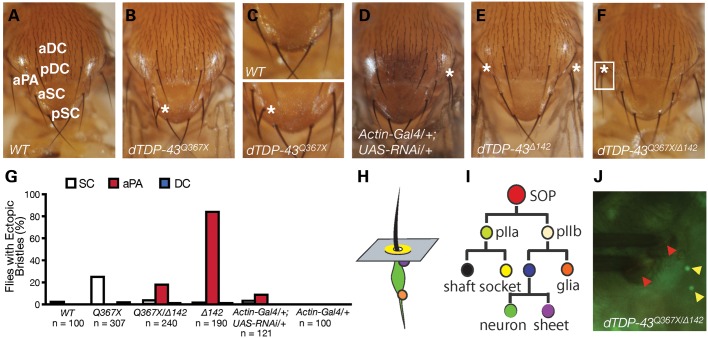
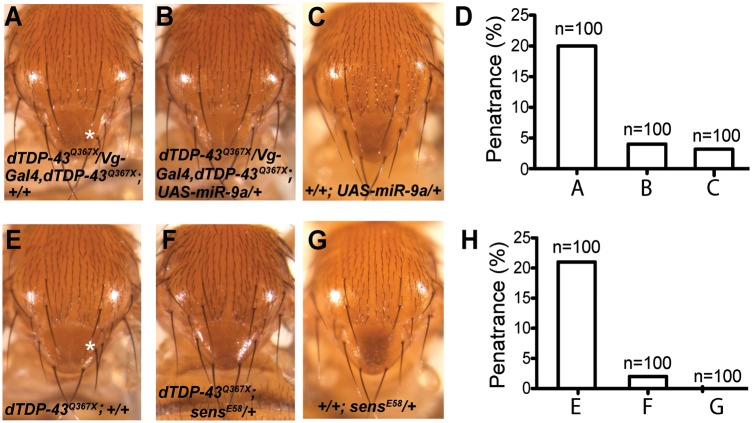
Figure 1.
Ectopic production SOPs in dTDP-43 mutant flies. (A) The notum of a wild-type (w1118) fly. Some macrochaetes are labeled at the right of the bristles: aSC, anterior scutellar; pSC, posterior scutellar; aDC, anterior dorsocentral; pDC, posterior dorsocentral; aPA, anterior postalar. (B) The asterisk indicates an ectopic SC bristle in a dTDP-43Q367X mutant fly. (C) Enlarged images of notum of wild-type or dTDP-43 mutant flies from panels A and B. The asterisk indicates the ectopic bristle. (D) The asterisk indicates an ectopic aPA bristle in a fly expressing dTDP-43 RNAi. (E) Two ectopic aPA bristles in a dTDP-43Δ142 mutant fly. (F) An ectopic aPA bristle in a dTDP-43Q367X/dTDP-43Δ142 transheterozygous fly. (G) Penetrance of the ectopic bristle phenotype in dTDP-43 mutant flies with various combinations of alleles. The number of flies examined for each genotype is listed that included roughly equal numbers of male and female flies. (H) Schematic representation of a sensory organ. Black: sensory bristle. Green: sensory neuron. Orange: glia cell. Purple: sheet cell. Yellow: socket cell. (I) The SOP lineage that gives rise to different cell types in a sensory organ. (J) Immunostaining with anti-embryonic lethal abnormal vision (ELAV) antibody revealed that ectopic macrochaetes in dTDP-43 mutants were accompanied with ectopic sensory neurons, indicating ectopic production of SOPs. In this image, an ectopic bristle is shown at the aPA position in a dTDP-43Q367X/dTDP-43Δ142 transheterozygous mutant fly. Red arrows indicate socket cells; yellow arrows indicate sensory neurons underneath each sensory bristle.
Among dTDP-43Q367X homozygous mutant flies, 26% had at least one extra macrochaete on the notum, and among them 90% had only one extra bristle (Fig. 1B and G). The positions of ectopic bristles might differ in different flies. Eighty-seven percent were near any one of the four SC bristles, and a few flies had ectopic bristles at one of the two aPA or four DC positions. dTDP-43Q367X homozygous mutant flies were analyzed here. To further confirm this developmental phenotype, we expressed a dTDP-43-specific RNAi construct (Vienna Drosophila RNAi Center) under the control of the actin-Gal4 driver. About 7% of flies with RNAi knockdown had ectopic bristles (Fig. 1D), whereas the flies with the actin-Gal4 alone (Fig. 1G) or UAS-RNAi alone (data not shown) had no bristle phenotype.
We also examined dTDP-43 mutant fly lines generated independently by Feiguin et al. (21) (Supplementary Material, Fig. S1). Among dTDP-43Δ142 homozygous mutant flies, 81% had ectopic macrochaetes and 56% of those flies had only one extra bristle and 44% had two (Fig. 1E and G). Surprisingly, 81% of ectopic bristles were at one of the two aPA positions, in contrast to our findings in dTDP-43Q367Xflies. When we examined mutant flies with dTDP-43Q367X and dTDP-43Δ142 alleles in trans, we found that the penetrance of the ectopic bristle phenotype was 21% (Fig. 1F and G), similar to that in dTDP-43Q367X flies, but most of the ectopic bristles were at the aPA position (Fig. 1G), more similar to the bristle location in dTDP-43Δ142 flies. Moreover, the ectopic bristle phenotype at the aPA position in dTDP-43Q367X/Δ142mutants could be rescued by dTDP-43 expression. The differences in the penetrance and location of ectopic bristles between dTDP-43Q367Xand dTDP-43Δ142 alleles remain after several generations of outcross with w1118 flies, raising the possibility that dTDP-43 function may be modified by other genetic factors near the dTDP-43 locus.
Each sensory bristle is accompanied by a sensory neuron and a few other support cells (Fig. 1H) that are derived from a single SOP through two rounds of asymmetric division (Fig. 1I). To examine whether the ectopic bristles we observed in dTDP-43 mutant flies are a reflection of increased SOP production, we performed immunostaining experiment using the embryonic lethal abnormal vision (ELAV) antibody and found that ectopic bristles (indicated by the asterisk in the white square in Fig. 1F) are accompanied by ectopic sensory neurons (Fig. 1J). Thus, the precise specification of neural progenitor cells is altered by the loss of dTDP-43 function in Drosophila.
Ectopic expression of TDP-43 results in a mixed SOP phenotype
To further examine TDP-43 function, we performed overexpression studies. Overexpression of dTDP-43 with Vg-Gal4, which drives gene expression in the thin strip of dorsal–ventral boundary of wing imaginal discs (22), led to the loss of SOPs at one of the four SC positions (Fig. 2B and C). This phenotype is consistent with the finding that the loss of endogenous dTDP-43 function leads to increased production of SOPs (Fig. 1). Interestingly, some flies with dTDP-43 overexpression also had ectopic bristles at one of the four SC or DC positions (Fig. 2B and C), probably reflecting a dominant-negative effect induced by overexpression due to well-documented tendency of the protein to aggregates under non-physiological conditions (4,5). When we expressed hTDP-43 using Vg-Gal4, we only observed ectopic SOP production on the notum (Fig. 2D), consistent with a previous report that hTDP-43 overexpression in flies mimics a loss of function in locomotor activity and survival (23). Previously, we generated transgenic fly lines that express ALS-associated mutant hTDP-43, hTDP-43M337V and hTDP-43Q331K, at comparable levels as hTDP-43WT (20). Expression of hTDP-43M337V and hTDP-43Q331K resulted in a less severe SOP phenotype than hTDP-43WT (Fig. 2E–G), further supporting the notion that some ALS-associated pathogenic mutations reduce the normal activity of TDP-43 and partial loss of TDP-43 function caused by genetic mutations may contribute to disease process (20,23).
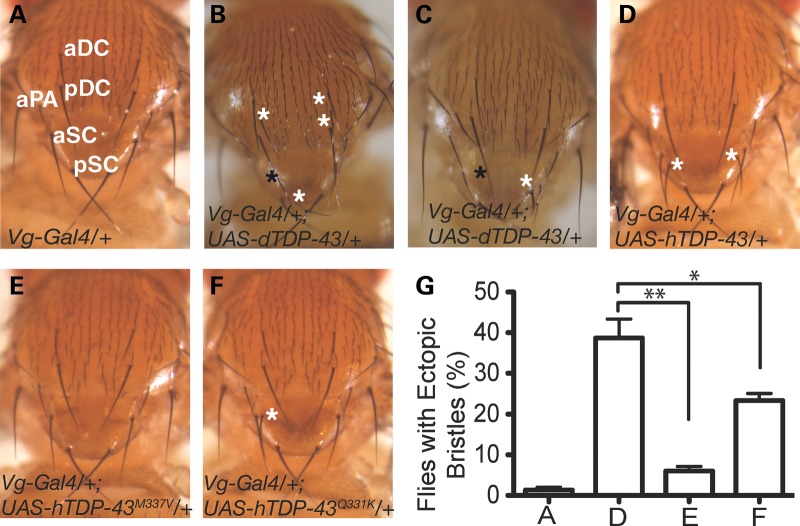
Figure 2.
Ectopic expression of TDP-43 affects SOP specification. (A) The notum of a Vg-Gal4/+ fly with no ectopic sensory bristles. (B and C) Ectopic expression of dTDP-43 with Vg-Gal4 leads to not only the loss of SC bristles (black asterisk) but also ectopic production of DC or SC bristles (white asterisks). (D) Ectopic expression of hTDP-43 with Vg-Gal4 results in the ectopic production of SC bristles only. (E and F) The effects of ectopic expression of hTDP-43M337V (E) or hTDP-43Q331K (F) on ectopic SC bristle production is less potent than hTDP-43. The expression levels of wild-type and mutant hTDP-43 were comparable (Lu et al., 2009) (20). (G) Quantification of the penetrance of the ectopic SC bristle phenotype caused by expression of wild-type or mutant hTDP-43. Fifty flies were counted for each genotype and the average of results from three countings are presented here. *P< 0.05; **P< 0.01. The letters A, D, E and F indicate the genotypes of the flies presented in Panels A, D, E and F.
The effects of dTDP-43 on SOP specification are influenced by gender and environment
When we compared female and male flies, we found that in all dTDP-43 mutants with different allele combinations, the penetrance of ectopic SOP production was consistently higher in females (Fig. 3A). Environmental fluctuation can also affect the penetrance and expressivity of the ectopic SOP phenotype. The precision of SOP specification of wild-type flies is not affected by environmental fluctuation. No difference was observed when wild-type flies were grown at 18, 25 or 30°C. In contrast, when grown at 18°C, 50% of dTDP-43Q367X/Δ142 mutant flies had one or more ectopic bristles, about twice the rate in flies grown at 25 or 30°C. Furthermore, ectopic bristles occurred preferentially at the aPA positions at 18 and 25°C, but at the SC positions at 30°C (Fig. 3B). Thus, dTDP-43 functions as a ‘gatekeeper’ to ensure the robustness of SOP specification, despite fluctuating environmental conditions. Without dTDP-43 function, SOP specification becomes more prone to environmental influence.
Figure 3.
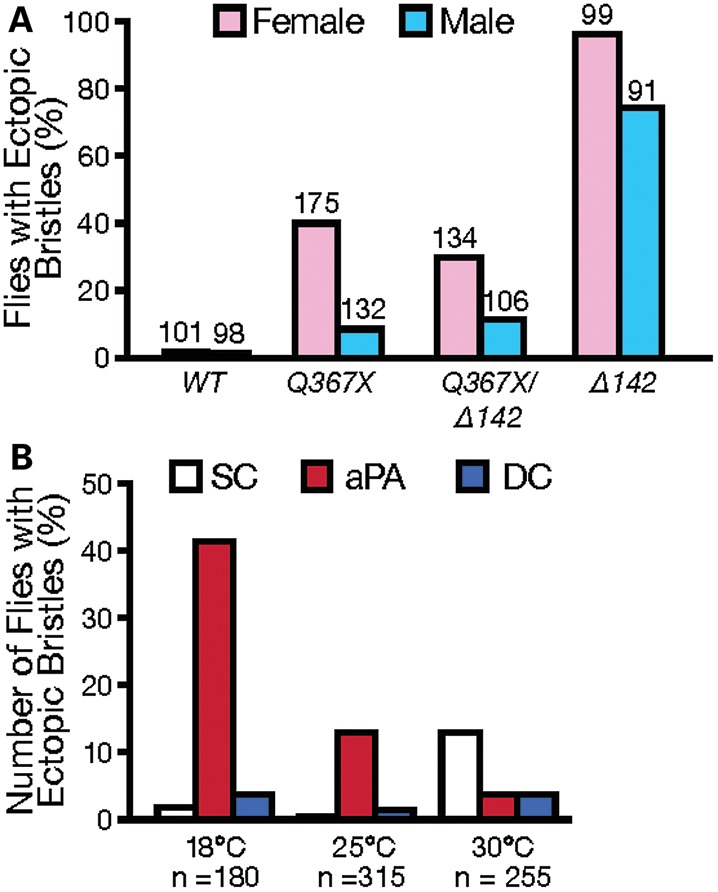
The effects of gender and environment on dTDP-43 function in SOP specification. (A) Gender strongly influences penetrance of the ectopic bristle phenotype in dTDP-43 mutants. The number of flies examined for each genotype is shown on the top of each column. (B) Effect of temperature on the penetrance of the phenotype and the location of ectopic bristles in male and female dTDP-43Q367X/dTDP-43Δ142 transheterozygous mutant flies.
MiR-9a is substantially downregulated in dTDP-43 mutant larvae
The SOP phenotype in dTDP-43 mutant flies is remarkably similar to that in miR-9a knockout flies as a consequence of misregulated SOP specification (14,15). Examination of the anterior margin of the wing revealed increased sensory bristle production in 8 of 52 dTDP-43Q367X mutants (Fig. 4A–D). In some hemisegments of dTDP-43Q367X mutant embryos, ectopic sensory neurons were also produced (Fig. 4E and F). Both the wing and embryo phenotypes also resembled those in miR-9a knockout flies (14,15). We, therefore, determined whether miR-9a is downregulated in dTDP-43 mutants.
Figure 4.
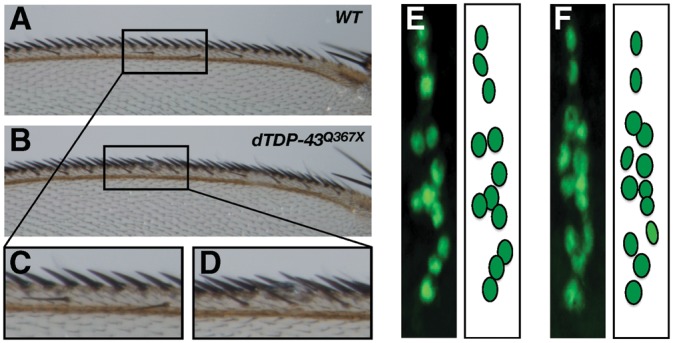
SOP phenotypes on the anterior margin of the wing and in embryos. (A) The anterior margin of a wild-type wing showing sensory bristles assembled in a regular pattern. (B) A dTDP-43Q367X mutant with ectopic production of sensory bristles on the anterior wing margin. (C and D) Enlarged images of the areas highlighted in (A) and (B). (E) The dorsal cluster in an abdominal hemisegment of a wild-type embryo contains 12 sensory neurons that can be labeled with anti-ELAV antibody (green). (F) In some hemisegments of dTDP-43Q367X mutant embryos, an ectopic sensory neuron is produced.
Because TDP-43 overexpression in the notum resulted in a complex SOP phenotype (Fig. 2), we focused our biochemical analysis on dTDP-43 loss of function mutant flies. Indeed, the expression of miR-9a, but not of the muscle-specific miR-1, was reduced in dTDP-43Q367X mutant larvae, as shown by northern blot analysis (Supplementary Material, Figs. S2 and 5A). Similar results were obtained and confirmed in dTDP-43 mutant larvae with other allele combinations (Fig. 5A). Of note, miR-9a expression was not completely abolished in dTDP-43 mutants. Indeed, unlike miR-9a knockout flies (14,15,24), dTDP-43 mutants did not have a notching defect at the posterior margin of the wing. To examine how dTDP-43 regulates miR-9a biogenesis, we measured the levels of pri-miR-9a and pre-miR-9a and found that both were reduced as shown by quantitative real-time PCR (qRT-PCR) (Fig. 5B). Thus, the lower level of miR-9a in dTDP-43 mutants is partially due to a function of dTDP-43 in pri-miR-9a transcription and/or stability. Indeed, it has been reported that the complex containing TDP-43 and overexpressed Drosha in cultured cells does not possess miRNA processing activity (25).
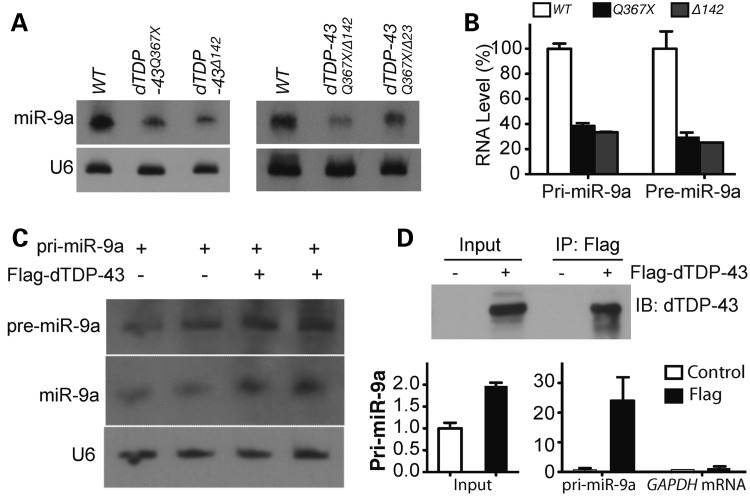
Figure 5.
MiR-9a is downregulated in dTDP-43 mutants. (A) miR-9a expression level is decreased in dTDP-43 mutant third instar larvae with combinations of different alleles. (B) Downregulation of pri-miR-9a and pre-miR-9a in dTDP-43 mutants. (C) Overexpression of dTDP-43 in HEK293 cells increases the levels of pre-miR-9a and mature miR-9a. (D) dTDP-43 binds to pri-miR-9a as shown by co-immunoprecipitation experiments and increases the steady-state level of pri-miR-9a.
To further examine the role of dTDP-43 in miR-9a biogenesis, we co-transfected pri-miR-9a and dTDP-43 into HEK293 cells. dTDP-43 overexpression increased the production of both pre-miR-9a and mature miR-9a (Fig. 5C and Supplementary Material, Fig. S3); and the level of pri-miR-9a was also higher and a biochemical interaction between TDP-43 and pri-miR-9a was detected with GAPDH mRNA as the negative control (Fig. 5D). Thus, these results raise the possibility that dTDP-43 regulates the transcription and/or stability of pri-miR-9a.
dTDP-43 regulates SOP specification in part through miR-9a
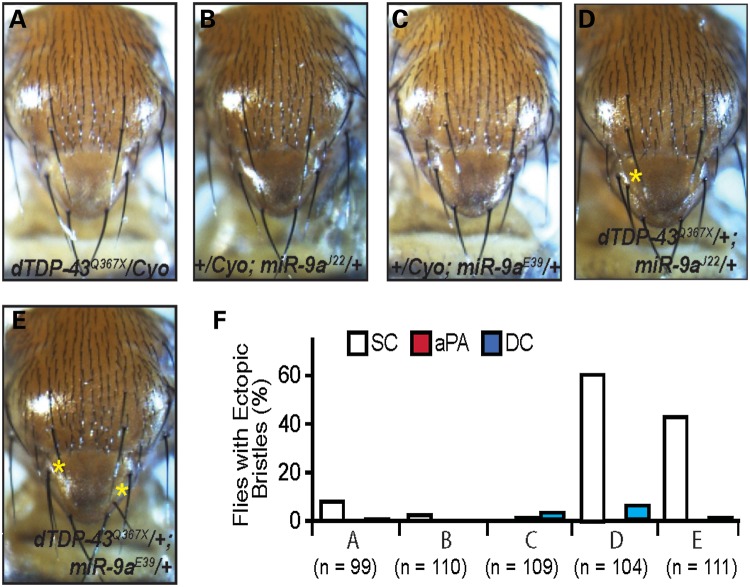
To further demonstrate the involvement of miR-9a in the dTDP-43 genetic pathway controlling the robustness of SOP specification, we first performed genetic interaction experiments using only female flies because their SOP phenotype is stronger than that in males. dTDP-43 heterozygous female flies exhibited an ectopic bristle phenotype at a low penetrance (Fig. 6A and F), indicating that dTDP-43 itself functions in a dose-dependent manner in SOP specification. Loss of one copy of miR-9a, either the miR-9aJ22 or miR-9aE39 allele, significantly enhanced the SOP phenotype of dTDP-43 heterozygous flies (Fig. 6D–F). Only one extra bristle was observed in dTDP-43, miR-9a transheterozygous flies. We repeated this experiment using both male and female flies and obtained a similar result (Supplementary Material, Fig. S4). This result supports the notion that the two genes function in the same pathway.
Figure 6.
Genetic interactions between dTDP-43 and miR-9a. (A–E) Representative images of female flies with different genotypes as indicated in each panel. (F) Penetrance of the ectopic bristle phenotype in flies with the genotypes listed in (A–E). Only female flies for each genotype were used in this analysis; thus, the penetrance of the phenotype in dTDP-43Q367X mutant flies is higher than that in Figure 1.
The reduced miR-9a expression in dTDP-43 mutant larvae (Fig. 5) raises the possibility that dTDP-43 regulates SOP specification in part through miR-9a. To further support this hypothesis, we performed rescue experiment and expressed miR-9a in dTDP-43 mutant background. To this end, we recombined Vg-Gal4 and dTDP-43Q367X onto the same chromosome. Vg-Gal4/+ (Fig. 2A) or UAS-pre-miR-9a/+ by itself had no SOP phenotype (Fig. 7C). Expression of UAS-pre-miR-9a by Vg-Gal4 in the dTDP-43Q367X mutant background could largely rescue the SOP phenotype (Fig. 7A and B). Moreover, our previous studies showed that sens mRNA is a key target of miR-9a in SOP specification and loss of one copy of sens could partially rescue miR-9a mutant phenotype (14). SensE58/+ by itself had no SOP phenotype (Fig. 7G) but loss of one copy of sens markedly suppressed the ectopic SOP phenotype in dTDP-43 mutant flies (Fig. 7E and F), further supporting the importance of miR-9a in the regulation of SOP specification by dTDP-43.
Figure 7.
dTDP-43 regulation of SOP specification is mediated through miR-9a and sens. (A) Vg-Gal4 and dTDP-43Q367X were recombined onto the same chromosome and dTDP-43Q367X/Vg-Gal4, dTDP-43Q367X flies show ectopic production of SC bristles. (B) Expression of miR-9a rescues the dTDP-43Q367X mutant phenotype. (C) UAS-pre-miR-9a/+ flies have no bristle phenotype. (D) Quantification of the ectopic SC bristle phenotype observed in (A–C). (E) A representative image of the notum of a dTDP-43Q367X/dTDP-43Q367X; +/+ fly. Asterisk indicates an ectopic macrochaete at the SC position. (F) An image of the notum of a dTDP-43Q367X/dTDP-43Q367X; sensE58/+ fly. (G) SensE58/+ flies have no bristle phenotype. (H) Penetrance of the ectopic bristle phenotype in flies presented in (E–G). Equal numbers of male and female flies were used.
DISCUSSION
In this study, we identified a novel function for the evolutionarily conserved RNA-binding protein TDP-43 in neural specification in Drosophila. Unlike several well-studied transcription factors in the Notch-Delta lateral inhibition pathway (e.g. 9,26), TDP-43 seems to function as a ‘robustness’ factor in a manner similar to that of miR-9a (14,16). In the absence of TDP-43 activity, ectopic bristles appear at one of several locations and SOP specification becomes more sensitive to environmental influences. Thus, TDP-43 seems to serve as a ‘gatekeeper’ to ensure the reproducible execution of genetic control programs and to canalize developmental phenotypes.
Since mRNA synthesis can fluctuate (27), regulation of mRNA metabolism plays a central role in gene expression. miRNAs are a key class of regulatory molecules that ensure the robustness of developmental programs and may play a key role in canalization (18,19), which is partly due to the modest effect of each miRNA on expression levels of multiple mRNA targets (13). Hundreds of RNA-binding proteins are encoded by the Drosophila genome (28) or in other species, and many simultaneously regulate multiple mRNAs (29). It is likely some other RNA-binding proteins also serve as robustness factors through direct interactions with target mRNAs.
Our findings show that the ‘robustness’ function of TDP-43 is mediated in part by miR-9a in one specific neural developmental process. The molecular functions of TDP-43 in development as revealed here may provide a new perspective on its endogenous role in neurodegeneration. Although the loss of TDP-43 leads to an early lethal phenotype in mouse embryos (30–32), loss of nuclear TDP-43 does not necessarily result in rapid neuronal cell death (33). Gene expression in individual cells is tightly regulated and also varies significantly, in part due to stochastic biochemical events (34). Chronic loss of nuclear TDP-43 or compromise of its buffering function by genetic mutations may cause an imbalance in protein homeostasis in human neurons before eventual neurodegeneration, during which miRNAs may play an essential role downstream of TDP-43 (35). Both TDP-43 and miR-9 are highly conserved through evolution and their interaction may occur in mammalian cells as well. It is interesting to note that miR-9 is significantly downregulated in Huntington's disease (36) and a mouse model of spinal motor neuron disease (37). Thus, downregulation of miR-9 and possibly other miRNAs as well in stressed neurons in which TDP-43 has been depleted from the nucleus may be a common contributing factor in different neurodegenerative disorders.
MATERIALS AND METHODS
Fly strains and genetics
All flies were maintained at 25°C on standard medium except for those in the experiment in Figure 3B. The w1118 strain served as a wild-type control. The following dTDP-43 mutant lines were studied: dTDP-43Q367X/Cy0 (20), dTDP-43Δ142/Cy0 (21). miR-9aJ22, miR-9aE39 and sensE58 flies were as described (14). For dTDP-43 overexpression, vg-Gal4 (on the second chromosome) and UAS-dTDP-43 (on the third chromosome) lines were crossed to generate vg-Gal4/+; UAS-dTDP-43/+ flies. For dTDP-43 knockdown, actin-Gal4/Cy0 flies were crossed with UAS-dTDP-43 RNAi line 38377 (Vienna Drosophila RNAi Center) to generate actin-Gal4/+; 38377/+ and Cy0/+; 38377/+ flies. UAS-hTDP-43WT, UAS-hTDP-43M337V and UAS-hTDP-43Q331K were described before (20).
Northern blot and qRT-PCR analysis
Northern blot analysis was performed as described (38), using digoxygenin-labeled probes from Exiqon as recommended by the manufacturer. Total RNA was extracted from third instar larvae with the miRNAeasy mini kit (Qiagen). For northern blot, 25 μg of total RNA for each sample was separated on a 12% polyacrylamide gel (Sequagel, National Diagnostics) and transferred to a Nytran SuperCharge Signal membrane (Schleicher & Schuell). miRNAs were detected with 5’-digoxigenin-labeled mercury-LNA probes (Exiqon) specific for individual miRNAs according to the manufacturer's protocol. Alternatively, miRNAs were detected with CDP-Star substrate according to the manufacturer's protocol (Roche). The membrane was then incubated with anti-digoxigenin-AP (1:100 000 in blocking buffer) and washed multiple times. U6 probe served as a loading control.
The following miRNA probe sequences were used: miR-9a: TCATACAGCTAGATAACCAAAGA; miR-1: CTCCATACTTCTTTACATTCCA; miR-8: GACATCTTTACCTGACAGTATTA; bantam: AATCAGCTTTCAAAATGATCTCA. The membrane was stripped by boiling in stripping buffer (0.1% SDS and 5 mm EDTA) at 85°C for 1 h and reprobed with different miRNA probes.
For qRT-PCR, total RNA was used in reverse transcription reaction using Taqman reverse transcription reagent (Applied Biosystems). The first-strand cDNAs were used as templates for qRT-PCR in a final volume of 20 μl. A standard curve was run in each PCR reaction. Individual values were normalized to the value of the gene encoding the ribosomal protein RP-49. The primers used are listed in Supplementary Material, Table S1. Relative expression of RNAs was calculated with the standard curve and ΔCt–ΔCt methods.
Generation of expression constructs
Drosophila miR-9a pri-miRNA (>1.6 kb) was cloned into pSuper-GFP vector from the wild-type genomic DNA with primers 5′-TATAAGCTTGCCGATGCAGGTCCGAGTCC-3′ and 5′-TATCTCGAGAGCTGTTTTGTTTAAATATGCCGT-3′. cDNA of dTDP-43 protein coding region was cloned into p3 × Flag-CMV-7.1 vector.
Cell culture and immunoprecipitation assay
HEK293 cells were maintained in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and penicillin/streptomycin and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Immunoprecipitation was performed as described (38) with some modifications. Briefly, 36 h after transfection with pri-miR-9a and Flag-dTDP-43 or control vector, HEK293 cells were lysed in cold RNase-free lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 0.5% NP-40, 1 mm dithiothreitol, protease inhibitor and RNase inhibitor). The protein extracts were centrifuged for 10 min at 4°C. The supernatant was incubated with protein-G-agarose beads conjugated with anti-Flag antibody (Sigma) at 4°C for 4 h. The precipitated complex was washed three times with RNase-free phosphate-buffered saline. The associated RNA was isolated with Trizol (Invitrogen) and detected with primers specific for pri-miR-9a (5′-TGTGCGTGAGTGGGCGACAA-3′ and 5′-TGTCTCGCTGGGCAGACGCTA-3′).
Immunostaining
In Figure 1, aPA bristles in late pupae were immunostained with antibodies against the pan-neuronal marker ELAV (Developmental Hybridoma Study Bank; 1:20) as described (14). The pupae were dissected and fixed in 4% formaldehyde in PBT (0.3% Triton in PBS) overnight at 4°C.
Standard protocols were used to immunostain Drosophila embryos. Briefly, Drosophila stage-15 embryos were collected, dechorionated in 50% sodium hypochlorite and fixed in 100% methanol for 1–2 h at room temperature. The primary antibody as anti-ELAV (1:200; Developmental Hybriodoma Study Bank) and the second antibody was Alexa Fluor 488 goat anti-rat IgG (1:400, A11006, Invitrogen). All samples with fluorescent signals were imaged by confocal microscopy (Nikon, D-Eclipse C1).
Western blot analysis
Western blot analysis was performed according to the standard protocol. Briefly, protein was extracted from the heads of 60 adult flies (30 females and 30 males) of each genotype and homogenized in 200 μl of lysis buffer (0.137 M NaCl, 20 mm Tris, pH 8.0, 10% glycerol, 1% NP-40, 0.1% SDS, 0.1% sodium deoxycholate, 1 mm dithiothreitol and Pierce protease inhibitor). Protein (10 μg), as measured with the Bio-Rad reagent, was separated by SDS-PAGE with 10% gels, transferred to PVDF membranes and immunoblotted with anti-dTDP-43 rabbit polyclonal antibody (1:3000; a gift of Dr Baralle, Italy) as described (21). The second antibody was horseradish peroxidase-conjugated IgG antibody (Jackson Laboratory). β-actin served as a loading control and was detected with antibodies from Cell Signaling (1:5000).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (R21NS066901, RO1NS066586 and RO1NS057553 to F.-B.G.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Bellen, F. Feiguin and the Bloomington Drosophila Stock Center for fly lines and reagents. We also thank S. Ordway for editorial assistance, A. Wilson for graphics help and Gao lab members for discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 2.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 3.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Bennett C.F., Cleveland D.W., Yeo G.W. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012;1462:3–15. doi: 10.1016/j.brainres.2012.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee E.B., Lee V.M., Trojanowski J.Q. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waddington C.H. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. [Google Scholar]
- 7.Jan Y.N., Jan L.Y. Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu. Rev. Genet. 1994;28:373–393. doi: 10.1146/annurev.ge.28.120194.002105. [DOI] [PubMed] [Google Scholar]
- 8.Modolell J. Patterning of the adult peripheral nervous system of Drosophila. Perspect. Dev. Neurobiol. 1997;4:285–296. [PubMed] [Google Scholar]
- 9.Barad O., Hornstein E., Barkai N. Robust selection of sensory organ precursors by the Notch–Delta pathway. Curr. Opin. Cell Biol. 2011;23:663–667. doi: 10.1016/j.ceb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Nolo R., Abbott L.A., Bellen H.J. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 11.Jafar-Nejad H., Acar M., Nolo R., Lacin H., Pan H., Parkhurst S.M., Bellen H.J. Senseless acts as a binary switch during sensory organ precursor selection. Genes. Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambros V. MicroRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Wang F., Lee J.A., Gao F.-B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes. Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejarano F., Smibert P., Lai E.C. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 2010;338:63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S.M., Brennecke J., Stark A. Denoising feedback loops by thresholding–a new role for microRNAs. Genes. Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 17.Li X., Cassidy J.J., Reinke C.A., Fischboeck S., Carthew R.W. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornstein E., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38(suppl.):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 19.Wu C.I., Shen Y., Tang T. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome. Res. 2009;19:734–743. doi: 10.1101/gr.084640.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y., Ferris J., Gao F.-B. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol. Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feiguin F., Godena V.K., Romano G., D'Ambrogio A., Klima R., Baralle F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Calleja M., Moreno E., Pelaz S., Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- 23.Estes P.S., Boehringer A., Zwick R., Tang J.E., Grigsby B., Zarnescu D.C. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum. Mol. Genet. 2011;20:2308–2321. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biryukova I., Asmar J., Abdesselem H., Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev. Biol. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 25.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 26.Schweisguth F., Posakony J.W. Suppressor of hairless, the Drosophila homolog of the mouse recombination signal-binding protein, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 27.Raj A., Peskin C.S., Tranchina D., Vargas D.Y, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasko P. Translation factors and RNA binding proteins. J. Cell Biol. 2000;150:51–56. doi: 10.1083/jcb.150.2.f51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keene J.D. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang P.M., Ling J., Jeong Y.H., Price D.L., Aja S.M., Wong P.C. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl. Acad. Sci. USA. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P., 3rd, Herz J., Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L.S., Cheng W.C., Hou S.C., Yan Y.T., Jiang S.T., Shen C.K. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 33.Sato T., Takeuchi S., Saito A., Ding W., Bamba H., Matsuura H., Hisa Y., Tooyama I., Urushitani M. Axonal ligation induces transient redistribution of TDP-43 in brainstem motor neurons. Neuroscience. 2009;164:1565–1578. doi: 10.1016/j.neuroscience.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 34.Raser J.M., O'Shea E.K. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gascon E., Gao F.-B. Cause or effect: misregulation of microRNA pathways in neurodegeneration. Front. Neurosci. 2012;6:48. doi: 10.3389/fnins.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Packer A.N., Xing Y., Harper S.Q., Jones L., Davidson B.L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haramati S., Chapnik E., Sztainberg Y., Eilam R., Zwang R., Gershoni N., McGlinn E., Heiser P.W., Wills A. M., Wirguin I., et al. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X.L., Zong R., Li Z., Biswas M.H., Fang Z., Nelson D.L., Gao F.-B. FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J. Neurosci. 2011;13:13705–13709. doi: 10.1523/JNEUROSCI.2827-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.