Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most frequent known cause of late-onset Parkinson's disease (PD). To explore the therapeutic potential of small molecules targeting the LRRK2 kinase domain, we characterized two LRRK2 kinase inhibitors, TTT-3002 and LRRK2-IN1, for their effects against LRRK2 activity in vitro and in Caenorhabditis elegans models of LRRK2-linked neurodegeneration. TTT-3002 and LRRK2-IN1 potently inhibited in vitro kinase activity of LRRK2 wild-type and mutant proteins, attenuated phosphorylation of cellular LRRK2 and rescued neurotoxicity of mutant LRRK2 in transfected cells. To establish whether LRRK2 kinase inhibitors can mitigate pathogenesis caused by different mutations including G2019S and R1441C located within and outside of the LRRK2 kinase domain, respectively, we evaluated effects of TTT-3002 and LRRK2-IN1 against R1441C- and G2019S-induced neurodegeneration in C. elegans models. TTT-3002 and LRRK2-IN1 rescued the behavioral deficit characteristic of dopaminergic impairment in transgenic C. elegans expressing human R1441C- and G2019S-LRRK2. The inhibitors displayed nanomolar to low micromolar rescue potency when administered either pre-symptomatically or post-symptomatically, indicating both prevention and reversal of the dopaminergic deficit. The same treatments also led to long-lasting prevention and rescue of neurodegeneration. In contrast, TTT-3002 and LRRK2-IN1 were ineffective against the neurodegenerative phenotype in transgenic worms carrying the inhibitor-resistant A2016T mutation of LRRK2, suggesting that they elicit neuroprotective effects in vivo by targeting LRRK2 specifically. Our findings indicate that the LRRK2 kinase activity is critical for neurodegeneration caused by R1441C and G2019S mutations, suggesting that kinase inhibition of LRRK2 may represent a promising therapeutic strategy for PD.
INTRODUCTION
Parkinson's disease (PD) is a common neurodegenerative disorder with the pathological hallmark of progressive loss of dopamine (DA) neurons in affected brains. There is no cure for PD and current therapy does not halt the underlying degenerative process. Recent discovery of genetic causes of PD has uncovered potentially novel targets for therapeutic interventions that may prevent or slow down the progression of the disease. Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most frequent known cause of late-onset familial PD (1,2). The G2019S substitution occurs within the kinase domain of LRRK2, and it is the most common mutation observed across a majority of populations. Patients carrying this mutation present with clinical features indistinguishable from those of idiopathic PD (3). G2019S has been found to enhance kinase activity in vitro and to promote toxicity in neuronal cells in culture (4,5). These findings provide strong support for the notion that G2019S may play a pathogenic role through a ‘gain-of-function’ mechanism, prompting development of small molecule LRRK2 kinase inhibitors to counteract hyperactive LRRK2 signaling. However, PD-associated mutations in LRRK2 occur throughout the protein; and several of them are outside the kinase domain, including R1441C/G and Y1699C. Although the prevalence of R1441C/G in PD is lower than G2019S in many populations (6,7), the reverse is also true in specific ethnic groups such as those from the Basques and southern Italy (8–11). Biochemical studies by different groups on the effect of R1441C/G and Y1699C on LRRK2 kinase activity have not provided consistent results (12). It remains to be established whether all LRRK2 mutations initiate pathogenesis through a gain-of-function, and whether LRRK2 kinase-specific inhibition will be effective against neurodegeneration caused by different LRRK2 mutations.
Several commercially available kinase inhibitors, originally targeted against other kinases, have also been found to suppress LRRK2 kinase activity in vitro (13,14). These include H-1152, sunitinib and GW-5074. GW-5074 has also been reported to be protective against G2019S-induced neurodegeneration in Caenorhabditis elegans, Drosophila and mice (14,15). However, the low potency and poor selectivity of these compounds toward LRRK2 raise a concern about side effects, and make it difficult to assess whether inhibition of LRRK2 kinase alone is sufficient to confer neuroprotection in vivo. Recently, the first and highly selective LRRK2 inhibitor, LRRK2-IN1, was described (16). In the present study, we have utilized a combined pharmacological and genetic approach to examine whether LRRK2-specific inhibition confers disease-modifying protection against neurodegeneration caused by different LRRK2 mutations. We have utilized LRRK2-IN1 and TTT-3002, a potent inhibitor with a relatively high degree of specificity for LRRK2, to evaluate in vivo potency, LRRK2 selectivity and treatment time-dependent effectiveness relative to onset and progression of behavioral symptoms and DA neurodegeneration caused by expression of either G2019S- or R1441C-LRRK2 in the C. elegans models of PD we have described previously (17). Both TTT-3002 and LRRK2-IN1 potently rescued the age-dependent dopaminergic behavioral deficit and the corresponding degeneration of DA neurons in transgenic R1441C and G2019S C. elegans. The inhibitors were effective when administered either before or after the pathological symptoms appeared, indicating they can also reverse dopaminergic degeneration. However, TTT-3002 and LRRK2-IN1 were ineffective in treating neurodegeneration caused by dual-mutants of LRRK2, containing a kinase inhibitor-resistant mutation (A2016T), in addition to either the G2019S or the R1441C mutations, indicating that TTT-3002 and LRRK2-IN1 are effective in vivo by specifically targeting LRRK2. These results demonstrate that the LRRK2-kinase activity is critical for neurodegeneration caused by the R1441C and G2019S mutations of LRRK2, suggesting that kinase-targeted inhibition of LRRK2 may represent a promising strategy for therapy of PD.
RESULTS
Selective inhibition of LRRK2-dependent phosphorylation in vitro and in intact cells
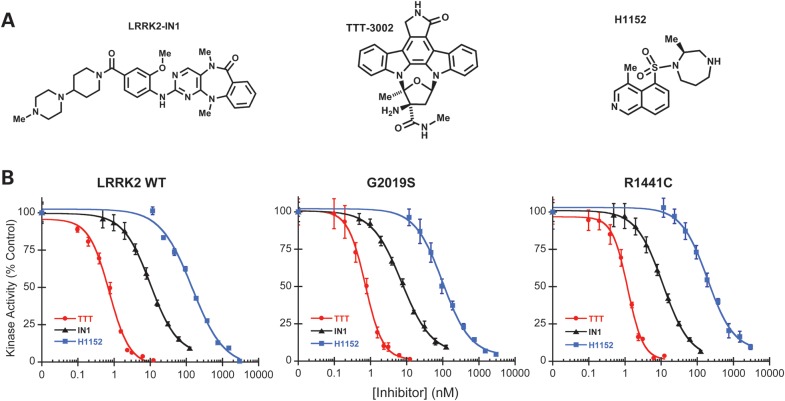
The availability of potent and selective LRRK2 inhibitors is highly desirable to pharmacologically interrogate LRRK2 kinase activity and its role in PD, and to explore the therapeutic potential of LRRK2 inhibition. We evaluated several small molecules with unrelated structures (Fig. 1A) for their inhibitory efficacy against LRRK2 in vitro. These compounds include LRRK2-IN1, the first reported LRRK2-selective kinase inhibitor with excellent potency and selectivity profile (16); TTT-3002, a potent LRRK2 inhibitor used in the present study and H-1152, a previously reported non-selective kinase inhibitor of LRRK2 (13). These inhibitors were tested for concentration-dependent inhibition of LRRK2-catalyzed phosphorylation of the specific peptide substrate LRRKtide, using three recombinant LRRK2 variants. Namely, GST-tagged LRRK2 fragment encompassing residues 970–2527, corresponding to LRRK2 wild-type (WT) and two LRRK2 mutants containing PD-causing substitutions either within (G2019S) or outside (R1441C) the kinase domain. All three compounds showed dose-dependent inhibition of the kinase activities of the three LRRK2 variants (Fig. 1B), displaying the same rank order of inhibitory potencies against WT, R1441C and G2019S, as measured by their half-maximal inhibitory concentrations (IC50 values) (Table 1). LRRK2-IN1 had nanomolar inhibitory potency with IC50 of 10 and 7.3 nm for LRRK2 WT and G2019S, respectively, in good agreement with the previous report under essentially the same assay conditions (16). The IC50 of LRRK2-IN1 for LRRK2 R1441C is 11 nm, similar to that for LRRK2 WT. In comparison, TTT-3002 showed the highest potency with IC50 values of 0.7, 0.7 and 1.2 nm for LRRK2 WT, G2019S and R1441C, respectively. H-1152 displayed the lowest potency with IC50 values of 140, 99 and 190 nm for LRRK2 WT, G2019S and R1441C, respectively. These data identified TTT-3002 and LRRK2-IN1 as potent LRRK2 kinase inhibitors with subnano- to nano-molar potencies against in vitro phosphorylation by all three LRRK2 variants. LRRK2-IN1 has already been shown to be quite specific for LRRK2 according to extensive selectivity profiling against many different protein kinases (16). The characterization of TTT-3002, however, has previously not been presented. Although an indolocarbazole, a priori assumed to be not very selective, it exhibited an unexpected degree of specificity for LRRK2 when profiled against a panel of 23 other kinases (Supplementary Material, Table S1), many of which are normally expressed in the brain or are members of the mitogen-activated protein kinase superfamilies to which LRRK2 belongs. More extensive analysis of TTT-3002 selectivity against a panel of 140 kinases revealed a pattern of concentration-dependent selectivity profile (Supplementary Material, Table S2). At a low concentration (1 nm), TTT-3002 only inhibited the activities of a few kinases to >70% of the vehicle (DMSO) control. However, the selectivity of TTT-3002 decreased at higher concentrations (above 10 and 100 nm). These results suggest that TTT-3002 could have a good LRRK2 selectivity at very low but not high concentrations when used in vitro.
Figure 1.
Kinase inhibitory effects of LRRK2-IN1, TTT-3002 and H-1152 on recombinant LRRK2 and mutants in vitro. (A) Chemical structures of LRRK2-IN1, TTT-3002 and H-1152. (B) Inhibition curves of LRRK2-IN1, TTT-3002 and H-1152 for LRRK2 WT, G2019S and R1441C. Inhibitors were assayed in triplicate using 8 nm Invitrogen LRRK2, R1441C and G2019S separately in the presence of 100 μm ATP and 100 μm LRRKtide. The results are presented as percentage of kinase activity relative to the vehicle control from three independent experiments performed in triplicate. Data are expressed as mean ± SEM.
Table 1.
IC50 values for LRRK2-IN1, TTT-3002 and H-1152 toward WT-, R1441C- and G2019S-LRRK2a
| IC50 (nm) |
|||
|---|---|---|---|
| WT | R1441C | G2019S | |
| LRRK2-IN1 | 10.0 | 11.0 | 7.3 |
| TTT-3002 | 0.70 | 1.2 | 0.70 |
| H-1152 | 140 | 190 | 99 |
aDerived from inhibition curves in Figure 1.
Notably, the nominal IC50 values of TTT-3002 for all LRRK2 variants were below the concentrations of the respective enzyme (3–8 nm) in the in vitro assays, indicating that TTT-3002 has a high affinity for LRRK2 and its measured potencies were limited by the kinase concentration. Thus, its actual inhibitory potencies and thereby the specificity for LRRK2, would be even higher if suitable assay formats allowed for lower kinase concentrations. Taken together, the results indicate LRRK2-IN1 and TTT-3002 are relatively potent and selective LRRK2 inhibitors. Since LRRK2-IN1 and TTT-3002 differ chemically, display 10-fold different relative potencies for LRRK2 and have off-target effects on different non-LRRK2 kinases, their parallel use should be helpful in the analysis of LRRK2-specific inhibition in cells.
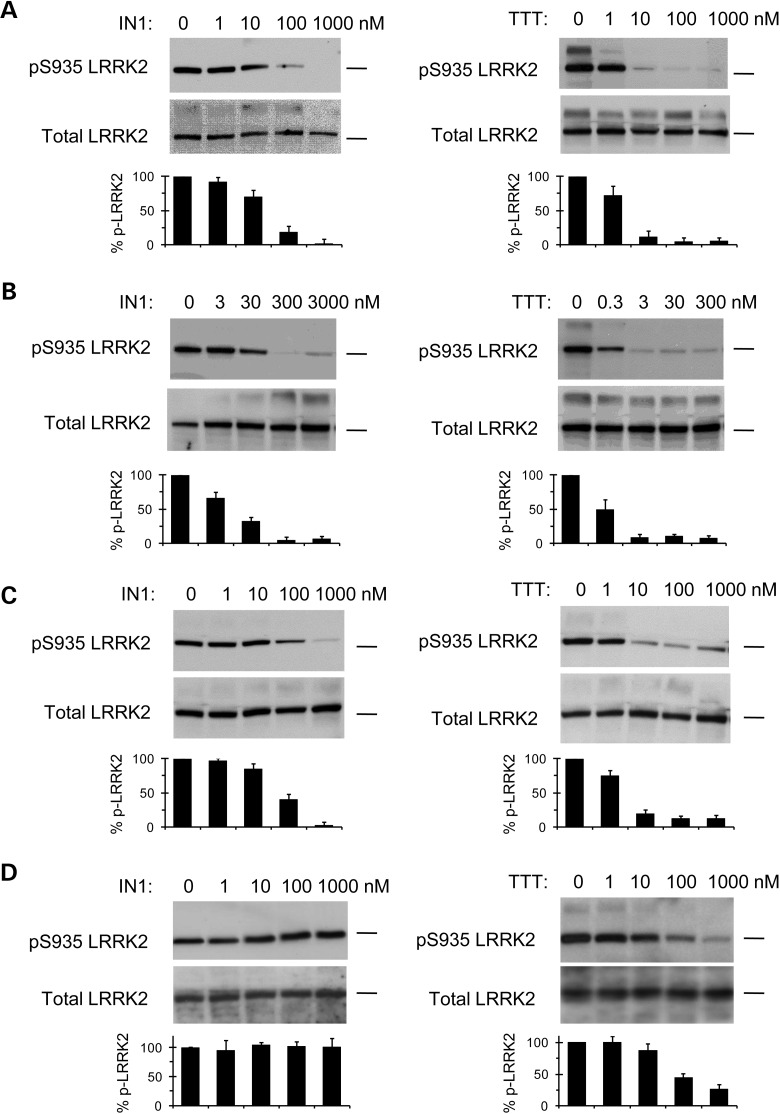
We next examined the inhibitory effects of LRRK2-IN1 and TTT-3002 on LRRK2-dependent phosphorylation in human cell lines. LRRK2 is constitutively phosphorylated on serine 935 (S935), and inhibition of LRRK2 kinase activity results in diminished S935 phosphorylation in cultured cells (13). This allows for a robust cellular assay for small molecule inhibitors using S935 phosphorylation as a readout. We probed S935 phosphorylation of LRRK2 in human macrophage cells and lymphocytes that express high levels of endogenous LRRK2 (18,19), obviating the need for transfection in these cell lines. In human THP-1 macrophage cells, LRRK2-IN1 and TTT-3002 dose-dependently inhibited endogenous S935 phosphorylation of LRRK2 (Fig. 2A). In an immortalized lymphoblastoid cell line derived from a PD patient carrying the G2019S mutation, LRRK2-IN1 inhibited S935 phosphorylation of LRRK2 at concentrations of 30 nm and higher while TTT-3002 abolished the phosphorylation at a concentration of 3 nm (Fig. 2B). In human SH-SY5Y neuroblastoma cell line transiently transfected with G2019S-LRRK2, LRRK2-IN1 inhibited S935 phosphorylation of LRRK2 at concentrations of 100 nm and higher while TTT-3002 showed prominent inhibition at concentrations 10 nm and higher (Fig. 2C). Overall, TTT-3002 was about one order of magnitude more potent than LRRK2-IN1 in inhibiting S935 phosphorylation of LRRK2 in cells, in general agreement with their relative potency in inhibiting kinase activity of recombinant LRRK2 (Fig. 1B, Table 1).
Figure 2.
Effect of LRRK2-IN1 and TTT-3002 on S935 phosphorylation of LRRK2 in human cell lines. LRRK2-IN1 and TTT-3002 inhibited S935 phosphorylation of LRRK2 in human THP-1 macrophages (A), human lymphoblastoid cell line derived from a PD patient carrying the G2019S mutation (B) and human SH-SY5Y neuroblastoma cell line transfected with G2019S-LRRK2 plasmid (C). The inhibitory effects of LRRK2-IN1 and TTT-3002 on S935 phosphorylation of LRRK2 were attenuated in cells transfected with G2019S/A2016T plasmid (D). All cell lines were treated with varying concentrations of LRRK2-IN1 and TTT-3002 for 1.5 h. Cell lysates were subjected to immunoblotting for detection of S935 phosphorylation (pS935 LRRK2) using a rabbit mAb UDD2 against phospho-serine 935 LRRK2, and for detection of total LRRK2 using a mouse mAb against LRRK2 amino acids 970–2527. The position of the 250 kDa molecular weight marker was indicated on each blot. The bar graphs represent percent pS935 LRRK2 normalized to total LRRK2 in untreated and inhibitor-treated samples, using band intensities determined from scanned blot images with the ImageJ software. The results were obtained from three independent experiments and values are expressed as mean ± SEM.
TTT-3002 and LRRK2-IN1 attenuate toxicity induced by G2019S-LRRK2 in human SH-SY5Y neuroblastoma cell line
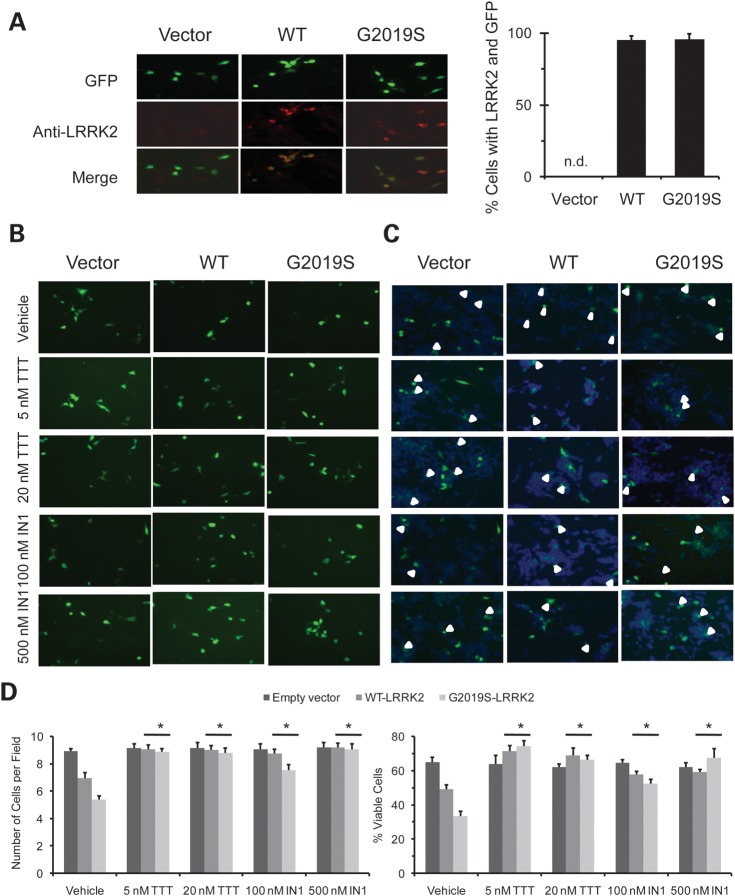
Overexpression of mutant G2019S-LRRK2 causes cytotoxicity in transfected cells, and such toxicity has been shown to correlate with the enhanced kinase activity of the G2019S mutation (20,21). We transfected human SH-SY5Y neuroblastoma cell line with WT- or G2019S-LRRK2 along with the GFP marker. We verified co-expression of LRRK2 and GFP in most of the transfected cells (Fig. 3A). We confirmed that overexpression of G2019S-LRRK2 caused significantly higher toxicity than overexpression of WT-LRRK2, as evidenced by reduced cell survival (Fig. 3B–D, vehicle-treated), consistent with previous reports of a higher percentage of apoptotic cells for G2019S overexpression (20,21). We tested if treatment with TTT-3002 and LRRK2-IN1 could protect against G2019S-associated cellular toxicity and found that they significantly improved cell survival in the G2019S transfectants (Fig. 3B–D). In direct comparison, TTT-3002 was again more potent than LRRK2-IN1 in rescuing G2019S-induced cell death. These results suggest that inhibition of LRRK2 protects neuronal cells from toxicity induced by overexpression of mutant LRRK2.
Figure 3.
TTT-3002 and LRRK2-IN1 attenuate cytotoxicity induced by overexpression of G2019S-LRRK2 and WT-LRRK2 in SH-SY5Y cells. SH-SY5Y cells were transiently transfected with WT-LRRK2, G2019S-LRRK2 and empty vector along with GFP at 15:1 ratio. (A) Images of GFP, LRRK2 immunostaining (Anti-LRRK2) and merges were made from SH-SY5Y cells transfected with empty vector, WT-LRRK2 and G2019S-LRRK2 plasmids. Cells were stained 48 h post-transfection with LRRK2 rabbit mAb UDD3 and Alexa Fluor 488 labeled goat anti-rabbit antibody. Cell images were taken at ×40 magnification. There was little LRRK2 staining in the empty vector control group. The cells positive for LRRK2 staining were analyzed for their overlap with co-expressed GFP signals in WT-LRRK2 and G2019S-LRRK2 transfection groups using 30 randomly imaged fields (n = 120–150 cells for each group), as shown in the bar graph. The results were obtained from three independent experiments and values are expressed as mean ± SEM. n.d., not detected. (B) GFP images of cells treated with vehicle, TTT-3002 and LRRK2-IN1. At 48 h post-transfection, cell images were taken at 30 randomly selected fields at ×20 magnification per treatment group by using GFP channel. (C) Merged images of GFP and Hoechst 3332 staining. At 48 h post-transfection, cells were treated with Hoechst 3332 and images were taken at 30 randomly selected fields at ×20 magnification per group by using both GFP channel and DAPI channel. The GFP image and DAPI image were superimposed for identifying the viable GFP positive cells. (D) Quantitative analysis of GFP positive and viable cells. WT-LRRK2 and G2019S-LRRK2 transfection caused a reduction in the number of GFP positive (left panel) and percentage (right panel) of viable GFP positive cells when compared with empty vector (vehicle controls). Both TTT-3002 and LRRK2-IN1 treatments significantly increased the number of GFP positive cells (left panel) and percentage of viable GFP positive cells (right panel) in WT-LRRK2 and G2019S-LRRK2 transfectants. TTT-3002 and LRRK2-IN1 did not affect number of GFP positive cells transfected with empty vector. The results were obtained from three independent experiments and values are expressed as mean ± SEM. *P < 0.05 by one-way ANOVA, inhibitor-treated vs. vehicle-treated samples in WT-LRRK2 and G2019S-LRRK2 transfectants.
Treatment of C. elegans models of PD with LRRK2 inhibitors at different stages of neuronal impairment
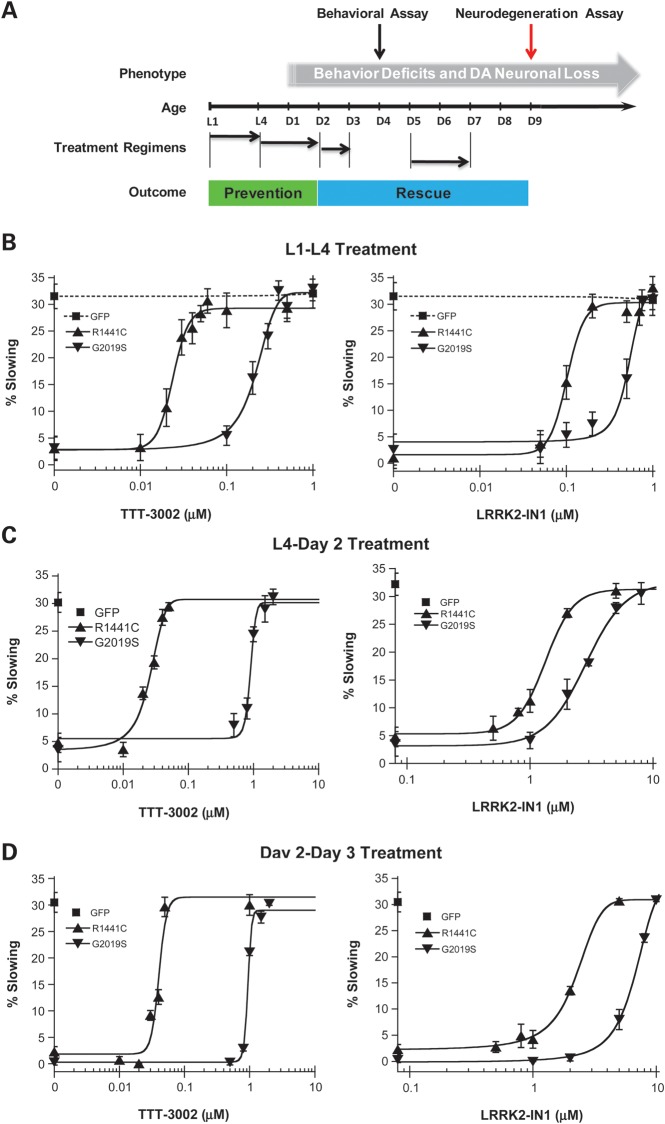
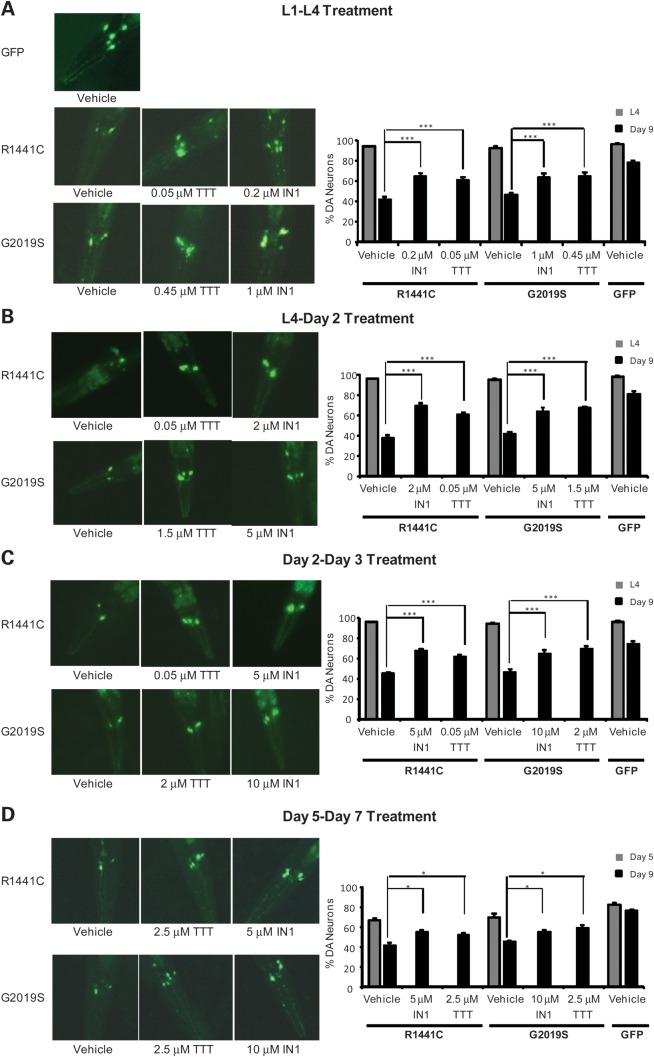
We previously characterized C. elegans models of LRRK2-linked PD in which transgenic expression of human R1441C and G2019S LRRK2 in dopaminergic neurons caused adult-onset and progressive behavioral deficits and neurodegeneration (17). We have devised a liquid-culture method that allows for treatment with small molecule inhibitors for discrete periods at different stages of worm development and manifestation of neurodegeneration. This approach permits us to evaluate therapeutic outcomes in terms of prevention or rescue of PD-relevant phenotypes in C. elegans (Fig. 4A). We sought to validate whether TTT-3002 and LRRK2-IN1 could protect against mutant LRRK2-induced dopaminergic behavioral deficits and neurodegeneration in vivo, and to determine if these compounds are effective both pre- and post-symptomatically.
Figure 4.
Treatment with TTT-3002 and LRRK2-IN1 during different life stages prevents or rescues behavioral deficit in transgenic R1441C and G2019S worms. (A) Treatment regimens of C. elegans with small molecule inhibitors. For assessing the ‘preventive’ effect of LRRK2 inhibitors against C. elegans behavioral deficits and DA neuronal loss, R1441C and G2019S transgenic C. elegans were treated with TTT-3002 and LRRK2-IN1 in liquid culture over the periods L1–L4 and L4 to adult Day 2, respectively. These are early stages of life span prior to overt LRRK2-linked behavioral abnormality and DA neuronal loss. For assessing the ‘rescue’ effect of LRRK2 inhibitors against C. elegans behavior deficit and DA neuronal loss, R1441C and G2019S transgenic C. elegans were treated with TTT-3002 and LRRK2-IN1 in liquid culture for the periods of adult Day 2 to Day 3, and adult Day 5 to Day 7, respectively. Both transgenic R1441C and G2019S lines manifested severe behavioral impairment since adult Day 2 and progressive DA neurodegeneration starting approximately at adult Day 5. (B) Treatment during L1–L4: dose–response curves for TTT-3002 and LRRK2-IN-1 in preventing R1441C and G2019S transgenic C. elegans behavior abnormality. The behavioral response for GFP control worms (dotted lines) was not altered by either TTT-3002 or LRRK2-IN1. (C) Treatment during L4 to adult Day 2: shown are dose–response curves for TTT-3002 and LRRK2-IN-1 in rescuing behavior deficit of R1441C and G2019S transgenic C. elegans. (D) Treatment during adult Day 2 to Day 3: shown are dose–response curves for TTT-3002 and LRRK2-IN1 in reversing the behavioral deficit of R1441C and G2019S transgenic C. elegans. Results were obtained from three independent experiments, each with 10–15 worms per genotype for a given compound concentration. Values are expressed as mean ± SEM.
TTT-3002 and LRRK2-IN1 prevent and rescue mutant LRRK2-induced dopaminergic behavioral dysfunction
The basal slowing response (also called food-sensing response) is a well-established behavioral assay for dopamine-specific function in C. elegans, in which well-fed worms slow down their locomotive activity after encountering bacterial food (22). As we described previously (17), transgenic expression of R1441C- and G2019S-LRRK2 causes dopamine-specific locomotive dysfunction in C. elegans manifested as adult-onset and progressive loss of the basal slowing response beginning on adult Day 2, with a complete loss of the dopamine-dependent behavioral response on adult Day 4. To test whether inhibition of LRRK2 kinase activity could protect against dopaminergic behavioral dysfunction caused by PD-linked mutations, we treated R1441C- and G2019S-LRRK2 transgenic C. elegans with TTT-3002 and LRRK2-IN1 at different developmental stages and assayed the behavioral response on adult Day 4.
We first incubated worms with the chemicals during the first (L1) to the last (L4) larval stage. As shown in Figure 4B, the vehicle-treated R1441C and G2019S worms were defective in basal slowing response (i.e., low % slowing). However, treatment with TTT-3002 and LRRK2-IN1 restored the basal slowing response in both R1441C and G2019S transgenic LRRK2 worms in a dose-dependent manner. The control worms expressing the GFP marker in DA neurons showed normal behavior that was unaltered by the treatments with either compound. The EC50 (half maximal effective concentration) values for TTT-3002 were 0.02 and 0.22 μm for preventing the dopaminergic behavioral deficit in transgenic worms expressing R1441C and G2019S, respectively (Table 2). The corresponding EC50 values for behavioral rescue by LRRK2-IN1 were 0.10 and 0.51 μm in transgenic worms expressing R1441C and G2019S, respectively. In contrast, H-1152 was ineffective even at concentrations as high as 10 μm, and showed only a partial effect at 100 μm in transgenic R1441C worms (Supplementary Material, Fig. S1). Therefore, TTT-3002 and LRRK2-IN1 are highly effective in early prevention of dopaminergic behavioral impairment caused by R1441C and G2019S expression in C. elegans DA neurons.
Table 2.
Quantification of in vivo EC50 values in different treatment regimens for TTT-3002 and LRRK2-IN1 in rescuing the behavioral deficit displayed by R1441C- and G2019S-LRRK2 transgenic C. elegansa
| Treatment | Inhibitor | R1441C (EC50, μm) | G2019S (EC50, μm) |
|---|---|---|---|
| L1–L4 | LRRK2-IN1 | 0.10 | 0.51 |
| TTT-3002 | 0.02 | 0.22 | |
| L4–Day 2 | LRRK2-IN1 | 1.33 | 2.83 |
| TTT-3002 | 0.03 | 0.90 | |
| Day 2–Day 3 | LRRK2-IN1 | 2.22 | 6.80 |
| TTT-3002 | 0.04 | 0.94 |
aDerived from Figure 4.
We examined LRRK2 expression levels in transgenic worms expressing G2019S- and R1441C-LRRK2. We were unable to detect LRRK2 proteins in total lysates of LRRK2 transgenic worms by western blotting, likely due to the expression of LRRK2 constructs only in dopaminergic neurons (eight per animal). Following affinity pull-down of LRRK2 from large populations of worms, we confirmed the expression of LRRK2 proteins in transgenic worms expressing R1441C and G2019S before and after treatment with inhibitors (Supplementary Material, Fig. S2). To accurately determine LRRK2 expression in individual worms, we examined LRRK2 mRNA expression in single worms using quantitative real-time PCR (qPCR). We verified amplification of LRRK2 transcripts in R1441C and G2019S transgenic worms from RNA extracted from single animals using an LRRK2-specific primer. Data from qPCR analysis revealed a higher LRRK2 mRNA level in transgenic R1441C worms relative to transgenic G2019S worms (Supplementary Material, Fig. S2).
We next treated worms from L4 (adult Day 0) to adult Day 2 when the initial behavioral symptoms began to develop in transgenic worms expressing mutant LRRK2. Again, TTT-3002 and LRRK2-IN1 were able to prevent the dopaminergic behavioral deficit caused by mutant LRRK2 (Fig. 4C). Compared with the L1-L4 treatment, TTT-3002 maintained similar potency (EC50 = 0.03 μm) while LRRK2-IN1 showed a reduction in potency (EC50 = 1.33 μm) in behavioral rescue of R1441C transgenic worms (Table 2). For G2019S transgenic worms, both chemicals exhibited considerably reduced potency when compared with the L1–L4 treatment, with EC50 values of 0.90 μm for TTT-3002 and 2.83 μm for LRRK2-IN1.
Finally, when treatment occurred from adult Day 2 to Day 3, during which severe behavioral phenotype already was manifest in transgenic worms expressing R1441C and G2019S, both TTT-3002 and LRRK2-IN1 also showed dose-dependent rescue of the behavioral deficit in these worms (Fig. 4D). EC50 values were 0·04 μm for TTT-3002 and 2.22 μm for LRRK2-IN1 in behavioral rescue of R1441C transgenic worms, and 0.94 μm for TTT-3002 and 6.80 μm for LRRK2-IN1 in treating G2019S transgenic worms (Table 2). Taken together, our results demonstrate that TTT-3002 and LRRK2-IN1 potently prevent and rescue the dopamine-dependent behavioral phenotype induced by LRRK2 mutants in C. elegans during pre- and post-symptomatic periods, respectively.
TTT-3002 and LRRK2-IN1 arrest mutant LRRK2-induced neurodegeneration in vivo
We evaluated whether rescue of the dopamine-dependent behavioral phenotype by TTT-3002 and LRRK2-IN1 correlated as expected with protection against dopaminergic neurodegeneration in transgenic R1441C and G2019S worms. We conducted treatment regimens analogous to those used for behavioral rescue (treatments during L1–L4, L4–Day 2 and Day 2–Day 3), as well as treatment during adult Day 5 to Day 7 when there was already marked neuronal loss in untreated animals. Following treatments with TTT-3002 and LRRK2-IN1, we monitored the surviving dopamine (DA) neurons in the head (CEP and ADE neurons according to nomenclature for C. elegans) in aged worms on adult Day 9. As shown for the L1–L4 treatment period (Fig. 5A), only about 40% of DA neurons remained on adult Day 9 in transgenic worms expressing either R1441C or G2019S that were treated with vehicle only, while about 80% of DA neurons survived in the control worms expressing the GFP marker. Treatment with TTT-3002 and LRRK2-IN1 at minimal dosages required for full behavioral rescue resulted in significant improvement of neuronal survival in transgenic R1441C and G2019S worms. Consistent with the findings from behavioral rescue, transgenic worms expressing R1441C were more sensitive to LRRK2 inhibitors, requiring lower dosages for arresting neurodegeneration than those expressing G2019S. Likewise, treatments during L4 to Day 2 (Fig. 5B) and Day 2 to Day 3 (Fig. 5C) with TTT-3002 and LRRK2-IN1, each at a concentrations required for full behavioral rescue, resulted in significant protection against neurodegeneration in transgenic R1441C and G2019S worms. We observed similar findings even for the later treatment during Day 5 to Day 7, albeit with higher concentrations of inhibitors than used in younger animals (Fig. 5D). Taken together, our data indicate that treatment of TTT-3002 and LRRK2-IN1 during both pre- and post-symptomatic stages significantly protect DA neurons against neurodegeneration caused by R1441C and G2019S.
Figure 5.
Treatment with TTT-3002 and LRRK2-IN-1 arrests neurodegeneration in transgenic R1441C and G2019S worms. Transgenic C. elegans lines were treated with vehicle, TTT-3002 and LRRK2-IN-1 during L1–L4 stage (A), L4 to adult Day 2 (B), adult Day 2 to adult Day 3 (C) and adult Day 5 to adult Day 7 (D). When left untreated (vehicle control) and examined on adult Day 9, only ∼40% of ADE/CEP head DA neurons remained in both R1441C and G2019S transgenic C. elegans, while ∼80% of ADE/CEP head DA neurons were present in GFP control C. elegans. (A) Representative fluorescence images taken on adult Day 9 following treatments in liquid culture during L1–L4 stage with vehicle control, LRRK2-IN1 and TTT-3002 of transgenic C. elegans expressing GFP marker only or additionally R1441C- and G2019S-LRRK2 in DA neurons (left panel), and quantitation of DA neurons in the head region monitored at L4 stage (adult Day 0) and on adult Day 9 (right panel). (B) Representative fluorescence images taken on adult Day 9 following treatments in liquid culture from L4 to adult Day 2 with vehicle control, LRRK2-IN1 and TTT-3002 of transgenic C. elegans expressing GFP marker only or additionally R1441C- and G2019S-LRRK2 in DA neurons (left panel), and quantitation of DA neurons in the head region monitored at L4 stage (adult Day 0) and on adult Day 9 (right panel). (C) Representative fluorescence images taken on adult Day 9 following treatments in liquid culture from adult Day 2 to Day 3 with vehicle control, LRRK2-IN1 and TTT-3002 of transgenic C. elegans expressing GFP marker only or additionally R1441C- and G2019S-LRRK2 in DA neurons (left panel), and quantitation of DA neurons in the head region monitored at L4 stage (adult Day 0) and on adult Day 9 (right panel). (D) Representative fluorescence images taken on adult Day 9 following treatments in liquid culture from adult Day 5 to Day 7 with vehicle control, LRRK2-IN1 and TTT-3002 of transgenic C. elegans expressing GFP marker only or additionally R1441C- and G2019S-LRRK2 in DA neurons (left panel), and quantitation of DA neurons in the head region monitored on adult Day 5 and on adult Day 9 (right panel). In all the treatment regimens, both TTT-3002 and LRRK2-IN1 significantly enhanced the percentage of surviving DA neurons assayed on adult Day 9. DA neurons in the head region were monitored at the L4 stage, on adult Day 5 or adult Day 9 using a fluorescence microscope. Results were obtained from three independent experiments, each with about 30 worms per genotype for a given compound concentration. Values are expressed as mean ± SEM. ***P < 0.01 and *P < 0.05 by one-way ANOVA.
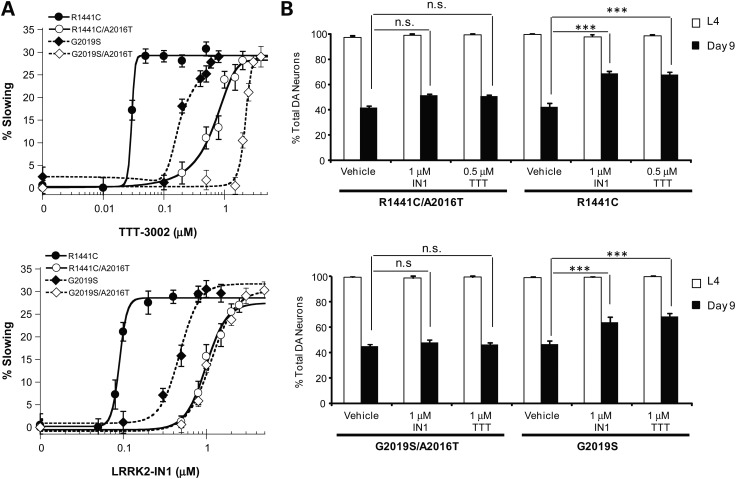
TTT-3002 and LRRK2-IN1 specifically target LRRK2 in vivo
To validate that the therapeutic outcomes of TTT-3002 and LRRK2-IN1 against dopaminergic behavioral deficit and neurodegeneration in transgenic worms expressing R1441C- and G2019S-LRRK2 are due to specific inhibition of the LRRK2 kinase activity, we took advantage of a rationally designed inhibitor-resistant A2016T mutation of LRRK2. As shown previously, the A2016 residue in the kinase domain of LRRK2 is required for its sensitivity to LRRK2 kinase inhibitors (13), and the A2016T mutation renders LRRK2 wild-type and G2019S resistant to LRRK2-IN1 in vitro (16). We confirmed that SH-SY5Y cells overexpressing G2019S/A2016T were more resistant to inhibition of S935 LRRK2 phosphorylation by LRRK2-IN1 and TTT-3002 (Fig. 2D) when compared with those overexpressing G2019S (Fig. 2C). However, the effects of the A2016T mutation on LRRK2 activity and its sensitivity to inhibitors have not been examined in vivo. Therefore, we constructed transgenic C. elegans lines expressing R1441C/A2016T- or G2019S/A2016T-LRRK2 double mutants in DA neurons. As shown in Figure 6, transgenic R1441C/A2016T- or G2019S/A2016T-LRRK2 worms manifested the same phenotype of dopaminergic behavioral deficit and neurodegeneration as transgenic single mutant R1441C- and G2019S-LRRK2 lines (vehicle treatment), indicating that the A2016T mutation does not alter the pathogenicity of R1441C- and G2019S-LRRK2 in vivo. This is consistent with the in vitro finding that A2016T mutation does not affect the intrinsic kinase activity of LRRK2 (13). When treated with TTT-3002 and LRRK2-IN1, transgenic double mutant R1441C/A2016T and G2019S/A2016T worms were much more resistant to the rescuing effects on behavioral defects by both inhibitors than their respective single-mutant R1441C and G2019S worms (Fig. 6A). When treated with TTT-3002 and LRRK2-IN1 at concentrations that fully rescue the behavioral deficits in transgenic R1441C and G2019S worms but with no or partial effects on the R1441C/A2016T and G2019S/A2016 worms, both inhibitors significantly enhanced the survival of DA neurons in transgenic worms expressing R1441C and G2019S, but failed to rescue neurodegeneration in transgenic R1441C/A2016T and G2019S/A2016T worms (Fig. 6B). Hence, the A2016T mutation desensitizes LRRK2 in response to kinase inhibitors. These results fully establish transgenic R1441C/A2016T and G2019S/A2016T worms as useful in vivo models for validating direct targeting of LRRK2 by small molecule inhibitors. Our findings suggest that TTT-3002 and LRRK2-IN1 arrest the PD-relevant pathogenesis by specifically inhibiting the kinase activity of R1441C and G2019S in vivo.
Figure 6.
Transgenic worms expressing R1441C/A2016T- and G2019S/A2016T-LRRK2 exhibit dopaminergic behavioral deficit and neurodegeneration but are resistant to treatment with LRRK2 inhibitors. When left untreated, R1441C/A2016T- and G2019S/A2016T-LRRK2 transgenic C. elegans manifested similar age-dependent dopaminergic behavioral deficit (A) and neurodegeneration (B) as the R1441C- and G2019S-LRRK2 transgenic lines. (A) Transgenic C. elegans lines expressing R1441C, G2019S, R1441C/A2016T and G2019S/A2016T in DA neurons were treated during L1–L4 in liquid culture with vehicle and different concentrations of TTT-3002 and LRRK2-IN1. Transgenic C. elegans lines expressing R1441C/A2016T and G2019S/A2016T were less sensitive to behavioral rescue by TTT-3002 and LRRK2-IN1 than those expressing R1441C and G2019S. (B) TTT-3002 at 0.5 and 1 μm, and LRRK2-IN1 at 1 μm significantly enhanced DA neuron survival on adult Day 9 in R1441C- and G2019S-LRRK2 transgenic C. elegans lines, respectively. Little or no improvement in DA neuron survival was observed for either of the R1441C/A2016T- and G2019S/A2016T-LRRK2 lines treated with TTT-3002 and LRRL2-IN1. DA neurons in the head region were monitored at the L4 stage or on adult Day 9 using a fluorescence microscope. Results were obtained from three independent experiments, each with about 30 worms per genotype for a given compound concentration. Values are expressed as mean ± SEM. ***P < 0.01, and n.s. (not significant) by one-way ANOVA.
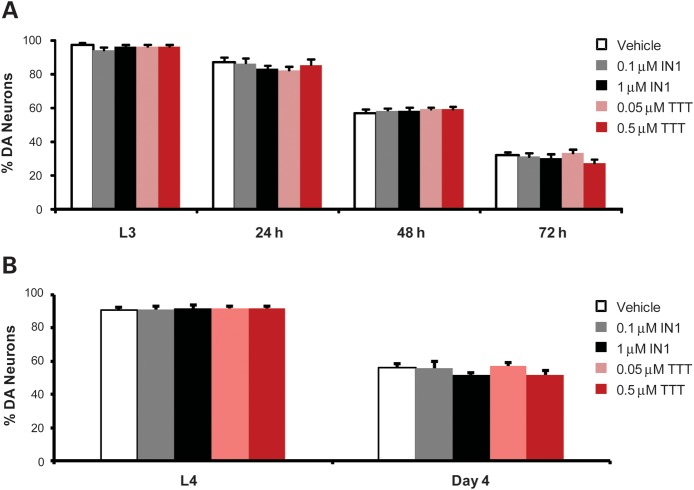
TTT-3002 and LRRK2-IN1 have no effect on neurodegeneration induced by exogenous neurotoxin or other factors
We also examined whether TTT-3002 and LRRK2-IN1 had any protective effects on other worm models of neurodegeneration unrelated to LRRK2. We exposed worms expressing GFP marker in DA neurons to the neurotoxin 6-hydroxy-dopamine (6-OHDA) which is selectively transported into DA neurons of both mammals and worms and induces degeneration (23). We incubated the GFP worms with vehicle or LRRK2 inhibitors from L1 until L3, exposed L3 worms to 6-OHDA for 1 h and re-incubated with vehicle or LRRK2 inhibitors for an additional 24 h. Then DA neurons were monitored at 24, 48 and 72 h post 6-OHDA exposure. As shown in Figure 7A, 6-OHDA caused progressive loss of DA neurons 48 and 72 h post exposure, a reproducible phenotype, as described previously (23). We also considered whether TTT-3002 and LRRK2-IN1 inhibit LRK-1 in vivo, the C. elegans homolog of mammalian LRRK2. As reported previously, loss of LRK-1 in worms render the DA neurons hypersensitive to 6-OHDA (24). We treated lrk-1 deletion worms in the absence or presence of TTT-3002 (0.5 µM) and LRRK2-IN1 (1 µM), and assayed vulnerability of DA neurons to 6-OHDA. As shown in Supplementary Material, Figure S3, we confirmed that the lrk-1 null worms were more vulnerable to neurodegeneration caused by 6-OHDA. However, TTT-3002 and LRRK2-IN1 did not seem to alter 6-OHDA induced toxicity in either the control GFP or lrk-1 mutant animals when compared with the vehicle control. Therefore, it is likely that these two inhibitors did not affect endogenous LRK-1, at least in the concentrations used for our study. Taken together, TTT-3002 and LRRK2-IN1 had no effect on neurodegeneration in this model.
Figure 7.
TTT-3002 and LRRK2-IN1 have no effect on neurodegeneration induced by exposure to 6-OHDA or overproduction of DA. (A) GFP worms were treated with different concentrations of TTT-3002 and LRRK2-IN-1 from L1 to L3, exposed to 6-OHDA for 1 h, and re-treated with TTT-3002 and LRRK2-IN1 for another 24 h. The DA neurons were monitored from 0 h (L3), 24, 48 and 72 h after 6-OHDA exposure. No significant improvement of DA neuron survival in TTT-3002 and LRRK2-IN1 treatment groups were observed. (B) The CAT-2 overexpressing C. elegans were treated with different concentrations of TTT-3002 and LRRK2-IN-1 from L1–L4. CAT-2 overexpression generates excessive dopamine and leads to DA neuron degeneration in C. elegans on adult Day 4. The DA neurons were monitored at L4 stage and on adult Day 4. No change in DA neuron survival was observed between vehicle controls and groups treated with TTT-3002 or LRRK2-IN1.
We next evaluated neurodegeneration induced by overproduction of DA in transgenic worms overexpressing CAT-2 (the C. elegans homolog of tyrosine hydroxylase) in DA neurons. The transgenic CAT-2 overexpression line exhibited marked neurodegeneration on adult Day 4 (Fig. 7B), consistent with previous results (17). Treatment with TTT-3002 and LRRK2-IN1 showed no protective effect on these worms.
We also treated the GFP control worms with inhibitors during L1–L4 and monitored DA neurons in old animals on adult Day 9 (Supplementary Material, Fig. S4). The GFP worms showed about 20% loss of DA neurons on adult Day 9 due to aging in vehicle-treated controls. TTT-3002 and LRRK2-IN1 had no protective effect on DA neuron survival, indicating that these LRRK2 inhibitors do not affect DA neuron integrity in normal animals. We also observed that TTT-3002 and LRRK2-IN1 had no effect on lifespan in either worms expressing GFP marker alone or G2019S-LRRK2 (Supplementary Material, Fig. S5), consistent with the fact that DA neurons per se in worms are dispensable for organism survival. Since changes in signaling through post-synaptic dopamine receptors DOP-1 and DOP-3 expressed in other neurons can also affect the basal slowing response of the worms (25), we tested whether rescuing effects of TTT-3002 and LRRK2-IN1 on R1441C-induced behavioral deficits depend on DOP-1 or DOP-3. We generated R1441C transgenic worms in the dop-1 null background, and assayed effects of TTT-3002 and LRRK2-IN1 in this model. As shown in Supplementary Material, Figure S6, loss of DOP-1 did not change the neurotoxicity of R1441C, and the LRRK2 inhibitors remained effective in rescuing the behavioral deficits in the dop-1/R1441C worms. In addition, the LRRK2 inhibitors did not alter the basal slowing response in either dop-1 or dop-3 mutant worms when compared with the vehicle controls. Therefore, it seemed unlikely that TTT-3002 and LRRK2-IN1 rescued R1441C-induced behavioral deficits through a DOP-1/DOP-3-dependent mechanism. Finally, the C. elegans lines expressing R1441C, G2019S, R1441C/A2016T and G2019S/A2016T all displayed prominent dopaminergic behavioral deficits that can be symptomatically rescued with exogenous L-DOPA (Supplementary Material, Fig. S7; and data not shown). Hence, they are useful models for studying LRRK2-relevant pathogenesis and for screening small molecules with neuroprotective effects. Taken together, these results indicate that TTT-3002 and LRRK2-IN1 do not affect DA neurodegeneration induced by factors other than LRRK2, which is consistent with the interpretation that TTT-3002 and LRRK2-IN1 specifically target LRRK2 kinase activity to confer neuroprotection.
DISCUSSION
In the present study, we take advantage of potent and selective LRRK2 inhibitors to interrogate LRRK2 kinase activity and elucidate its role in dopaminergic neurodegeneration caused by different LRRK2 mutations using C. elegans models. We have been able to pharmacologically rescue the pathological phenotype manifested by the expression of mutant LRRK2 G2019S and R1441C in vivo. Although it is still being debated whether and how mutations outside kinase domain such as R1441C affect the intrinsic LRRK2 kinase activity (12), the results from our study are consistent with a toxic ‘gain of function’ mechanism for G2019S- and R1441C-linked PD, likely due to aberrant kinase activity that can be suppressed by select small molecule inhibitors.
We have shown that both TTT-3002 and LRRK2-IN1 potently inhibit in vitro kinase activities of LRRK2 wild-type, R1441C and G2019S at nanomolar concentrations, and have confirmed their ability to inhibit endogenous LRRK2 in a cellular environment based on detection of S935 phosphorylation of LRRK2 at potencies that reflect those determined in the enzymatic assays. The inhibitory effects of TTT-3002 and LRRK2-IN1 on LRRK2 kinase activity also correlated well with their protective effect on neurotoxicity induced by transfected G2019S-LRRK2 in SH-SY5Y neuroblastoma cells. Furthermore, we have evaluated the effects of both inhibitors on dopaminergic behavioral and neurodegenerative phenotypes in our previously established transgenic C. elegans lines overexpressing G2019S- and R1441C-LRRK2. TTT-3002 and LRRK2-IN1 prevented and rescued G2019S- and R1441C-linked behavioral abnormality when treated during both pre- and post-symptomatic stages, and arrested DA neurodegeneration in aged G2019S- and R1441C-LRRK2 transgenic C. elegans. In all of these studies, TTT-3002 was typically about 10-fold more potent than LRRK2-IN1. Various control experiments indicated that the rescue of behavior abnormality and DA neurodegeneration in R1441C and G2019S transgenic lines are not the result of non-specific or off-target effects of either inhibitor: (i) TTT-3002 and LRRK2-IN1 were much less effective in rescuing phenotypes of inhibitor-resistant R1441C/A2016T and G2019S/A2016T C. elegans lines, suggesting that both inhibitors directly target LRRK2. Like the R1441C and G2019S transgenic worms, R1441C/A2016T and G2019S/A2016T displayed dopaminergic defects that could be rescued by treatment with exogenous L-DOPA. Therefore, the R1441C/A2016T and G2019S/A2016T models are highly relevant in testing the specificity of LRRK2 inhibitors and (ii) both inhibitors had no effect on DA neuron survival of wild-type animals expressing GFP marker alone, or on neurodegeneration caused by factors other than mutant LRRK2. These results suggest that kinase activity is critical for both R1441C- and G2019S-linked pathogenesis in C. elegans, and support the notion that pharmacological inhibition of LRRK2 may have disease-modifying effects on PD associated with aberrant LRRK2-dependent signaling. TTT-3002 is particularly promising for further evaluation in mammalian models of PD as it can be dosed orally in mice to sustain brain levels of 300–400 nm over a period of several months without notable toxicity or any other detectable adverse effects on behavior and learning and memory (H. Roder, unpublished data).
Our liquid-culture treatment regimens developed in the present study have allowed for sensitive and quantitative assessment of in vivo efficacies and kinetics of rescue by LRRK2 inhibitors at different stages of development and manifestation of neurodegenerative phenotypes in C. elegans models. Consequently, we are able to make several observations that reveal interesting differences in efficacy and pharmacodynamics of different LRRK2 inhibitors, and in sensitivity of transgenic C. elegans carrying different LRRK2 mutations toward compound treatments. First, the potencies of both TTT-3002 and LRRK2-IN1 in the C. elegans models (Fig. 4 and Table 2) were lower than in the in vitro enzyme assays (Fig. 1 and Table 1) or in the cellular model systems (Figs 2 and 3). It is possible that the availability of the inhibitors to its target (mutant LRRK2) expressed in DA neurons of C. elegans is limited by tissue penetration and other regulatory or metabolic mechanisms, which also affect whole-animal pharmacology in mammals. Second, TTT-3002 was not only more potent but also had a remarkably steeper dose–response curve than LRRK2-IN1 across the treatment regimens and in both R1441C and G2019S transgenic C. elegans lines (Fig. 4 and Table 2). It is also noteworthy that increasing concentrations are required the later the treatment is administered, more so for the less potent LRRK2-IN1 than for TTT-3002. These results correlate with the better in vitro potency of TTT-3002 but also suggest higher affinity of TTT-3002 for LRRK2 proteins than that of LRRK2-IN1. Unexpectedly, transgenic worms expressing R1441C were dramatically more sensitive to either inhibitor in rescue of behavior deficit (Fig. 4) and neurodegeneration (Fig. 5) than those expressing G2019S, with EC50 values differing across treatment regimens by 20- to 100-fold for TTT-3002 and 2- to 5-fold for LRRK2-IN1, respectively (Table 2). This is intriguing considering that in vitro kinase assays revealed similar potency of inhibition toward R14411C- and G2019S-LRRK2 by either TTT-3002 or LRRK2-IN1 (Table 1). Conceivably, there could be several explanations for the observed differences in the response of R1441C- and G2019S transgenic worms to treatment by LRRK2 inhibitors. Considering the high affinity of TTT-3002 and LRRK2-IN1 for LRRK2, their apparent EC50 values would become dependent on LRRK2 concentration at levels approaching or exceeding the dissociation constant (Ki). Accordingly, the two inhibitors, especially the more potent TTT-3002, would display different apparent potencies that correlate with the expression levels of R1441C- and G2019S-LRRK2 proteins in the respective transgenic C. elegans models. However, we consider this scenario less likely since relative LRRK2 mRNA levels in transgenic R1441C and G2019S worms do not correlate directly with their sensitivities to the inhibitors (Supplementary Material, Fig. S2). Another possibility is that binding of the different LRRK2 inhibitors to the R1441C- or G2019S-LRRK2 proteins may induce different conformational changes in the mutant proteins and differentially affect downstream signaling. Consistent with the concept of differential conformation in LRRK2 mutants, it was previously reported that the R1441C mutation alters the folding properties of the GTPase domain of LRRK2 and also lowers the thermodynamic stability in vitro (26). Adding to the complexity of the interpretation is the observation that LRRK2 proteins can undergo dimerization and association with membrane structures and likely with other proteins (27–32), which may modify local concentrations of LRRK2 proteins. However, it is unknown whether these interactions would be impacted differentially by the different LRRK2 mutations and LRRK2 inhibitors. Finally, different degrees of inhibition may be required to rescue the equivalent phenotype caused by the R1441C and G2019S mutations. Further research is clearly required to resolve these issues. Nevertheless, our pharmacological approach that integrates different treatment regimens and sensitive phenotypic assays, combined with genetic models with an engineered mutation affecting drug sensitivity should be generally applicable to screening and characterization of other bioactive compounds in C. elegans models.
In summary, our study has provided strong evidence that specific inhibition of LRRK2 kinase activity can protect DA neurons from degeneration induced by pathogenic LRRK2 mutations. Our findings on the anti-neurodegenerative potential of LRRK2-selective inhibitors should encourage further validation of this therapeutic strategy in mammalian models, and ultimately clinical evaluation in PD.
MATERIALS AND METHODS
Chemicals
Small molecule kinase inhibitors used were LRRK2-IN1 (16), obtained from Michael J. Fox Foundation for Parkinson's Research; TTT-3002, supplied by TauTaTis, Inc. (San Diego, CA, USA); and H-1152, purchased from Calbiochem. All other chemicals were from Sigma unless otherwise indicated.
Determination of in vitro IC50 and kinase selectivity
We used the ADP-Glo kinase assay kit (Promega) to determine the IC50 values for LRRK2 inhibitors. All inhibitors were stored at −80°C as 10 mm stocks dissolved in dimethyl sulfoxide (DMSO). Prior to each assay, they were diluted with Kinase Buffer A (Invitrogen), with the final DMSO concentration kept at 1% for all samples. Other assay components were also prepared in Kinase Buffer A. Assays were conducted in triplicate in separate wells of a Corning 384-well low-volume white plate in a final volume of 5 μl, containing 8 nm recombinant LRRK2 wild-type [GST-LRRK2 (970–2547), Invitrogen], 100 μm LRRKtide (LRRK2 peptide substrate, Invitrogen), and varying concentrations of inhibitors (LRRK2-IN1, TTT-3002 and H-1152). The vehicle control contained 1% DMSO in place of inhibitor. The reactions were initiated with 100 μm ATP (Promega), and incubated at room temperature (22°C) for 1 h. The reactions were quenched and detection of the ATP turnover was accomplished by using the ADP-Glo system according to the manufacturer's instructions (Promega). Briefly, 5 μl of ADP-Glo reagent was added to each well and incubated for 40 min. Then, 10 μl of kinase detection substrate was added to each well and incubation was continued for an additional 30 min at room temperature (22°C). Luminescence was measured with a TECAN Genios plate reader. Analogous reactions were conducted with recombinant G2019S-LRRK2 or R1441C-LRRK2 (Invitrogen).
Independent assays for inhibitory potency of LRRK2-IN1 for LRRK2 wild-type and G2019S, but not R1441C, as well as selectivity profiling of LRRK2-IN1 against many different kinases were reported previously (16). Kinase selectivity profiling for TTT-3002 was described in the Supplemental Experimental Procedures.
Detection of S935 phosphorylation of cellular LRRK2
Immortalized primary human lymphoblastoid cells propagated from leukocytes from one PD patient carrying the G2019S mutation were obtained from the Coriell Institute for Medical Research (Camden, NJ). Human lymphoblastoid cells were maintained in RPMI 1640 with 10% FBS, 2 mm glutamine, 1× penicillin/streptomycin solution (Invitrogen) at a cell density of 1 × 106 cells per milliliter. Two million cells were plated into each well of a six-well plate, with 5 ml medium per well. Various working solutions of LRRK2-IN1 and TTT-3002 (at 1000 times the final concentrations) were made by serial dilutions with water from stock solutions made in DMSO and adjusting the DMSO content to 10%. Vehicle controls were made by diluting DMSO with water in a parallel fashion. A volume of 2 μl of each diluted inhibitor solution was added to 2 ml of the culture medium in separate wells of the culture plate; and equivalent volumes of the corresponding vehicle controls were added to the medium in a parallel set of wells. The final concentration of DMSO for all groups was 0.01%. Cells were incubated at 37°C for 1.5 h in 5% CO2. Cells were then washed with PBS and collected by brief centrifugation. Cells were lysed on ice for 15 min after adding 300 μl of ice-cold buffer containing PBS, 1% Triton X-100 and a cocktail of protease and phosphatase inhibitors (Roche). Cell lysates were centrifuged at 16 000g for 15 min at 4°C and the supernatant was collected for immediate use or storage at −80°C. Human THP-1 monocyte cells were differentiated into macrophages by treatment with 400 nm phorbol 12-myristate 13-acetate for 24 h. THP-1 macrophage cells were then stimulated with human interferon-gamma (100 ng/ml) for 48 h to induce expression of endogenous LRRK2 (18), followed by treatment with LRRK2 kinase inhibitors as described earlier. Cleared cell lysates from human lymphoblastoid cells and THP-1 macrophages were resolved on 7.5% SDS–PAGE gels and transferred to PVDF membranes for western blot analysis of LRRK2 phosphorylated at S935 (33) using a rabbit monoclonal antibody against phospho-serine 935 LRRK2 (clone UDD2, 1:6000; Epitomics). The blot was re-probed with a mouse monoclonal antibody against LRRK2 amino acids 970–2527 (clone N241A/34, 1:15 000; NeuroMab) to detect total LRRK2. Western blots were developed with enhanced ECL reagents (Amersham) on X-ray films, which were scanned into image files and analyzed for intensities of protein bands using ImageJ.
LRRK2 immunostaining, cell toxicity and rescue assay
SH-SY5Y neuroblastoma cells were cultured in Opti-MEM (Invitrogen) with 10% fetal bovine serum (FBS, Atlanta Biologicals) and 1% penicillin (Invitrogen) and incubated in 5% CO2 at 37°C. One day prior to transfection, SH-SY5Y cells were split and placed into six-well plates at a density of 3 × 105/ml in 2 ml of medium per well. SH-SY5Y cells were transfected with pCEP4 empty vector (Invitrogen), pCEP4 WT-LRRK2-FLAG and pCEP4 G2019S-LRRK2-FLAG (34), all mixed with PCMV-GFP (Addgene) at 15:1 ratio. The culture was changed to serum-free medium 1 h before transfection and to complete medium 6 h post-transfection. At 48 h post transfection, cells were fixed with freshly made 4% paraformaldehyde for 30 min. Cells were permealized with phosphate-buffered saline (PBS) containing 0.2% Triton X-100 on ice for 10 min and were blocked with 3% BSA for 1 h at room temperature. Cells were then incubated with 1:100 diluted rabbit mAb primary anti-LRRK2 (clone UDD3, Epitomics) at 4°C overnight, washed four times (each for 15 min) in PBS and incubated with 1:300 diluted Alexa Fluor488 goat anti-rabbit antibody for 2 h at room temperature in the dark. Images were taken by using Zeiss Axiovert 200M microscope with 1.5 s exposure time at 20 or 40× magnifications. LRRK2 inhibitors and vehicle controls were added to the cell medium from the start of transfection until image collection. GFP positive cell images were captured from 40 randomly selected fields 12 and 48 h post transfection. Viable cells were quantified using Hoechst 33 342 stain added to each well at 12 and 48 h post-transfection and images of 40 random fields for each well were captured using a fluorescence microscope with the GFP and DAPI channels. GFP and corresponding DAPI images were superimposed and GFP positive viable cells were counted as having intact nuclei with dispersed chromatin versus total GFP positive cells.
Nematode strains and maintenance
Transgenic C. elegans lines expressing green fluorescent protein (GFP) either alone or together with G2019S- or R1441C-LRRK2 specifically in dopaminergic neurons using the promoter of dopamine transport dat-1 have been described previously (17). As all of the transgenic lines carry GFP as the fluorescent marker for DA neurons, they were simply designated (with transgene specified in parentheses) as the GFP only line (Pdat-1::GFP), G2019S line (Pdat-1::G2019S-LRRK2; Pdat-1::GFP) and R1441C line (Pdat-1::R1441C-LRRK2; Pdat-1::GFP), respectively. The A2016T inhibitor-resistant mutation of LRRK2 (13) was introduced into pFXneEGFP plasmid containing Pdat-1::R1441C (17) by site-directed mutagenesis using QuikChange II XL kit (Stratagene), generating a Pdat-1::R1441C/A2016T construct. The Pdat-1::G2019S/A2016T construct was made by subcloning the insert from pCMV-FLAG-LRRK2 G2019S/A2016T (13) into pFXneEGFP plasmid containing Pdat-1 using BamHI and NotI sites. All constructs were confirmed by DNA sequencing. For generating transgenic C. elegans lines expressing LRRK2 dual mutant R1441C/A2016T or G2019S/A2016T in dopaminergic neurons, young adult C. elegans hermaphrodites with lin-15(765ts) allele (strain MT1898) were injected with a mixture of DNA constructs consisting of Pdat-1::GFP (20 ng/μl) together with Pdat-1::R1441C/A2016T or Pdat-1::G2019S/A2016T (20 ng/μl), as well as lin-15+ selection marker (50 ng/μl; pJM23, a gift from L. Avery, University of Texas Southwestern Medical Center) and an empty vector pSL1180 (20 ng/μl; Amersham). To construct dop-1/R1441C C.elegans lines, a mixture of Pdat-1::R1441C (20 ng/ul), Pdat-1::GFP (20 ng/ul) and an empty vector pBluescript (70 ng/ul) were injected into C. elegans with a dop-1 null allele (vs101). Stable extra-chromosomal transgenic lines were selected based on the absence of the lin-15 Muv phenotype and presence of both GFP marker and LRRK2 transgene, as previously described (17). Corresponding lines with chromosomally integrated transgene were made by treatment with UV and trimethyl psoralen, followed by backcrossing with the wild-type strain (N2 Bristol) for at least three times. At least three C. elegans lines were characterized for each genotype, and data from a representative line were presented. The CAT-2 overexpression line (Pdat-1::CAT-2; Pdat-1::GFP, strain UA57, generated by G. Caldwell, University of Alabama) was obtained from the Caenorhabditis Genetics Center (CGC). A list of C. elegans strains used in this study is described in Supplementary Material, Table S3.
C. elegans strains were cultured on standard nematode growth medium (NGM) agar plates seeded with E. coli OP50 as a food source according to established protocols (35). NGM was made with Bacto agar and Bacto peptone from BD Biosciences and other chemicals from Sigma. Mixed stage animals were maintained as bulk culture on NGM agar at room temperature (22°C). Prior to each experiment, animals were age-synchronized by standard bleaching and washing protocol to obtain embryos, from which developmental stages were followed.
Behavioral assay
Well-fed worms with intact dopaminergic neural circuitry move slower in the presence of bacterial food than in its absence. This basal slowing response (or food-sensing response) is solely controlled by endogenous dopamine levels and was assayed as described (22). Briefly, food-containing assay plates were prepared by spreading the bacterial food, E. coli (OP50), in a ring on NGM agar, and together with non-coated assay plates, were incubated at 37°C overnight and cooled to room temperature prior to assay. Age-synchronized worms (about 10 worms of each strain) were transferred from their food-containing culture plate to a non-food plate and washed twice in S basal buffer (100 mm NaCl, 10 μg/ml cholesterol, 50 mm potassium phosphate, pH 6.0) to get rid of the remaining food on their bodies. Worms were then transferred to the food-containing or non-food assay plates prepared as described earlier, settled for 5 min, and body bends in 20 s intervals were recorded for each of the worms.
L-DOPA treatment
C. elegans to be tested was placed on NGM agar plates containing 20 mm 3,4-dihydroxyphenylalanine (L-DOPA) for 6 h prior to assay for basal slowing response.
Neurodegeneration assay
Degeneration of dopamine (DA) neurons in live C. elegans was analyzed essentially as described (36,37). Briefly, age-synchronized worms were paralyzed by treatment with 3 mm levamisole and mounted on glass slides. The DA neurons in the head regions [four cephalic (CEP) neurons and two anterior dierdic (ADE) neurons] were visualized for GFP fluorescence under a Zeiss Axiovert 200M microscope and monitored at different ages (L4/adult Day 0 through Day 9). The total numbers of CEPs and ADEs were scored for the presence of intact neurons. DA neurons missing most of the cell body and neurites were counted as degenerated. For each strain, about 30 worms were analyzed in at least three independent experiments, with a total of ∼90–120 animals scored in the same manner. The percent of intact neurons survived was calculated as number of intact ADE/CEP neurons observed in all animals divided by total number of ADE/CEP neurons expected if no degeneration occurred (six in each animal times the number of animals tested), times 100. Fluorescent images of DA neurons in the head region of worms were taken with a Zeiss Axiovert 200M microscope using 1 s exposure time at 20× magnification.
Time course of worm treatment with LRRK2 inhibitors
Worm treatment with LRRK2 inhibitors was done in liquid culture to achieve better drug exposure. For treatment of C. elegans, stage L1–L4, with LRRK2 inhibitors, young adult worms were age-synchronized by soaking in bleach and sodium hydroxide to destroy worms but not eggs; the resulting eggs were left in 1 ml M9 buffer [For 1 l: KH2PO4, 3 g; Na2HPO4, 6 g; NaCl, 5 g; MgSO4 (1 m), 1 ml] and shaken at 100 r.p.m. at room temperature overnight to generate synchronized L1 worms. The M9 buffer containing L1 worms was distributed into 12-well plates together with E. coli OP50 to make a total volume of 900 μl (300 μl M9 + 600 μl OP50) in each well. Each well contained roughly 100 L1 worms. LRRK2 inhibitors were added to achieve the desired concentrations. The 12-well plates were maintained in a water chamber at room temperature and shaken at 100 r.p.m. Worms were monitored every day and placed onto agar plates with OP50 when most of them reached L4 stage, according to morphological assessment (roughly 60 h). L4 worms were picked, settled in agar plates with OP50 for 48 h and tested by the behavioral assay as described earlier. For treatment of worms at stage L4 to Day 2 with LRRK2 inhibitors, synchronized L1 worms were left growing on agar plates with OP50 until reaching L4 stage. L4 worms were washed with M9 buffer and distributed into 12-well plates together with OP50 and LRRK2 inhibitors (each well contained ∼50 worms). The plates were left in a water chamber at room temperature (22oC) and shaken at 100 r.p.m. for 48 h and then the worms were transferred onto agar plates with OP50 and settled for 24 h for the food sensing test. For treatment from adult Day 2 to Day 3 and Day 5 to Day 7, synchronized worms at Day 2 and Day 5 were treated essentially the same as for L4 to Day 2 treatment, except Day 2 to Day 3 treatment only lasted 24 h in liquid culture.
6-OHDA treatment
For testing whether the LRRK2 inhibitors afforded protection from 6-OHDA-induced DA neuron death in C. elegans, synchronized L1 Pdat-1::GFP worms were treated in liquid culture for 36 h with LRRK2 inhibitors, or vehicle control, as described earlier until they reached the L3 stage. The L3 worms were collected into 1.5 ml tubes and centrifuged at 1000 r.p.m. to remove supernatant. Five hundred microliters of 50 mm 6-OHDA and 10 mm ascorbic acid were added to each tube and mixed gently every 15 min for 1 h at room temperature (22°C). The worms were then washed with M9 three times and re-placed into liquid culture with E. coli OP50 and LRRK2 inhibitors for another 24 h. After that, worms were transferred onto agar plates with OP50 for monitoring DA neuron death.
Worm protein extraction, affinity pull-down and western blotting
Five 10 cm NGM plates full of adult transgenic worms expressing Flag-tagged G2019S- or R1441C-LRRK2 were used to obtain eggs by alkaline bleaching and subsequently age-synchronized L1 larva. Worms were grown in liquid culture until L4 stage, harvested and homogenized in ice-cold lysis buffer containing 0.5% Triton X-100 in 20 mm Tris, pH 7.5 supplemented with protease inhibitors (Roche). Total lysates were clarified by centrifugation at 16 000g for 15 min at 4°C. Affinity pull-down of Flag-tagged LRRK2 proteins was achieved by incubation of 20 mg lysate proteins with anti-Flag M2 mAb conjugated beads (Sigma). The beads were washed in lysis buffer and bound proteins were eluted with 2× SDS sample buffer heated at 95°C. Proteins were separated on 7.5% SDS–PAGE gels and western blotting was performed using anti-LRRK2 rabbit mAb (Epitomics, clone MJFF-2). An aliquot of lysate was analyzed by western blotting as an input control using anti-tubulin mouse mAb E7 (Developmental Studies Hybridoma Bank).
RT–PCR and quantitative PCR (qPCR)
Age-synchronized adult transgenic worms expressing GFP, G2019S-LRRK2 and R1441C-LRRK2 were picked individually into PCR tubes (one worm per tube) and washed twice in water. RNA was extracted from each worm using PicoPure RNA Isolation Kit (Invitrogen), followed by DNase treatment according to the manufacturer's instructions. First strand cDNA was synthesized from RNA using SuperScript II Reverse Transcriptase (Invitrogen). One-tenth of the synthesized cDNA was used as template for conventional PCR with primers specific to LRRK2 and an internal reference gene act-1 to amplify unique products for visualization on agarose gel. Another aliquot of cDNA was used as template for quantitative real-time PCR reactions with Fast SYBR Green Master Mix and appropriate primers using StepOne qPCR system (Applied Biosystems). The cycling was performed at 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and at 60°C for 1 min. Data were processed with StepOne software and analyzed using the comparative cycle threshold method. Relative LRRK2 mRNA levels in individual worms were calculated as the values normalized to the internal control gene act-1. Primer sequences (5′ to 3′) specific for human LRRK2 and worm act-1 genes were listed as follows:
LRRK2: forward-TTT GCT GGT TCC AAG CAG TT, reverse-ACA TTC TGT TTG GGC GAA GT;
act-1: forward-GCT GGA CGT GAT CTT ACT GAT TAC C, reverse-GTA GCA GAG CTT CTC CTT GAT GTC
Worm lifespan assay
Worms were treated with different concentrations of inhibitors for the L1–L4 duration in liquid culture and then transferred to OP50 agar plates and settled for 24 h. Fifteen post L4, Day 1 worms were transferred into one OP50 agar plate with 1.25 mg/ml 5-fluorodeoxyuridine. The worms were monitored every day until all worms were dead in each plate. The life span curves were generated by plotting percentage of surviving worms at each time point. Percent surviving was determined by scoring 10 plates of worms at each inhibitor concentration.
Statistics
Data were presented as mean ± SEM. Statistical significance of differences between results was analyzed using the Student's t-test for single comparisons or one-way ANOVA followed by the Turkey–Kramer post hoc test for multiple comparisons. A P-value <0.05 was considered significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by grants from the National Institutes of Health (S.G.C. and A.L.W.), Michael J. Fox Foundation for Parkinson's Research (S.G.C., A.L.W. and D.R.A.) and UK Medical Research Council (D.R.A.), and by a Department of Veterans Affairs Merit Review Grant (J.J.M.). Some strains were obtained from the Caenorhabditis Genetics Center, which is supported by National Center for Research Resources, NIH.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the anonymous reviewers for their helpful suggestions on the initial version of this manuscript.
Conflict of Interest statement: H.R. is Chairman and CEO, and a major shareholder of TauTaTis, Inc.
REFERENCES
- 1.Gandhi P.N., Chen S.G., Wilson-Delfosse A.L. Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson's disease. J. Neurosci. Res. 2009;87:1283–1295. doi: 10.1002/jnr.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cookson M.R. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldwurm S., Zini M., Di Fonzo A., De Gaspari D., Siri C., Simons E.J., van Doeselaar M., Tesei S., Antonini A., Canesi M., et al. LRRK2 G2019S mutation and Parkinson's disease: a clinical, neuropsychological and neuropsychiatric study in a large Italian sample. Parkinsonism Relat Disord. 2006;12:410–419. doi: 10.1016/j.parkreldis.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Smith W.W., Pei Z., Jiang H., Dawson V.L., Dawson T.M., Ross C.A. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 5.West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A., Dawson V.L., Dawson T.M. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl Acad. Sci. USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira J.J., Guedes L.C., Rosa M.M., Coelho M., van Doeselaar M., Schweiger D., Di Fonzo A., Oostra B.A., Sampaio C., Bonifati V. High prevalence of LRRK2 mutations in familial and sporadic Parkinson's disease in Portugal. Mov. Disord. 2007;22:1194–1201. doi: 10.1002/mds.21525. [DOI] [PubMed] [Google Scholar]
- 7.Di Fonzo A., Tassorelli C., De Mari M., Chien H.F., Ferreira J., Rohe C.F., Riboldazzi G., Antonini A., Albani G., Mauro A., et al. Comprehensive analysis of the LRRK2 gene in sixty families with Parkinson's disease. Eur. J. Hum. Genet. 2006;14:322–331. doi: 10.1038/sj.ejhg.5201539. [DOI] [PubMed] [Google Scholar]
- 8.Mata I.F., Taylor J.P., Kachergus J., Hulihan M., Huerta C., Lahoz C., Blazquez M., Guisasola L.M., Salvador C., Ribacoba R., et al. LRRK2 R1441G in Spanish patients with Parkinson's disease. Neurosci. Lett. 2005;382:309–311. doi: 10.1016/j.neulet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 9.Simon-Sanchez J., Marti-Masso J.F., Sanchez-Mut J.V., Paisan-Ruiz C., Martinez-Gil A., Ruiz-Martinez J., Saenz A., Singleton A.B., Lopez de Munain A., Perez-Tur J. Parkinson's disease due to the R1441G mutation in Dardarin: a founder effect in the Basques. Mov. Disord. 2006;21:1954–1959. doi: 10.1002/mds.21114. [DOI] [PubMed] [Google Scholar]
- 10.Gorostidi A., Ruiz-Martinez J., Lopez de Munain A., Alzualde A., Masso J.F. LRRK2 G2019S and R1441G mutations associated with Parkinson's disease are common in the Basque Country, but relative prevalence is determined by ethnicity. Neurogenetics. 2009;10:157–159. doi: 10.1007/s10048-008-0162-0. [DOI] [PubMed] [Google Scholar]
- 11.Criscuolo C., De Rosa A., Guacci A., Simons E.J., Breedveld G.J., Peluso S., Volpe G., Filla A., Oostra B.A., Bonifati V., et al. The LRRK2 R1441C mutation is more frequent than G2019S in Parkinson's disease patients from southern Italy. Mov. Disord. 2011;26:1733–1736. doi: 10.1002/mds.23735. [DOI] [PubMed] [Google Scholar]
- 12.Greggio E., Cookson M.R. Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro. 2009;1 doi: 10.1042/AN20090007. art:e00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols R.J., Dzamko N., Hutti J.E., Cantley L.C., Deak M., Moran J., Bamborough P., Reith A.D., Alessi D.R. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson's disease. Biochem. J. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B.D., Shin J.H., VanKampen J., Petrucelli L., West A.B., Ko H.S., Lee Y.I., Maguire-Zeiss K.A., Bowers W.J., Federoff H.J., et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat. Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z., Hamamichi S., Lee B.D., Yang D., Ray A., Caldwell G.A., Caldwell K.A., Dawson T.M., Smith W.W., Dawson V.L. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson's disease models. Hum. Mol. Genet. 2011;20:3933–3942. doi: 10.1093/hmg/ddr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X., Dzamko N., Prescott A., Davies P., Liu Q., Yang Q., Lee J.D., Patricelli M.P., Nomanbhoy T.K., Alessi D.R., et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat. Chem. Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao C., El Khoury R., Wang W., Byrd T.A., Pehek E.A., Thacker C., Zhu X., Smith M.A., Wilson-Delfosse A.L., Chen S.G. LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson's disease. Neurobiol. Dis. 2010;40:73–81. doi: 10.1016/j.nbd.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardet A., Benita Y., Li C., Sands B.E., Ballester I., Stevens C., Korzenik J.R., Rioux J.D., Daly M.J., Xavier R.J., et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J. Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maekawa T., Kubo M., Yokoyama I., Ohta E., Obata F. Age-dependent and cell-population-restricted LRRK2 expression in normal mouse spleen. Biochem. Biophys. Res. Commun. 2010;392:431–435. doi: 10.1016/j.bbrc.2010.01.041. [DOI] [PubMed] [Google Scholar]
- 20.Smith W.W., Pei Z., Jiang H., Moore D.J., Liang Y., West A.B., Dawson V.L., Dawson T.M., Ross C.A. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc. Natl Acad. Sci. USA. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M.P., Beilina A., Blackinton J., Thomas K.J., et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Sawin E.R., Ranganathan R., Horvitz H.R. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 23.Nass R., Hall D.H., Miller D.M., III, Blakely R.D. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2002;99:3264–3269. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y., Cao P., Smith M.A., Kramp K., Huang Y., Hisamoto N., Matsumoto K., Hatzoglou M., Jin H., Feng Z. Dysregulated LRRK2 signaling in response to endoplasmic reticulum stress leads to dopaminergic neuron degeneration in C. elegans. PLoS One. 2011;6:e22354. doi: 10.1371/journal.pone.0022354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chase D.L., Pepper J.S., Koelle M.R. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat. Neurosci. 2004;7:1096–1103. doi: 10.1038/nn1316. [DOI] [PubMed] [Google Scholar]
- 26.Li Y., Dunn L., Greggio E., Krumm B., Jackson G.S., Cookson M.R., Lewis P.A., Deng J. The R1441C mutation alters the folding properties of the ROC domain of LRRK2. Biochim. Biophys. Acta. 2009;1792:1194–1197. doi: 10.1016/j.bbadis.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen S., Webber P.J., West A.B. Leucine-rich repeat kinase 2 (LRRK2) kinase activity: dependence on dimerization. J. Biol. Chem. 2009;284:36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biskup S., Moore D.J., Celsi F., Higashi S., West A.B., Andrabi S.A., Kurkinen K., Yu S.W., Savitt J.M., Waldvogel H.J., et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 29.Gandhi P.N., Wang X., Zhu X., Chen S.G., Wilson-Delfosse A.L. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J. Neurosci. Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dachsel J.C., Taylor J.P., Mok S.S., Ross O.A., Hinkle K.M., Bailey R.M., Hines J.H., Szutu J., Madden B., Petrucelli L., et al. Identification of potential protein interactors of Lrrk2. Parkinsonism Relat Disord. 2007;13:382–385. doi: 10.1016/j.parkreldis.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J.M., Daniels V., Lewis P., Jain S., Ding J., Syed A., et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger Z., Smith K.A., Lavoie M.J. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry (Mosc) 2010 doi: 10.1021/bi100157u. , 49, 5511–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dzamko N., Deak M., Hentati F., Reith A.D., Prescott A.R., Alessi D.R., Nichols R.J. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14–3–3 binding and altered cytoplasmic localization. Biochem. J. 2010;430:405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo L., Gandhi P.N., Wang W., Petersen R.B., Wilson-Delfosse A.L., Chen S.G. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp. Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkowitz L.A., Hamamichi S., Knight A.L., Harrington A.J., Caldwell G.A., Caldwell K.A. Application of a C. elegans dopamine neuron degeneration assay for the validation of potential Parkinson's disease genes. J. Vis. Exp. 2008 doi: 10.3791/835. 17, pii: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao S., Gelwix C.C., Caldwell K.A., Caldwell G.A. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J. Neurosci. 2005;25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.