Abstract
The microbial community of a fermented molasses-fed sequencing batch reactor (SBR) operated under feast and famine conditions for production of polyhydroxyalkanoates (PHAs) was identified and quantified through a 16 S rRNA gene clone library and fluorescence in situ hybridization (FISH). The microbial enrichment was found to be composed of PHA-storing populations (84% of the microbial community), comprising members of the genera Azoarcus, Thauera and Paracoccus. The dominant PHA-storing populations ensured the high functional stability of the system (characterized by high PHA-storage efficiency, up to 60% PHA content). The fermented molasses contained primarily acetate, propionate, butyrate and valerate. The substrate preferences were determined by microautoradiography-FISH and differences in the substrate-uptake capabilities for the various probe-defined populations were found. The results showed that in the presence of multiple substrates, microbial populations specialized in different substrates were selected, thereby co-existing in the SBR by adapting to different niches. Azoarcus and Thauera, primarily consumed acetate and butyrate, respectively. Paracoccus consumed a broader range of substrates and had a higher cell-specific substrate uptake. The relative species composition and their substrate specialization were reflected in the substrate removal rates of different volatile fatty acids in the SBR reactor.
Keywords: polyhydroxyalkanoates (PHA), mixed microbial cultures (MMC), culture selection, sequencing batch reactor (SBR), substrate preferences, microautoradiography-fluorescence in situ hybridization (MAR-FISH)
Introduction
Polyhydroxyalkanoates (PHAs) are biologically synthesized polymers that have gained increasing attention for their possible use as eco-efficient bioplastics. These intracellularly stored polyesters are both bio-based and biodegradable, allowing for a closed loop carbon cycle. Furthermore, they display a wide range of thermoplastic properties, thus providing a high technical replacement potential versus conventional oil-based plastics (Crank and Patel, 2005). The implementation of PHAs as commodity plastic material has so far been limited by their high production cost, given that, commercially available PHAs are produced by pure microbial culture fermentations (with high operating costs) and from raw materials with high market price (Choi and Lee, 1997; Crank and Patel, 2005). Consequently, finding less expensive feedstocks are decisive for the development of economically effective PHA-production processes.
Wastewaters and industrial effluents have been suggested as an economically viable and environmentally sustainable alternative to the current industrial PHA-production processes (Gurieff and Lant, 2007), because PHA storage has been demonstrated to have a physiological role for microorganisms in biological wastewater treatment. Enhanced PHA storage was found in activated sludge systems subjected to alternate carbon substrate availability, also designated as feast and famine (FF) (Majone et al., 1996; van Loosdrecht et al., 1997). Under FF, microorganisms that can rapidly store the substrate have a competitive advantage. Thus, imposing FF conditions allows for the selection of cultures enriched in organisms with enhanced PHA-storage capacity.
PHA production by mixed microbial cultures from waste or surplus-based feedstocks is usually carried out in a three-stage process, comprising: (1) acidogenic fermentation, where the organic content of the feedstock is biologically converted into volatile fatty acids (VFAs) under anaerobic conditions; (2) culture selection, typically carried out in sequencing batch reactors (SBRs) operated under FF conditions; and (3) PHA production, designed to maximize PHA accumulation in cells harvested from the enrichment bioreactor.
The key to optimizing mixed microbial culture-PHA-production processes is the maximization of the selective pressure imposed on culture enrichment. Extensive research has been carried out on the impact of different SBR-operating conditions (Dias et al., 2006; Serafim et al., 2008). However, sufficient knowledge on all factors governing microbial competition has not yet been gathered to support comprehensive mathematical models that allow the design of a strategy able to couple a high selective pressure for PHA storage with a high biomass-production rate. Indeed, both factors are crucial to maximize the productivity of the process.
Recent studies (Johnson et al., 2009; Jiang et al., 2011a, 2011b) demonstrated the possibility to obtain highly selected enrichments, through the use of FF SBRs operated under highly controlled conditions (chemically defined media with only one or two carbon sources) and using a low number of cycles per sludge retention time (SRT). The cultures thus selected had very narrow community structures (virtually monocultures) and were capable of reaching very high PHA contents (84–92% PHA cell content). These results show that the imposition of a high selective pressure through a low feast-to-famine length ratio and a low number of cycles per SRT will select microorganisms based on their efficiency to store PHA. However, because these studies used only a limited number of carbon sources, it cannot be anticipated whether the same high FF-selective pressure would yield the same culture simplification if in the presence of multiple substrates. Because the mixed culture-PHA-production process is only economically viable when using VFA-rich feedstocks arising from wastewater treatment processes (for example, fermented industrial effluents) containing numerous carbon sources, it is important to assess not only the impact of the FF-selective pressure but also the impact of this pressure when combined with the presence of multiple substrates.
In order to encompass the goal of population selection and that of biomass production, it is necessary to better understand the relation between factors influencing microbial selection and the resultant community structure. Substrate composition is one of the major factors influencing the microbial population structure and performance of PHA-production systems. The relationship between feed composition and microbial community selection is likely conditioned by the specific carbon source preferences of each population, which is ultimately reflected on the overall substrate-uptake performance observed in the SBR. The aim of this study was therefore to investigate the effect of mixed substrates on a PHA-storing community, by assessing the carbon source preferences of the PHA-storing organisms, and, in the long run, on microbial community changes when variations in the VFA composition in the feed took place.
In a previous study, Albuquerque et al. (2010a) operated a FF SBR fed with fermented molasses, comprising four VFAs: acetate, propionate, butyrate and valerate. The optimization of this system resulted in a microbial enrichment able to reach a PHA cell content of 78%. In the present study, the microbial community selected in the same system, after undergoing oscillations in the substrate composition, was characterized through clone library studies and quantitative fluorescence in situ hybridization (FISH) in order to identify the dominant microbial groups, particularly those with PHA-storage properties. The link between the phylogenetic classification of the different populations and their functionality was done through microautoradiography (MAR)-FISH, which also enabled detection of the substrate preference of each of the probe-defined populations. To the best of our knowledge, this last aspect has never been reported for PHA-producing mixed culture systems.
Materials and methods
Experimental setup
The experimental setup consisted of three bench-scale reactors and a hollow fiber membrane filtration module (Albuquerque et al., 2007). The molasses acidogenic fermentation (stage 1) was carried out in a continuous stirred tank reactor operated under anaerobic conditions. The conditions used to operate the acidogenic continuous stirred tank reactor are described in detail in Albuquerque et al. (2007). The fermented molasses was clarified by microfiltration and used as a feedstock for subsequent culture selection and PHA accumulation. Selection of a PHA-accumulating culture (stage 2) was carried out in a SBR subjected to FF conditions. PHA accumulation (stage 3) took place in a batch reactor inoculated with sludge from the culture-enrichment SBR and fed with clarified fermented molasses. The SBR was inoculated with a PHA-accumulating mixed culture acclimatized to the fermented molasses feedstock (Albuquerque et al., 2010a).
Culture selection in SBR
A SBR (working volume of 800 ml) was operated using the conditions reported in Albuquerque et al., 2010a: 10-day SRT, 1-day hydraulic retention time (HRT), 12 h cycle length, organic loading rate of 90 Cmmol VFA l−1 d−1. The SBR 12-h cycles consisted of four discrete periods: fill (5 min); aerobiosis (FF) (11 h); settling (45 min); and draw (10 min). The SBR was fed with clarified fermented molasses produced in stage 1 (200 ml per cycle) at an initial SBR substrate concentration of 45 Cmmol l−1 VFA, which corresponded to 80–85% of the soluble chemical oxygen demand. A mineral nutrient solution containing both ammonia (NH4Cl) and phosphate (KH2PO4) was also added during the fill phase (200 ml per cycle) so as to make up initial reactor concentrations of 3.75 mmol N l−1 and 0.48 mmol P l−1, thereby keeping the C/N/P molar ratios at 100/8/1. Thiourea was added to inhibit nitrification. At the end of the reaction phase, a purge of mixed liquor (40 ml) was withdrawn in order to keep the SRT at 10 days. Following the settling phase, the exhaust supernatant was withdrawn (400 ml per cycle). Air was supplied through a ceramic diffuser. Stirring was kept at 400 r.p.m. pH was left uncontrolled. Pumping (fill and draw), aeration and mixing were automatically controlled by a software program. In addition, the software was also used to acquire pH and dissolved oxygen data. The reactor was placed in a temperature-controlled room (23–25 °C).
Batch PHA-production assays
PHA-accumulation assays were carried out to determine kinetic and stoichiometric PHA-storing parameters of the SBR-selected culture. These assays were carried out in batch mode, feeding the clarified fermented molasses produced in stage 1 to SBR excess sludge (that is, without nutrient suplementing). Air was supplied through a ceramic diffuser and mixing was provided by magnetic stirring (400 r.p.m.). Feed pH was adjusted to 8 before reactor feeding and the pH in the reactor was left uncontrolled during the reaction phase. Temperature was kept at 23–25 °C.
The sludge PHA content is given as a percentage of volatile suspended solids (VSSs) on a mass basis. Active biomass (X) is defined as the VSS concentration minus the intracellular PHA content. The maximum specific substrate uptake (−qS in Cmol VFA/(Cmol X.h)) and PHA-storage rates (qP in Cmol PHA/(Cmol X.h)) are specific rates of substrate consumed or PHA stored per amount of active biomass per time. The yields of PHA (YP/S in Cmol PHA/Cmol VFA) and active biomass (YX/S in Cmol X/Cmol VFA) on substrate consumed are respectively defined as the ratios between the amount of product produced (either PHA or active biomass) and the amount of substrate consumed. These stoichiometric and kinetic parameters were calculated as described in Albuquerque et al. (2010a, 2010b).
Analytical procedures
Biomass concentration was determined using the VSS procedure described in the standard methods (APHA, 1998).
Volatile fatty-acid (acetate, propionate, butyrate and valerate) concentrations were determined by high-performance liquid chromatography using a Merck–Hitachi chromatographer (Merck–Hitachi, Manchester, UK) equipped with a refractive index detector and Aminex HPX-87 H pre-column (Aminex, Hercules, CA, USA) and column from Bio-Rad (Hercules, CA, USA). Sulphuric acid 0.01 M was used as an eluent at a flow rate of 0.6 ml min−1 and 50 °C. The organic-acid concentrations were calculated using a standard curve of 25–1000 mg l−1.
PHAs were determined by gas chromatography using the method described in Albuquerque et al. (2010b). Hydroxybutyrate (HB) and hydroxyvalerate (HV) concentrations were calculated using the P(HB-HV) (88%/12%) (Sigma) standards and corrected using an heptadecane internal standard.
Ammonia concentration was determined using an ammonia gas-sensing combination electrode ThermoOrion 9512 (ThermoOrion, Karlsruhe, Germany). A calibration curve was obtained with the NH4Cl standards (0.01–10 mmol N l−1).
Microbial characterization
16 S rRNA gene cloning and sequencing
Bacterial DNA was extracted from SBR biomass using the Ultraclean Soil DNA isolation kit (MO BIO Laboratories Inc., Carlsbad, CA, USA), following the manufacturer's instructions. DNA quantification was performed by spectrophotometric readings at 260 nm (NanoDrop 1000, ThermoScientific, Rockford, IL, USA). Bacterial 16 S rRNA gene fragments were amplified by PCR using primers 27f and 1492rBac (Lane, 1991). The reaction mixture (25 μl) contained 2 ng μl−1 of template DNA, 1 mℳ MgCl2, 1 × PCR buffer, 2.2 mℳ of each dNTP, 2.2 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) and 1 μℳ of each primer. The cycle program comprised: 5 min at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 48 °C and 2 min at 72 °C, followed by a final extension at 72 °C for 5 min. The PCR products were cleaned with the QIAquick PCR Purification Kit (Qiagen GmbH, Hilden, Germany) before proceeding with cloning.
Cloning was performed using the pGem-T easy vector system I (Promega, Madison, WI, USA) and the JM109 High efficiency competent cells (Promega). The 16 S rRNA gene fragments from selected clones (71) were amplified and grouped into operational taxonomic unit (OTU) according to restriction fragment length polymorphism analysis. The restriction enzymes used were AluI and Sau3 AI, following the manufacturer conditions (Jena Bioscience GmbH, Löbstedter, Germany). Representatives from each OTU were amplified with SP6 and T7 primers and the PCR products were purified with the QIAquick PCR Purification Kit (Qiagen GmbH) and sequenced (STAB VIDA Lda, Lisbon, Portugal) using the 27f as the sequencing primer. The sequences were deposited in the GenBank database under accession numbers JQ658298–JQ658336.
Phylogenetic position of the clones (amplicons of 713 bp in average) was performed with the MEGA5 software (Tamura et al. 2011) using reference sequences obtained from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Cluster analysis was performed using the neighbor joining method with bootstrap test of 1000 replicates (significant clustering considered >70%). Similarity matrices were obtained in MEGA5. Clones with ⩾97% sequence similarity were considered to belong to the same species.
FISH
Fixation of SBR biomass samples in 4% paraformaldehyde and FISH analysis were performed according to Amann (1995). The fluorescently labeled oligonucleotide probes used were as follows: EUBmix, for all Bacteria (a mixture of probes EUB338 (Amann et al., 1990), EUB338-II and EUB338-III (Daims et al., 1999), Azo644 for most members of the Azoarcus cluster (Hess et al., 1997), THAU832 for Thauera (Loy et al., 2005), PAR651 for Paracoccus (Neef et al., 1996), CF319a for most Flavobacteria, some Bacteroidetes and some Sphingobacteria (Manz et al., 1996) and ALF968 for most bacteria belonging to the class Alphaproteobacteria (Neef et al., 1997). Hybridized samples were viewed with a Zeiss LSM 510 Meta confocal laser scanning microscope (Zeiss, Oberkochen, Germany). FISH quantification of Cy3-labeled Azoarcus, Thauera and Paracoccus in respect to all Bacteria (Cy5-labeled) was done by image analysis (30 images of each sample) with the Daime software (Daims et al., 2006), which determines the biovolume fraction of the specifically labeled target population relative to the biovolume of the total bacteria. The s.e.m. was calculated as the s.d. divided by the square root of the number of images.
Post-FISH staining with Nile blue
PHA was stained by Nile blue after FISH was carried out by FLUOS-labeled probes on biomass in suspension and spread out on gelatine-coated slides as described by Thomsen et al., 2004. Examination of probe-positive bacteria (PAR651 and ALF968) was done using a confocal laser scanning microscope with automatic stage controller. Different probe-defined bacteria of interest were detected on the slide and their exact position on were marked. The slides were stained according to the protocol by Ostle and Holt, 1982, and the automatic stage controller enabled the relocation of the probe-defined bacteria following the Nile blue staining.
MAR and FISH
Biomass was harvested from the SBR reactor (Portugal), and kept at 4 °C while transported to Aalborg, Denmark, within 72 h. The biomass first was acclimatized at 20 °C for 2 h in mineral nutrient solution (used to feed the SBR, see above) without carbon sources. Prior to the MAR studies, a washing step was done to remove any residual substrate and the biomass was resuspended in fresh medium that mimicked that in the SBR, consisting of a mixture of exhausted SBR supernatant, collected at the end of the famine phase and mineral nutrient solution (ratio 0.75:0.25). The MAR experiments were carried out slightly modified from the protocol by Lee et al. (1999).
In total, four substrates radiolabeled with either 3H or 14C were investigated: acetate, butyrate, propionate and valerate, to determine which substrates alone and in combination resulted in substrate uptake for probe-defined Azoarcus, Thauera, Paracoccus and other unidentified heterotrophs present. All the experiments were conducted under aerobic conditions, with minor pH variation (average pH increase in single substrate tests was 0.07 and in mixed substrate tests was 0.05). Final radioactivity per vial was 5–20 μCi. The amount of substrate added at the biomass concentration used (approximately 1 g l−1) ensured that all substrates were available during the entire 2-h incubation time and no substrate limitation occurred. After fixation, a thorough homogenization of the biomass was needed in order to separate the very dense flocs into individual cells, so that uptake could be monitored as silver grain formation on single cells after the MAR procedure. Silver grains indicate cell uptake of the labeled substrate, and results are from the cumulative activity during the 2-h experiment. MAR results were thus interpreted in a scale from no uptake to strong uptake, based on the density of the MAR signal (Supplementary Figure S1).
Two types of experiments were conducted: (1) determination of uptake of single substrates and (2) determination of uptake of multiple substrates. For the experiments with multiple substrates, the relative proportion between substrates mimicked that of the SBR. The composition of labeled and non-labeled substrates in the experiments is shown in Table 1. All experiments were carried out in duplicates. The length of all the experiments was 2 h and they were terminated using 4% paraformaldehyde. The samples were washed three times to remove excess tracer, homogenized and plated out on gelatin-coated cover slips (approximately 20 μl from each sample). After the FISH procedure, slides were coated with film emulsion and exposed for 3 and 6 days before development and examined under the microscope. Silver grain density was assessed using light microscopy, and epifluorescence microscopy (Zeiss AxioScope 2) was simultaneously used for the FISH identification by examining at least 20 fields of view. The ALF968 probe was used for examination of the MAR slides instead of the PAR651 probe to identify Paracoccus due to problems with probe precipitation. This was possible because nearly all ALF968-positive bacteria were also targeted with the probe specific for Paracoccus (data not shown).
Table 1. Overview of incubation setup with single and multiple substrates, corresponding concentrations of cold substrates and final radioactivity in incubation vials.
|
Experiments with single substrates |
Experiments with multiple substrates |
||
|---|---|---|---|
| Tracer | Substrate concentration and final radioactivity | Tracer | Substrate concentrationa and final radioactivity |
| 3H-Acetate | 4 mℳ, 20 μCi | 3H-Acetate | Ace (4 mℳ, 20 μCi)+Prop (2 mℳ)+But (2 mℳ)+Val (1 mℳ) |
| 14C-Propionate | 2 mℳ, 10 μCi | 14C-Propionate | Prop (2 mℳ, 10 μCi)+Ace (4 mℳ)+But (2 mℳ)+Val (1 mℳ) |
| 14C-Butyrate | 2 mℳ, 10 μCi | 14C-Butyrate | But (2 mℳ, 10 μCi)+Ace (4 mℳ)+Prop (2 mℳ)+Val (1 mℳ) |
| 14C-Valerate | 1 mℳ, 5 μCi | 14C-Valerate | Val (1 mℳ, 5 μCi)+Ace (4 mℳ)+Prop (2 mℳ)+But (2 mℳ) |
Ace, acetate; But, n-butyrate; Prop, propionate; Val, valerate.
Results
Characterization of SBR performance
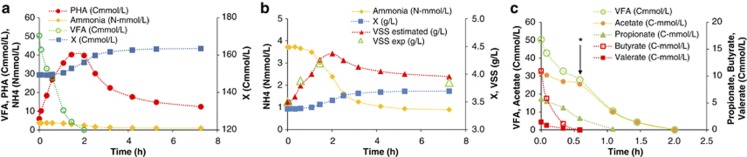
The SBR fed with fermented molasses was operated under FF conditions using the operation conditions optimized by Albuquerque et al. (2010a). A typical SBR cycle of operation is shown in Figure 1 and average values of kinetic and stoichiometric parameters for the time period of the microbial characterization and MAR-FISH studies are shown in Table 2. Typical initial VFA concentrations of acetate, propionate, butyrate and valerate fed to the SBR were: 14.6, 1.6, 2.8 and 0.2 mℳ, respectively (as can be seen in Figure 1c).
Figure 1.
SBR cycle of operation: (a) VFA and ammonia uptake, PHA storage and active biomass (X) growth; (b) ammonia uptake as estimate of cell growth, estimated VSS and active biomass concentrations and experimental VSS values; (c) VFA-uptake profile during the feast phase. *Marks the point of inflexion of the acetate-uptake curve.
Table 2. PHA-storage efficiency in the enrichment SBR and the batch accumulation assay.
| VFA initial (Cmmol l−1) | VFA profile Ace/Prop/But/Val (Cmol per 100 Cmol VFA) | NH4i (mmol N l−1) | PHAi (%) | PHAmax (%) | PHA composition (Cmol HB:Cmol HV) | YPHA/VFA (Cmol PHA/Cmol VFA) | YX/VFA(Cmol X/Cmol VFA) | |
|---|---|---|---|---|---|---|---|---|
| SBR | 47.7 (±1.4) | 63/11/24/2 | 3.7 (±0.3) | 5.7 (±2.2) | 21.5 (±1.5) | 80 (±2):20 (±2) | 0.68 (±0.03) | 0.13 (±0.04) |
| Batch | 59.0±4.6 (5 pulses of ∼60) | 62/11/21/6 | 2.5 | 4.0 | 60.5 | 87:13 | 0.62 | 0.10 |
Abbreviations: Ace, acetate; But, n-butyrate; HB, hydroxybutyrate; HV, hydroxyvalerate; Prop, propionate; PHA, polyhydroxyalkanoate; SBR, sequencing batch reactor; Val, valerate; VFA, volatile fatty acid.
The selected culture had a high PHA storage and low growth response during the feast period of the SBR operation cycles: a high PHA-storage yield on substrate of 0.68 Cmol PHA/Cmol VFA and a cell growth yield of 0.13 Cmol X/Cmol VFA (Figure 1a and Table 2). The high polymer yield on substrate was directly correlated to the internal growth limitation induced by the low feast-to-famine length ratio of 0.22, as can be infered from the low cell growth yield on substrate, despite excess nutrient availability. This is also confirmed by the lag phase observed in the ammonia-uptake curve (Figure 1b), and suggests, as already observed by Albuquerque et al. (2010a), that following the long famine period, cells undergo physiological adaptation before reaching their maximum growth rate. Thus, under these conditions, substrate was mainly diverted to PHA storage.
The observed storage yield is consistent with the fast substrate depletion (qVFA of 0.34 Cmol VFA/(Cmol X.h)) and PHA-storage rate observed (qPHA of 0.22 Cmol PHA/(Cmol X.h)), particularly in the beginning of the SBR cycle (first 40-min period) (Figure 1c and Table 3). After about 40 min of feast phase, these rates decreased to about 0.19 Cmol VFA/(Cmol X.h) and 0.10 Cmol PHA/(Cmol X.h), respectively, either denoting a loss of the physiological adaptation effect (which decays after replenishement of intracellular growth requirements allowing cells to resume their maximum growth rates) or a lower substrate consumption and PHA-storage efficiencies from the carbon sources for which the uptake was slower. Although decreased substrate uptake and PHA-storage rates can also be observed as a result of proximity to the saturation level (because the PHA synthesis reaction is inhibitted by high intracellular PHA concentrations), this cannot be the case in the SBR cycles discussed here because the maximum PHA content reached per cycle is in the range of 25% (gPHA/g TSS), whereas the saturation level was shown by the batch accumulation studies to be above 60%.
Table 3. Kinetic parameters (with s.d. in between brackets) determined for one SBR operation cycle and the first pulse of the batch accumulation study.
| VFA initial(Cmmol l−1) |
Substrate-uptake rates (Cmol/(Cmol X.h)) |
PHA-storage rates (Cmol/(Cmol X.h)) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| qVFA | qHAce | qHProp | qHBut | qHVal | qHB | qHV | qPHA | ||
| SBR (Albuquerque et al., 2010a) | — | — | 0.12 (0.01) | 0.03 (0.01) | 0.06 (0.03) | 0.02 (0.003) | — | — | — |
| SBR (this study) | 50 | 0.34 (0.01)/0.19 (0.006)a | 0.07 (0.003)/0.17 (0.02)a | 0.03 (0.001) | 0.18 (0.02) | 0.02 (0.01) | 0.19 (0.004)/ 0.09 (0.002) | 0.03 (0.002)/ 0.01 (0.001) | 0.22 (0.003)/ 0.10 (0.001)a |
| Batch | 64 | 0.64 (0.03)/0.29 (0.01)a | 0.10 (0.007)/0.29 (0.04)a | 0.05 (0.001) | 0.33 (0.01) | 0.11 (0.01) | 0.21 (0.02)/0.12 (0.004) | 0.06 (0.004)/0.007(0.003)a | 0.27 (0.007)/0.13 (0.001)a |
Abbreviations: Ace, acetate; But, n-butyrate; HB, hydroxybutyrate; HV, hydroxyvalerate; Prop, propionate; PHA, polyhydroxyalkanoate; SBR, sequencing batch reactor; Val, valerate; VFA, volatile fatty acid.
First rate is the initial rate; second rate is the rate determined from the point of inflexion of the acetate-uptake curve onward.
In fact, the substrate-uptake profile (Figure 1c) seems to indicate that there are clear substrate preferences by the SBR-selected culture. Butyrate was consumed at the highest rate (0.18 Cmol/(Cmol X.h)), followed by acetate (0.07 Cmol/(Cmol X.h)), then propionate (0.03 Cmol/(Cmol X.h)) and valerate (0.02 Cmol/(Cmol X.h)) (Table 3).
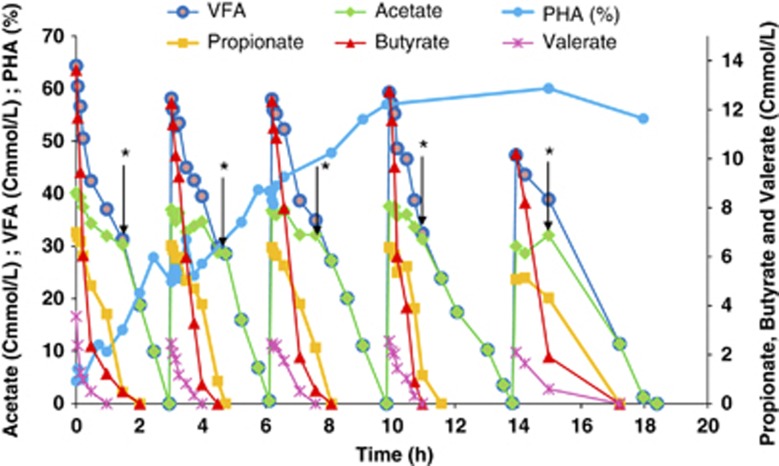
The high PHA-storage capacity of the enriched culture was demonstrated in a batch accumulation study, using a pulse-feeding addition of the fermented molasses (5 × 60 Cmmol l−1 VFA) as carbon source, in which a maximum intracellular PHA content of 60% was attained (Table 2). Growth was prevented because ammonia concentration in the feed was limiting. The substrate-uptake profile observed for each pulse (Figure 2 and Table 3) was similar to that observed in each SBR operation cycle (Figure 1 and Table 3), where butyrate was consumed at the highest rate and the acetate-uptake rate showed a sudden rise when other VFA substrates were exhausted. A copolymer P(HB-co-HV) with a composition of 87:13 (Cmol HB:Cmol HV) was obtained at the end of the accumulation assay.
Figure 2.
PHA batch accumulation study using the SBR-selected culture fed with fermented molasses using a pulse-wise feeding strategy. *Marks the point of inflexion in the acetate-uptake curve.
Characterization of the microbial community
A clone library was generated and 15 different species were detected (Table 4). The phylogenetic groups comprising the highest number of clones from the phylum Proteobacteria belonged to the genera Paracoccus (two species, 14% of clones) and Thauera (one species, 13% of clones), and another group was identified as belonging to either Thauera or Azoarcus (14% of clones). Two other microbial groups had a high number of clones (Sphingobacteria and Flavobacteria). However, FISH assessment of these communities using the CF319a probe showed that they were present in low abundance (<5%), suggesting that the high representativeness in the clone library is due to artifacts of PCR amplification (Kanagawa, 2003) or any other artifact of the cloning process.
Table 4. Phylogenetic position of the clones retrieved from the SBR.
| Phylum | Class | Family | Genus | Number of speciesa | Clones (% of total) | Boostrap valueb |
|---|---|---|---|---|---|---|
| Proteobacteria | Alphaproteobacteria | Rhodobacteraceae | Paracoccus | 2 | 14% | 100% |
| Phyllobacteriaceae | — | 1 | 1% | 95% | ||
| Betaproteobacteria | Rhodocyclaceae | Thauera | 1 | 13% | 100% | |
| Thauera/Azoarcus | 1 | 14% | 86% | |||
| Comamonadaceae | Lampropedia | 1 | 3% | 98% | ||
| — | — | 1 | 1% | 88% | ||
| — | — | — | 1 | 1% | 92% | |
| Bacteroidetes | Sphingobacteria | Sphingobacteriaceae | — | 2 | 12% | 92% |
| Flavobacteria | Flavobacteriaceae | Flavobacterium | 1 | 33% | 72% | |
| — | — | — | 3 | 4% | 84% | |
| Acidobacteria | — | — | — | 1 | 1% | 100% |
(—) not possible to classify to this level.
Clones with similarity <97% were considered to belong to different species.
Bootstrap values supporting the inclusion of the clones in the phylogenetic group.
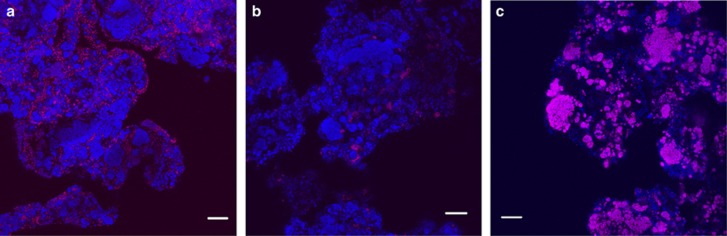
Quantitative FISH showed that Azoarcus represented about 26% of the microbial community, and Thauera constituted approximately 8% (Table 5), which was less than previously found in this system, where the sum of the two populations added up to 88% (Albuquerque et al., 2010a). Paracoccus constituted 50% of the community (Table 5). These three probe-defined populations covered 84% of all bacteria (Figure 3). Post-FISH staining with Nile Blue on the PAR651-hybridized samples showed that Paracoccus were PHA-storing organisms.
Table 5. Quantification of microbial populations in the SBR reactor by FISH and corresponding s.e.m.
| Abundance (% of EUBmix)a | s.e.m. (% of EUBmix) | Congruency (%)a | |
|---|---|---|---|
| Azoarcus | 26% | 2% | 99% |
| Thauera | 8% | 1% | 98% |
| Paracoccus | 50% | 2% | 97% |
Abundance determined as biovolume fraction of total bacteria and congruency between the population and general probes, as per the software Daime.
Figure 3.
Microbial community characterization by FISH. Specific probes for Azoarcus (a), Thauera (b) and Paracoccus (c) are in magenta and other bacteria in blue. Bar=20 μm. The colour reproduction of this figure is available at The ISME Journal online.
The changes in microbial population structure observed in this study (corresponding to a period of operation between 600 and 630 days after inoculation) in respect to Albuquerque et al. (2010a) (between days 120–150 of enrichment) were probably the result of a temporary variation in the VFA composition of the feed. Before the current study, the concentrations of the two major carbon species present (acetate and butyrate) varied in the fermented molasses composition during a period of about 60 days, where butyrate increased to up to 50% of the Cmol/Cmol VFA at the expense of acetate. After that period, the VFA composition of the fermented molasses shifted back (over a period of approximately 20 days) to a higher acetate than butyrate concentration. These two consecutive shifts in substrate composition resulted in a substantial rearrangement in the PHA-storing population structure, with an increase of the Paracoccus from <1% to 50%, and a decrease in Azoarcus and Thauera to less than half and about one-third of their initial numbers, respectively, clearly demonstrating the direct impact of carbon source composition on population selection in PHA-storing systems.
Substrate-uptake preferences by probe-defined populations
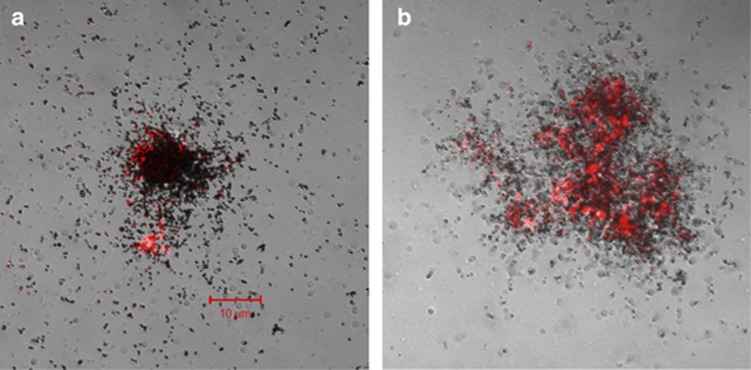
Substrate-uptake preferences by the probe-defined populations were investigated by the use of MAR-FISH (Figure 4). The potential for uptake of single substrates is shown in Table 6a. Interestingly, propionate was taken up efficiently by all probe-defined populations, whereas other substrates could only be consumed by some populations. In addition to propionate, Azoarcus showed also a moderate-to-high uptake of acetate and valerate, but not butyrate. Thauera did not take up any acetate and only moderately took up butyrate and valerate. Paracoccus was able to take up all substrates tested, although the uptake of acetate was only moderate compared with other substrates.
Figure 4.
MAR-FISH images: (a) Azoarcus and labeled propionate and (b) Paracoccus and labeled butyrate.
Table 6a. Overview of single substrate uptake by probe-defined populations determined by MAR-FISH.
| Tracer | Azoarcus | Thauera | Paracoccus |
|---|---|---|---|
| 3H-Acetate | + | − | (+) |
| 14C-Propionate | ++ | ++ | ++ |
| 14C-Butyrate | − | (+) | ++ |
| 14C-Valerate | + | (+) | + |
−, no substrate uptake (approximately 10–30% of the population showed a small uptake but most were negative); (+), moderate substrate uptake (minimum 50% took up a small amount); +, moderate-to-high substrate uptake (80–90% took up substrates); ++, strong uptake (80–90% very positive).
The uptake of specific labeled substrates in the presence of multiple unlabeled substrates was carried out to mimic their simultaneous presence in the FF reactor. The results showed a slightly different uptake pattern compared with the experiments performed with a single-labeled substrate (Table 6b). All the populations were able to take up several substrates simultaneously, as indicated by the positive MAR signal for different specific labeled substrates in the presence of all other unlabeled substrates. However, the level of uptake of the different substrates as assessed by the silver grain density on top of the single cells was different from the relative uptake when taken up as single substrates. A certain niche differentiation or substrate specialization could be observed, where acetate was primarily taken up by Azoarcus and Paracoccus, propionate by Paracoccus, butyrate by Thauera and Paracoccus and valerate by Paracoccus.
Table 6b. Overview of substrate uptake by probe-defined populations determined by MAR-FISH together with multiple unlabeled substrates.
| Tracer | Azoarcus | Thauera | Paracoccus |
|---|---|---|---|
| 3H-Acetate+ULa | ++ | − | + |
| 14C-Propionate+UL | (+) | (+) | ++ |
| 14C-Butyrate+ UL | + | ++ | ++ |
| 14C-Valerate+UL | (+) | (+) | ++ |
−, no substrate uptake; (+), moderate substrate uptake; +, moderate-to-high substrate uptake; ++, strong uptake.
UL, all unlabeled substrates added (acetate, propionate, butyrate and valerate).
Discussion
Microbial population characterization
PHA-storing organisms
The clone library detected the presence of organisms belonging to the Azoarcus and Thauera genera, which were also confirmed by FISH. Both genera have been shown to be responsible for PHA storage in FF-operated microbial enrichments fed with acetate, propionate and/or lactate (Lemos et al., 2008). Bacteria from the genus Thauera were also reported as PHA-storing organisms in a FF-operated SBR fed with a mixture of acetate, propionate and lactate (Dionisi et al., 2005) or a mixture of acetate and lactate (Jiang et al., 2011b).
Furthermore, the clone library and quantitative FISH allowed identifying Paracoccus as the dominant microorganism in the present SBR-enriched culture. Though the genus Paracoccus has not been identified in other FF systems, Paracoccus denitrificans is a known PHA producer in nitrogen-limited pure culture fermentations (Yamane et al., 1996; Gao et al., 2001; Maehara et al., 2001; Ueda et al., 2002; Kojima et al., 2004). Another Paracoccus sp. has been identified as denitrifying polyphosphate accumulating organism, capable of taking up acetate and producing PHA (Lee and Park, 2008). The PHA-storing capacity of other members of the Paracoccus genus is still unknown, but Nile blue staining confirmed that most of the PAR651-targeted bacteria stored PHA in this study, although not all with the same capacity.
Microbial diversity and function
FISH quantification identified PHA-storing organisms as covering at least 84% of the community, which correlated well with a relatively high PHA-accumulation efficiency (maximum 60% PHA content). On the other hand, the number of different species (15) included in the clone library suggests a certain microbial diversity. A fairly diverse microbial population is not contrary to a high functional specialization (apparent from the high PHA-storage efficiency). Diversity can result from multiple PHA-storing organisms, because there are multiple VFA precursors for PHA storage, and/or from the presence of a significant flanking population. FF conditions are designed to ensure VFA exhaustion during the feast phase, but nonetheless other carbon sources (non-VFA organic compounds present in the feed, exopolymeric material, lysed cells or hydrolysis products) could be taken up more slowly during the remaining cycle length by non-PHA-storing organisms.
FF systems operated with a low number of cycles per SRT (for example, two cycles per SRT, Jiang et al., 2011a, 2011b, 2011c) and single substrates have shown to be a very high selective pressure for PHA storage because this strategy forces the microorganisms to grow exclusively on the stored PHA. The operating conditions used in our system, with a high number of cycles per SRT (20) and the use of a complex fermented feed, loosens the selective pressure for PHA-storing organisms, allowing for a flanking population to establish.
Substrate-uptake preferences
Substrate preferences by probe-defined populations
The substrate-uptake capabilities of probe-defined Azoarcus and Thauera were largely in agreement with those present in full-scale water treatment plants, consuming acetate and propionate, and propionate but not acetate, respectively (Hagman et al., 2008; Thomsen et al., 2007). An additional difference was that Azoarcus did not uptake butyrate as a single substrate. On the other hand, Paracoccus seemed to be more of a generalist, showing a strong uptake of all but one VFA, acetate, which was consumed moderately. Interestingly, when multiple substrates were added, this differentiation of substrate-uptake pattern was more pronounced. Possibly, when a single substrate was supplied, all the populations that were able to take it up competed for the same substrate. However, in the presence of other substrates, each population will likely show a stronger uptake of its preferred substrate, leaving the remaining carbon sources available for the other microbial groups. This could explain the increased acetate uptake by Azoarcus in the presence of other substrates, for which the other acetate-consuming population, Paracoccus, showed a stronger preference (propionate, butyrate and valerate). Similarly, more butyrate would be available for Thauera when the other population capable of taking it up, Paracoccus, extended their uptake to other carbon sources (propionate and valerate) for which they demonstrated equal preference.
Another plausible explanation to the differences observed when feeding one or multiple substrates could be the activation of PHA-storage by co-substrate metabolism. For example, Azoarcus seemed to be incapable of taking up butyrate unless it was in the presence of other substrates. This suggests that butyrate was used as a co-substrate by Azoarcus, requiring the uptake of another carbon source to activate butyrate consumption.
When butyrate or valerate are added as sole carbon sources, they have to undergo β-oxidation to acetylCoA (or acetylCoA and propionylCoA) to be available for the cell growth through the TCA cycle, whereas the remaining fraction can be used for PHA storage (either through β-oxidation or be activated directly to hydroxybutyrylCoA and hydroxyvalerylCoA). When other substrates are available for cell growth (such as acetate), microorganisms can use butyrate and valerate for PHA synthesis more efficiently by not going through the β-oxidation pathways but only through direct activation followed by polymerization (Dias et al., 2006). For instance, if both butyrate and acetate are available and consumed by the microorganisms, and both are used for cell growth as well as PHA storage, the metabolic flux from butyrate toward the TCA cycle has to go through the formation of acetoacetylCoA and then acetylCoA. The metabolic flux from acetate toward PHA storage has to go through acetylCoA to form acetoacetylCoA and then hydroxybutyrylCoA to form PHB. This means that if both substrates are used for both ends, the enzymatic reaction between acetylCoA and acetoacetylCoA will run in both directions. It can be suggested that the acetate will be preferably used for cell growth, whereas butyrate can be more efficiently stored as PHB directly. This hypothesis has been suggested by Pardelha et al. (2010) to explain the gradual increase of external substrate-uptake rates of butyrate and valerate (in the simultaneous presence of acetate and propionate) concurrently with increased HB and HV synthesis rates.
Little is known about uptake of multiple substrates of uncultured bacteria in natural or engineered systems, so the observations here are novel and seem to demonstrate a highly flexible physiology allowing them to coexist in the community.
Substrate-uptake pattern in SBR as a function of microbial composition
SBR operation cycles show that the mixed culture selected in this study demonstrated a preference for butyrate, which was consumed at the highest rate, followed by acetate, then propionate and valerate (Table 3). A similar substrate-uptake pattern was observed during the pulse-wise feeding batch accumulation assay (Figure 2 and Table 3). Though the low uptake rates of propionate and valerate could be associated with limiting concentrations of these carbon sources (as suggested in Albuquerque et al. (2010b)), the same argument cannot justify the lower uptake rate of acetate, because this was the most abundant carbon compound (Table 2). Furthermore, a sudden increase of the acetate- and propionate-uptake rates was observed once the remaining VFA substrates were depleted (Figures 1c and 2), suggesting that the lower acetate- and propionate-uptake rates were not associated to inhibiting concentrations of these carbon sources but that, while present, other carbon sources were preferred in detriment of acetate and propionate.
Given the presence of multiple VFA substrates in different concentrations, each SBR cycle starts with a mixture of substrates and gradually a reduction in number of VFA takes place, so all bacteria experience a shift from multiple to fewer or a single substrate. Thus, the increase in propionate uptake rate when butyrate and valerate are depleted (Figures 1 and 2) could be explained by a shift by both Azoarcus and Thauera from butyrate to propionate because both can consume this substrate fast (as demonstrated in the single substrate MAR experiment with propionate). Then, as propionate becomes exhausted, Azoarcus and Paracoccus speed up the acetate consumption by shifting to acetate.
Overall, the VFA-uptake profile observed in the SBR is consistent with the substrate-uptake pattern observed in the MAR study in the presence of multiple substrates. The population in the highest abundance, Paracoccus (50% of the bacteria), was found to prefer butyrate, valerate and propionate over acetate, which justifies the lower uptake rate of acetate in the initial part of the SBR cycle. Furthermore, Thauera did not take up acetate at all. Only Azoarcus preferred acetate over the remaining VFA, thus the rate of acetate uptake measured at the beginning of the cycle may be attributed primarily to the activity of Azoarcus. Considering that acetate was the most abundant VFA and that Azoarcus only represented a quarter of the bacterial population, it is not surprising that the uptake rate of this substrate was slower than the others. The increase of the acetate-uptake rate following the exhaustion of the remaining substrates indicates a substrate shift of Paracoccus, caused by unavailability of their preferred substrates. This also explains that in Albuquerque et al., 2010a, acetate was consumed at the highest specific uptake rate as compared with the other substrates (0.12 Cmol/CmolX.h), whereas the present biomass initially consumed acetate slower (0.07 Cmol/CmolX.h, about 60% of the rate observed in that study), because Azoarcus abundance was reduced from 65% to 26% (that is, about 40% of the numbers in Albuquerque et al., 2010a) and the other populations preferred to take up other carbon sources first (Table 6b).
Impact of substrate-uptake preferences on microbial population
The microbial population structure in the PHA-production system in this study differed remarkably from the community in Albuquerque et al. (2010a), despite the same operating conditions being used: Albuquerque et al. (2010a) reported that Azoarcus (65%) and Thauera (25%) covered 88% of the bacterial population, whereas in the current study, those two populations covered only 34% of the bacterial population, with Paracoccus making up another 50%. This different microbial enrichment was likely a direct consequence of an important variation in the feed composition that took place in between the two studies, during which butyrate was the most abundant carbon source.
Interestingly, in the MAR-FISH experiments, the Paracoccus population showed a rather strong substrate uptake for several substrates, as well as a broader range of substrates, as compared with Azoarcus and Thauera, which seemed to be more specialized in acetate and butyrate, respectively. This can justify the increase of the Paracoccus fraction, because variations in the VFA composition fed to the SBR may have created a greater advantage to generalists like Paracoccus rather than very specialized populations, which would be less fit to adapt when their preferred substrate became scarce.
The change in community structure was likely the cause for the differences in substrate-uptake pattern observed in the SBR in this study as compared with that reported by Albuquerque et al. (2010a), because the feed supplied to the SBR had by then shifted back to a composition very similar to that study. In Albuquerque et al. (2010a), all four carbon substrates were simultaneous taken up, and acetate (the most abundant carbon species) was consumed at the highest rate, which can be explained by the higher abundance of Azoarcus in that period. Indeed, Azoarcus was shown by MAR to have a strong preference for acetate. The simultaneous uptake of butyrate reported in that study is also consistent with the MAR results obtained in the present study, because both Azoarcus and Thauera were shown to take up butyrate in the presence of other VFA.
It is also interesting to note that the copolymer P(HB-co-HV) composition obtained in the batch accumulation study carried out with the current enrichment (87:13 CmolHB:CmolHV) is quite distinct from the composition obtained by Albuquerque et al. (2010a) of 80:20 CmolHB:CmolHV. Because the fermented molasses feedstock supplied in both studies had a similar VFA profile, the difference in HV content is most likely associated with the observed difference in community structure (decreased Azoarcus and Thauera fractions and increased Paracoccus). However, the causes for such a cause-effect correlation are not obvious and should be the subject of further research.
Future prospects
Information regarding the substrate preferences of each dominant microbial population will in the future allow the development of segregated metabolic models taking into account the microbial composition. Previously developed metabolic models for PHA production by mixed cultures considered average metabolic activity (Dias et al., 2008; Pardelha et al., 2010). Improved models could now be developed, incorporating the fraction of each PHA-producing population and their corresponding substrate preferences, which would allow more specific metabolic flux analysis. Furthermore, the link between microbial population structure and polymer composition could be a valuable additional asset for the selection of operating conditions that result in a polymer with specific characteristics.
Acknowledgments
We thank M Stevenson for technical assistance with MAR experiments. We thank Fundação para a Ciência e a Tecnologia (Portugal) for funding through SFRH/30800/2006, PEst-C/EQB/LA0006/2011 and PEst-OE/EQB/LA0004/2011.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albuquerque MGE, Concas S, Bengtsson S, Reis MAM. mixed culture polyhydroxyalkanoates production from sugar molasses: the use of a 2-stage CSTR system for culture selection. Bioresource Technol. 2010b;101:7112–7122. doi: 10.1016/j.biortech.2010.04.019. [DOI] [PubMed] [Google Scholar]
- Albuquerque MGE, Eiroa M, Torres C, Nunes BR, Reis MAM. Strategies for the development of a side stream process for polyhydroxyalkanoate (PHA) production from sugar cane molasses. J Biotechnol. 2007;130:411–421. doi: 10.1016/j.jbiotec.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Albuquerque MGE, Torres C, Reis MAM. Polyhydroxyalkanoate (PHA) production by a mixed microbial culture using sugar molasses: effect of the influent substrate concentration on culture selection. Water Res. 2010a;44:3419–3433. doi: 10.1016/j.watres.2010.03.021. [DOI] [PubMed] [Google Scholar]
- Amann RI.1995In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probesIn: Akkermans ADL, van Elsas JD, de Bruijn FJ, (eds).Molecular Microbial Ecology Manual Kluwer Academic Publications: Dordrecht, Holland; 1–15. [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl Environ Microb. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA . Standard methods for the examination of water and wastewater. American Public Health Association: Washington; 1998. [Google Scholar]
- Choi J, Lee SY. Process analysis and economic evaluation for poly-(3-hydroxybutyrate) production by fermentation. Bioprocess Eng. 1997;17:335–342. [Google Scholar]
- Crank M, Patel M. Techno-economic Feasibility of Largescale Production of Bio-based Polymers in Europe. European Commission: Brussels, Belgium; 2005. [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Daims H, Lücker S, Wagner M. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- Dias JML, Lemos PC, Serafim LS, Oliveira C, Eiroa M, Albuquerque MGE, et al. Recent advances in polyhydroxyalkanoateproduction by mixed aerobic cultures: from the substrate to the final product. Macromol Biosci. 2006;6:885–906. doi: 10.1002/mabi.200600112. [DOI] [PubMed] [Google Scholar]
- Dias JML, Oehmen A, Serafim LS, Lemos PC, Reis MAM, Oliveira R. Metabolic modelling of polyhydroxyalkanoate copolymers production from different volatile fatty acids by mixed microbial cultures. BMC Systems Biology. 2008;2:59. doi: 10.1186/1752-0509-2-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisi D, Carucci G, Petrangeli M, Papini P, Riccardi C, Majone M, et al. Olive oil mill effluents as a feedstock for production of biodegradable polymers. Water Res. 2005;39:2076–2084. doi: 10.1016/j.watres.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Gao D, Maehara A, Yamane T, Ueda S. Identification of the intracellular polyhydroxyalkanoate depolymerase gene of Paracoccus denitrificans and some properties of the gene product. FEMS Microbiol Lett. 2001;196:159–164. doi: 10.1111/j.1574-6968.2001.tb10558.x. [DOI] [PubMed] [Google Scholar]
- Gurieff N, Lant P. Comparative life cycle assessment and financial analysis of mixed culture polyhydroxyalkanoate production. Bioresource Technol. 2007;98:3393–3403. doi: 10.1016/j.biortech.2006.10.046. [DOI] [PubMed] [Google Scholar]
- Hagman H, Nielsen JL, Nielsen PH, Jansen JC. Mixed carbon sources for nitrate reduction in activated sludge—identification of bacteria and process activity studies. Wat Res. 2008;42:1539–1546. doi: 10.1016/j.watres.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Hess A, Zarda B, Hahn D, Haner A, Stax D, Hohener P, et al. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl. Environ. Microbiol. 1997;63:2136–2141. doi: 10.1128/aem.63.6.2136-2141.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Hebly M, Kleerebezem R, Muyzer G, van Loosdrecht MCM. Metabolic modeling of mixed substrate uptake for polyhydroxyalkanoate (PHA) production. Water Res. 2011c;45:1309–1321. doi: 10.1016/j.watres.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Marang L, Kleerebezem R, Muyzer G, Van Loosdrecht MCM. Effect of temperature and cycle length on microbial competition in PHB-producing sequencing batch reactor. ISME J. 2011a;5:896–907. doi: 10.1038/ismej.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Marang L, Kleerebezem R, Muyzer G, van Loosdrecht MCM. Polyhydroxybutyrate production from lactate using a mixed microbial culture. Biotechnol Bioeng. 2011b;108:2022–2035. doi: 10.1002/bit.23148. [DOI] [PubMed] [Google Scholar]
- Johnson K, Kleerebezem R, Muyzer G, van Loosdrecht MCM. Enrichment of a mixed bacterial culture with a high polyhydroxyalkanoate storage capacity. Biomacromolecules. 2009;10:670–676. doi: 10.1021/bm8013796. [DOI] [PubMed] [Google Scholar]
- Kanagawa T. Bias and artifacts in multitemplate PCR. J Biosci Bioeng. 2003;96:317. doi: 10.1016/S1389-1723(03)90130-7. [DOI] [PubMed] [Google Scholar]
- Kojima T, Nishiyama T, Maehara A, Ueda S, Nakano H, Yamane T. Expression profiles of polyhydroxyalkanoate synthesis-related genes in Paracoccus denitrificans. J Biosci and Bioeng. 2004;97:45–53. doi: 10.1016/S1389-1723(04)70164-4. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M, (eds).Nucleic acid techniques in bacterial systematic John Wiley & Sons: New York; 115–175. [Google Scholar]
- Lee N, Nielsen PH, Andreasen K, Juretschko S, Nielsen JL, Schleifer K-H, et al. Combination of fluorescent in situ hybidization and microautoradiography - a new tool for structure-function analysis in microbial ecology. Appl. Environ. Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Park YK. Characterizations of denitrifying polyphosphate-accumulating bacterium Paracoccus sp. Strain YKP-9. J Microbiol Biotechnol. 2008;18:1958–1965. [PubMed] [Google Scholar]
- Lemos PC, Levantesi C, Serafim LS, Rossetti S, Reis MAM, Tandoi V. Microbial characterisation of polyhydroxyalkanoates storing populations selected under different operating conditions using a cell-sorting RT-PCR approach. Appl Microbiol Biotechnol. 2008;78:351–360. doi: 10.1007/s00253-007-1301-5. [DOI] [PubMed] [Google Scholar]
- Loy A, Schulz C, Lücker S, Schöpfer-Wendels A, Stoecker K, Baranyi C, et al. 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order Rhodocyclales. Appl Environ Microbiol. 2005;71:1373–1386. doi: 10.1128/AEM.71.3.1373-1386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara A, Doi Y, Nishiyama T, Takagi T, Ueda S, Nakano H, et al. PhaR, a protein of unknown function conserved among short-chain-length polyhydroxyalkanoic acids producing bacteria, is a DNA-binding protein and represses Paracoccus denitrificans phaP expression in vitro. FEMS Microbiol Lett. 2001;200:9–15. doi: 10.1111/j.1574-6968.2001.tb10685.x. [DOI] [PubMed] [Google Scholar]
- Majone M, Masanisso P, Carucci A, Lindrea K, Tandoi V. Influence of storage on kinetic selection to control aerobic filamentous bulking. Wat Sci Technol. 1996;34:223–232. [Google Scholar]
- Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiol. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- Neef A.1997Anwendung der in situ Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen BiozönosenDoctoral thesis (Technische Universität München).
- Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K-H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostle AG, Holt JG. Nile blue A as a fluorescent stain for poly-b-hydroxybutyrate. Appl Environ Microbiol. 1982;44:238–241. doi: 10.1128/aem.44.1.238-241.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardelha F, Albuquerque MGE, Reis MAM, Oliveira R, Dias JML. Metabolic modeling of polyhydroxyalkanoates (PHA) production from complex mixtures of competing volatile fatty acids (VFA) by mixed microbial cultures (MMC) J Biotechnol. 2010;150:565. [Google Scholar]
- Serafim LS, Lemos PC, Albuquerque MGE, Reis MAM. Strategies for PHA production by mixed cultures and renewable waste materials. Appl Microbiol Biotechnol. 2008;81:4. doi: 10.1007/s00253-008-1757-y. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen TR, Kong Y, Nielsen PH. Ecophysiology of dominant denitrifying bacteria in activated sludge. FEMS Microbiol Ecol. 2007;60:370–382. doi: 10.1111/j.1574-6941.2007.00309.x. [DOI] [PubMed] [Google Scholar]
- Thomsen TR, Nielsen JL, Ramsing NB, Nielsen PH. Micromanipulation and further identification of FISH-labelled microcolonies of a dominant denitrifying bacterium in activated sludge. Environ Microbiol. 2004;6:470–479. doi: 10.1111/j.1462-2920.2004.00580.x. [DOI] [PubMed] [Google Scholar]
- Ueda S, Sano K, Gao D, Tomihari N, Yamane T, Endo I. Purification and properties of (−)-3-hydroxybutyrate oligomer hydrolase of Paracoccus denitrificans. FEMS Microbiol Lett. 2002;206:179–184. doi: 10.1111/j.1574-6968.2002.tb11006.x. [DOI] [PubMed] [Google Scholar]
- van Loosdrecht MCM, Pot MA, Heijnen JJ. Importance of bacterial storage polymers in bioprocesses. Wat Sci Technol. 1997;35:41–47. [Google Scholar]
- Yamane T, Chen X-F, Ueda S. Polyhydroxyalkanoate synthesis from alcohols during the growth of Paracoccus denitrificans. FEMS Microbiol Lett. 1996;135:207–211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.