Abstract
The microbial mats of Guerrero Negro (GN), Baja California Sur, Mexico historically were considered a simple environment, dominated by cyanobacteria and sulfate-reducing bacteria. Culture-independent rRNA community profiling instead revealed these microbial mats as among the most phylogenetically diverse environments known. A preliminary molecular survey of the GN mat based on only ∼1500 small subunit rRNA gene sequences discovered several new phylum-level groups in the bacterial phylogenetic domain and many previously undetected lower-level taxa. We determined an additional ∼119 000 nearly full-length sequences and 28 000 >200 nucleotide 454 reads from a 10-layer depth profile of the GN mat. With this unprecedented coverage of long sequences from one environment, we confirm the mat is phylogenetically stratified, presumably corresponding to light and geochemical gradients throughout the depth of the mat. Previous shotgun metagenomic data from the same depth profile show the same stratified pattern and suggest that metagenome properties may be predictable from rRNA gene sequences. We verify previously identified novel lineages and identify new phylogenetic diversity at lower taxonomic levels, for example, thousands of operational taxonomic units at the family-genus levels differ considerably from known sequences. The new sequences populate parts of the bacterial phylogenetic tree that previously were poorly described, but indicate that any comprehensive survey of GN diversity has only begun. Finally, we show that taxonomic conclusions are generally congruent between Sanger and 454 sequencing technologies, with the taxonomic resolution achieved dependent on the abundance of reference sequences in the relevant region of the rRNA tree of life.
Keywords: Guerrero Negro, rRNA phylogeny, microbial mats, microbial ecology, QIIME
Introduction
Extensive hypersaline microbial mats occur in the evaporation lagoons at the Exportadora de Sal SA saltern, in Guerrero Negro (GN), Baja California Sur, Mexico (Risatti et al., 1994; Des Marais, 1995; Des Marais, 2003). The GN mats consist of a macroscopically stratified biomass of microorganisms embedded in extracellular polymeric and mineralized materials, typically several centimeter thick, that underlies ∼100 km2 of shallow evaporative lagoons. The mats are supported ultimately by photosynthesis, and so offer a relatively closed system for study of the interplay between the energy influx available to a complex natural microbial community and the carbon flow that drives the community. Consequently, the GN mats have served for decades as a model system for microbial ecology and life on the early Earth (Cohen et al., 1984; D'Amelio D'Antoni et al., 1989; Canfield and Des Marais, 1993; Des Marais, 1995; Bebout et al., 2002; Des Marais, 2003; Spear et al., 2003; Ley et al., 2006; Feazel et al., 2008; Dillon et al., 2009; Fike et al., 2009; Robertson et al., 2009).
Extensive biogeochemical studies have shown that the overall metabolism of the GN mats is complex. Light penetrates only a few millimeters into the mats and oxygen produced by photosynthesis rapidly diminishes with depth (Jorgensen and Des Marais, 1986; Jorgensen, 1992). Turnover of carbon fixed in the upper portion of the mats is effected by fermentations and sulfate reduction deeper in the mat, the sulfate from seawater that fills the evaporation lagoons (Ley et al., 2006). Thus, the sulfur cycle also has an important role in the overall bioenergetics of the GN mats. An additional complexity is that the chemical properties of the mats fluctuate daily and seasonally. During the day, oxygenic photosynthesis operates in the uppermost layers. At night, however, the mats and even the water column above become anoxic and high in hydrogen sulfide concentrations, a consequence of ongoing sulfate reduction in the absence of photosynthesis.
The chemical complexity of the GN mats might be expected to indicate microbiological complexity as well. Early microscopic and culture-based studies suggested instead that the microbiology of the mats was relatively simple, dominated by cyanobacteria and sulfate-reducing bacteria (Des Marais et al., 1989). However, recent rRNA gene sequence surveys markedly altered the view of the GN mats as a simple ecosystem and showed instead that these mats are among the most diverse, complex and novel microbial ecosystems so far encountered (Spear et al., 2003; Ley et al., 2006; Lozupone and Knight, 2007; Feazel et al., 2008; Robertson et al., 2009). A preliminary study of only ∼1500 rRNA gene sequences (Ley et al., 2006; Lozupone and Knight, 2007) revealed a bewildering complexity of rRNA phylotypes, including several novel, phylum-level groups of bacteria. Similar gene complexity was seen in a limited shotgun Sanger-sequencing analysis of the GN mat (Kunin et al., 2008). Because of the extreme sequence complexity, however, and the under-sampling of genes in the mat indicated by collectors curves, the distributions of different phylotypes in mat strata were unclear and the diversity was predicted to be under-represented. Data based on these prior sequencing efforts predicted that the richness of the GN bacterial community analyzed was on the order of 104 species-level rRNA sequence types, and that a sampling effort on the order of 105 sequences would be required to capture even a representative sample of the most abundant phylotypes.
To test these statistical predictions and to further explore the phylogenetic diversity and stratification in this unprecedentedly complex ecosystem, we determined an additional ∼147 000 rRNA gene sequences from a 10-layer depth profile of the mat, using both Sanger and 454 FLX sequencing technologies. Although we focus our analysis primarily on the full-length Sanger data, we show that similar biological conclusions are drawn from the partial-length 454 sequence data. The results provide a far more comprehensive view of the phylogenetic makeup and structure of the mat than previously known. The results also significantly expand the known rRNA sequence diversity of bacteria.
Materials and methods
Sample collection and sequencing
Microbial mat samples were collected as cores (∼1 × 5 cm) at ∼1400 hours from pond 4 (near pond 5), the site of our previous study (Ley et al., 2006), at the Exportadora de Sal Saltworks, GN, Baja California Sur, Mexico. Cores were field sectioned unfrozen into 23 layers, 1-mm sections in the uppermost 1 cm and 3-mm sections in the lower mat. Sections from four cores taken within∼0.25 m2 were pooled to minimize influence of patchiness on the stratification profile. Dissected samples were frozen in liquid nitrogen for transport to the laboratory. Genomic DNA was extracted from each layer by phenol:chloroform as described (Ley et al., 2006). Each DNA sample was used to construct two independent libraries. PCR reactions to amplify 16S rRNA genes were conducted (30 cycles with annealing temperature of 52 °C) with the bacterial domain-specific primers 27F and 1492 R (Weisburg et al., 1991). Eight independent PCR reactions were run with each template and four each were pooled for gel purification and cloning with the TopoTA sequencing kit (Invitrogen, Grand Island, NY, USA) as specified by the manufacturer. Clone colonies were robotically picked and sequenced.
DNA sequencing
Sanger sequencing was performed at the Joint Genome Institute (under the Community Sequencing Program) using standard protocols for the MegaBace4000 capillary electrophoresis platform. Sequences were assembled by PHRAP using XplorSeq (Frank, 2008). 454 amplicon production was performed as previously described using barcoded 27F/338R primers targeting the V1/2 region of the bacterial 16S rRNA gene (Hamady et al., 2008). Sequencing was performed per manufacturer's protocols using the 454 FLX system (Branford, CT, USA).
Data analysis
All data analysis was performed using the Quantitative Insights Into Microbial Ecology software package, version 1.3.0-dev (QIIME) (Caporaso et al., 2010b). Sanger reads were required to be ⩾1250 and <1550 bases in length to be included in the analysis, and 454 reads were required to be ⩾200 bases and <500 bases in length to be included in the analysis. Any reads containing N characters were discarded.
Operational taxonomic unit (OTU) picking was performed using an ‘open-reference' OTU picking protocol, where sequences are clustered against the Greengenes database (DeSantis et al., 2006) pre-clustered at 97% identity using uclust (Edgar, 2010). Reads that matched a Greengenes reference sequence at ⩾97% identity were clustered to an OTU defined by that reference sequence. Reads that did not match a reference sequence were subsequently clustered de novo. OTU picking (and all subsequent steps) were performed independently for both 454 and Sanger reads. OTUs observed in only one of the two replicate PCR and sequencing runs for each sample were discarded.
The centroid sequence in each cluster was chosen to represent the cluster and aligned with the Greengenes core set using PyNAST (Caporaso et al., 2010a). Chimeric sequences, identified with ChimeraSlayer (Haas et al., 2011) (run independently for the 454 and Sanger data sets), and reads that failed to align with PyNAST were excluded from subsequent analyses. PyNAST version 1.1 was used through QIIME using default parameters. Phylogenetic trees were constructed using FastTree (Price et al., 2010), and taxonomic assignments were made for each representative sequences using tax2tree (McDonald et al., 2011). Taxonomies were additionally assigned using the RDP Classifier (Wang et al., 2007) retrained on the Greengenes database. RDP taxonomy assignments were largely consistent with the tax2tree taxonomy assignments, although fewer sequences were unassigned with tax2tree (data not shown).
Beta diversity between 16S rRNA samples was computed using unweighted UniFrac (Lozupone and Knight, 2005). Metagenomic data were obtained from MG-RAST (Meyer et al., 2008), and distances between metagenomes were computed as Bray–Curtis distances using QIIME based on counts of all metagenome reads that were classified in the Kyoto Encyclopedia of Genes and Genomes. Principal coordinates (PCoA) plots were generated independently for the 454, Sanger, and metagenome data and compared using Procrustes analysis (Gower, 1975), using as input the distance matrix of unweighted UniFrac distances for 16S rRNA and the distance matrix of Bray–Curtis distances for KO groups from the Kyoto Encyclopedia of Genes and Genomes (as in Muegge et al., 2011). Phylogenetic and OTU gain were computed using the unifrac_g and binary_otu_gain metrics in QIIME.
Detrending of PCoA plots was performed by fitting PC2 values to a quadratic function of PC1, projecting each point onto the fitted curve, and reporting the positions of the points along the curve as the transformed PC1 values, and the residuals as the transformed PC2 values. This process was iterated until the average change in PC1 value was>0.001% of the range (five iterations). This novel but simple method allows detrending of even extreme curvature exhibiting a ‘horseshoe' effect while generally preserving inter-sample distances, in contrast to well-known but controversial previous approaches to detrending ordination coordinates (Hill and Gauch, 1980; Wartenberg et al., 1987; Peet et al., 1988). To justify our approach, we show that the analysis preserves the correlation of PC1 with the expected dominant gradient, sample depth (Spearman's rank correlation coefficient=0.976, P<1 × 10−23), while removing most of the correlation between PC2 and depth (mean Spearman's rank correlation coefficient before=0.9511, P<1 × 10−8, after=0.2579, P>0.05; correlation is calculated separately below and above the numerical midpoint of the gradient, then averaged). We also show that this approach generally preserves the original ordination distances (Spearman's rank correlation coefficient=0.9019 (P<1 × 10−230).
Results
Physiological and phylogenetic stratification was expected in the GN mat based on the properties of the mat and earlier investigations. However, the previous under-sampling limited specific conclusions regarding phylogenetic makeup and structure in the mat (Ley et al., 2006, Spear et al., 2003). To gain better perspective on the composition, cores of a ∼5-cm thick mat were taken as outlined in Materials and Methods from Exportadora de Sal's GN Pond Four near Pond Five (with 90 ppt salinity; ∼1.5 M Na+ and ∼80 mM sulfate), an historically studied site in the system. Cores were sliced into sections in the field to obtain a depth profile of specimens. Equivalent sections from four different cores taken within ∼0.25 m2 were pooled to accommodate potential patchiness in the strata. DNA was extracted from the sections (Materials and Methods), and rRNA genes for cloning and sequence analysis were obtained by PCR using primer sets specific for the bacterial phylogenetic domain. Previous studies have shown that archaeal and eukaryotic sequences in the mat are far less abundant than bacterial sequences (Robertson et al., 2009). Approximately 119 000 near full-length Sanger sequences and ∼28 000 454 FLX sequences were determined (Materials and Methods).
Microbial community composition is stratified
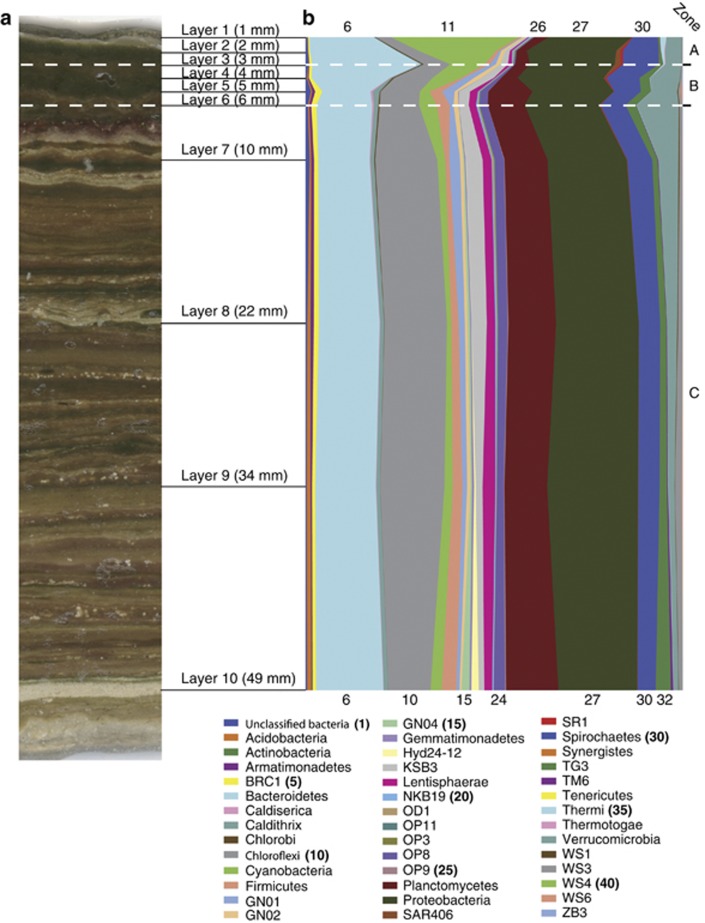
Figure 1 juxtaposes the intricate macroscopic striations of the ∼50-mm-thick mat (Figure 1a) with a summary of the phylum-level relatedness groups encountered throughout the mat (Figure 1b), and indicates the main (known) chemical zones: the photic/oxic (0–3 mm depth, zone A in the Figure), low sulfide (3–6 mm depth, zone B) and high sulfide (6 mm and deeper, zone C) zones seen with light intensity and microelectrode measurements taken previously at the site (Jorgensen and Des Marais, 1986; Jorgensen, 1992; Ley et al., 2006). These boundaries intrinsically are not sharp and in addition migrate with light intensity (Jorgensen and Des Marais, 1986; Jorgensen, 1992). The mat is highly complex throughout; sequences representing >40 phyla were detected in each of the 10 layers analyzed (Figure 1 legend, Supplementary Table S1). Overall, only ∼10 phyla are the main contributors to the makeup of this system throughout the mat, although each of those phylum-level groups is represented by broad intra-phylum diversity. Phylogenetic stratification is evident even at the phylum level, as previously determined (Ley et al., 2006) and seen in Figure 1. Conspicuous gradients in phylum-level variation are seen particularly in the upper 5–6 mm of the mat (Figure 1), which includes the photic zone (zone A in Figure 1) and oxic/anoxic interface (zone B). The distribution of the sequences among the phyla is more uniform deeper in the mat (zone C), although subphylum variation occurs throughout.
Figure 1.
The GN microbial mat (a) photograph and (b) taxonomic composition by depth. Taxonomic assignments were made with tax2tree (Materials and methods). (a) and (b) are consistent in scale. Numeric values on top and bottom of (b) correspond to the phylum legend to aid orientation. As discussed in the text, the dashed white lines indicate approximate depths of the photic/oxic (0–3 mm depth, zone A), low sulfide (3–6 mm depth, zone B) and high sulfide (6 mm and deeper, zone C) zones seen with light intensity and microelectrode measurements (Jorgensen and Des Marais, 1986; Jorgensen, 1992; Ley et al., 2006).
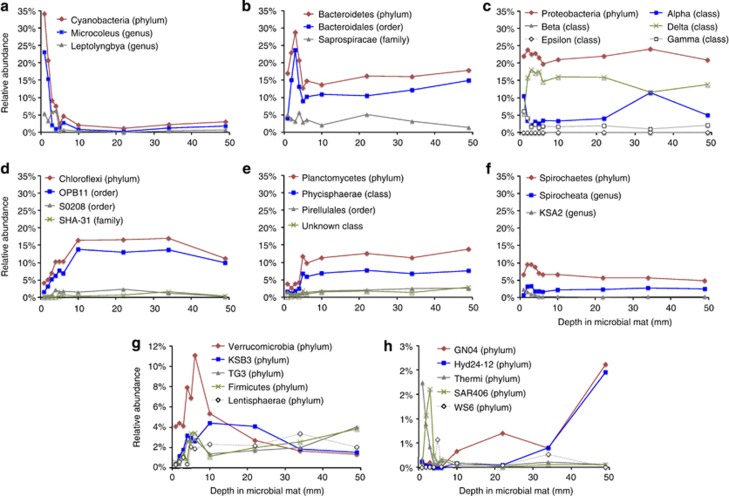
There are many possible correlations between the depths of different phylotypes in the mat and physiological attributes of known, albeit distant relatives of the GN microbes. Figure 2 illustrates the distribution of some of the most abundant (>5% overall) subphylum groups and a few of the more rare phylum-level groups of sequences. The upper ∼5 mm, which includes the photic zone and oxic/sulfide boundary (zones A and B in Figure 1), clearly demarcates the main gradient of variation. All the groups of cyanobacterial sequences (Figure 2a), for instance, are highly enriched, ∼35% of total, in the upper few millimeters and diminish drastically below that. We note that rRNA gene abundance does not translate directly into cellular abundance because of variable gene contents, growth rates, and so on. Subgroups of other phyla, for instance Bacteroidetes or Proteobacteria (Figures 2b and c), are abundant in both zones and so consist of more physiologically cosmopolitan organisms than the obligately photosynthetic Cyanobacteria. Many of the representatives of Bacteroidetes seen deeper in the mat have relatives among human fecal bacteria, consistent with the expected fermentative nature of the mat physiology below the photic zone. The persistence of Delta-proteobacteria throughout the deep mat (Figure 2c) including relatives of organisms known for hydrogen metabolism and sulfate reduction, also is consistent with a fermentative environment deep in the mat. Many groups with unknown physiological properties are restricted to the anoxic portion of the mat (for example, Chloroflexi (Figure 2d), Planctomycetes (Figure 2e)). Although the deeper mat seems more uniform in composition than the upper portion, some stratification is seen even at the phylum level, including candidate phyla with abundance peaks at various positions in the lower portion of the mat (Figures 2g and h). These gradients of variation deep in the mat may represent unidentified biochemical gradients or intrinsic patchiness in the organismal structure of the mat. Overall, however, the deeper portion of the mat, >10 mm, is more homogeneous in composition than the upper few millimeters.
Figure 2.
Abundance of some prominent phyla and taxonomic subgroups by depth in the mat. (a) Cyanobacteria, (b) Bacteroidetes, (c) Proteobacteria, (d) Chloroflexi, (e) Planctomycetes, (f) Spirochetes, (g) and (h) other low-abundance phyla.
By far, most of the organisms represented by the sequences have no known physiological attributes and are not sufficiently closely related to known organisms to infer more than general properties. Nonetheless, the positions of some sequences in the mat suggest more specific physiological properties for the corresponding, otherwise unknown organisms. For example, the enrichment of sequences representing Thermi or the candidate phylum SAR406 in the upper portion of the mat (Figure 2h) may suggest a photosynthetic lifestyle, or close associations with phototrophs, for the otherwise uncharacterized organisms that correspond to these sequences.
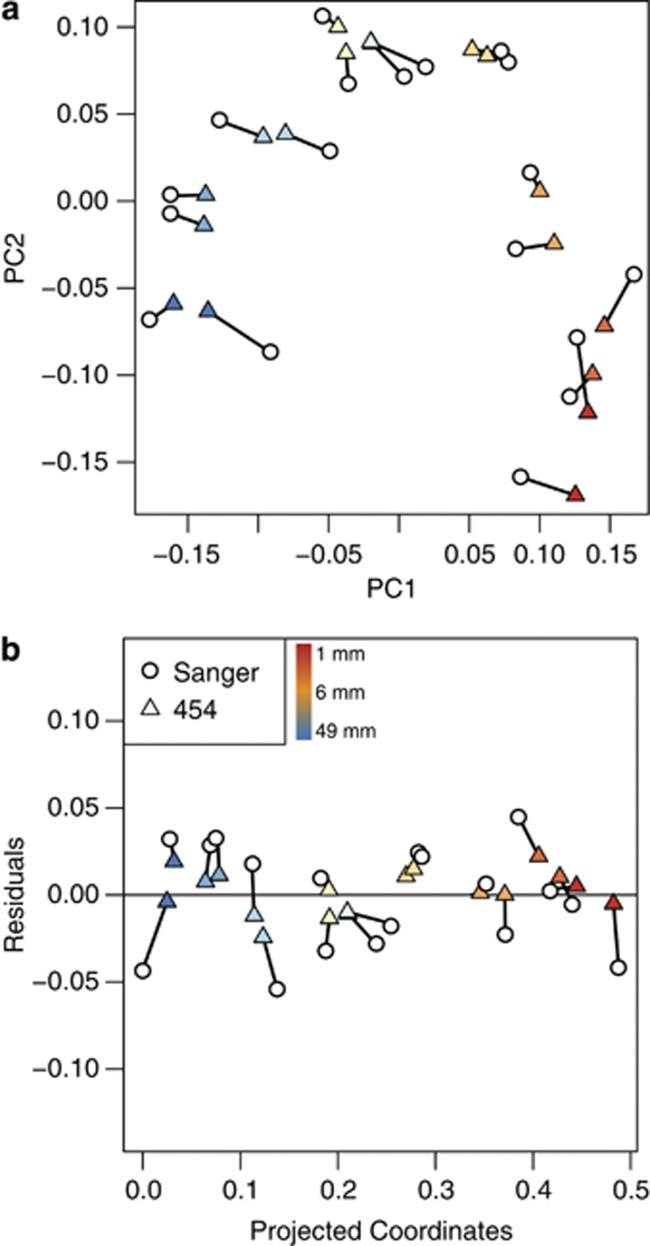
Community clustering is not affected by sequencing technology
Despite differences in sequencing technology (Sanger vs 454) and read length (Sanger reads: 1389±53 nt; 454 read lengths: 236±11 nt), the same general biological conclusions are drawn with regard to the stratified distribution of the many phylogenetic types of organisms. The similarities of the Sanger and 454 results, as well as the phylogenetic gradient, are seen by PCoA analysis clustering of phylogenetic variation (unweighted UniFrac distances) in both sets of data from the different depths of the mat as shown in Figure 3a. PC1 (Figure 3a) correlates with depth in the mat (Spearman's correlation: ρ=0.9455, P=3.9 × 10−18). PC2 reflects phylogenetic variation in the makeup of the sample constituents and the arc-like distribution indicates a gradient in variation with depth. This correlation is even more apparent after detrending as shown in Figure 3b (Spearman's correlation: ρ=0.9756, P=5.9 × 10−24). Further, a Procrustes comparison of the 454 and Sanger PC matrices (the linked points in Figure 3) show that the clustering patterns overlap nearly perfectly (M2=0.024; P<0.001 based on 1000 Monte Carlo iterations).
Figure 3.
(a) Procrustes plot comparing PCoA analysis results from unweighted UniFrac distances for 454 and Sanger sequences. (b) Detrended Procrustes plot (Methods and materials) illustrates the correlation between PC1 and depth in the microbial mat. Points are colored by depth in the microbial mat with red indicating the top layers, yellow indicating the middle layers and blue indicating the bottom layers.
Novel biodiversity in the GN mat
Previous studies of the GN mat identified sequences with no specific affiliation with known phylum-level clades, thus indicating novel candidate phyla (Ley et al., 2006). The new sequences substantially increase the representation of most of these new phylum-level phylogenetic groups: hundreds or thousands of sequences now represent some groups indicated by only a few rRNA sequences in the original study (Supplementary Table S2). For example, the candidate phylum GN01 was represented by 22 sequences in the original study and ∼1300 here; GN02 was represented by 14 sequences in the original study and ∼800 collected here; and GN04, seen as 14 sequences is now represented by ∼300 sequences. These additional representative sequences solidly establish the groups as novel phylum-level clades and will provide the basis for exploring further their phylogenetic structure.
In total, 50 phylogenetically novel, but poorly represented (rare), Sanger OTUs could not be unambiguously assigned to currently recognized phyla in Greengenes or other databases, or in the GN sequence set (Supplementary Table S3). Many of these likely represent undocumented candidate phyla, but additional sequences will be required to confirm their phylogenetic novelty.
To estimate the contribution of the total GN sequence set to known rRNA sequence diversity, we analyzed a combined phylogenetic tree of all Sanger GN sequences and 97% identity-bin OTUs from the Greengenes database using Faith's G measure (gain in phylogenetic diversity). This measure assesses the overall branch length in a phylogenetic tree that is contributed by a particular sequence set (Faith, 1992). Approximately 7.4% of the total branch length in the combined Greengenes and GN tree is represented only among GN sequences (that is, G=7.4% for the GN set); these branches thus represent novel diversity with respect to the Greengenes database.
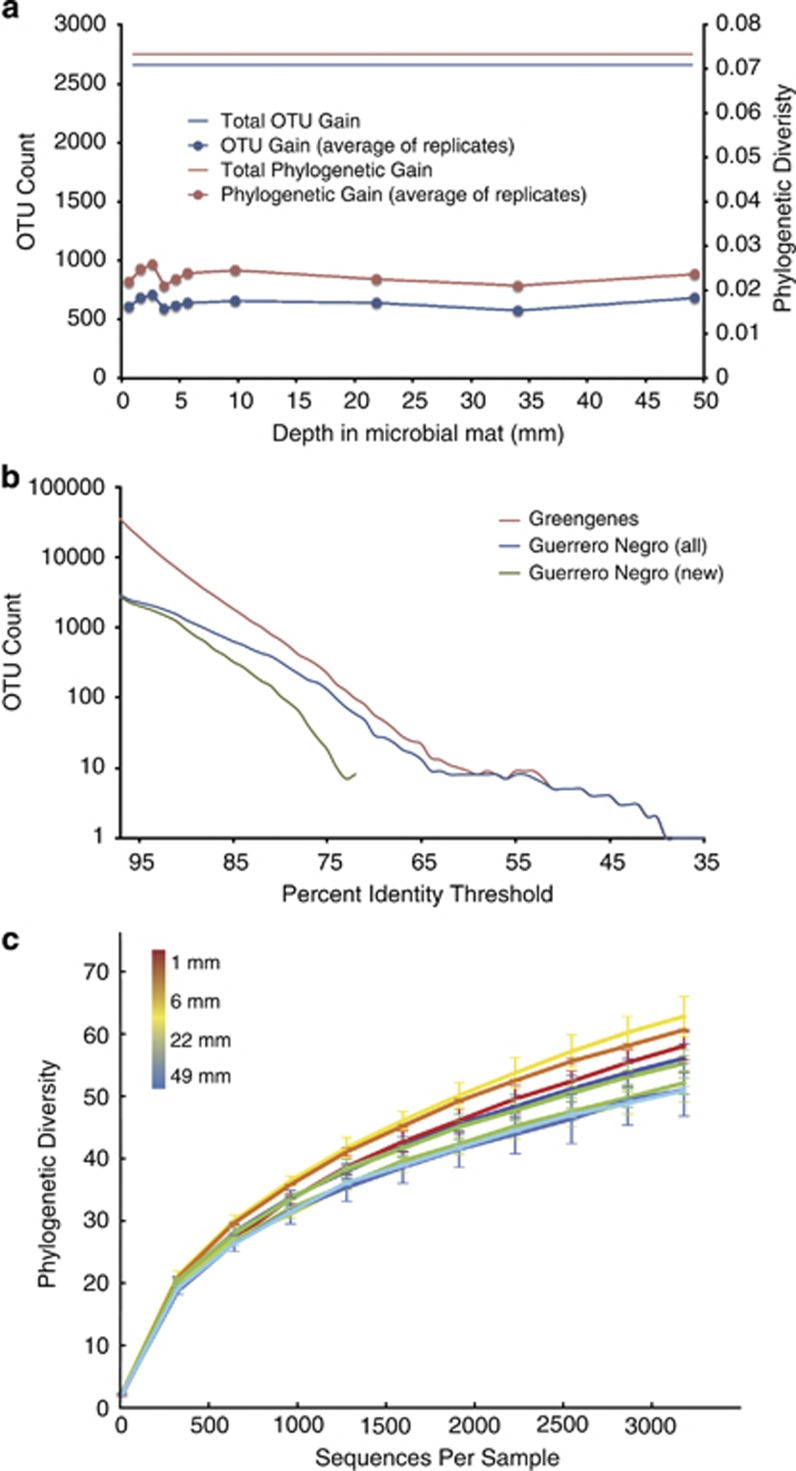
We additionally compared the phylogenetic gain in GN novelty relative to the Greengenes database for each individual layer in the GN mat. Each of the 10 samples was subsampled to even depth (3042 sequences/sample for Sanger; 995 for 454), to correct for the effect that samples with more sequences would automatically be expected to cover more diversity. With both Sanger (Figure 4a) and 454 (Supplementary Figure 1a) sequences, the phylogenetic gain was largely consistent across different depths in the mat, ∼2–3% novel branch length contribution per layer assessed. As seen in Figure 4b, a comparison of OTU numbers vs sequence identities, the phylogenetic novelty in the GN sequence collection is seen at all levels of relatedness, from the OTU levels of the species (⩾97% identity) to the phylum (∼70% identity). Similar results were obtained with the 454 sequence set (Supplementary Figure 1b).
Figure 4.
(a) Phylogenetic gain (new phylogenetic tree branch length, right axis) and OTU gain (left axis) by layer in the GN mat for Sanger reads with respect to the Greengenes database. (b) OTU counts from 97% identity to 35% identity for Sanger sequences. The red line shows the number of OTUs in Greengenes at a given OTU identity threshold; the blue line shows the number of OTUs in the GN data set at a given OTU identity threshold; and the green line shows the number of OTUs in the GN but not Greengenes data base, that is, the number of novel OTUs in the GN data set, at a given identity threshold. The y axis is log scale, so lines are not present when y=0. (c) Alpha rarefaction plots for Sanger reads. Plots are color-coded by depth in the mat according to the inset: red indicates top layers, yellow and green indicate middle layers and blue indicates bottom layers.
Although substantial new phylogenetic diversity is documented in this survey, considerable diversity remains to be revealed. Collector's curves for the accumulation of novelty in the different mat layers provide some assessment of the completeness of our survey of the microbial diversity in the GN mat to date. As seen in Figure 4c, the phylogenetic diversity of the GN mat, as assessed by Faith's Phylogenetic Diversity (PD) measure, remains incompletely sampled and the vectors of the curves show little sign of plateau. The upper portion of the mat contains a somewhat broader phylogenetic diversity than the deeper strata (Figure 4c), but further sampling at all depths is expected to yield still further diversity. Based on the above analyses, however, additional phylogenetic diversity is expected to occur mainly at lower taxonomic levels, at the species and genus levels of phylogenetic variation.
A main contribution of this study is the large number of sequences that populate poorly described regions of the bacterial 16S rRNA tree. To gain some perspective on the taxonomic breakdown of the novel GN sequences relative to the taxonomy of the Greengenes database, we selected a representative Sanger sequence for each of 2898 OTUs identified by UCLUST (Edgar, 2010) at 97% identity (Materials and Methods). We then constructed a tree with FastTree (Price et al., 2010) to infer taxonomic associations based on ancestral nodes with neighboring branches using tax2tree (McDonald et al., 2011). This procedure provides an assessment of the taxonomic specificity with which the GN OTUs can be identified with those described in the Greengenes database. This analysis, reported in Table 1, shows that 2848/2898 (>98%) of the GN OTUs, are affiliated with 41 specific phyla. (50 OTUs, identified in Supplementary Table S2, could not be assigned to a specific phylum as described above.) However, because the Greengenes database is poorly populated by sequences related to those encountered in the GN mat, the level of taxonomic resolution of the GN sequence collection is low. For example, only <1% of the GN OTUs match a Greengenes reference sequence at the species level; only ∼30% and ∼10% of the Sanger OTUs could be classified at the family and genus levels, respectively (Table 1).
Table 1. Counts and fractions of taxonomically classified OTUs by taxonomic level for (a) Guerrero Negro Sanger reads, (b) Guerrero Negro 454 reads, (c) Guerrero Negro Sanger reads trimmed to 454 read lengths and (d) human fecal 454 reads. All classification was performed against Greengenes using tax2tree.
| Taxonomic rank | No. of unclassified OTUs | No. of classified OTUsa | % classified OTUs |
|---|---|---|---|
| (a) GN Sanger OTUs vs Greengenes | |||
| Domain | 0 | 2898 | 100.0 |
| Phylum | 43 | 2855 | 98.5 |
| Class | 388 | 2510 | 86.6 |
| Order | 1109 | 1789 | 61.7 |
| Family | 2019 | 879 | 30.3 |
| Genus | 2598 | 300 | 10.4 |
| Species | 2876 | 22 | 0.8 |
| (b) GN 454 OTUs vs Greengenes | |||
| Domain | 0 | 1298 | 100.0 |
| Phylum | 7 | 1291 | 99.5 |
| Class | 177 | 1121 | 86.4 |
| Order | 322 | 976 | 75.2 |
| Family | 734 | 564 | 43.5 |
| Genus | 1097 | 201 | 15.5 |
| Species | 1264 | 34 | 2.6 |
| (c) GN Sanger OTUs trimmed to 454 length vs Greengenes | |||
| Domain | 5 | 2893 | 99.8 |
| Phylum | 92 | 2806 | 96.8 |
| Class | 390 | 2508 | 86.5 |
| Order | 1128 | 1770 | 61.1 |
| Family | 1967 | 931 | 32.1 |
| Genus | 2479 | 419 | 14.5 |
| Species | 2853 | 45 | 1.6 |
| (d) Human Fecal OTUs (454)b vs Greengenes | |||
| Domain | 0 | 1314 | 100.0 |
| Phylum | 0 | 1314 | 100.0 |
| Class | 7 | 1307 | 99.5 |
| Order | 9 | 1305 | 99.3 |
| Family | 194 | 1120 | 85.2 |
| Genus | 421 | 893 | 68.0 |
| Species | 1037 | 277 | 21.1 |
Abbreviation: OTU, Operational taxonomic unit.
Classification by tax2tree with Greengenes including candidate phyla.
From Costello et al. (2009).
We conducted a similar analysis with the GN 454 OTUs and observed minor disagreement with the Sanger sequence results; in some taxonomic groups assignments of the 454 OTUs seemed more successful than with the full-length sequences (Table 1b). For instance, the full-length and 454 sequences have similar frequencies of classified OTUs at the class level, ∼16%. However, at other taxonomic levels successful assignment of full-length Sanger sequences (Table 1a) was less than seen with the shorter 454 sequences. For example, ∼75% of 454 OTUs, but only ∼60% of Sanger OTUs, could be classified at the order level. To test whether this variation was an effect of sequence length or some intrinsic difference between the Sanger and 454 data sets, we compiled a similar analysis with Sanger sequences pared to match the lengths of the 454 sequences, shown in Table 1c. These classification results match well with those obtained with the full-length sequences (Table 1a). Thus, the incongruities in classifications with the GN Sanger and 454 sequences are not a consequence of sequence length, but may instead lie in differences in methodologies of sequence acquisition or data processing. We do not understand this variation at this time.
In contrast to the relatively low levels of taxonomic identification seen with either the Sanger or the 454 GN OTUs, those from more familiar and frequently studied environments receive more detailed taxonomic assignments, a consequence of their comparatively high representation in the Greengenes database. This is illustrated by an analysis, as above, of human fecal 454 rRNA OTUs (Table 1d). In this case, ∼70% of OTUs could be classified at the genus level, considerably better than the 10–15% success with GN OTUs at the same level (Table 1a–c). This comparison between the resolution of identification possible with more (fecal) or less (GN) representation in the Greengenes database illustrates the considerable extent of novel diversity contributed by this study of the GN mat.
Overall, this study determined ∼119 000 Sanger sequences from 20 libraries from the most-studied site in the GN mat system. Our estimates from previous studies of GN microbial diversity suggested that alpha diversity would be on the order of 104 unique SSU rRNA sequences and that a sampling of 105 sequences would be required to capture a representative sample of this diversity. We observed 5909 species-level (⩾97% sequence identity) OTUs (including singletons, which were filtered for all other analyses presented) and estimated total alpha diversity (Chao1) at 12 227 OTUs. Thus, from the perspective of this deep level of sequencing at only one site in the ∼100 km2 portion of the GN saltern containing microbial mats, it is clear that unrealistic effort would be required to fully analyze the microbial mats of GN. Nonetheless, we have characterized the main microbial components of the community studied and provided new perspective on the complex phylogenetic structure of the mat.
Discussion
Knowledge of the kinds and quantities of organisms that constitute particular ecosystems is a foundation of any ecological understanding. Previous sequence results indicated high complexity in the GN mat ecosystem, but the limited sampling of the survey, only ∼1500 rRNA sequences, provided only a glimpse of the makeup of the mat. These much deeper sequencing results provide a greatly expanded view of the bacterial diversity of the GN microbial mat and a rough phylogenetic stratigraphy to orient with the known properties of the mat. Most of the sequences are from little-studied clades and so richly populate the bacterial phylogenetic tree in poorly documented areas. The GN sequences will serve as a resource for future environmental correlations.
The gross physiological properties of the mat have been studied extensively (Cohen et al., 1984; D'Amelio D'Antoni et al., 1989; Canfield and Des Marais, 1993; Des Marais, 1995; Bebout et al., 2002; Des Marais, 2003; Spear et al., 2003; Ley et al., 2006; Feazel et al., 2008; Dillon et al., 2009; Fike et al., 2009; Robertson et al., 2009). Cyanobacteria are generally considered the main source of the photosynthates that fuel the mat and the high abundance of cyanobacterial rRNA genes in the upper portion of the mat is consistent with that role. However, sequences of other phyla known for photosynthesis, for instance representatives of Proteobacteria, as well as phyla not known for photosynthesis, for example, Bacteroidetes, also are conspicuous in the upper portion of the mat, the photic zone. Because of their abundance, we would predict these phylotypes not known for photosynthesis are phototrophic (Figures 1 and 2). Also because of their relatively high abundance, such organisms likely have a role in primary productivity in this ecosystem and in others where similar sequences might be encountered. If enrichment in the photic zone of the mat suggests photosynthetic capacity, then some of the poorly known GN phyla that are particularly abundant in the upper portion of the mat, because of their considerable evolutionary distance from known organisms, may yield novel photosynthetic pathways upon further study.
The abundances of the different phylotypes shown in the figures correspond only to the particular depth section and are not normalized to biomass distribution in the mat. The actual distribution of active biomass, complicated by many factors, is not known currently. Bulk ATP concentrations, one surrogate for active biomass (Karl, 1980), are 5–10-fold higher in the upper portion of the mat than deeper than a depth of ∼10 mm, suggesting that biomass, hence phylotype (rRNA gene) abundance, diminishes with depth in the mat (Ley et al., 2006); the deeper mat appears to be a starved environment. As of the high biomass load of the mat, probably not much fermentable reduced carbon reaches the depths by diffusion and the bioenergetics deep in the mat may depend mainly on metabolic hydrogen, which perfuses the mat (Hoehler et al., 2001). Nonetheless, many phylotypes representative of the uppermost microbiota also are seen deep in the mat (Figures 1 and 2). Obligately phototrophic cyanobacteria, for example, would seem unlikely to occur deep in the mat unless by migration (gliding motility is common among cyanobacteria) or transportation. Slow stirring of the mat also could directly transport nutrients and microbes from the photic zone into the deeper mat. One mechanism for stirring the mat might be the activities of abundant nematodes, which constitute ∼1% of rRNA genes obtained from the mat with universal PCR primers (Ley et al., 2006; Feazel et al., 2008).
The phylogenetic diversity of the GN mat studied is remarkable. More than 40 of the ∼100 bacterial phyla have so far been encountered in the mat. In contrast, other complex systems examined so far are much simpler. The human gut microbiota, for instance, includes representatives of only ∼8 phyla. Why is the GN microbiota so diverse? The answer likely lies in the complex chemical basis of the GN mat. Rather than a ‘simple' ecosystem, the GN mat presents many energetic opportunities for exploitation by microbial diversity: photosynthesis, including oxygenic photosynthesis; oxygen-dependent oxidative reactions; fermentations; sulfate reduction coupled to oxidation of fermentation products; and so on. Gut microbiota, in contrast, have mainly fermentative capacity and so present more limited complexity than seen in the GN mat. Moreover, the metabolic properties of the upper mat are in continuous flux, dependent on incident light and oxygen production. Another consideration in the context of the complexity of the GN mat is its relative structural stability over time. Whereas gut microbiology is continuously churned and turns over rapidly with excretion, the setting of the GN microbiology, in undisturbed shallow ponds, persists over time. This might allow development in the GN mat of a more extensive repertoire of resources than supports gut microbiota and similar flow through assemblages, for instance through formation of local assemblages, symbioses and other community functions.
The natural microbial world is complex and any understanding of its molecular biology requires high-volume sequencing, currently best achieved with short-read sequencing. Here, we sequence with both long-read Sanger and short-read 454 technologies, and confirm that the same general conclusions are obtained with both sequencing platforms as we, and others, have found previously. With this large and diverse data set we note, however, some differences in classifications obtained with 454 vs full-length sequences (Table 1); the 454 data seemed somewhat more successful at taxon assignment in some cases than the longer Sanger sequences. The reason(s) for any incongruent results is not known, but for instance possibly lies in PCR biases in full-length or 454 libraries, or data processing differences. Nonetheless, short sequencing technologies are appropriate alternatives to longer read technologies for many applications. An exception is the placement of novel diversity into a phylogenetic context; short reads cannot do so as accurately as long sequences. Although short reads can detect new diversity or identify sequences with previously described sequences, longer sequences are best for phylogenetic reconstructions because of the greater information content. At this time, large portions of the bacterial phylogenetic tree are poorly sampled, which, as we show, can limit accurate placement of novel sequences, short or long, into low-level taxonomic groups such as species or genera.
Based on rRNA gene sequences the GN mat clearly is phylogenetically structured. Functional profiles of the GN mat based on a comparatively limited shotgun metagenomic sequence analysis (MG-RAST metagenomes 4440963.3–4440972.3) of the same samples as categorized by rRNA sequences here also correlate with depth in the mat (Kunin et al., 2008). This indicates not only that different taxonomic groups dominate different layers, but also the metagenomic makeup differs by layer. Thus, the phylogenetic (rRNA) and metagenomic results paint a similar picture of biological gradients in the GN mat (Sanger 16S reads vs metagenomes: Procrustes M2=0.040, P<0.001; 454 16S reads vs metagenomes: Procrustes M2=0.038; P<0.001; based on 1000 Monte Carlo iterations). As correlations of functional genes and phylogenetic diversity accumulate it will become possible to predict many phenotypic properties of environmental organisms on the basis of their phylogenetic types.
Acknowledgments
We thank the Exportadora de Sal SA for access and assistance in working on-site. We are grateful for the enabling support of David DesMarais, Brad Bebout and Tori Hoehler, NASA Ames, Moffat Field, CA. Sanger sequencing was provided by the Joint Genome Institute Community Sequencing Program. JRS was supported by an NSF Microbial Biology Postdoctoral start-up fund. Work in the Knight lab was supported in part by the National Institutes of Health and the Howard Hughes Medical Institute. Work in the Pace laboratory was supported by grants from the NASA Astrobiology Institute. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract no. DE-AC02-05CH11231.
Data deposition: the sequences were assigned GenBank accession numbers JN427016–JN539989.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bebout BM, Carpenter SP, Des Marais DJ, Discipulo M, Embaye T, Garcia-Pichel F, et al. Long-term manipulations of intact microbial mat communities in a greenhouse collaboratory: simulating earth's present and past field environments. Astrobiology. 2002;2:383–402. doi: 10.1089/153110702762470491. [DOI] [PubMed] [Google Scholar]
- Canfield DE, Des Marais DJ. Biogeochemical cycles of carbon, sulfur, and free oxygen in a microbial mat. Geochim Cosmochim Acta. 1993;57:3971–3984. doi: 10.1016/0016-7037(93)90347-y. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010a;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010b;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Castenholz RW, Halvorson HO.1984The Solar Lake cyanobacterial mats: strategies of photosynthetic life under sulfideIn: Liss AR, (ed)Microbial Mats: Stromatolites New York; 133–148. [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amelio D'Antoni E, Cohen Y, Des Marais DJ.1989Comparative functional ultrastructure of two hypersaline submerged cyanobacterial mats: Guerrero Negro, Baja California Sur, Mexico, and Solar Lake, Sinai, EgyptIn: Cohen Y, Rosenberg E, (eds)Microbial Mats: Physiological Ecology of Benthic Microbial Communities American Society for Microbiology: Washington DC; 97–113. [Google Scholar]
- Des Marais DJ, Cohen Y, Nguyen H, Cheatham T, Mu–Oz E.Carbon isotopic trends in the hypersaline ponds and microbial mats at Guerrero Negro, Baja California Sur, Mexico: Implications for Precambrian stromatolites 1989. In: Cohen Y, Rosenberg E, (eds)Microbial Mats, Physiological Ecology of Benthic Microbial Communities American Society for Microbiology; 191–203. [Google Scholar]
- Des Marais DJ. The biogeochemistry of hypersaline microbial mats. Adv Microb Ecol. 1995;14:251–274. doi: 10.1007/978-1-4684-7724-5_6. [DOI] [PubMed] [Google Scholar]
- Des Marais DJ. Biogeochemistry of hypersaline microbial mats illustrates the dynamics of modern microbial ecosystems and the early evolution of the biosphere. Biol Bull. 2003;204:160–167. doi: 10.2307/1543552. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon JG, Miller S, Bebout B, Hullar M, Pinel N, Stahl DA. Spatial and temporal variability in a stratified hypersaline microbial mat community. FEMS Microbiol Ecol. 2009;68:46–58. doi: 10.1111/j.1574-6941.2009.00647.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;60:1–10. [Google Scholar]
- Feazel LM, Spear JR, Berger AB, Harris JK, Frank DN, Ley RE, et al. Eucaryotic diversity in a hypersaline microbial mat. Appl Environ Microbiol. 2008;74:329–332. doi: 10.1128/AEM.01448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fike DA, Finke N, Zha J, Blake G, Hoehler TM, Orphan VJ. The effect of sulfate concentration on (sub)millimeter-scale sulfide δ34S in hypersaline cyanobacterial mats over the diurnal cycle. Geochim Cosmochim Acta. 2009;73:6187–6204. [Google Scholar]
- Frank DN. XplorSeq: a software environment for integrated management and phylogenetic analysis of metagenomic sequence data. BMC Bioinform. 2008;9:420. doi: 10.1186/1471-2105-9-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower JC. Generalized Procrustes Analysis. Psychometrika. 1975;40:33–51. [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill O, Gauch HG. Detrended correspondence analysis: an improved ordination technique. Plant ecology. 1980;42:47–58. [Google Scholar]
- Hoehler TM, Bebout BM, Des Marais DJ. The role of microbial mats in the production of reduced gases on the early Earth. Nature. 2001;412:324–327. doi: 10.1038/35085554. [DOI] [PubMed] [Google Scholar]
- Jorgensen B. Spectral light measurements in microbenthic phototrophic communities with a fiber-optic microprobe coupled to a sensitive diode array detector. Limnol Oceanog. 1992;37:1813–1823. [Google Scholar]
- Jorgensen BB, Des Marais DJ. Competition for sulfide among colorless and purple sulfur bacteria in cyanobacterial mats. FEMS Microbiol Ecol. 1986;38:179–186. doi: 10.1111/j.1574-6968.1986.tb01727.x. [DOI] [PubMed] [Google Scholar]
- Karl DM. Cellular nucleotide measurements and applications in microbial ecology. Microbiol Rev. 1980;44:739–796. doi: 10.1128/mr.44.4.739-796.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, et al. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microbio. 2006;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, Desantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2011. [DOI] [PMC free article] [PubMed]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet RK, Knox RG, Case JS, Allen RB. Putting things in order: the advantages of detrended correspondence analysis. Am Nat. 1988;131:924–934. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risatti JB, Capman WC, Stahl DA. Community structure of a microbial mat: the phylogenetic dimension. Proc Natl Acad Sci USA. 1994;91:10173–10177. doi: 10.1073/pnas.91.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson CE, Spear JR, Harris JK, Pace NR. Diversity and stratification of archaea in a hypersaline microbial mat. Appl Environ Microbiol. 2009;75:1801–1810. doi: 10.1128/AEM.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear JR, Ley RE, Berger AB, Pace NR. Complexity in natural microbial ecosystems: the Guerrero Negro experience. Biol Bull. 2003;204:168–173. doi: 10.2307/1543553. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartenberg D, Ferson S, Rohlf FJ. Putting things in order: a critique of detrended correspondence analysis. Am Nat. 1987;129:434–448. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.