Abstract
Mixed microbial communities are complex, dynamic and heterogeneous. It is therefore essential that biomolecular fractions obtained for high-throughput omic analyses are representative of single samples to facilitate meaningful data integration, analysis and modeling. We have developed a new methodological framework for the reproducible isolation of high-quality genomic DNA, large and small RNA, proteins, and polar and non-polar metabolites from single unique mixed microbial community samples. The methodology is based around reproducible cryogenic sample preservation and cell lysis. Metabolites are extracted first using organic solvents, followed by the sequential isolation of nucleic acids and proteins using chromatographic spin columns. The methodology was validated by comparison to traditional dedicated and simultaneous biomolecular isolation methods. To prove the broad applicability of the methodology, we applied it to microbial consortia of biotechnological, environmental and biomedical research interest. The developed methodological framework lays the foundation for standardized molecular eco-systematic studies on a range of different microbial communities in the future.
Keywords: biomolecules, eco-systems biology, extraction, isolation, metabolomics, sample heterogeneity
Introduction
Natural microbial communities play fundamental roles in the Earths biogeochemical cycles as well as in human health and disease, and provide essential services to mankind. They represent highly complex, dynamic and heterogeneous systems (Denef et al., 2010). The advent of high-resolution molecular biology methodologies, including genomics (Tyson et al., 2004), transcriptomics (Frias-Lopez et al., 2008), proteomics (Ram et al., 2005) and metabolomics (Li et al., 2008), is facilitating unprecedented insights into the structure and function of microbial consortia in situ. Beyond isolated biomolecular characterization, integrated omics combined with relevant statistical analyses offer the ability to unravel fundamental ecological and evolutionary relationships, which are indiscernible from isolated omic data sets (Wilmes et al., 2010). Furthermore, space- and time-resolved integrated omics have the potential to uncover associations between distinct biomolecules, which allows for discovery of previously unknown biochemical traits of specific microbial community members (Fischer et al., 2011). Such linkages will, however, only be discernible if representative biomolecular fractions are obtained. Thus, within the emerging field of molecular eco-systems biology (Raes and Bork, 2008), molecule-level systematic characterizations of microbial consortia will only fulfill their full potential if biomolecular fractions (DNA, RNA, proteins and small molecules) are obtained from single unique samples (Kitano, 2001). Only then will the subsequent integration of high-resolution omic data enable true systems-level views of community-wide, population-wide and individual-level processes. To our knowledge, no methodology currently exists for the isolation of all concomitant small molecules (metabolites) and biomacromolecules (DNA, RNA and proteins) from biological systems.
A well-established method for the isolation of concomitant biomacromolecules only is based around the simultaneous addition of a monophasic mixture of phenol and guanidine isothiocyanate, commercially available as TRIzol and TRI Reagent (TR), and chloroform to biological samples to obtain an aqueous phase containing primarily RNA, an interphase containing DNA and an organic phase containing proteins (Chomczynski, 1993; Hummon et al., 2007; Chey et al., 2011; Xiong et al., 2011). Individual biomacromolecular fractions are purified from the respective phases and may be subjected to specialized downstream analyses. Adaptation of the standard TR protocol for the additional extraction of small molecules was carried out on plant material by Weckwerth et al. (2004). For this, a solvent mixture of methanol, chloroform and water is first used for the fractionation of small molecules into polar and non-polar metabolites, and the precipitation of biological macromolecules. Polar and non-polar metabolites are retrieved from the aqueous and organic phases, respectively. RNA and proteins are isolated from the remaining pellet following extraction in dedicated buffers and phenol, respectively. However, no genomic DNA fraction was obtained using this method, a need that is particularly important in microbial communities that exhibit extensive genetic heterogeneity (Wilmes et al., 2009) and for which genomic context is thus essential for meaningful interpretation of functional omic data.
Owing to the hazardous chemicals involved and the methods being unsuited for routine high-throughput laboratory use, chromatographic spin column-based procedures have been introduced for the isolation of concomitant biomacromolecules (Morse et al., 2006; Tolosa et al., 2007; Radpour et al., 2009). These methods rely on the pH- and salt-concentration-dependent adsorption of nucleic acids and proteins to solid phases such as silica or glass. The solid phases are washed and the biomacromolecules of interest are sequentially eluted. Such methods are now available as commercial kits from Qiagen (AllPrep DNA/RNA/Protein Mini kit, QA) and Norgen Biotek (All-in-One Purification kit for large RNA, small/microRNA, total proteins and genomic DNA; NA). The latter has the advantage of offering the ability to deplete the total RNA fraction of small RNA (<200 nt), which can be analyzed separately. Chromatographic spin column-based biomacromolecular isolation has yet to be combined with the extraction of concomitant small molecules.
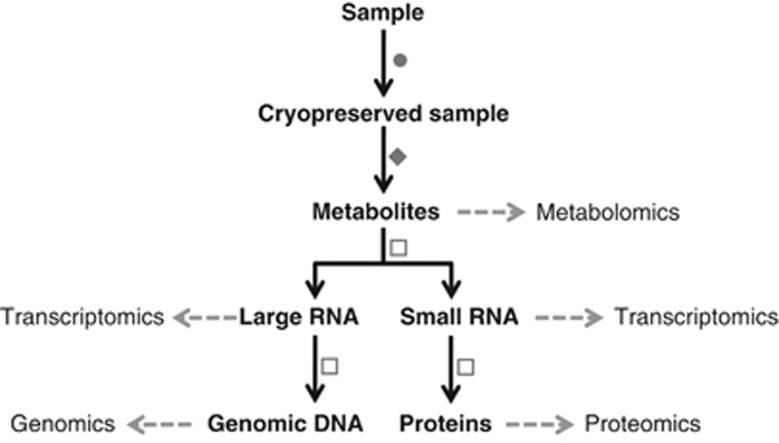
Here, we combine sample processing, sample cryopreservation, cell disruption by cryomilling, small molecule extraction and biomacromolecular isolation based on chromatographic spin columns to result in a universal methodological framework that allows the standardized isolation of extracellular and intracellular polar and non-polar metabolites, genomic DNA, RNA (divided into large and small RNA fractions) and proteins from single microbial community samples (Figure 1). We validated the performance of the methodology by comparison to widely used exclusive and simultaneous biomolecular isolation methods. Furthermore, we proved its general applicability to diverse mixed microbial communities of biotechnological, environmental and biomedical research interest, that is, lipid-accumulating organisms (LAOs), that may be exploited for the reclamation of energy-rich lipids from wastewater, river water filtrate and human feces. The framework may also find broader appeal for integrated omics on other heterogeneous and/or precious biological samples.
Figure 1.
The developed biomolecular isolation framework highlighting the NA-based methodological workflow. Legend: ( ) Sample processing and preservation: immediate snap-freezing by immersion in liquid nitrogen for LAO-enriched mixed microbial communities; concentration by tangential flow filtration followed by high-speed centrifugation, then snap-freezing of the resulting cell pellet for freshwater mixed microbial communities; homogenization with RNAlater followed by centrifugation steps before snap-freezing for fresh human fecal samples (Materials and methods). (
) Sample processing and preservation: immediate snap-freezing by immersion in liquid nitrogen for LAO-enriched mixed microbial communities; concentration by tangential flow filtration followed by high-speed centrifugation, then snap-freezing of the resulting cell pellet for freshwater mixed microbial communities; homogenization with RNAlater followed by centrifugation steps before snap-freezing for fresh human fecal samples (Materials and methods). ( ) Cryomilling and metabolite extraction: cryomilling of cell pellets and solvent extraction of the intracellular polar and non-polar metabolite fractions (extracellular fractions were only prepared for the LAO-enriched microbial communities; Materials and methods). (
) Cryomilling and metabolite extraction: cryomilling of cell pellets and solvent extraction of the intracellular polar and non-polar metabolite fractions (extracellular fractions were only prepared for the LAO-enriched microbial communities; Materials and methods). ( ) Physicochemical biomacromolecular isolation: use of sequential physicochemical separation based around chromatographic spin columns following bead-beating in the modified NA-lysis buffer, resulting in the isolation of high-purity biomacromolecular fractions (Materials and methods).
) Physicochemical biomacromolecular isolation: use of sequential physicochemical separation based around chromatographic spin columns following bead-beating in the modified NA-lysis buffer, resulting in the isolation of high-purity biomacromolecular fractions (Materials and methods).
Materials and methods
For more details see Supplementary Materials and methods.
Sampling and sample processing
LAO-enriched mixed microbial community
Floating LAO biomass was sampled from the air–water interface of the anoxic activated sludge tank at the Schifflange wastewater treatment plant (Esch-sur-Alzette, Luxembourg; 49°30′48.29″N; 6°1′4.53″E). For each sampling date, four different ‘islets' (I1-I4, defined herein as biological replicates; Supplementary Figure 1) were sampled using a levy cane of 500 ml. Samples were collected in 50 ml sterile Falcon tubes and then immediately snap-frozen by immersion in liquid nitrogen and stored at −80 °C.
For the determination of sample heterogeneity by metabolomics, four replicates of 200 mg of LAO biomass (technical replicates) were obtained from four different islets (biological replicates) for four different dates, leading to an overall sample set of 64. The different sampling dates were chosen to reflect different LAO microbial community compositions. Samples were taken on 4 October 2010 (date 1, D1; wastewater temperature 20.7 °C), 25 October 2010 (date 2, D2; 18.9 °C), 25 January 2011 (date 3, D3; 14.5 °C) and 23 February 2011 (date 4, D4; 13.9 °C). For the development and assessment of the biomolecular extraction protocols, a single representative islet sample taken on 13 December 2010 was used. For the comparative metabolomic analysis of the three microbiomes, three technical replicates from D4 islet 4 were used. For each extraction, technical replicates of 200 mg were subsampled from the collected islet using a sterile metal spatula while at all times guaranteeing that the samples remained in the frozen state.
Following weighing out, 200 mg of LAO-enriched microbial community samples were briefly thawed on ice followed by centrifugation at 18 000 g for 10 min at 4 °C to separate the supernatant (∼150 μl; extracellular fraction) from the biomass (intracellular fraction). The intracellular fraction was then immediately refrozen in liquid nitrogen before homogenization by cryomilling (Figure 1). In contrast, the extracellular fraction immediately underwent metabolite extraction.
Freshwater mixed microbial community
Forty liters of river water were collected at a depth of around 1 m from the Alzette river (Schifflange, Luxembourg; 49°30′28.04″N; 6°0′11.48″E). Cells were concentrated by tangential flow filtration and centrifugation (Supplementary Materials and methods). Resulting cell pellets were snap-frozen and stored at −80 °C until the cryomilling step (Figure 1).
Human fecal samples
Three fresh human fecal samples, 300 mg each, were collected from a young healthy individual and placed immediately on ice. Samples were treated with RNAlater and cell pellets were obtained following centrifugation (Supplementary Materials and methods). Pellets were stored at −80 °C until the cryomilling step (Figure 1).
Cryomilling
Each of the three different microbial community samples were homogenized by cryomilling for 2 min at 30 Hz using two 5 mm and five 2 mm stainless steel milling balls (Retsch, Haan, Germany) in a Mixer Mill MM 400 (Retsch; Figure 1).
Metabolite extractions
Extracellular metabolite extractions were only carried out on supernatant from the LAO-enriched microbial communities. For the river water filtrate and human fecal samples, supernatants were not obtainable because of the need for concentrating the river water sample by tangential flow filtration and the very limited liquid content in the human fecal samples, respectively.
Briefly, small molecules were cold (4 °C) solvent extracted by bead-beating (2 min at 20 Hz in a Retsch Mixer Mill MM400) the samples in defined mixtures of polar (methanol and water) and non-polar solvents (chloroform) using the same stainless steel balls as used previously for sample cryomilling. For a detailed description of the respective metabolite extraction protocols, see Supplementary Materials and methods. Following centrifugation at 14 000 g for 10 min at 4 °C, metabolite extractions resulted in an upper phase comprising polar metabolites, an interphase pellet comprising genomic DNA, large and small RNA, proteins and non-lysed cells, and a lower phase containing non-polar metabolites. Defined volumes of both polar and non-polar metabolites extracts were dried in specific gas chromatography (GC) glass vials before metabolomic analyses (Supplementary Materials and methods).
Sample processing and biomacromolecular isolations
After removal of the respective metabolite fractions, the interphase pellet (along with the steel milling balls) was kept on ice for the subsequent total RNA (enriched in large RNA), genomic DNA, small RNA and protein sequential isolations and purifications using the different methods as specified below. As the described metabolite extraction is a major modification of the typical extraction workflow, the interphase pellet was lysed in the respective lysis buffers by bead-beating at 30 Hz for 30 s at 4 °C (Retsch Mixer Mill MM 400) with stainless steel balls (the same as previously used for sample cryomilling and metabolite extraction steps).
The Norgen Biotek All-in-One Purification kit-based biomacromolecular isolation method (NA, Labomics S.A., Nivelles, Belgium; Figure 1) was applied to the interphase pellet according to the manufacturer's instructions with a few important modifications (Supplementary Materials and methods).
The Qiagen AllPrep DNA/RNA/Protein Mini kit-based method (QA, Qiagen, Venlo, The Netherlands) was applied to the interphase pellet according to the manufacturer's instructions (Supplementary Materials and methods).
The TRI Reagent-based method (TR, Sigma-Aldrich, Bornem, Belgium) was directly applied to the interphase pellet according to the manufacturer's instructions with a few important modifications (Supplementary Materials and methods).
Reference methods
To qualitatively and quantitatively assess the biomolecular fractions obtained through the sequential and simultaneous biomolecular isolation protocols, widely used dedicated biomolecular extraction and purification methods were used as reference methods. In each case, the reference methods were applied to 200 mg of LAO-enriched biomass (a single islet sampled on 13 December 2010).
DNA extraction was performed using the PowerSoil DNA isolation kit (MO BIO Laboratories, Brussels, Belgium) according to the manufacturer's instructions (Supplementary Materials and methods). RNA extraction was performed with an RNeasy Mini kit (Qiagen) including the optional DNase treatment to eliminate contaminating genomic DNA according to the manufacturer's instructions. Protein extractions were carried out using a metaproteomic extraction method for activated sludge (Wilmes and Bond, 2004; Supplementary Materials and methods).
Metabolomics
Identical procedures were used for the analysis of both intracellular and extracellular metabolite fractions.
For the determination of sample heterogeneity by comparative metabolomics, gas chromatography (GC) coupled to mass spectroscopy (MS) measurements were performed on intracellular polar extracts from the LAO-enriched microbial communities obtained as specified above. A pool, from which analytical replicates were obtained, was also prepared by combining 100 μl of each of the 64 polar extracts. Aliquots of 40 μl of both intracellular polar extracts and of the pool were dried under vacuum and analyzed following resuspension and derivatization (Supplementary Materials and methods). Each pool aliquot represented an analytical replicate.
For the metabolomic analyses of the representative LAO-enriched microbial community samples, 50 μl of the polar and non-polar phase extracts of the extracellular and intracellular compartments were dried down and analyzed following resuspension and derivatization (Supplementary Materials and methods).
For the comparative metabolomic analysis of the different microbial communities, 50, 10 and 5 μl of the intracellular polar and non-polar phase extracts of the LAO-enriched microbial community, river water filtrate and diluted human fecal samples were dried and analyzed, respectively. To prevent overloading of the GC column, a 1 in 10 dilution in a 1:1 (v:v) methanol:water mixture was previously carried out on the raw polar phase extracts derived from the human fecal samples.
Biomacromolecular quality and quantity assessment
Isolated genomic DNA was separated by electrophoresis on agarose gels (Supplementary Materials and methods). PCR amplifications were carried out for each of the triplicate DNA fractions obtained using the NA-based method from the different microbial community samples, without any additional DNA purification steps (Supplementary Materials and methods).
For RNA quality assessment and quantification, an Agilent 2100 Bioanalyser (Agilent Technologies, Diegem, Belgium) was used. The Agilent RNA 6000 Nano kit and Agilent Small RNA kit for prokaryotes were used. To compare different RNA fraction traces obtained using the Agilent 2100 Bioanalyser, fluorescent units were normalized (Supplementary Materials and methods).
Genomic DNA and RNA fractions were quantified and assessed using a NanoDrop Spectrophotometer 1000 (Thermo Scientific, Erembodegem, Belgium).
Protein extracts were separated using 1D sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad Laboratories, Nazareth, Belgium; Supplementary Materials and methods). The protein fractions were quantified using a 2-D Quant kit (GE Healthcare, Diegen, Belgium).
Peptide tandem mass spectra were obtained on a Matrix-Assisted Laser Desorption Ionization-Time of Flight mass spectrometer (MALDI-ToF/ToF, 4800 Proteomics Analyzer, Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands) after tryptic digestion of a dominant protein band excised from a SDS-PAGE gel on which an NA-based LAO-enriched microbial community protein extract had been separated.
Determination of intact cells versus cells with a compromised cell membrane
To evaluate the efficiency of cell lysis and variation between biomolecular extraction protocols on representative LAO-enriched microbial community samples, the percentage of damaged cells was determined by epifluorescent microscopic quantification after staining using the Live/Dead BacLight Bacterial Viability kit (Invitrogen, Merelbeke, Belgium; Supplementary Materials and methods).
Data treatment and statistical analyses
The data sets resulting from the individual analyses were processed and analyzed using appropriate statistical methods (Supplementary Materials and methods).
Results
Analysis of sample heterogeneity by comparative metabolomics
The metabolome represents the final output that results from the cellular interactions of the genome, transcriptome, proteome and environment and, thus, is the most sensitive indicator of cellular activity and sample-to-sample variation. To assess the extent of heterogeneity within microbial communities, we carried out an initial metabolomics experiment on spatially and temporally resolved LAO-enriched microbial communities from the air–water interface of a biological wastewater treatment plant (Supplementary Figure 1). These LAO-enriched microbial communities allow spatially and temporally defined samples to be taken, exhibit medial species richness (α diversity of around 900 species; unpublished information) and are typically dominated by organisms belonging to the genus Microthrix spp. (Levantesi et al., 2006).
For the four different sampling dates, we extracted intracellular polar metabolites from four different biological replicates, that is, different islets (Supplementary Figure 1), and per islet we extracted four technical replicates (different subsamples derived from the same islet). In addition, an analytical reference sample for quality control and normalization, consisting of a pool of all polar metabolite extracts (Dunn et al., 2011), was prepared using the remaining polar extracts from all the samples (Materials and methods).
The samples were analyzed by GC coupled to mass spectrometry (GC–MS) and the peak intensities of detected metabolite features integrated using specialized software (Supplementary Materials and methods; Supplementary Table 1). The data obtained for 16 analytical pool replicates highlights the high reproducibility of the metabolomics method (mean concordance correlation coefficient (Lin, 1989) =0.993±0.006; mean CCC±s.d.). Owing to the apparent unevenness of total integrated peak intensities in the metabolomics data (Supplementary Table 1) that results from sample heterogeneity, we chose a unit vector normalization strategy analogous to that commonly used for shotgun proteomics experiments (Florens et al., 2006). This involves dividing each metabolite peak intensity by the total peak intensity of the sample (Wilmes et al., 2010). To further exclude experimental variation, the metabolomics data set was filtered according to the presence in all analytical pool replicates. Finally, to account for instrument drift, which was observed for a very small fraction of metabolites, we normalized the data using the pool replicate data (Dunn et al., 2011) and finally re-normalized the data to the total intensity (Supplementary Materials and methods).
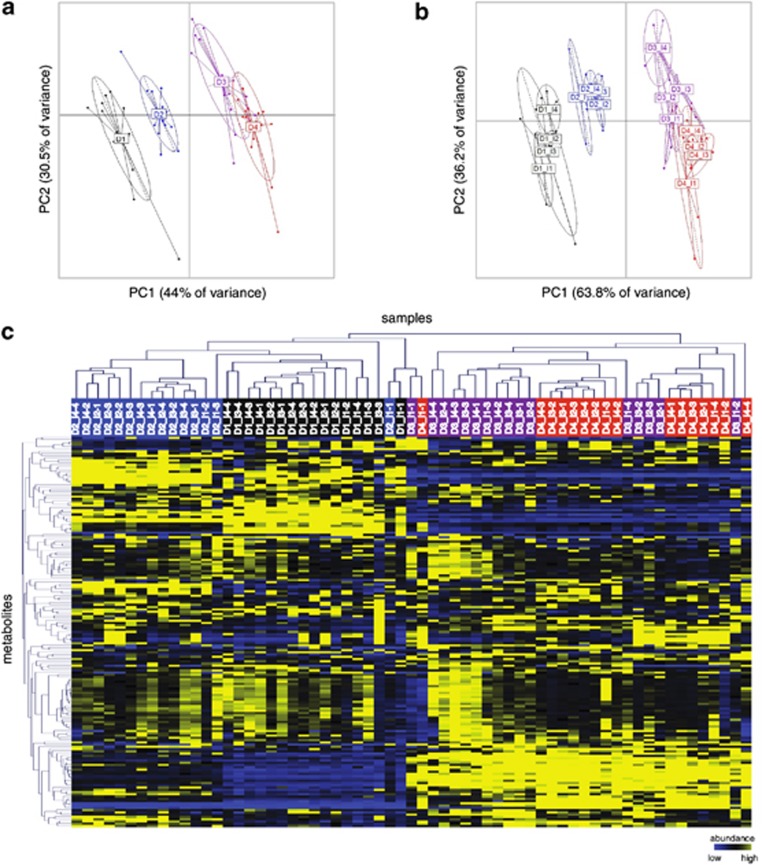
Using the normalized metabolomics data, extensive sample-to-sample variation is apparent for both biological (islets) and technical replicates. Although most samples from specific dates are distinguishable by their metabolomic profiles (Figure 2a and Supplementary Figure 2), extensive overlap between biological and technical replicates is apparent (Figure 2b), with numerous samples clustering outside of their respective replicate groups (Figure 2c). These results indicate that sample heterogeneity is an important consideration for integrated omic studies of natural microbial consortia.
Figure 2.
Metabolome heterogeneity within LAO-enriched microbial community samples. (a) Scatter plot of the two first principal components obtained using principal component analysis (PCA) of the normalized metabolomics data derived from the four biological replicates (islets; I1-I4), for each of the four distinct sampling dates (D1-D4). Each microbial community metabolome is indicated by a dot and color-coded according to sampling date. (b) Between-class PCA of the individual technical replicates for each biological replicate (islets; I1-I4). (a and b) The center of gravity for each date/islet cluster is marked by a rectangle and the colored ellipse covers 67% of the samples belonging to the cluster. (c) Hierarchical clustering of the normalized metabolomics data using the Pearson product moment correlation coefficient.
Conceptualization of a biomolecular isolation framework
In the context of dealing with extensive sample-to-sample and endogenous sample heterogeneity, understanding of these complex systems' structure and dynamics requires integrated analyses of their parts, which in our case demands the isolation of concomitant metabolites, nucleic acids and proteins from single unique microbial community samples before the individual fractions are able to be subjected to specialized metabolomic, genomic, transcriptomic and proteomic analyses (Figure 1). Major considerations to allow comprehensive and reproducible extraction and purification of biomolecules from mixed microbial communities are: (i) standardized and representative sample preservation, (ii) indiscriminate cell lysis and (iii) retrieval of high-quality biomolecular fractions. These constraints were taken into account when devising the biomolecular isolation framework and resulted in a methodology, which is based on (i) immediate snap-freezing of mixed microbial community samples in liquid nitrogen following sampling (or after sample-specific cold preprocessing) and preservation of the biomass at a minimum of −80 °C, (ii) mechanical (cryogenic) and chemical cell lysis, and (iii) reliance on robust methods for biomolecular extraction and purification. The resultant methodological framework offers a fully integrated workflow that can easily be adjusted for specific samples and which allows flexibility for use of sequential or dedicated biomolecular extraction and purification protocols. We developed and validated the methodology on representative LAO-enriched microbial community sample (Materials and methods).
Cell lysis efficiency
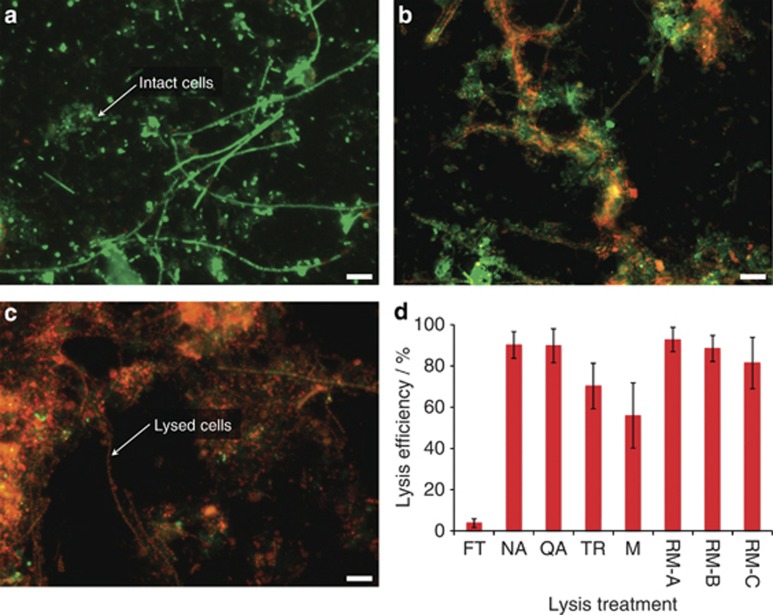
Conservation of cell integrity before sample processing and representative cell lysis before biomolecular extraction are essential considerations for the methodology to result in reproducible and representative biomolecular fractions. Following snap-freezing in liquid nitrogen, sample preservation at −80 °C and thawing on ice, almost all cells are left intact (Figures 3a and d). The conceptualized methodological framework relies on an initial cryomilling homogenization and lysis step followed by polar and non-polar metabolite extractions. These steps result in indiscriminate and comprehensive cell disruption and lysis (Figures 3b and d). Following the further addition of dedicated lysis buffers, the vast majority of cells (90.20±6.46% NA-based method mean±s.d.) are lysed (Figures 3c and d). Importantly, as highlighted in Figure 3d, the lysis efficiencies of the methods allowing sequential biomolecular isolations (NA and QA) compare favorably to widely used reference methods for exclusive biomacromolecular isolation, that is, no statistically significant differences are apparent (Kruskal–Wallis, P=0.189, n=10), and they significantly outperform the standard TR-based simultaneous biomolecular isolation method (Kruskal–Wallis, P=0.002, n=10). These results validate the chosen approach for sample preservation, homogenization and cell lysis.
Figure 3.
Efficiencies of different cell lysis methods. (a–c) Representative epifluorescence micrographs of microbial cells from a representative LAO-enriched microbial community sample stained with the Live/Dead stain (intact cells highlighted in green, disrupted cells with a compromised cell membrane in red). Scale bar is equivalent to 10 μm. (a) Sample having undergone a single freeze-thaw cycle. (b) Sample having undergone polar and non-polar metabolite extractions. (c) Sample having undergone the additional mechanical and chemical lysis step using the NA kit's modified lysis buffer. (d) Bar chart highlighting the percentages of cells disrupted by the different treatments (n=10; error bars represent s.d.). x-axis legend: FT, sample having undergone a single freeze-thaw cycle, reflecting panel a; NA, sample having been subjected to cell lysis in the modified NA lysis buffer, reflecting panel c; QA, QA lysis buffer; TR, TR-based lysis; M, sample after polar and non-polar metabolite extractions reflecting panel b; RM-A, sample having been subjected to the dedicated DNA extraction reference method; RM-B, dedicated RNA extraction method; RM-C, dedicated protein extraction method.
Quality and quantity of obtained biomolecular fractions
In addition to the need for efficient cell lysis, the most important consideration is the requirement for obtaining representative high-quality biomolecular fractions. As metabolites are extracted first, the methodology results in identical results for metabolomic analyses if subsequent biomacromolecular isolations are carried out or not. Metabolomic analysis of the representative LAO-enriched microbial community sample resulted in clear and reproducible total ion chromatograms (Supplementary Figure 3). These allowed the robust detection and semi-quantification of 267 polar and 242 non-polar intracellular metabolites as well as 268 polar and 176 non-polar extracellular metabolites (Supplementary Table 2). Using a spectral reference library, this enabled the combined identification of 62 polar and 30 non-polar metabolites. Consequently, the vast majority (>85%) of detected metabolites were not identified in line with earlier metabolomics results obtained on mixed microbial communities (Wilmes et al., 2010).
The nucleic acid fractions obtained using the different extraction protocols were firstly analyzed by a NanoDrop spectrophotometer, the absorbance ratios at 260/280 nm in particular reflecting purity of the respective fractions obtained (Supplementary Table 3). The mean absorbance ratios at 260/280 nm for the DNA fractions are between 1.9 and 2.1 for the methods used, generally accepted as ‘pure' (Manchester, 1995). The only exception is the DNA fraction obtained using the simultaneous biomolecular isolation using the TR-based method for which a poor mean ratio of 1.5 was measured, highlighting the superior performance of the chromatographic spin column-based methods.
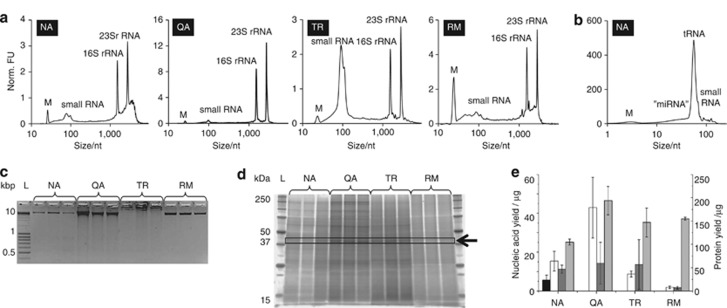
The absorbance ratios at 260/280 nm of all RNA fractions are between 1.8 and 2.1 (Supplementary Table 3), indicating that overall high-quality RNA was extracted using the different protocols. The Agilent Bioanalyzer 2100 electropherograms (Figure 4a) show distinct peaks between 100 and 4500 nt representing the total RNA fractions obtained using the different methods. The 23S/16S rRNA ratios vary depending on the RNA extraction method used from 0.7 to 1.5 (Supplementary Table 3). However, integrity of the isolated RNA was mainly assessed using the RNA integrity number (RIN) score, which is now commonly accepted as a better RNA quality indicator (Fleige and Pfaffl, 2006; Jahn et al., 2008). The RIN scores obtained with the NA-based method (7.03±1.20) are similar to those obtained using the Qiagen RNeasy Mini kit-based reference method (6.60±0.88; Kruskal–Wallis, P=0.806, n=5) and the sequential biomolecular extraction based on the TR-based method (7.44±0.28; Kruskal–Wallis, P=0.352, n=5) but lower than those obtained for the QA-based method (9.68±0.05; Kruskal–Wallis, P=0.011, n=5), which had a very high and consistent score. The RNA fraction obtained using the TR-based method is particularly enriched in small RNAs, indicative of extensive RNA degradation resulting in a non-representative RNA fraction (Figure 4a). Overall, the RNA fractions derived using the simultaneous and sequential biomolecular isolation methods are of sufficient quality for downstream ribosomal RNA removal, reverse transcription and high-throughput cDNA sequencing (He et al., 2010).
Figure 4.
Quality and quantity of biomacromolecular fractions isolated from the representative LAO-enriched microbial community sample using either the NA-, QA- and TR-based method (following prior metabolite extractions) or using the reference methods (no metabolite extractions were carried out before the respective extractions). (a and b) Representative Agilent Bioanalyzer 2100 electropherograms of the total RNA and small RNA fractions, respectively. (c) Agarose gel image highlighting representative genomic DNA fractions obtained (Mean amount (n=3) loaded in μg±s.d., from right to left; NA: 0.35±0.17; QA: 0.35±0.08; TR: 0.83±0.85; RM: 0.08±0.04) and (d) SDS-PAGE image of representative protein fractions (Mean amount (n=3) loaded in μg±s.d., from right to left; NA, first elution: 3.20±0.19; QA: 5.44±1.06; TR: 3.88±0.30; RM: 4.62±0.09). The arrow and the box represent the dominant gel band which was submitted to tryptic digestion and MALDI-ToF/ToF analysis. (e) Biomacromolecular yield obtained for the  small RNA, □ total and large RNA,
small RNA, □ total and large RNA,  DNA and
DNA and  protein (first elution) fractions (n=5, error bars represent s.d.). FU, fluorescent unit; L, ladder; M, marker; NA, NA-based method; Norm., normalized; nt, nucleotides; RM, reference methods; TR, TR-based method; QA, QA-based method.
protein (first elution) fractions (n=5, error bars represent s.d.). FU, fluorescent unit; L, ladder; M, marker; NA, NA-based method; Norm., normalized; nt, nucleotides; RM, reference methods; TR, TR-based method; QA, QA-based method.
The NA-based method allows for the additional subfractionation of the total RNA extracted into a small RNA fraction (Figure 4b). As expected, the major components of the small RNA fractions are transfer RNAs with a mean size of around 60 nt and other small RNAs of larger size, represented by multiple peaks around 120 nt, for example, 5S rRNA. A ‘miRNA'-like region is observed as a broad peak ranging from 10 to 40 nt (Figure 4b). The ‘miRNA' region relative to small RNA content (‘miRNA'/small RNA ratio) is 26.43±1.97% for the NA-based method. Owing to the important regulatory roles fulfilled by small bacterial regulatory RNAs, the small RNA fraction, only provided by the NA-based method, adds an important additional level of information to the integrative molecular analyses that can be carried out using the reported methodology.
The size, the quality (degraded versus intact) and semi-quantitative amount of DNA extracted were determined by gel electrophoresis (Figure 4c). Rather expectably, the dedicated reference method results in the best DNA quality extract (Figure 4c, lane RM). The genomic DNA fraction isolated using the sequential biomolecular extraction protocol based on NA provides the most similar results to the reference method. Sequential biomolecular isolation based on QA also results in DNA extracts exhibiting intense and large bands. Importantly, the TR-based extraction method results in poor quality DNA extracts (Figure 4c), in concordance with the low-absorbance ratios measured at 260/280 nm (Supplementary Table 3) and, thus, is rather ill suited for comprehensive biomolecular isolations.
Gel electrophoresis of protein fractions (SDS-PAGE) provides a visual representation of the community metaproteomes derived from the representative LAO-enriched microbial community samples (Figure 4d). Importantly, in terms of band diversity and clarity, the efficiency of protein extraction is superior for the sequential and simultaneous isolation protocols (Figure 4d, lanes NA, QA and TR) compared to those obtained using the reference method (Figure 4d, lane RM). This is most likely due to the removal of ‘contaminant' biomolecular fractions (for example, small molecules, nucleic acids) from the protein fraction during the sequential or simultaneous isolation methods. In contrast, some of these biomolecules may be retained in the extracts obtained with the reference method. In summary, the developed methodology results in higher quality protein extracts than the dedicated protein extraction method.
To further assess the compatibility of the protein fractions obtained with the NA-based method with subsequent downstream analyses, a protein band of ∼45 kDa was excided from an SDS-PAGE gel (Figure 4d) and analyzed by MALDI-ToF/ToF following tryptic in-gel digestion. A de novo peptide sequence was derived from a clear tandem mass spectrum (Supplementary Figure 4). The peptide (de novo sequence: VESITAPVVVTEDQTQR) has been putatively identified as a possible cell surface protein (Corynebacterium glucuronolyticum spp.; GI: 227541404, e-value=3.7, score=31.6, BLASTP against NCBInr). Overall, these results highlight the ability to carry out proteomic analyses using either gel- or liquid chromatography-based separation followed by mass spectrometry on the obtained protein fractions.
In terms of yields, larger quantities of nucleic acids were obtained using the chromatographic spin column-based methods (NA and QA) compared with the reference methods (Kruskal–Wallis, P=0.009, n=5; Figures 4c and e and Supplementary Table 3). An analogous finding was previously highlighted in a comparative analysis of RNA yields obtained using a dedicated RNA extraction method and the simultaneous biomacromolecular extraction method based on TR (Chomczynski, 1993). In terms of protein quantity (Figures 4d and e), the overall yield is highest for the QA-based method, followed by the exclusive isolation method. However, extensive variation in terms of the quantities of obtained biomolecular fractions is apparent for the QA-based isolation methodology. In contrast, the NA-based method results in the most consistent results for the tested sequential/simultaneous extraction methods.
Broad applicability of the methodological framework
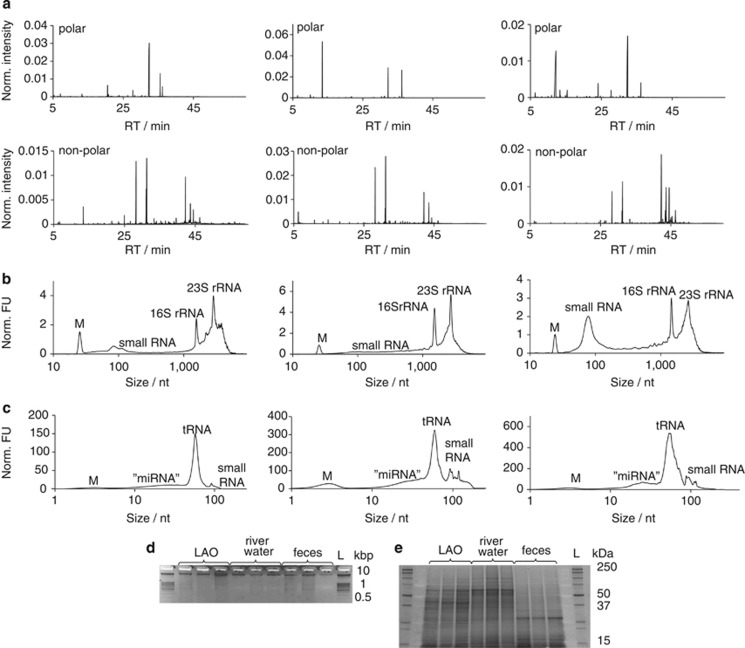
The broad applicability of the developed methodology was demonstrated by applying the NA-based method to two additional mixed microbial community samples of environmental and biomedical research interest, that is, river water filtrate and human feces. Some minor sample-specific pretreatments, for example, RNAlater treatment of the fecal samples, were necessary to guarantee sufficient recovery of biomolecules. Analyses of the respective biomacromolecular fractions' quality and quantity resulted in no apparent differences compared with those obtained from the LAO-enriched microbial community (Kruskal–Wallis, 0.054<P<0.729, n=5, Figure 5, Supplementary Table 4 and Supplementary Table 5). To ascertain the quality of the obtained DNA for further downstream analyses, each DNA fraction from the three environmental samples was subjected to PCR amplification with primers targeting the 16S rRNA gene. For each microbial community DNA extract, clear and distinct bands with the correct molecular size were obtained (Supplementary Figure 5), highlighting the possibility for specialised downstream analyses without the need for additional purification steps.
Figure 5.
Application of the developed biomolecular isolation methodology to a LAO-enriched microbial community (n=3), river water filtrate (n=3) and human feces (n=3). (a–c) Left-hand panels represent LAO-enriched microbial communities, middle panels represent river water filtrate and right-hand panels represent human feces. (a) Representative GC–MS total ion chromatograms of polar and non-polar metabolite fractions. Representative Agilent Bioanalyzer 2100 electropherograms of the (b) total RNA fractions and (c) small RNA fractions. (d) Agarose gel electrophoresis image of the genomic DNA fractions (Mean amount loaded in μg±s.d., right to left; LAO: 0.63±0.28; river water: 0.35±0.03; feces: 0.61±0.26) for each of the three technical replicates considered. (e) SDS-PAGE image of protein fractions, first elution (Mean amount loaded in μg±s.d., right to left; LAO: 1.19±0.40; river water: 1.70±0.63; feces: 1.19±1.21) for each of the three technical replicates considered. L, ladder; RT, retention time.
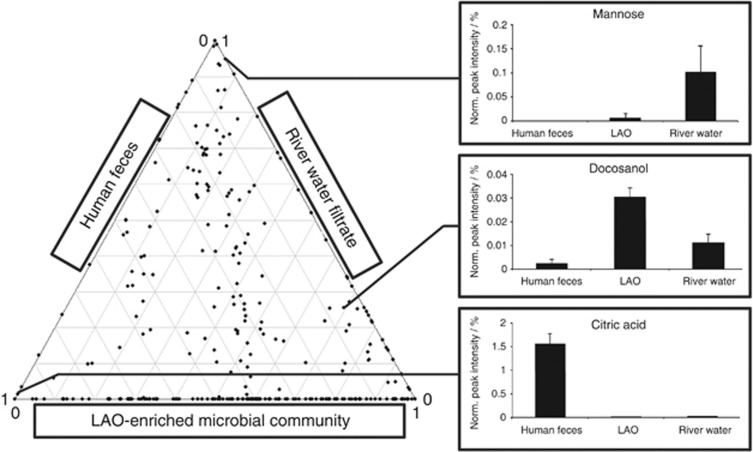
Use of the developed standardized methodology further allows comparative analysis of different microbiomes. For example, it has not escaped our notice that an apparent enrichment in small RNA exists in human fecal samples (Figure 4b). Furthermore, to highlight potential signature metabolites for the three analyzed microbial communities, we contrasted the relative abundances of detected metabolites for each microbiome. For this, the metabolomic data sets from the respective microbiomes were subjected to a three-way comparative analysis (Supplementary Materials and methods). Clear differences in the metabolite composition of the three microbiomes are apparent, allowing the identification of specific signature metabolites for three microbiomes (Figure 6). For example, the enrichment of docosanol, a saturated fatty alcohol, in the LAO-enriched microbial community from the wastewater treatment plant may be explained by the wide use of this molecule as an emollient, emulsifier and thickener in cosmetics, nutritional and pharmaceutical products. Consequently, its presence may be expected in domestic wastewater and through its chemical properties would be enriched in LAOs. The pronounced enrichment of mannose, a simple sugar, in river water filtrate may be a direct result of photosynthesis by the dominant phototrophic organisms. Finally, a comparatively strong enrichment in citric acid was found in the human fecal samples. An increase of all metabolites involved in energy metabolism including citric acid was previously found in the serum of mice conventionally raised versus germ-free mice (Velagapudi et al., 2010). Our present results indicate that this elevated level may directly result from the metabolic activities of microbial communities in the gastrointestinal tract.
Figure 6.
Three-way comparison of microbial community metabolomes obtained from a LAO-enriched microbial community, river water filtrate and human feces. Each dot represents the relative abundance of a metabolite in the three different environmental samples. Exemplary metabolite signatures for each microbiome are highlighted with their respective abundance levels on the right. Error bars represent s.d.
By applying the NA-based workflow to these two additional mixed microbial communities, we demonstrate that the methodological framework can be applied to a range of different samples and, thus, represents a standardized biomolecular isolation procedure for comparative eco-systems biology investigations on a range of different microbial communities in the future.
Discussion
With the advent of high-throughput omic technologies, powerful and sensitive methods are available for the analysis of nucleic acids (DNA and RNA), proteins and small molecules obtained from biological samples including microbial communities (Jansson et al., 2012). Molecular eco-systems biology investigations, centered around the integration of omic data from microbial communities, are faced with major challenges arising from the complexity, dynamics and heterogeneity of microbial consortia. In the present work, we demonstrate that microbial community metabolomes do not allow certain biological and technical replicates to be discriminated, which is a direct consequence of the extensive heterogeneity encountered in these systems. Consequently, major artifacts may be introduced into coupled high-resolution omic experiments by sample splitting before dedicated isolation of the respective individual biomolecular fractions. Such approaches will result in disparate omic results and, thus, will not fulfill the premise of systematic measurements. Furthermore, they will not allow meaningful in silico reconstructions and modeling of community-wide processes and interactions.
Here, we describe a methodological framework that allows for the sequential extraction and purification of all known biomolecular fractions from single unique samples. The methodology is based on the combination of chromatographic spin column-based biomacromolecular isolation with prior extraction of concomitant polar and non-polar metabolite fractions. We demonstrate that the devised methodology yields equivalent or even better results compared with those obtained with dedicated extraction protocols, and that it is applicable to a range of different microbial community samples. Furthermore, the methodology may prove successful on a range of other biological samples, in particular those that are precious or which are characterized by extensive within-sample heterogeneity, for example, human tumor samples. The utilization of spin column chromatography for biomacromolecular separation minimizes the risk of exposure to harmful chemicals and, thus, allows routine laboratory use. Finally, the methodological framework is potentially compatible with sequential biomacromolecular isolation methods other than those used in the present work, for example, the GE Healthcare illustra triplePrep kit.
The reported methodological framework provides the basis for conducting molecular eco-systems biology investigations on a range of different mixed microbial communities in the future. The methodology will facilitate meaningful data integration, analysis and modeling of metabolomic, transcriptomic, genomic and proteomic data. Furthermore, it will allow exploitation of the inherent heterogeneity through spatial and temporal sampling of microbial communities to allow deconvolution of community-wide, population-wide and individual-level processes using statistical approaches. Consequently, it will allow the determination of associations between distinct biomolecules that may provide pointers towards the discovery of previously unknown metabolic processes linked to specific community members, a need which is highlighted by the relatively large number of uncharacterized metabolites detected in the metabolomic experiments carried out as part of the present work. Finally, by providing a standardized workflow, the methodology lays the foundation for future comparative molecular studies of different microbial ecosystems leading to further discovery of functional microbiome-specific biomolecular signatures.
Acknowledgments
The scientists and technical staff of the Centre de Recherche Public Gabriel-Lippmann, particularly Dr Christian Penny, Cécile Walczak and Sébastien Planchon, and from the Luxembourg Centre for Systems Biomedicine, particularly Dr Pranjul Shah and Laura Lebrun, are thanked for their assistance and support. Mr Bissen and Mr Di Pentima from the Syndicat Intercommunal à Vocation Ecologique (SIVEC) are thanked for their permission to collect samples and gain access to the monitoring platform of the Schifflange wastewater treatment plant. Rudi Balling and Anders Andersson are thanked for helpful comments on the manuscript. The present work was supported by an ATTRACT programme grant to PW (ATTRACT/A09/03) and Aide à la Formation Recherche (AFR) grants to HR (PHD-MARP-04) and EM (PRD-2011-1/SR), all funded by the Luxembourg National Research Fund (FNR).
PW and HR designed the research; HR, PW, TC and KH planned the experiments; HR, EM, PW, JR and TC carried out the analyses; HR, PW and EM analyzed the data; and HR, EM and PW wrote the manuscript.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Chey S, Claus C, Liebert UG. Improved method for simultaneous isolation of proteins and nucleic acids. Anal Biochem. 2011;411:164–166. doi: 10.1016/j.ab.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–536. [PubMed] [Google Scholar]
- Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Fischer CR, Wilmes P, Bowen BP, Northen TR, Banfield JF.2011Deuterium-exchange metabolomics reveals N-methyl lyso phosphatidylethanolamines as abundant lipids in acidophilic mixed microbial communities Metabolomicsadvance online publicationdoi: 10.1007/s11306-011-0344-x [DOI]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wurtzel O, Singh K, Froula JL, Yilmaz S, Tringe SG, et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Meth. 2010;7:807–812. doi: 10.1038/nmeth.1507. [DOI] [PubMed] [Google Scholar]
- Hummon AB, Lim SR, Difilippantonio MJ, Ried T. Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42:467–472. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn CE, Charkowski AO, Willis DK. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods. 2008;75:318–324. doi: 10.1016/j.mimet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Jansson JK, Neufeld JD, Moran MA, Gilbert JA. Omics for understanding microbial functional dynamics. Environ Microbiol. 2012;14:1–3. doi: 10.1111/j.1462-2920.2011.02518.x. [DOI] [PubMed] [Google Scholar]
- Kitano H. Foundations of Systems Biology. The MIT Press: Cambridge, Massachusetts, London, England; 2001. [Google Scholar]
- Levantesi C, Rossetti S, Thelen K, Kragelund C, Krooneman J, Eikelboom D, et al. Phylogeny, physiology and distribution of ‘Candidatus Microthrix calida', a new Microthrix species isolated from industrial activated sludge wastewater treatment plants. Environ Microbiol. 2006;8:1552–1563. doi: 10.1111/j.1462-2920.2006.01046.x. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- Manchester KL. Value of A260/A280 ratios for measurement of purity of nucleic acids. Biotechniques. 1995;19:208–210. [PubMed] [Google Scholar]
- Morse S, Shaw G, Larner S. Concurrent mRNA and protein extraction from the same experimental sample using a commercially available column-based RNA preparation kit. Biotechniques. 2006;40:54–58. doi: 10.2144/000112100. [DOI] [PubMed] [Google Scholar]
- Radpour R, Sikora M, Grussenmeyer T, Kohler C, Barekati Z, Holzgreve W, et al. Simultaneous isolation of DNA, RNA, and proteins for genetic, epigenetic, transcriptomic, and proteomic analysis. J Proteome Res. 2009;8:5264–5274. doi: 10.1021/pr900591w. [DOI] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Micro. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- Tolosa JM, Schjenken1 JE, Civiti TD, Clifton VL, Smith R. Column-based method to simultaneously extract DNA, RNA, and proteins from the same sample. Biotechniques. 2007;43:799–804. doi: 10.2144/000112594. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Wenzel K, Fiehn O. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics. 2004;4:78–83. doi: 10.1002/pmic.200200500. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Bond PL. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol. 2004;6:911–920. doi: 10.1111/j.1462-2920.2004.00687.x. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Bowen BP, Thomas BC, Mueller RS, Denef VJ, VerBerkmoes NC, et al. Metabolome-proteome differentiation coupled to microbial divergence. mBio. 2010;1:e00246–00210. doi: 10.1128/mBio.00246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P, Simmons SL, Denef VJ, Banfield JF. The dynamic genetic repertoire of microbial communities. FEMS Microbiol Rev. 2009;33:109–132. doi: 10.1111/j.1574-6976.2008.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Yang Q, Kang J, Sun Y, Zhang T, Margaret G, et al. Simultaneous isolation of DNA, RNA, and protein from Medicago truncatula L. Electrophoresis. 2011;32:321–330. doi: 10.1002/elps.201000425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.