Abstract
Very little is known about growth rates of individual bacterial taxa and how they respond to environmental flux. Here, we characterized bacterial community diversity, structure and the relative abundance of 16S rRNA and 16S rRNA genes (rDNA) using pyrosequencing along the salinity gradient in the Delaware Bay. Indices of diversity, evenness, structure and growth rates of the surface bacterial community significantly varied along the transect, reflecting active mixing between the freshwater and marine ends of the estuary. There was no positive correlation between relative abundances of 16S rRNA and rDNA for the entire bacterial community, suggesting that abundance of bacteria does not necessarily reflect potential growth rate or activity. However, for almost half of the individual taxa, 16S rRNA positively correlated with rDNA, suggesting that activity did follow abundance in these cases. The positive relationship between 16S rRNA and rDNA was less in the whole water community than for free-living taxa, indicating that the two communities differed in activity. The 16S rRNA:rDNA ratios of some typically marine taxa reflected differences in light, nutrient concentrations and other environmental factors along the estuarine gradient. The ratios of individual freshwater taxa declined as salinity increased, whereas the 16S rRNA:rDNA ratios of only some typical marine bacteria increased as salinity increased. These data suggest that physical and other bottom-up factors differentially affect growth rates, but not necessarily abundance of individual taxa in this highly variable environment.
Keywords: ribosome, growth, estuaries, pyrosequencing, 16S rRNA
Introduction
The structure of bacterial communities and the abundances of individual bacterial taxa have been examined extensively in aquatic ecosystems over the past 30 years as have bulk properties of bacterial communities, such as biomass production and community-level growth rates (Ducklow, 2000; Morris et al., 2005; Gilbert et al., 2009; Fouilland and Mostajir, 2010). However, few studies have examined growth rates of specific taxa in natural microbial communities. We need to know growth properties in order to gage the contribution of bacteria to carbon cycling and other biogeochemical processes. Growth and general levels of metabolic activity may be explored by examining cell properties that change with growth rate, such as the number of ribosomes per cell. In cultures of non-marine and marine bacteria (Kemp et al., 1993; Bremer and Dennis, 1996; Fegatella et al., 1998; Kerkhof and Kemp, 1999; Deutscher, 2006), ribosome numbers increase with faster growth and decrease during starvation, although the levels and rates of decline may differ in different taxa (Kramer and Singleton, 1992; Nilsson et al., 1997; Fegatella et al., 1998).
Recently, we and others used 16S rRNA relative abundance and the ratio of 16S rRNA to rRNA genes (rDNA) as indexes of activity and potential growth rate of specific taxa in natural communities (Campbell et al., 2009; Jones and Lennon, 2010; Gaidos et al., 2011). There was a positive correlation between 16S rRNA and rDNA frequencies in both coral sediments and coastal surface bacterial communities (Campbell et al., 2011; Gaidos et al., 2011), suggesting that activity followed abundance in these cases. However, rare bacteria were found to have higher levels of 16S rRNA (ribosomes) per rDNA (cell number) in both lakes and in surface marine communities, suggesting that growth rates were higher (Jones and Lennon, 2010; Campbell et al., 2011). These data suggest that although the most abundant bacteria often did not have the highest growth rates, they still contributed the most to production or ‘activity' on a per volume basis in the few systems examined so far. Clearly, more work is needed to understand the relationship between abundance, activity and potential growth rates and how these relationships are affected by environmental factors.
Estuaries are ideal systems for exploring these questions because they are dynamic with gradients in salinity and other environmental factors, as well as in microorganisms (Kirchman et al., 2005; Sharp et al., 2009; Telesh and Khlebovich, 2010; Fortunato et al., 2012). One well-studied estuary, the Delaware Bay, has the biggest salinity gradient of the three largest urban estuaries in the United States (Sharp et al., 2009). Bacterial community structure at the phylum and subphylum level changes predictably in the Delaware Bay and in other estuaries. At the freshwater end of the estuarine gradient, bacterial communities are dominated by Betaproteobacteria and Actinobacteria, while marine communities are dominated by Alphaproteobacteria; levels of the Bacteroidetes phylum are fairly constant along the estuary (Bouvier and del Giorgio, 2002; Crump et al., 2004; Kirchman et al., 2005; Kan et al., 2008; Fortunato et al., 2012; Herlemann et al., 2011). These changes in community structure may be driven by changes in growth rates, but the relationships between abundance and growth are complex, even at the phylum or subphylum levels (Cottrell and Kirchman, 2004; Yokokawa et al., 2004). Although these studies point to differences in growth at the phylum or class level, nothing is known about activity or growth rates at the operational taxonomic unit (OTU) level along estuarine gradients.
Estuaries often have large numbers of particle-attached bacteria, sometimes making up >50% of the total community, more than that observed in other marine systems (Kirchman, 1993; Crump et al., 1998; Simon et al., 2002). These bacteria probably disproportionally contribute to bacterial production and particulate organic matter degradation as suggested by studies that explored per cell rates by normalizing bulk activity measures by cell abundance of particle-attached and free-living bacteria (Crump and Baross, 2000; Ghiglione et al., 2007). It remains to be seen if a more direct method also shows that particle-attached bacteria grow faster than free-living bacteria.
The goals of this study were to learn more about potential growth rates and activity of individual bacterial taxa in a dynamic aquatic environment and, in particular, to see if the positive correlation between 16S rRNA and rDNA that was observed at the coastal Delaware site (Campbell et al., 2011) held over a large environmental gradient. We examined diversity, composition and activity of bacteria along the Delaware Bay using a tagged 16S rRNA/rDNA pyrosequencing approach. This method allowed a detailed characterization of bacterial richness, evenness, compositional changes and activity of the bacterial communities at the OTU level. Samples were taken over 2 days in July 2009 along a 200 km transect of the Bay, spanning both freshwater and marine sites. The relative abundance and activity of taxa, defined by 16S rDNA and rRNA sequences sharing 97% similarity, were characterized for both whole water and free-living communities. We found marked changes in diversity, composition and 16S rRNA:rDNA ratios of taxa along the salinity gradient.
Materials and methods
Site description, sample collection and characterization
Surface water samples were collected along the Delaware Bay and nearby at a coastal ocean observatory site termed FB in July 2009 (Supplementary Table S1). Standard oceanographic properties, including water temperature, salinity, light intensity (photosynthetically active radiation), light attenuation, abundance of total bacterioplankton, leucine incorporation, chlorophyll a concentration and nutrients (NO3, NH4, PO4, Si), were measured as described previously (Preen and Kirchman, 2004; Kirchman et al., 2005; Cottrell et al., 2006; Michelou et al., 2007). Samples for nucleic acids were either directly collected on 0.22 μm Durapore membranes or prefiltered through 0.8 μm pore size filters before collection on 0.22 μm membranes. Filters were frozen at −80 °C in 1 ml of CTAB buffer (Dempster et al., 1999) until nucleic acids were extracted.
Nucleic acid extractions and high-throughput sequencing
Total nucleic acids were extracted using a modified CTAB extraction protocol (Dempster et al., 1999) with two chloroform extractions instead of one. DNA was separated from RNA via RNAase I digestion, and RNA was isolated from the total nucleic acid preparation by DNAase I digestion of the sample as outlined in the manufacturer's protocol (Ambion/Life Technologies, Grand Island, NY, USA). DNA and RNA were quantified via a picogreen or ribogreen assay, respectively, as recommended by the manufacturer (Invitrogen/Life Technologies, Grand Island, NY, USA). RNA samples did not have contaminating DNA, according to a polymerase chain reaction test for 16S rRNA genes. Approximately 50 ng of RNA from each sample were reverse transcribed into cDNA with random primers using the Superscript First-Strand synthesis system for reverse transcription-polymerase chain reaction (Invitrogen). Nucleic acids (DNA at 10 ng μl−1 and cDNA at 1 ng μl−1) were sent to the Research and Testing Laboratory (http://www.researchandtesting.com/, Lubbock, TX, USA) for high-throughput sequencing on the Roche 454 platform with titanium chemistry. Primers used spanned the V1–V3 region (28F-519R) of the 16S rRNA gene.
Sequences are available in the NCBI short-read archive database, accession number SRA052537. A MIMARKS compliant table of sample metadata is included in the Supplementary Table S1.
Sequence analyses
Sequences were run through the AmpliconNoise/Perseus pipeline (Quince et al., 2011) to remove noise and chimeras from the original 493 031 SFF files. The resulting 250 478 sequences were then passed through the mothur program to further reduce errors as outlined previously (Schloss et al., 2009; Campbell et al., 2011). Briefly, after trimming, pre-clustering, removal of chloroplast sequences and alignments, a total of 138 532 sequences remained. These were clustered with the average neighbor algorithm at a 0.03 distance, which resulted in 2446 OTUs. Before comparisons, samples were normalized to a total of 1675 sequences each by randomly resampling the sequence data 100 times using the sample function in R (http://www.r-project.org/).
Alpha diversity measures (ChaoI, Sobs, Simpsoneven, Invsimpson) of the samples were calculated in mothur (Schloss et al., 2009). A representative sequence from each OTU was classified by several methods, including Silva, RDPII and greengenes web alignment and classification tools, as well as by BLAST analyses (DeSantis et al., 2006; Pruesse et al., 2007; Johnson et al., 2008; Cole et al., 2009). In general, the results from all classification schemes were consistent with each other (data not shown). Phylogenetic distances were calculated in MEGA5.05 using the maximum composite likelihood pairwise distance method after Clustal alignment (Tamura et al., 2011). Beta diversity measures (non-metric multidimensional scaling and the Theta (yc) similarity index) were also performed in mothur using the rarefied OTU table (Schloss et al., 2009). Sample clustering was also performed with the Bray–Curtis algorithm in PAST based on normalized abundances (Hammer et al., 2001).
Statistical analyses
Unless otherwise noted, data were square root or natural log transformed before statistical testing. Since assumptions of parametric regressions were not satisfied, nonparametric (Spearman and Kendall) regression analyses performed in R (http://www.r-project.org/) were used when all OTUs were analyzed. Reduced (or standard) major axis regression analysis was also performed in R with the lmodel2 function on individual OTU values.
Results
Environmental characterization of the Delaware Bay
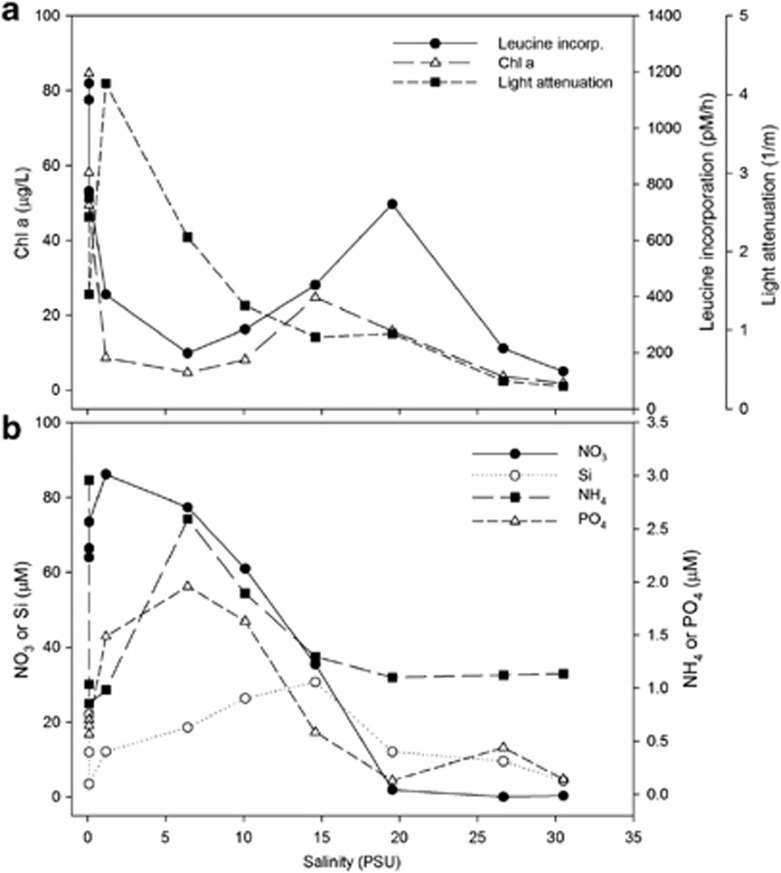
Surface water samples were examined for a variety of biogeochemical parameters, including chlorophyll a, leucine incorporation, light attenuation and inorganic nutrients in relation to salinity (Figure 1). There were two peaks of chlorophyll a concentration at the freshwater sites and mid-salinity sites (15–20 PSU; Figure 1a). Bacterial production was highest after the mesohaline chlorophyll a peak, while light attenuation was highest at about 1 PSU (Figure 1a). Most nutrients, except Si, peaked in the oligohaline sites (1–6 PSU) (Figure 1b). Si concentrations were highest at the same station as the second chlorophyll a peak (15 PSU).
Figure 1.
Environmental parameters along the Delaware Bay in relation to salinity in July 2009. (a) Leucine incorporation, chlorophyll a and light attenuation. (b) Concentrations of nitrate, silicate, phosphate and ammonium.
Bacterial diversity along the salinity gradient
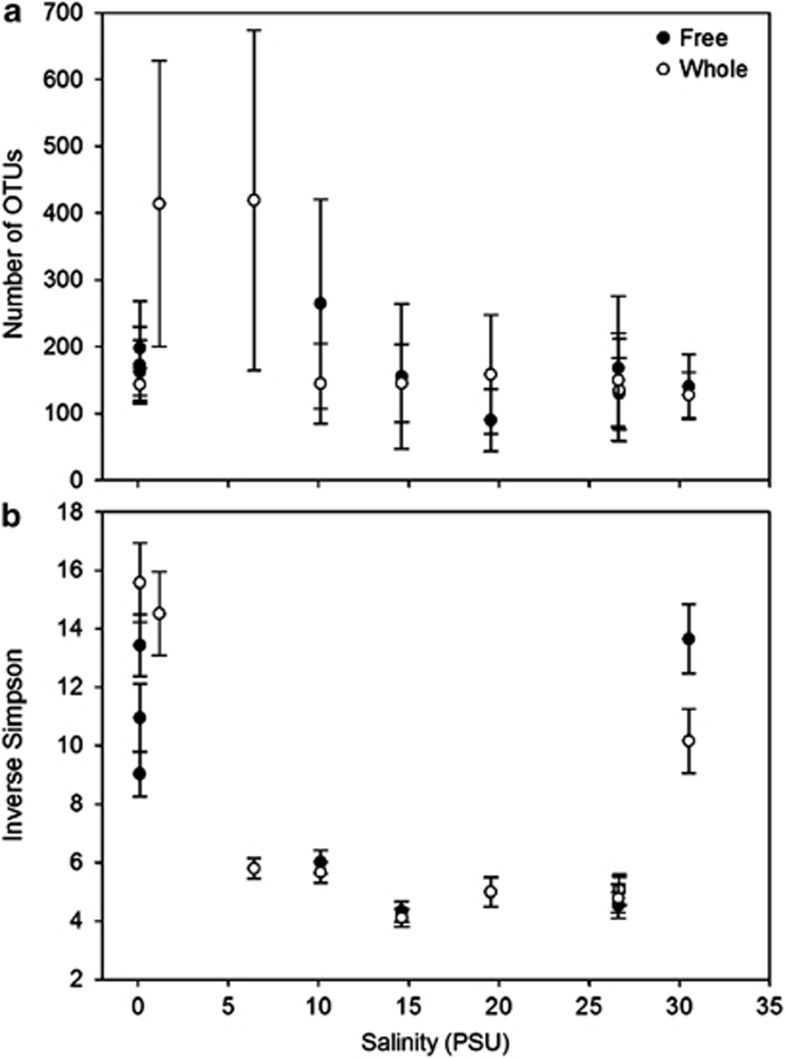
Species richness, evenness and alpha diversity of free-living (<0.8 μm) and of the entire (>0.22 μm) bacterial communities were assessed at nine sites along the Delaware Bay after trimming and normalization of the sequence data. Species richness according to the nonparameteric Chao1 index varied similarly between the two fractions along the salinity gradient (Figure 2a). The richness of the entire bacterial community was highest at 1.2 and 6.4 PSU, and declined to less than half that in the lowest and highest salinity samples. Although not statistically significant, similar trends in the Chao1 index were observed for the free-living community, and with richness estimated by the number of observed species for both communities (data not shown). In contrast to the Chao1 index, the lowest and highest salinity samples had significantly higher overall diversity (Inverse Simpson) than the other samples for both communities (Figure 2b). Identical patterns to the diversity values were observed in evenness as measured by the Simpson evenness calculator (data not shown).
Figure 2.
Bacterial richness (a) and diversity (b) along the Delaware Bay in relation to salinity. OTUs were defined at a 97% similarity level. Error bars indicate 95% confidence interval calculated in mothur. Closed circles indicate bacteria from the free-living fractions, whereas open circles indicate bacteria from the whole water fraction.
Overall bacterial community structure in the Delaware Bay
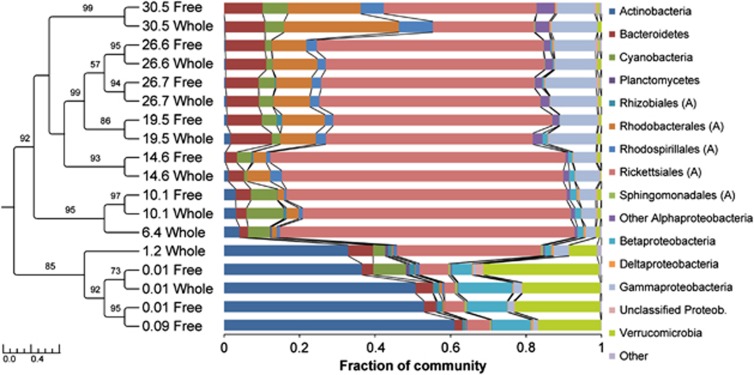
As expected, bacterial community composition changed greatly along the salinity gradient, from a community dominated by Actinobacteria, Verrucomircobia and Betaproteobacteria in fresh waters to a typical marine community dominated by SAR11 taxa (∼40%), Rhodobacterales (∼20%), Gammaproteobacteria (∼15%) and Bacteroidetes (∼15%) in the highest salinity, coastal waters (Figure 3). The variation in composition was mirrored in the similarity dendrogram, where the freshwater and very low salinity samples (PSU 0.01–1.2) formed a group separate from the rest of the samples (Figure 3). There were also specific subclusters within the moderate to high salinity samples, which also grouped according to salinity and were driven mainly by relative abundance changes in Cyanobacteria and Rhodobacterales. Interestingly, the largest group of samples spanned a salinity range from 14.6 to 26.7 PSU. The coastal ocean sample, which differed in salinity only slightly from the Bay mouth samples, formed a separate subcluster, probably due to the higher relative abundance of the Rhodobacterales. In all cases where paired samples could be compared, free-living and whole water communities clustered together and shared similar abundances of most groups (Figure 3).
Figure 3.
Bacterial composition along the Delaware Bay salinity gradient. Bray–Curtis dendrogram of 16S rDNA sequences clustered into OTUs at 97% similarity, followed by colored bars indicating the percentage of the designated group within each sample. (A)=Alphaproteobacteria. Numbers in dendrogram refer to salinity (PSU) of sample. Bootstrap values (>50) are listed. Scale bar indicates distance in length.
Some taxa were dominant in more than one salinity range. For instance, the three SAR11 taxa (OTUs 124, 125 and 267) comprised up to 70% of the community in both the mid-salinity and marine sites. Other taxa were most abundant within defined salinity ranges (Supplementary Table S2). The freshwater samples contained the greatest number of unique taxa, with a total of 41 OTUs that made up at least 0.5% of the community. Most of these OTUs belonged to the Verrucomicrobia (nine OTUs) and Actinomycetales (eight), with the other abundant taxa being within the Chloroflexi and Comamonadaceae groups (four each). The most selective salinity range was the mesohaline (8–10 PSU), where only two taxa, members of the Actinomycetales and Planctomycetales, were found in their highest abundance in this range. About the same number of taxa (10–12) reached their highest abundance in the mid-salinity (15–27 PSU) or marine sites. Four taxa in the mid-salinity range belonged to the Flavobacteriaceae, while three belonged to the Actinobacteria. The group with the largest number of taxa in the marine site was Gammaproteobacteria (five), followed by Flavobacteriaceae (three).
16S rRNA vs rDNA and 16S rRNA:rDNA ratios of bacteria in whole water and free-living communities
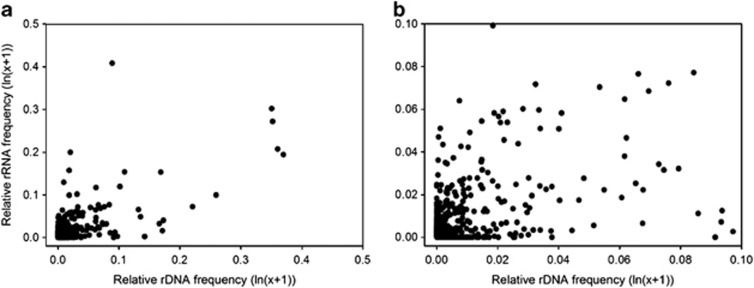
The relationship between 16S rRNA and rDNA frequency for each OTU and sample was examined in both whole water and free-living fractions (Figure 4 and Supplementary Figure S1). The correlations between rRNA and rDNA frequencies were low or even negative according to Kendall's nonparametric test (free-living τ=−0.04, P<0.001, n=1568; whole water τ=−0.10, P<0.001, n=1956) (Figure 4b and Supplementary Figure S1b), suggesting that overall, the contribution of all phylotypes to activity (rRNA frequency) did not follow their relative abundance (rDNA frequency).
Figure 4.
Relationship between 16S rRNA and 16S rDNA frequencies of bacterial OTUs along the Delaware Bay salinity gradient. The points are paired 16S rRNA and rDNA frequencies for each individual OTU in the free-living community at different salinities. (a) Entire data set and (b) <0.5% abundant OTUs.
We next asked if the 16S rRNA:rDNA ratios for the rare and abundant bacteria were different, as was the case in our previous study (Campbell et al., 2011). The average ratios for the whole water fraction were different, but not for the free-living fraction. The average 16S rRNA:rDNA ratios from the abundant (>1% rDNA) and rare (<1% rDNA) free-living populations were the same (average of about 1.2 for both communities). In contrast, the ratio for rare bacteria (average=1.6) was significantly higher than the ratio for abundant bacteria (average=1.1) in the whole water community (n=131, 1003, P<0.05, t-test).
16S rRNA vs rDNA and 16S rRNA:rDNA ratios of individual taxa
The relationship between the 16S rRNA and rDNA from individual OTUs was characterized to understand why there was no positive correlation in the entire data set. In all, 46 OTUs, defined at the family, order or phylum level, were chosen based on their relative abundance (frequency >0.5) and presence in at least three of the samples (Table 1 and Supplementary Table S3). About 40% of the taxa had a significant correlation between 16S rRNA and rDNA. Most of the significant relationships were within the Rhodobacteriaceae, Rhodospirillaceae, SAR11 and Gammaproteobacteria groups (Table 1 and Supplementary Table S3). Overall, more taxa had significant correlations in the free-living fraction than in the whole water fraction (Table 1), reflecting patterns in the entire community (Figure 4 and Supplementary Figure S1). The average correlation coefficients were higher in the Rhodobacteriaceae and Rhodospirillaceae and lower in the Gammaproteobacteria in the whole water communities compared to the free-living communities (Table 1). All other groups either had no or few representative OTUs with significant correlations between relative abundances of 16S rRNA and rDNA.
Table 1. Relationships of rRNA to rDNA of OTUs within the indicated group along the Delaware Bay salinity gradient using RMA regression analysis.
| Group |
Free
living |
Whole
community |
||||
|---|---|---|---|---|---|---|
| No. of OTUs | % Sig a | r | No. of OTUs | % Sig | r | |
| Actinobacteria | 6 | 0 | 5 | 20 | 0.97 | |
| Alphaproteobacteria | ||||||
| Rhodobacterales | 5 | 60 | 0.92±0.11 | 5 | 60 | 0.96±0.03 |
| Rhodospirillales | 6 | 33 | 0.87±0.05 | 6 | 66 | 0.94±0.06 |
| Rickettsiales | 4 | 100 | 0.93±0.02 | 4 | 75 | 0.93±0.02 |
| Betaproteobacteria | ||||||
| Burkholderiales | 3 | 0 | 3 | 0 | ||
| Deltaproteobacteria | 1 | 0 | 1 | 0 | ||
| Gammaproteobacteria | 11 | 45 | 0.92±0.06 | 11 | 55 | 0.89±0.05 |
| Chloroflexi | 1 | 0 | 0 | |||
| Cyanobacteria | 1 | 100 | 0.82 | 1 | 0 | |
| Flavobacteriales | 4 | 25 | 0.91±0.15 | 3 | 33 | 0.97 |
| Verrucomicrobia | 3 | 0 | 3 | 0 | ||
Abbreviations: OTU, operational taxonomic unit; RMA, reduced major axis.
The correlation coefficients (r) are averages±s.d. based on OTUs with significant correlations.
Percentage of OTUs with significant correlations between 16S rRNA and 16S rDNA.
As with the relative abundance estimates, 16S rRNA:rDNA ratios of some taxa were highest within certain salinity ranges (Supplementary Table S4). The ratios of OTUs within the Flavobacteriaceae (four), Rhodobacteriaceae (three) and Gammaproteobacteria (three) were highest within the 15–27 PSU range. The ratios of two Alphaproteobacteria within the Rhodobacteriaceae, closely related to Paracoccus aminovorans and P. alcaliphilus, were highest within the 8–10 and 15–27 PSU range, respectively. Notably, taxa related to SAR11 had their highest ratios at the marine site, whereas the ratios of Actinomycetales were highest in the freshwater sites.
Variation of 16S rRNA:rDNA ratios along the salinity gradient
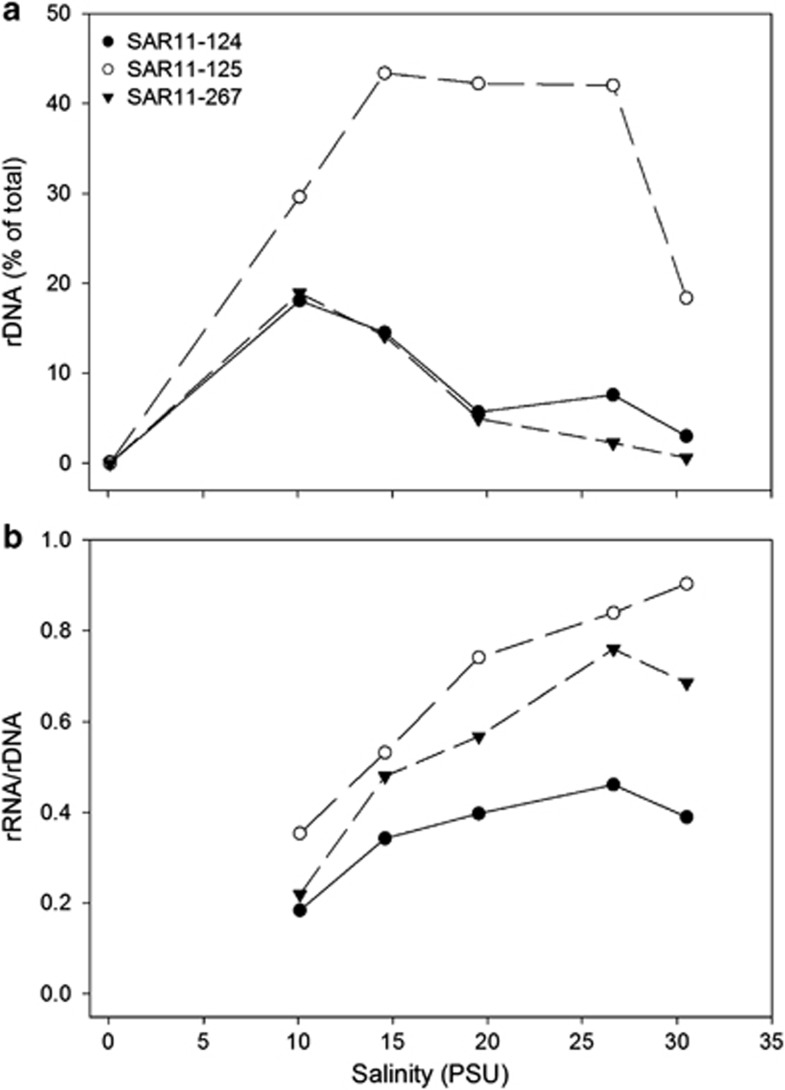
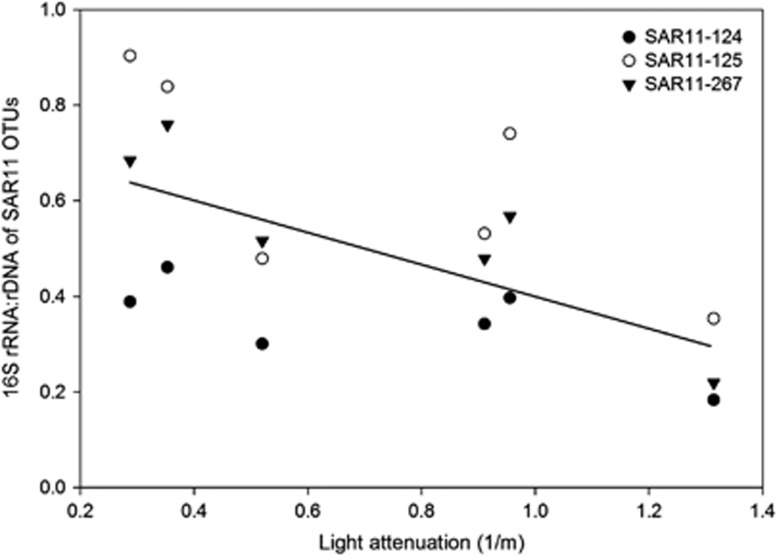
To understand how the potential growth rates varied along the transect, we analyzed the changes in relative abundance and 16S rRNA:rDNA ratios of the 46 taxa mentioned above (Supplementary Table S3) in relation to salinity or distance from the mouth of the bay. The relative abundance and 16S rRNA:rDNA ratios of the dominant SAR11 taxa varied along the transect. For instance, one SAR11 OTU (SAR11-807) was most prevalent in freshwater and low salinity waters, comprising 1–6% of the community (Supplementary Figure S2a). Its 16S rRNA:rDNA ratio did not change much; it was highest at 128 km from the bay mouth, or at 0.1 PSU, and it correlated with nitrate (r=0.95, P<0.05; Supplementary Figure S2b), although the sample size is small (n=4). The relative abundance of the other SAR11 taxa either peaked at the mid-salinity site (10 PSU) or had a broader abundance peak in higher salinity waters (15–26 PSU) (Figure 5a). The 16S rRNA:rDNA ratios of these SAR11 taxa steadily increased along the salinity gradient and peaked at either the bay mouth (26.6 PSU) or the coastal ocean site (OTU-125, 31 PSU) (Figure 5b). The ratios of all three increased when light attenuation decreased (Figure 6), and, as a group, were negatively correlated with light attenuation (r=−0.60, 0.82, n=18, P <0.01). OTU-125 was 97% identical to Pelagibacter ubique in this region of the 16S rRNA gene, whereas the other OTUs were only 90% similar to P. ubique (Supplementary Table S3). The 16S rRNA:rDNA ratios of the three SAR11s were negatively correlated with nitrate, ammonium and phosphate levels (r range from −0.51 to −0.61, n=18, P<0.05). In contrast, there was no significant correlation between abundance (16S rDNA) of the SAR11 OTUs as a group and any environmental parameter (data not shown).
Figure 5.
Relative abundance (a) and ratio of 16S rRNA to rDNA (b) of the indicated SAR11 OTU in relation to salinity along the Delaware Bay.
Figure 6.
Relationship between light attenuation and 16S rRNA:rDNA ratios of three marine SAR11 OTUs. The linear regression is indicated by the solid line.
The peak 16S rRNA:rDNA ratio of OTU-104, closely related to Roseovarius aestuarii, corresponded to the peak in leucine production (Table 1, Supplementary Table S2 and Supplementary Figure S3c), although the ratio was not significantly correlated to leucine production. The 16S rRNA:rDNA ratios of OTU-270, whose closest BLAST hit was P. alcaliphilus, was significantly correlated to chlorophyll a levels (r=0.89, n=6, P<0.01). Although abundances increased in two Rhodospirillaceae (OTUs 102 and 298, related to Nisaea denitrificans; Supplementary Table S3 and Supplementary Figure S3b) along the salinity gradient, their ratios declined at the highest salinity (Supplementary Figure S3d). As a group, the 16S rRNA:rDNA ratios of the Rhodospirillaceae were negatively correlated to photosynthetically active radiation (r=−0.63, n=11, P<0.05). However, there were no significant correlations between abundance (16S rDNA) of the Rhodospirillaceae group and any environmental parameter (data not shown).
It was more difficult to compare relative abundance and 16S rRNA:rDNA ratios of the taxa abundant at freshwater sites because of their low occurrence in our data set. The abundances of two Betaproteobacteria taxa (OTUs 34 and 1370) increased along the transect at the freshwater sites, but their 16S rRNA:rDNA ratios remained relatively constant or declined slightly (Supplementary Figure S2). The abundances of three taxa within the Verrucomicrobia phylum (Opitutae family) peaked at the second or third freshwater station (either at 128 or 136 km from the mouth of the bay; Supplementary Figure S4a). However, the 16S rRNA:rDNA ratios of two OTUs declined more than twofold from the most freshwater site to the high salinity waters (Supplementary Figure S4c). Other taxa had either (1) fewer than three locations where their abundances were >0.05% (2) ratios that changed directly with their abundance; (3) little variation in their ratios; or (4) no discernible trends in their ratios along the transect (data not shown).
Discussion
Surface coastal marine bacterial communities vary over time, reflecting broad changes in the environment (Fuhrman et al., 2006; Gilbert et al., 2009). Bacterial communities in estuaries and some coastal margins also greatly vary in space because of sharp gradients in salinity, nutrients and microbial communities, among other properties (Kirchman et al., 2005; Sharp et al., 2009; Fortunato et al., 2012; Herlemann et al., 2011). Although several studies have shown that these gradients lead to large variation in bulk bacterial production and biomass (Hoch and Kirchman, 1993; Goosen et al., 1999; Puddu et al., 1998; Preen and Kirchman, 2004) and community structure at the phylum and subphylum level (Bouvier and del Giorgio, 2002; Kirchman et al., 2005), few studies have examined the abundance of OTUs at the 97% level in this type of ecosystem (Kan et al., 2008; Herlemann et al., 2011; Fortunato et al., 2012) and even fewer have examined activity or potential growth rates at the OTU level in any ecosystem (Campbell et al., 2011). Here, we assessed bacterial community diversity, structure and 16S rRNA:rDNA ratios as an index of potential growth rate along the salinity gradient in the Delaware Bay. Our data indicate that bacteria are least diverse in the transition zone between fresh and marine waters and that levels of 16S rRNA are not positively correlated with abundance. This was especially true in the whole water fraction compared to the free-living fraction. We did find, however, that 16S rRNA of several taxa positively correlated with rDNA, indicating that activity did follow their relative abundance in some cases. The 16S rRNA:rDNA ratios, but not relative abundance of several taxa, changed with the physical, biological or chemical gradients along the estuary. These data suggest that presumed growth rates were controlled by environmental factors such as salinity and may be a better indicator than abundance alone in understanding the interactions between individual taxa and their environment.
In this study, we explored how levels of 16S rRNA changed in relation to abundance (16S rDNA) along a salinity gradient. Unlike our previous study, which explored rRNA–rDNA relationships over time at the marine site also examined here (Campbell et al., 2011), we found few positive relationships between 16S rRNA and rDNA along the salinity gradient. It is possible that the sampling frequency contributed to these differences; samples from the prior study were taken monthly at one site and not from several sites over a day as in this study. Another part of the explanation is that the steep gradients in salinity and other parameters in the estuarine environment apparently resulted in large changes in growth conditions, resulting in the poor correlations between 16S rRNA and rDNA. Similar results were observed when examining growth rates or activity of bacteria at the phylum or subphylum level using other approaches (Cottrell and Kirchman, 2004; Yokokawa et al., 2004).
The poor relationship between 16S rRNA and rDNA was even more pronounced in the whole water fraction than the free-living fraction. While the community composition of the free-living and whole water fractions differed only slightly, the ratios of 16S rRNA per rDNA in the whole water fraction were much higher for rare than for abundant bacteria, and were also higher in the rare whole water fraction than the rare free-living fraction. These results suggest that bacteria found in the rare whole water community are most active. Similar to other studies of bulk properties, we interpret this difference as being due to attachment to particulate detritus, although we cannot rule out the possibility of a higher number of large bacteria in the whole fraction than in the free-living fraction. In any case, our results agree with previous studies that found that particle-associated bacteria are more active than free-living bacteria, especially in estuarine and other eutrophic marine environments (Kirchman, 1993; Crump and Baross, 1996; Ghiglione et al., 2007).
When analyzed individually, 16S rRNA and rDNA levels of several taxa were not correlated or were even negatively correlated. Generally, these taxa were found in Actinobacteria, Flavobacteriales, Burkholderiales and Verrucomicrobia, most of which are related to known freshwater taxa (Newton et al., 2011). Bacterial growth rates within these clades may be very sensitive to changes in environmental conditions, such as salinity, or rely on a narrow range of available organic compounds that vary in the estuary (Church, 1986; Sharp et al., 2009). Our data suggest that overall the Actinobacteria had lower than average growth rates, while the Verrucomicrobia and Betaproteobacteria had higher than average rates in some of the freshwater regions along the Delaware Bay transect. Although data on the freshwater taxa were generally limited to three sites, these results agree with growth rates of these groups estimated by other approaches in freshwater lakes (Šimek et al., 2006; Newton et al., 2011). Even with these few sites, the data make possible a much more in-depth resolution of how potential growth rate responds to environmental change than possible in previous studies.
Many individual marine-associated taxa within the SAR11, Rhodobacterales, Rhodospirillales and Gammaproteobacteria groups, did show a significant correlation between 16S rRNA and rDNA, but, in many cases, this ratio changed along the salinity gradient. For instance, although the marine-associated SAR11 taxa were abundant in low to mid-salinity waters, their 16S rRNA:rDNA ratios gradually increased to the mouth of the bay and open ocean sites and were positively correlated with light (negatively correlated with light attenuation), perhaps reflecting the presence of the light-harvesting proteorhodopsin gene and their potential use on light for extra energy (Béjà et al., 2001; Giovannoni et al., 2005; Gómez-Consarnau et al., 2010; Steindler et al., 2011). In addition, overall 16S rRNA:rDNA ratios of SAR11 were low in spite of its dominance within the estuary, perhaps reflecting a slow growing mode also observed in cultured SAR11 (Rappé et al., 2002). Variation in the 16S rRNA:rDNA ratios along the estuary was also observed for Rhodobacteriaceae and Rhodospirllaceae OTUs. Growth rates of these bacteria may be influenced by phytoplankton-produced compounds, as their 16S rRNA:rDNA ratios were highest at or near the peak in chlorophyll a levels.
Members of the SAR11 clade dominate most marine environments (Morris et al., 2002; Giovannoni and Stingl, 2005; Giovannoni and Vergin, 2012), including the Delaware Bay. Previous studies using metagenomic, clone library or fluorescence in situ hybridization analyses found that SAR11 made up about 50% of the community in the Bay (Kirchman et al., 2001; Rusch et al., 2007; Straza and Kirchman, 2011). SAR11 types varied along the salinity gradient, indicating that the Delaware Bay contains ecotypes adapted to different salinities and other environmental conditions. Large differences were also observed between estuarine, coastal and oligotrophic metagenomes dominated by SAR11 taxa (Rusch et al., 2007). In spite of these differences in abundance, our data suggest that growth patterns of these distinct SAR11 are similar, perhaps reflecting some genomic traits shared among SAR11 taxa that differ phylogenetically. Further studies are needed to understand the complex growth patterns of this important clade.
Other abundant typically marine taxa varied along the Delaware Bay, including members of the Rhodobacteriaceae, Rhodospirillaceae and Flavobacteraceae. Variations in abundance and phylogenetic makeup of these families were also observed in other 16S rRNA and whole genome phylogenetic analyses of various estuaries (Bouvier and del Giorgio, 2002; Heidelberg et al., 2002; Crump et al., 2004; Kirchman et al., 2005; Kan et al., 2006; Andersson et al., 2010; Herlemann et al., 2011). As with these other studies, we also found that the abundance of some taxa peaked in certain salinity ranges, such as Verrucomicrobia and Comamonadaceae (Betaproteobacteria) in fresh water and Gammaproteobacteria related to the OMG group at the marine site. Other taxa most abundant in the mid-salinity and marine waters of the Delaware Bay were in families represented by isolates known to respire nitrate or sulfur compounds, or are associated with phytoplankton (Buchan et al., 2005).
Although it is clear that microbial communities change in structure along salinity gradients (Kirchman et al., 2005; Herlemann et al., 2011; Fortunato et al., 2012), corresponding changes in bacterial richness and diversity are less well described. Unlike that of higher organisms (Telesh and Khlebovich, 2010), we found that bacterial community richness as measured by both Chao1 and the number of OTUs was highest in the very low salinity waters. In addition, bacterial diversity and evenness were different along the Delaware Bay transect. In contrast, no differences were seen in alpha diversity along the large Baltic Sea salinity gradient, even though the Baltic study (Herlemann et al., 2011) used the same high-throughput sequencing approach as in our Delaware study. The difference is most likely due to the long gradient in salinity in the Baltic vs the short salinity gradient in the Delaware Bay. Salinity is well known to be a major contributor to microbial community structure and function (Lozupone and Knight, 2007; Nemergut et al., 2011).
Many estuaries, such as the Delaware Bay, are in constant flux, with wide variations in environmental conditions, sometimes over very short time or spatial scales (Sharp et al., 2009; Telesh and Khlebovich, 2010). This variability was reflected in the low levels of bacterial community diversity in oligo- and mesohaline waters of the estuary, and weak correlations between individual bacterial taxon abundance and environmental factors, as seen in previous studies (Fuhrman et al., 2006; Gilbert et al., 2009). Our study indicates that 16S rRNA:rDNA ratios may be more informative than abundance alone in understanding how bacterial communities respond to variation in physical and other bottom-up factors along the estuarine gradient. Further work is needed to convert 16S rRNA:rDNA ratios to actual estimates of individual bacterial growth rates. These data are likely to be a powerful tool to help understand the dynamics and controls of bacterial communities and ultimately to gage the contributions of individual bacteria to biogeochemical cycling.
Acknowledgments
We thank Matt Cottrell and Sharon Grim for sampling assistance and Liying Yu for technical assistance. This work was supported by National Science Foundation grants to BJC and DLK (OCE-0825468) and to DLK (MCB-0453993 and OCE-1030306).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Bouvier TC, del Giorgio PA. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr. 2002;47:453–470. [Google Scholar]
- Bremer H, Dennis PP.1996Modulation of chemical composition and other parameters of the cell by growth rateIn: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al. (eds).Escherichia coli and Salmonella: Cellular And Molecular Biology ASM Press: Washington, DC; 1553–1569. [Google Scholar]
- Buchan A, Gonzalez JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA. 2011;108:12776–12781. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Straza TRA, Kirchman DL. Temporal changes in bacterial rRNA and rRNA genes in Delaware (USA) coastal waters. Aquat Microb Ecol. 2009;57:123–135. [Google Scholar]
- Church TM. Biogeochemical factors influencing the residence time of microconstituents in a large tidal estuary, Delaware Bay. Mar Chem. 1986;18:393–406. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Single-cell analysis of bacterial growth, cell size, and community structure in the Delaware estuary. Aquat Microb Ecol. 2004;34:139–149. [Google Scholar]
- Cottrell MT, Mannino A, Kirchman DL. Aerobic anoxygenic phototrophic bacteria in the Mid-Atlantic Bight and the North Pacific Gyre. Appl Environ Microbiol. 2006;72:557–564. doi: 10.1128/AEM.72.1.557-564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Baross JA. Particle-attached bacteria and heterotrophic plankton associated with the Columbia River estuarine turbidity maxima. Mar Ecol Progr Ser. 1996;138:265–273. [Google Scholar]
- Crump BC, Baross JA. Characterization of the bacterially-active particle fraction in the Columbia River estuary. Mar Ecol Progr Ser. 2000;206:13–22. [Google Scholar]
- Crump BC, Baross JA, Simenstad CA. Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol. 1998;14:7–18. [Google Scholar]
- Crump BC, Hopkinson CS, Sogin ML, Hobbie JE. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol. 2004;70:1494–1505. doi: 10.1128/AEM.70.3.1494-1505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ. Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques. 1999;27:66–68. doi: 10.2144/99271bm13. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducklow HW.2000Bacterial production and biomass in the oceansIn: Kirchman DL, (ed).Microbial Ecology of the Oceans Wiley: New York; 85–120. [Google Scholar]
- Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J. 2012;6:554–563. doi: 10.1038/ismej.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouilland E, Mostajir B. Revisited phytoplanktonic carbon dependency of heterotrophic bacteria in freshwaters, transitional, coastal and oceanic waters. FEMS Microbiol Ecol. 2010;73:419–429. doi: 10.1111/j.1574-6941.2010.00896.x. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13:1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- Ghiglione JF, Mevel G, Pujo-Pay M, Mousseau L, Lebaron P, Goutx M. Diel and seasonal variations in abundance, activity, and community structure of particle-attached and free-living bacteria in NW Mediterranean Sea. Microbial Ecol. 2007;54:217–231. doi: 10.1007/s00248-006-9189-7. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Vergin KL. Seasonality in ocean microbial communities. Science. 2012;335:671–676. doi: 10.1126/science.1198078. [DOI] [PubMed] [Google Scholar]
- Goosen NK, Kromkamp J, Peene J, van Rijswik P, van Breugel P. Bacterial and phytoplankton production in the maximum turbidity zone of three European estuaries: the Elbe, Westerschelde and Gironde. J Mar Syst. 1999;22:151–171. [Google Scholar]
- Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, et al. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol. 2010;8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- Heidelberg JF, Heidelberg KB, Colwell RR. Seasonality of Chesapeake Bay bacterioplankton species. Appl Environ Microbiol. 2002;68:5488–5497. doi: 10.1128/AEM.68.11.5488-5497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch MP, Kirchman DL. Seasonal and interannual variability in bacterial production and biomass in a temperate estuary. Mar Ecol Prog Ser. 1993;98:283–295. [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan J, Evans SE, Chen F, Suzuki MT. Novel estuarine bacterioplankton in rRNA operon libraries from the Chesapeake Bay. Aquat Microb Ecol. 2008;51:55–66. [Google Scholar]
- Kan JJ, Crump BC, Wang K, Chen F. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol Oceanogr. 2006;51:2157–2169. [Google Scholar]
- Kemp PF, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhof L, Kemp P. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol. 1999;30:253–260. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- Kirchman DL.1993Particulate detritus and bacteria in marine environmentsIn: Ford T, (ed).Aquatic Microbiology: An Ecological Approach Blackwell: Cambridge, MA; 321–341. [Google Scholar]
- Kirchman DL, Dittel AI, Malmstrom RR, Cottrell MT. Biogeography of major bacterial groups in the Delaware estuary. Limnol Oceanogr. 2005;50:1697–1706. [Google Scholar]
- Kirchman DL, Yu LY, Fuchs BM, Amann R. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat Microb Ecol. 2001;26:13–22. [Google Scholar]
- Kramer JG, Singleton FL. Variations in rRNA content of marine Vibrio spp. during starvation–survival and recovery. Appl Environ Microbiol. 1992;58:201–207. doi: 10.1128/aem.58.1.201-207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelou VK, Cottrell MT, Kirchman DL. Light-stimulated bacterial production and amino acid assimilation by cyanobacteria and other microbes in the North Atlantic Ocean. Appl Environ Microbiol. 2007;73:5539–5546. doi: 10.1128/AEM.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr. 2005;50:1687–1696. [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Bulow L, Wahlund KG. Use of flow field-flow fractionation for the rapid quantitation of ribosome and ribosomal subunits in Escherichia coli at different protein production conditions. Biotechnol Bioeng. 1997;54:461–467. doi: 10.1002/(SICI)1097-0290(19970605)54:5<461::AID-BIT6>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Preen K, Kirchman DL. Microbial respiration and production in the Delaware Estuary. Aquat Microb Ecol. 2004;37:109–119. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu A, La Ferla R, Allegra A, Bacci C, Lopez M, Oliva F, et al. Seasonal and spatial distribution of bacterial production and biomass along a salinity gradient (Northern Adriatic Sea) Hydrobiologia. 1998;363:271–282. [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinform. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JH, Yoshiyama K, Parker AE, Schwartz MC, Curless SE, Beauregard AY, et al. A biogeochemical view of estuarine eutrophication: seasonal and spatial trends and correlations in the Delaware Estuary. Estuar Coast. 2009;32:1023–1043. [Google Scholar]
- Šimek K, Horňàk K, Jezbera J, Nedoma J, Vrba J, Stralkrábová V, et al. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ Microbiol. 2006;8:1613–1624. doi: 10.1111/j.1462-2920.2006.01053.x. [DOI] [PubMed] [Google Scholar]
- Simon M, Grossart HP, Schweitzer B, Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 2002;28:175–211. [Google Scholar]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One. 2011;6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straza TRA, Kirchman DL. Single-cell response of bacterial groups to light and other environmental factors in the Delaware Bay, USA. Aquat Microb Ecol. 2011;62:267–U281. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesh IV, Khlebovich VV. Principal processes within the estuarine salinity gradient: a review. Mar Pollut Bull. 2010;61:149–155. doi: 10.1016/j.marpolbul.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Yokokawa T, Nagata T, Cottrell MT, Kirchman DL. Growth rate of the major phylogenetic bacterial groups in the Delaware estuary. Limnol Oceanogr. 2004;49:1620–1629. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.