Abstract
The residence of dinoflagellate algae (genus: Symbiodinium) within scleractinian corals is critical to the construction and persistence of tropical reefs. In recent decades, however, acute and chronic environmental stressors have frequently destabilized this symbiosis, ultimately leading to coral mortality and reef decline. Viral infection has been suggested as a trigger of coral–Symbiodinium dissociation; knowledge of the diversity and hosts of coral-associated viruses is critical to evaluating this hypothesis. Here, we present the first genomic evidence of viruses associated with Symbiodinium, based on the presence of transcribed +ss (single-stranded) RNA and ds (double-stranded) DNA virus-like genes in complementary DNA viromes of the coral Montastraea cavernosa and expressed sequence tag (EST) libraries generated from Symbiodinium cultures. The M. cavernosa viromes contained divergent viral sequences similar to the major capsid protein of the dinoflagellate-infecting +ssRNA Heterocapsa circularisquama virus, suggesting a highly novel dinornavirus could infect Symbiodinium. Further, similarities to dsDNA viruses dominated (∼69%) eukaryotic viral similarities in the M. cavernosa viromes. Transcripts highly similar to eukaryotic algae-infecting phycodnaviruses were identified in the viromes, and homologs to these sequences were found in two independently generated Symbiodinium EST libraries. Phylogenetic reconstructions substantiate that these transcripts are undescribed and distinct members of the nucleocytoplasmic large DNA virus (NCLDVs) group. Based on a preponderance of evidence, we infer that the novel NCLDVs and RNA virus described here are associated with the algal endosymbionts of corals. If such viruses disrupt Symbiodinium, they are likely to impact the flexibility and/or stability of coral–algal symbioses, and thus long-term reef health and resilience.
Keywords: coral reef, Heterocapsa circularisquama RNA virus (HcRNAV), nuclear cytoplasmic large DNA virus (NCLDV), Phycodnaviridae, Symbiodinium, virome
Introduction
Corals and their resident microbes form multi-domain symbiotic assemblages fundamental to the construction and persistence of coral reef ecosystems. Coral symbionts include bacteria and dinoflagellate algae (genus Symbiodinium), as well as archaea and fungi (for example, Baker, 2003; Knowlton and Rohwer, 2003). Combinations of host and symbiont genotypes have been shown to influence the phenotypes of individual coral colonies, including tolerance to thermal (Baker et al., 2004; Berkelmans and van Oppen, 2006; Jones et al., 2008) and light regimes (Rowan et al., 1997; Iglesias-Prieto et al., 2004), as well as colony growth rates (Little et al., 2004; Cantin et al., 2009) and disease resistance and susceptibility (Reshef et al., 2006; Rosenberg et al., 2007; Correa et al., 2009a). Although functional redundancy in symbiotic microbes may allow coral colonies to cope with acute stress events (Fine and Loya, 2002; Mieog et al., 2007; LaJeunesse et al., 2009; Correa et al., 2009b; Silverstein et al., 2012), extreme or persistent environmental challenge can destabilize coral–microbe symbioses, leading to colony bleaching or disease, and in some cases, mortality.
Coral bleaching is the loss of Symbiodinium and/or chlorophyll from coral tissues, and results in the pale or white appearance of colonies (Glynn, 1996; Baker et al., 2008). Bleaching most commonly results from the accumulation of oxidative stress within symbiont chloroplasts (Lesser, 1997; Downs et al., 2002) following damage to photosystem II (Lesser, 1996), which can trigger the expulsion of algae from coral tissues via unresolved mechanisms (Gates et al., 1992; Franklin et al., 2004). Bleaching induced by bacteria (Vibrio spp.), although controversial, also has been reported (Kushmaro et al., 1996; Ben-Haim et al., 1999; Kushmaro et al., 2001; Ben-Haim et al., 2003, but see Ainsworth et al., 2008), and viral infections have been hypothesized to be an unrecognized cause of some bleaching and/or responsible for some of the >18 coral diseases (described in Sutherland et al., 2004).
Support for viral-mediated coral bleaching and disease is currently inconclusive, but a broad diversity of virus-like particles (VLPs) has been described on and within reef-building (stony) corals using morphological and genomic approaches (reviewed in Vega Thurber and Correa, 2011). Viral infections of the coral meta-organism were inferred from elevated abundances of VLPs and virus-like genomic sequences within stressed or degraded coral tissues (Wilson et al., 2005; Davy et al., 2006; Marhaver et al., 2008; Vega Thurber et al., 2008; Wilson, 2011). For example, metagenomic analyses suggest that coral-associated viral consortia commonly include single-stranded (ss)- and double-stranded (ds) DNA viruses, particularly members of the Herpesviridae and Phycodnaviridae, as well as phages (Wegley et al., 2007; Marhaver et al., 2008; Vega Thurber et al., 2008; Littman et al., 2011). These different viral groups likely target distinct hosts such as corals, resident coral symbionts, or other reef-associated organisms (for example, corallivores). Vega Thurber et al. (2008) documented herpesvirus-like sequences in viromes generated from the stony coral, Porites compressa. These annotated herpes-like sequences were then identified within distantly related azooxanthellate coral relatives, including the genome of Nematostella vectensis and complementary DNA (cDNA) expressed sequence tag (EST) data sets of Hydra magnipapillata. These data demonstrated that herpes-like viral genes are associated with cnidarians, rather than solely corals. They further indicated that latent or endogenous herpes-like viruses target corals as hosts, rather than their resident Symbiodinium.
Although significant evidence supports the hypothesis that herpes-like viruses target the coral animal, less is known about viruses that infect other members of the holobiont, including the endosymbiotic dinoflagellate genus Symbiodinium. Viral groups previously have been shown to target free-living dinoflagellates (for example, dinornaviruses and DNA giruses, Tarutani et al., 2001; Tomaru et al., 2004; Nagasaki et al., 2005; Nagasaki, 2008; Ogata et al., 2009) or eukaryotic algae (for example, phycodnaviruses, Wilson et al. (2009)). Therefore, it is plausible that similar viral groups target the dinoflagellates of corals and that some reported bleaching signs are derived from viral infection and/or viral-induced lysis of Symbiodinium cells (Vega Thurber and Correa, 2011; Wilson, 2011).
One putative Symbiodinium-associated virus is ‘zooxanthellae filamentous virus 1', or ZFV1 (Lohr et al., 2007). Lohr et al. (2007) reported these VLPs from TEM images of a UV-irradiated Symbiodinium strain in culture. VLPs increased in abundance within the periphery of algal cells and were reminiscent of the Closteroviridae (Lohr et al., 2007), a family of +ssRNA viruses that commonly infect plants. In the absence of genomic or infectivity data, however, it is possible that the observed filamentous particles were degraded Symbiodinium cytoplasmic structures. In contrast, there is significant genomic and microscopic evidence to support the hypothesis that phycodnaviruses are present in corals, but whether these viruses infect Symbiodinium is unknown (Wegley et al., 2007; Marhaver et al., 2008; Vega Thurber et al., 2008; Littman et al., 2011). Next steps in the characterization of these putative coral-associated filamentous and phycodnavirus-like particles are to confirm their viral nature and target hosts, as well as to evaluate their effects on coral–algal symbioses.
To confirm the presence of RNA viruses and phycodnaviruses in coral colonies, and to develop inferences regarding their target hosts, we pyrosequenced metatranscriptomes from purified VLP fractions isolated from control and heat-stressed Montastraea cavernosa corals and compared them with independently generated cDNA libraries from Symbiodinium cultures. Based on multiple lines of evidence, we suggest here that at least two previously undescribed viruses, a dsDNA nucleocytoplasmic large DNA virus (NCDLV) and a +ssRNA dinornavirus, are associated with the dinoflagellate endosymbionts of corals. If viruses commonly infect Symbiodinium, this is likely to influence the standing pool of symbiont diversity available to coral hosts, as well as the stability of coral–microbial symbioses and reef health.
Materials and methods
Experimental treatments
Montastraea cavernosa was chosen for this experiment because it is a common Caribbean reef-building coral (Goodbody-Gringley et al., 2012) that is susceptible to a variety of coral diseases (Sutherland et al., 2004). M. cavernosa also has a thick tissue layer, which facilitates the isolation of coral- and coral symbiont-associated VLPs. Symbiodinium in clade C typically dominate Caribbean M. cavernosa colonies (Wilcox, 1997; Baker, 1999), but Symbiodinium in clade D have also been detected from these hosts at low abundances (Correa et al., 2009b). Fragments of M. cavernosa corals were obtained on August 24 2009 from the Coral Rescue and Protection Program (Key West, FL, USA, 24°33′2″ N, 81°48′32″ W), where they were being maintained in an open water nursery (mean water temperature for August of 2009: 29.8 °C±0.7 s.e.; http://www.ndbc.noaa.gov/). Following collection, coral fragments were kept in an aquarium at 28.0 °C with broad-spectrum lighting for ∼1 month before the experiment. To induce viral production, fragments were heated to 31.5 °C for 12 h (N=2), while controls (N=2) were maintained at 28.0 °C.
Coral-associated viral particle isolation and purification
Following treatment, the tissue layer from each fragment was removed using an airbrush and 0.02 μm filtered viral-free 1 × PBS (pH 7.8). Coral and coral symbiont homogenates were collected in sterile tri-pour containers and pre-filtered with an 1-μm nucleopore filter (Whatman, Piscataway, NJ, USA). VLPs were concentrated from 27 ml of homogenate using ultracentrifugation of four-layer (1.2, 1.35, 1.5 and 1.7 g ml−1) cesium chloride density gradients (Vega Thurber et al., 2009). A gray band formed in the 1.2 g ml−1 density layer (Supplementary Figure 1a) and was harvested using an 18-gage needle and sterile syringe. Before and following a 0.22-μm Sterivex (Millipore, Billerica, MA, USA) filtration step (for details, see Vega Thurber et al. (2009)), this fraction was visualized using epifluorescence microscopy and SYBR Gold (Invitrogen, Carlsbad, CA, USA) staining (Noble and Fuhrman, 1998; Vega Thurber et al., 2009). This approach confirmed that the filtered fraction contained abundant VLPs, yet relatively few contaminating eukaryotic and microbial cells. No nuclei or microbial cells were observed in the 0.22-μm sterivex filtrate Supplementary Figure 1b); this filtrate was prepared for pyrosequencing.
Viral RNA extractions
To remove free DNA and RNA, each 0.22-μm-filtered VLP fraction was digested using DNase 1 and RNase A/T1 for 3 and 8 h, respectively. Total RNA was extracted from 200 μl of concentrated VLPs using an RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's recommendations, which included a DNA removal step performed after viral capsids were broken open. For each treatment, random hexamers were used to shotgun amplify 2 ng of total RNA using a Transplex WTA2 kit (Sigma, St Louis, MO, USA). The resulting cDNA libraries were cleaned using a Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). The purity and concentration of cDNA was measured on a Nanodrop-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA). To determine the size range of cDNA fragments, 200 ng of each library was visualized on a 2% agarose gel containing 1 × gel red. The control and heat-stressed samples (N=1 each) that yielded the highest amounts and broadest size classes of cDNA were selected for pyrosequencing. To further test for contaminating eukaryotic and microbial cDNA, 18S and 16S PCRs were conducted on all samples before sequencing. No contamination was detected.
Approximately 2 μg of cDNA of each sample was pyrosequenced on a Roche Titanium 454 platform at the Broad Institute (Massachusetts Institute of Technology, Cambridge, MA, USA). SFF files were converted to FASTA and FASTAQ files, and each read was parsed and assigned an alphanumeric name. Reads were archived at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (ID SRA052068), as well as at CAMERA (IDs CAM_SMPL_000711 and CAM_SMPL_000712) (Sun et al., 2011) and MG-RAST (IDs 44551158.3 and 44551159.3).
Sequence homology analyses
A variety of step-wise bioinformatic analyses were performed using unassembled raw reads (Supplementary Figure 2). Initial sequence similarities were determined using batch BLASTx searches to the MG-RAST database (Meyer et al., 2008). Virome reads also underwent tBLASTx searches to a boutique viral database containing fully annotated viral genomes (∼2700), as well as to two Symbiodinium EST draft libraries (Bayer et al., 2012). A minimum of ⩽10−6 was used as the expected value (e-value) for these and all other analyses. Query sequences from the M. cavernosa viromes and the Bayer et al. (2012) Symbiodinium EST libraries that had the best similarity to a viral genome and had a quality score greater than 25 were selected for complete annotation (Supplementary Figure 2). This was conducted in SeqMan (DNASTAR Inc., Madison, WI, USA) via BLASTp searches to the non-redundant database at the NCBI.
Phylogenetic analysis
M. cavernosa virome reads with similarity to a given viral family and gene of interest were assembled using CAP3 (Huang and Madan, 1999), and contigs were subsequently used in phylogenetic reconstructions. For each phylogeny, sequences included were initially identified using tBLASTx searches to the NCBI viral database and Bayer et al. (2012) Symbiodinium EST draft libraries, selecting only subject sequences with e-values ⩽10−12 and query coverage ⩾60% (Supplementary Material). Sequences were aligned as codons in MUSCLE using the default parameters. Identical duplicate sequences were removed in MEGA5 based on pairwise distances. The reliability of each alignment was estimated based on a calculation of the average percent amino-acid identity and p-distance for each alignment. Trees were constructed from alignments with p-distances ⩽0.8 (Hall 2011). For each alignment, the most appropriate Maximum Likelihood model and rates among sites to be used in constructing the corresponding gene tree was determined based on the ‘Find Best DNA/Protein Models (ML)' function in MEGA5 (Supplementary Table 1). The robustness of each phylogenetic tree was assessed using 100 bootstrap replicates. The threshold of bootstrap support used to collapse polytomies for the trees was 50/100.
Results
M. cavernosa viromes contain homologs to DNA and RNA viruses
Pyrosequencing of the cDNA generated from M. cavernosa viral fractions yielded 175 044 reads with an average length of 277 bp (Table 1). Approximately 19% of reads were similar to known sequences based on BLASTx searches in MG-RAST; 4.4% of these known reads were similar to viruses (Supplementary Table 2). Reads similar to phages and archaeal viruses were present in the M. cavernosa viromes, but these data will be presented elsewhere. To increase analytical power in the identification of sequence similarities to RNA or DNA viruses, tBLASTx searches were used to compare sequence reads with a boutique genome database containing all annotated sequences from eukaryotic viruses (∼2700 genomes). Using this approach, ∼1590 sequences within the combined control and stressed viromes had significant sequence similarity to eukaryotic viruses (Table 2). The majority (∼90%) of these sequences were similar to dsDNA and ssDNA viruses, with only ∼10% of the sequences containing homology to viruses with RNA-based genomes.
Table 1. Summary statistics for the control and stressed Montastraea cavernosa viromes generated in this study.
| Libraries | Control | Stressed |
|---|---|---|
| No. of Reads | 60 485 | 114 559 |
| Average read length (bp) | 270±162 | 283±172 |
| Average GC content | 43±8% | 43±7% |
Table 2. Summary of unique sequence similarities to eukaryotic viruses in each library based on batch tBLASTx searches (e-value cutoff of ⩽10−6) to a boutique viral database.
| Similarities to | Control | Stressed | Both viromes |
|---|---|---|---|
| Eukaryotic viruses (% total library) | 526 (0.87%) | 1059 (0.92%) | 1585 (0.9%) |
| dsDNA viruses | 78.7% | 63.6% | 68.6% |
| NCLDV-like | 57.28% | 48.63% | 51.72% |
| ssDNA viruses | 14.1% | 25.3% | 21.6% |
| dsRNA viruses | 0.0% | 0.1% | 0.1% |
| ssRNA viruses | 0.4% | 0.9% | 0.8% |
| Retroviruses | 6.8% | 10.0% | 9.0% |
Abbreviations: ds, double-stranded; NCLDV, nuclear cytoplasmic large DNA virus; ss, single-stranded.
Details are provided on the relative percent of eukaryotic virus similarities that are dsDNA, ssDNA, dsRNA and ssRNA virus-like, as well as NCLDV-like.
Highly divergent, +ssRNA viral sequence similarities are present in the stressed virome
The majority (∼92%) of eukaryotic RNA viral similarities in the M. cavernosa viromes were retrovirus-like sequences (Figure 1a, Table 2). However, sequence-based support for the presence of most retrovirus genomes within the M. cavernosa fragments was relatively rare (that is, ⩽4 similarities detected for most genomes, Figure 1b); many retrovirus genomes were similar to only a single sequence in our cDNA libraries (data not shown). ss- and dsRNA viral similarities also were rare in the viromes, comprising <1% of eukaryotic viral similarities (Table 2) and 9% of RNA viral similarities (Figure 1a). There were, however, five sequence similarities (Figure 1a) to Heterocapsa circularisquama RNA virus (HcRNAV), a +ssRNA virus that infects free-living dinoflagellates. BLASTp searches of the NCBI non-redundant (nr) database found that three of these sequences (reads GAIR4WKO3F1XL6, GAIR4WKO3FYXY4 and GAIR4WK03GFJLN) were homologs to the diagnostic HcRNAV major capsid protein (mcp). Translated amino-acid versions of our three HcRNAV-like mcp homologs could be aligned to known HcRNAV mcp genes (Figure 2). Complete consensus among the aligned sequences most commonly occurred outside of the hypervariable mcp regions identified by Nagasaki et al. (2005) (dark gray highlights in Figure 2). For example, F1XL6 overlapped with hypervariable regions 2 and 3, whereas FYXY4 and GFJLN overlapped significantly with each other and extended into hypervariable region 1. These latter two sequences exhibited complete substitution, relative to known HcRNAV mcp sequences (AB218608-9), in various amino-acid positions (light gray highlights in Figure 2).
Figure 1.
Summary of the eukaryotic viral similarities in the combined (control and stressed) Montastraea cavernosa viromes. Unique similarities are based on batch tBLASTx searches (e-value cutoff of ⩽10−6) to a boutique viral database. (a) Similarities to eukaryotic viral RNA families. Percentages indicate the number of unique sequence similarities to a given viral family, relative to all eukaryotic viral RNA similarities in the M. cavernosa viromes. The ‘Other ssRNA families' category represents the combined sequence similarities for all ssRNA viral families that were represented by <5 unique similarities. (b) Breakdown of observed retrovirus similarities. Percentages indicate the number of unique sequence similarities to a given retrovirus, relative to all retrovirus similarities detected. The ‘Other retrovirus similarities' and ‘Other caulimovirus similarities' categories represent members of the Retroviridae and Caulimoviridae, respectively, for which fewer than five unique similarities were detected. (c) Summary of the most abundant sequence similarities to eukaryotic viral families in the combined M. cavernosa viromes. Percentages indicate the number of unique sequence similarities to a given viral family, relative to all eukaryotic similarities in the M. cavernosa viromes. The ‘Other' category represents the combined sequence similarities for all viral families that comprised <5% of all eukaryotic viral similarities. ‘Popped out' pie wedges are members of the NCLDV group.
Figure 2.
Amino-acid alignment of known HcRNAV major capsid gene fragments (GenBank Accession no. AB218608-9) with homologous transcripts identified in the Montastraea cavernosa-stressed virome. The alignment was performed by hand, based on the results of PSI-BLAST searches (e-value cutoff of ⩽10−6) to the NCBI nr database. The positions of highly variable regions 1 to 3 (identified by Nagasaki et al. (2005)) are depicted using black rectangles. Dark gray shading highlights completely conserved amino acids. Light gray shading indicates amino-acid positions exhibiting complete substitution in GAIR4WKO3FYXY4 and GAIR4WKO3GFJLN, relative to known HcRNAV mcp sequences (no. AB218608-9).
dsDNA NCLDV-like sequence similarities dominate the M. cavernosa viromes
Most (∼69%) eukaryotic viral sequences identified from the M. cavernosa viral metatranscriptomes were similar to dsDNA viruses (Table 2). Unlike previous reports on coral viruses, however, a majority (∼52%) of these dsDNA virus similarities were to nucleocytoplasmic large DNA virus (NCLDVs) genomes. Sequence similarities to all viral families (that is, Ascoviridae, Asfarviridae, Iridoviridae, Mimiviridae, Phycodnaviridae and Poxviridae; Iyer et al., 2006; Koonin and Yutin, 2010) currently recognized as NCLDVs were detected (Table 3), even though the 0.22-μm filtration step likely removed some of the larger NCLDVs before sequencing. The majority of these NCLDV-like sequences were highly similar (10−61 ⩽e-values ⩽10−6) to known mimi- or phycodnaviruses. A total of 186 transcripts from the control and stressed viromes were similar to phycodnaviruses that infect Chlorella, the green algal endosymbiont of various cnidarian and protistan hosts. Asfarvirus and poxvirus sequence similarities also were common (Table 3, Figure 1c). Iridovirus-like sequences were detected but comprised <5% of eukaryotic viral similarities (data not shown). Sequences similar to other dsDNA viral families, including the Baculo-, Circo-, Herpes-, Nano- and Parvoviridae additionally each comprised ⩾5% of unique eukaryotic viral similarities (Table 3, Figure 1c).
Table 3. Summary of unique sequence similarities to the viral families (Asfar-, Baculo-, Circo-, Herpes-, Mimi-, Nano-, Parvo-, Phycodna-, Pox- and Retroviridae) comprising ⩾5% of all eukaryotic viruses detected from the combined (control and stressed) Montastraea cavernosa viromes.
| Viral group | Nucleic acid | Unique similarities | Percent eukaryotic virus similarities (%) |
|---|---|---|---|
| Eukaryotic viruses | 1585 | 100.0 | |
| Mimiviridae* | dsDNA | 287 | 18.1 |
| Phycodnaviridae* | dsDNA | 217 | 13.7 |
| Herpesviridae | dsDNA | 139 | 8.8 |
| Parvoviridae | ssDNA | 132 | 8.3 |
| Poxviridae* | dsDNA | 122 | 7.7 |
| Asfarviridae* | dsDNA | 118 | 7.4 |
| Nanoviridae | ssDNA | 117 | 7.4 |
| Baculoviridae | dsDNA | 114 | 7.2 |
| Circoviridae | ssDNA | 93 | 5.9 |
| Retroviridae | retrovirus | 92 | 5.8 |
Abbreviations: ds, double-stranded; ss, single-stranded.
Similarities are based on batch tBLASTx (e-value cutoff of ⩽10−6) searches to a boutique viral database. An * indicates that a given viral family is a member of the nuclear cytoplasmic large DNA virus group.
Significant sequence similarities to phycodna- and mimivirus-like transcripts in the M. cavernosa libraries were annotated using BLASTp searches of the NCBI nr database (Table 4, Supplementary Figure 2). The mimivirus similarities ranged in e-value from 1.9e−79 to 9.7e−9, with query coverage from 59 to 98%. Similarly, phycodnavirus gene similarities ranged in e-value from 4.1e−77 to 5.3e−7, with query coverage between 58 and 93%. Similarities to DNA topoisomerases (N=10), major capsid proteins (N=5) and RNA polymerases (N=5) were detected, as well as other proteins previously identified from the Phycodnaviridae (for example, protein kinases, ribonucleotide reductases (Table 4)) (Dunigan et al., 2006). Some gene similarities (for example, ornithine decarboxylase, GDP-ℒ-fucose synthase) specific to Paramecium bursaria Chlorella virus-1 (PbCV-1, Phycodnaviridae) were observed (Dunigan et al., 2006) and were similar to early genes with known functionality in the Chlorella-virus system (for example, GDP-ℒ-fucose synthase, glucosamine-fructose-6-phosphate aminotransferase, potassium ion transporter protein, thymidylate synthase, topoisomerase II) (Kang et al., 2005). It is also noteworthy that similarities to MutS (a gene involved in DNA mismatch repair and recombination) were not detected. The presence/absence of MutS within viral data sets from corals is of interest because horizontal transfer between an ancestor of giruses (including the dinoflagellate-infecting Heterocapsa circularisquama DNA virus, HcDNAV), gorgonians and environmental epsilonproteobacteria is hypothesized to have occurred (Claverie et al., 2009a, 2009b; Ogata et al., 2011).
Table 4. Examples of gene similarities from mimivirus and phycodnavirus-like sequences in the combined (control and stressed) Montastraea cavernosa viromes.
| Viral similarity (read ID) | Read length (bp) | Gene annotation (Org.) | e-value | Query coverage (%) |
|---|---|---|---|---|
| M1 (F975P) | 460 | Esterase/Lipase/Thiosterase | 1.9e−79 | 88 |
| M2 (G35KQ) | 461 | Esterase/Lipase/Thiosterase | 2.1e−78 | 88 |
| M3 (GD1NB) | 525 | Chaperone protein DnaK | 1.7e−63 | 79 |
| M4 (FU6GZ) | 508 | Chaperone protein DnaK | 6.3e−62 | 87 |
| M5 (FMUTW) | 520 | DNA polymerase | 6.2e−58 | 85 |
| M6 (FNJIO) | 513 | Chaperone protein DnaK | 4.7e−58 | 94 |
| M7 (FZPI9) | 496 | Major capsid protein | 2.0e−58 | 89 |
| M8 (GHWP0) | 517 | Major capsid protein | 8.0e−57 | 85 |
| M9 (GHNLN) | 487 | RNA polymerase II | 2.8e−52 | 89 |
| M10 (F2D45) | 519 | Major capsid protein | 1.5e−50 | 92 |
| M11 (GF99F) | 520 | DNA-directed RNA polymerase | 1.6e−46 | 82 |
| M12 (FWPS6) | 508 | Major capsid protein | 2.9e−45 | 86 |
| M13 (HDMDS) | 488 | RNA polymerase II | 1.2e−45 | 90 |
| M14 (GOSHQ) | 513 | Major capsid protein | 1.7e−44 | 85 |
| M15 (G2MSW) | 507 | Chaperone protein DnaK | 1.7e−43 | 82 |
| M16 (GB6Z7) | 350 | ATP-dependent DNA helicase | 3.5e−42 | 84 |
| M17 (HDFNZ) | 545 | RNA polymerase II | 2.0e−42 | 92 |
| M18 (FPJGH) | 550 | Ribonucleoside-diphosphate reductase | 4.1e−41 | 94 |
| M19 (GKDGX) | 477 | DNA-directed RNA polymerase II | 8.0e−40 | 89 |
| M20 (GNZHE) | 534 | DNA polymerase family B | 9.9e−29 | 80 |
| M21 (GACVR) | 361 | DNA topoisomerase I | 1.1e−28 | 90 |
| M22 (GV1WZ) | 548 | Ribonucleoside-diphosphate reductase | 1.7e−24 | 98 |
| M23 (FN56M) | 343 | Ribonucleotide reductase | 8.7e−16 | 81 |
| M24 (FNDQM) | 407 | Glutamate dehydrogenase | 2.7e−15 | 69 |
| M25 (G6CQK) | 513 | Putative ankyrin repeat protein | 1.6e−15 | 82 |
| M26 (GOSUY) | 514 | Putative ankyrin repeat protein | 6.5e−13 | 87 |
| M27 (HB5YS) | 431 | Glutamate dehydrogenase | 2.8e−13 | 65 |
| M28 (F13CC) | 493 | DNA topoisomerase I | 5.1e−11 | 64 |
| M29 (FF8T4) | 438 | Hypothetical protein ATCV1_z279R | 9.7e−9 | 62 |
| M30 (FLF2W) | 507 | Hypothetical protein DICPUDRAFT_41242 | 3.3e−9 | 52 |
| P1 (FL4SB) | 477 | Potassium uptake ion transporter protein | 4.1e−77 | 84 |
| P2 (HH88H) | 492 | ABC transporter related protein | 1.3e−64 | 82 |
| P3 (G8IS6) | 533 | DNA topoisomerase II | 7.4e−58 | 88 |
| P4 (GIAK7) | 413 | ATPase copper transporter | 6.4e−50 | 79 |
| P5 (GDW8W) | 455 | DNA topoisomerase II | 7.9e−45 | 91 |
| P6 (GN760) | 494 | Phosphatase/phosphohexomutase | 6.5e−44 | 88 |
| P7 (GDOK5) | 457 | DNA topoisomerase II | 8.0e−42 | 78 |
| P8 (GNKAT) | 461 | DNA topoisomerase II | 1.9e−38 | 81 |
| P9 (GCV8W) | 563 | DNA topoisomerase II | 1.7e−38 | 90 |
| P10 (HCDAR) | 451 | DNA topoisomerase II | 3.0e−37 | 88 |
| P11 (HA44I) | 451 | DNA topoisomerase II | 1.9e−37 | 86 |
| P12 (FYAKA) | 533 | ABC transporter ATPase | 1.2e−35 | 70 |
| P13 (HAV73) | 546 | Phosphatase/phosphohexomutase | 1.2e−35 | 86 |
| P14 (GGB0Q) | 620 | Glucosamine-fructose-6-phosphate aminotransferase | 2.4e−33 | 86 |
| P15 (GB9BB) | 415 | Ribonucleotide reductase | 1.9e−33 | 90 |
| P16 (FNSCY) | 490 | Ankyrin repeat containing protein | 3.6e−32 | 87 |
| P17 (GNFSR) | 418 | Ribonucleotide reductase | 1.5e−32 | 86 |
| P18 (FVVQF) | 455 | DNA topoisomerase II | 1.6e−31 | 88 |
| P19 (F20P7) | 522 | DNA topoisomerase II | 3.5e−28 | 91 |
| P20 (F693D) | 321 | Phosphatase/phosphohexomutase | 1.7e−27 | 84 |
| P21 (F548L) | 527 | Putative thymidylate synthase | 1.5e−26 | 93 |
| P22 (GN6W3) | 469 | Ornithine decarboxylase | 3.0e−25 | 90 |
| P23 (FVOXB) | 529 | Hypothetical protein CNM01240 | 4.2e−21 | 85 |
| P24 (F16LN) | 536 | Hypothetical protein ATCV1_Z667L | 6.0e−17 | 58 |
| P25 (GWXIX) | 446 | Hypothetical protein COPCOM_02634 | 9.3e−16 | 64 |
| P26 (FOGBA) | 366 | Predicted protein | 8.8e−16 | 80 |
| P27 (GUAMA) | 449 | DNA topoisomerase II | 5.9e−16 | 75 |
| P28 (G8R2H) | 528 | GDP-ℒ-fucose synthase | 9.1e−14 | 72 |
| P29 (F4H6E) | 370 | Hypothetical protein PGAG_00149 | 1.6e−13 | 77 |
| P30 (FYR8O) | 524 | Protein kinase | 5.4e−10 | 61 |
Abbreviations: M, Mimivirus-like sequence similarity; P, Phycodnavirus-like similarity.
Similarities are based on BLASTp searches to the NCBI nr database (e-value cutoff of ⩽10−6). All sequences had an average quality score ⩾25.
Symbiodinium transcriptomes contain homologs of M. cavernosa NCLDV-like sequences
Given that a majority of the M. cavernosa virome similarities were to genes from the Phycodnaviridae, a family of viruses that typically infects eukaryotic algae, we hypothesized that these viruses must be infecting Symbiodinium algae within the experimental coral fragments. To confirm that the annotated phycodna- and other virus-like transcripts presented here are associated with Symbiodinium, we compared our sequences with EST libraries from several Symbiodinium cultures (Table 5). All unique similarities to a given viral group from the combined (control and stressed) M. cavernosa libraries were used as query sequences in tBLASTx comparisons with EST draft libraries constructed from two Symbiodinium sub-genera (that is, clades): KB8 (clade A, Genbank accession no. SRX076696), and Mf1.05b (clade B, no. SRX076709 and SRX076710) (Bayer et al., 2012). A total of 396 unique similarities were detected in these Symbiodinium EST libraries. Comparable numbers of similarities were detected from the Symbiodinium clade A versus clade B libraries for each set of query sequences (for example, mimivirus-like queries (Table 5)). The majority (∼79%) of the Symbiodinium EST homologies were to M. cavernosa virome sequences that were NCLDV-like (for example, annotated mimivirus- and phycodnavirus-like) transcripts (Table 6). Mimivirus-like similarities to Symbiodinium EST contigs ranged in e-value from 0.0 to 1.5e−86, with query coverage from 82 to 97%. Similarly, phycodnavirus gene homologies ranged in e-value from 0.0 to 5.5e−19, with query coverage between 18 and 97%. Overall, there was strong agreement between the gene annotations for a given M. cavernosa virome query sequence and its most similar contig from a given Symbiodinium EST library (Table 6). Relatively few contigs from the Symbiodinium EST libraries were similar to asfarvirus-like M. cavernosa query sequences, despite a previous demonstration that the DNA polymerase B genes of asfarviruses and HcDNAV are closely related (Ogata et al., 2009). Few to no similarities were detected in the Symbiodinium EST libraries to annotated retrovirus-, ssDNA-like (that is, Parvoviridae, Nanoviridae, or Circoviridae) transcripts, respectively. No similarities to the HcRNAV-like virus were detected in the Symbiodinium EST libraries (data not shown).
Table 5. Sequence similarities to asfarvirus-, baculovirus-, circovirus-, herpesvirus-, mimivirus-, nanovirus-, parvovirus-, phycodnavirus-, poxvirus- and retrovirus-like sequences detected from two Symbiodinium draft EST libraries: KB8 (clade A, Genbank Accession no. SRX076696) and Mf1.05b (clade B, no. SRX076709 and SRX076710) (Bayer et al., 2012).
| M. cavernosa virome |
Symbiodinium clade A (KB8) |
Symbiodinium clade B (Mf1.05b) |
|||
|---|---|---|---|---|---|
| Query sequences | Unique similarities | All similarities | Unique similarities | All similarities | |
| Mimiviridae* | 287 | 63 | 287 | 73 | 239 |
| Phycodnaviridae* | 217 | 52 | 236 | 48 | 177 |
| Herpesviridae | 139 | 20 | 45 | 14 | 40 |
| Parvoviridae | 132 | 0 | 0 | 0 | 0 |
| Poxviridae* | 122 | 35 | 123 | 30 | 117 |
| Asfarviridae* | 118 | 7 | 22 | 5 | 16 |
| Nanoviridae | 117 | 0 | 0 | 0 | 0 |
| Baculoviridae | 114 | 21 | 96 | 25 | 84 |
| Circoviridae | 93 | 0 | 0 | 0 | 0 |
| Retroviridae | 92 | 2 | 11 | 1 | 8 |
| NCLDVs % of total similarities | 744 | 157 (78.5%) | 668 (81.5%) | 156 (79.6%) | 549 (80.6%) |
| Total similarities | 1431 | 200 | 820 | 196 | 681 |
Abbreviation: NCLDV, nuclear cytoplasmic large DNA virus.
Similarities are based on batch tBLASTx searches (e-value cutoff of ⩽10−6). Query sequences used to search for each viral group were from the combined (control and stressed) Montastraea cavernosa viromes (that is, ‘Unique similarities' column in Table 3). An * indicates that a given viral family is a member of the NCLDV group. NCLDV calculations for this table do not include ascovirus or iridovirus sequence similarities because these constituted <5% of all eukaryotic viral similarities (see Table 3).
Table 6. Examples of gene similarities from mimivirus and phycodnavirus-like contigs in two Symbiodinium draft EST libraries (Bayer et al., 2012).
| Virome query (read ID) | Query gene annotation | Symbiodinium EST contig (ID) | Contig gene annotation | Read length (bp) | e-value | Query coverage (%) |
|---|---|---|---|---|---|---|
| M1 (HDFNZ) | RNA polymerase II | B1 (16882) | RNA polymerase | 1351 | 0.0 | 96 |
| M2 (HDFNZ) | RNA polymerase II | A1 (3434) | RNA polymerase | 3596 | 0.0 | 95 |
| M3 (GV1WZ) | Ribonucleoside-diphosphate reductase | A2 (10097) | Ribonucleoside-diphosphate reductase | 2537 | 0.0 | 88 |
| M4 (GV1WZ) | Ribonucleoside-diphosphate reductase | B2 (3609) | Ribonucleoside-diphosphate reductase | 1392 | 8.9e−145 | 85 |
| M5 (GD1NB) | Chaperone protein DnaK | B3 (47620) | Chaperone protein DnaK | 796 | 5.0e−119 | 92 |
| M6 (FNDQM) | Glutamate dehydrogenase | A3 (2623) | Glutamate dehydrogenase | 1882 | 7.1e−117 | 87 |
| M7 (GD1NB) | Chaperone protein DnaK | A4 (7292) | Glutamate dehydrogenase | 2046 | 7.4e−107 | 82 |
| M8 (F568L) | ATP-dependent metalloprotease FtsH | A5 (2286) | ATP-dependent metallopeptidase HflB | 1971 | 1.5e−86 | 97 |
| P1 (GGB0Q) | Glucosamine-fructose-6-phosphate aminotransferase | A1 (7236) | Glucosamine-fructose-6-phosphate aminotransferase | 1993 | 0.0 | 97 |
| P2 (G8IS6) | DNA topoisomerase II | A2 (968) | DNA topoisomerase II | 5668 | 0.0 | 63 |
| P3 (G8IS6) | DNA topoisomerase II | B1 (17340) | DNA topoisomerase II | 1783 | 1.2e−173 | 88 |
| P4 (FL4SB) | Potassium uptake ion transporter protein | A3 (10426) | Potassium uptake ion transporter protein | 2295 | 6.7e−109 | 94 |
| P5 (GCV8W) | DNA topoisomerase II | B2 (19936) | DNA topoisomerase II | 556 | 4.0e−75 | 95 |
| P6 (FVVQF) | DNA topoisomerase II | B3 (18169) | DNA topoisomerase II | 656 | 5.8e−63 | 86 |
| P7 (GB9BB) | Ribonucleotide reductase | B4 (30594) | Ribonucleoside-diphosphate reductase | 694 | 2.9e−63 | 85 |
| P8 (GDW8W) | DNA topoisomerase II | A4 (3814) | Hypothetical protein MT325_m550L | 6069 | 5.5e−19 | 18 |
Abbreviations: M, Mimivirus-like sequence similarity; P, Phycodnavirus-like sequence similarity.
Contig gene annotations are based on BLASTp searches to the NCBI nr database (e-value cutoff of <10−6); ‘A' contigs are Symbiodinium clade A, whereas ‘B' contigs are clade B.
Phylogenetic support for NCLDVs that target Symbiodinium
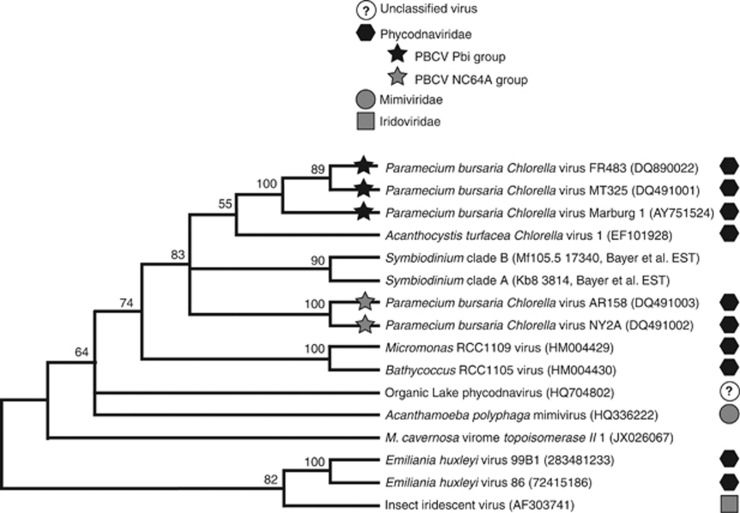
To further confirm the identity of these virus-like sequences, trees were generated for three phylogenetically informative genes common among NCLDVs: the major capsid protein, DNA polymerase family B and topoisomerase II (Figures 3, 4, 5). The M. cavernosa major capsid protein (mcp) sequences used in this phylogeny fell in their own clade, which was most closely related to a Chlorovirus (Phycodnaviridae) in the Pbi group (Figure 3). Therefore, these mcp-like sequences are interpreted to be viral genes belonging to a previously undescribed member of the Phycodnaviridae. In the DNA polymerase family B phylogeny, however, the M. cavernosa virome sequence is positioned in a clade with two unclassified giant viruses, but with minimal bootstrap support (Figure 4). In the topoisomerase II phylogeny, the M. cavernosa virome sequences also could not be assigned to a clade and may be relatively distinct from previously sequenced NCLDV topoisomerase II genes (Figure 5). Yet, the position of the topoisomerase II-like Symbiodinium EST contigs (Bayer et al., 2012) within this tree strongly suggests they are phycodnavirus sequences (Figure 5).
Figure 3.
Maximum Likelihood reconstruction of the mcp gene. The Montastraea cavernosa virome mcp sequences 1 and 2 are deposited in GenBank as Accession no. JX026064 and JX026065, respectively. The tree was rooted using iridovirus sequences (GenBank Accession no. M32799 and M33542).
Figure 4.
Unrooted Maximum Likelihood reconstruction of the DNA polymerase Family B (DNA pol B) gene. The Montastraea cavernosa virome DNA pol B sequence is deposited in GenBank as Accession no. JX026066.
Figure 5.
Unrooted Maximum Likelihood reconstruction of the DNA topoisomerase II gene. The Montastraea cavernosa virome topoisomerase II sequence is deposited in GenBank as Accession no. JX026067.
Discussion
Methods for generating and analyzing VLP cDNAs
To expand our knowledge of the diversity and host range of coral viruses, we generated and analyzed viral metatranscriptomes from control and thermally stressed specimens of M. cavernosa. Our experimental design was a modification of two previously published methods that analyzed DNA viruses from host corals (Vega Thurber et al., 2008) and RNA viruses from seawater (Culley and Steward, 2007). We modified these methods by adding DNase and RNase digests before the extraction of total viral RNA, as well as by shotgun amplifying the resulting total RNA using a commercial kit. Yet despite the RNase digest, our libraries contained 3 to 8% rRNA sequences (data not shown). Large subunit (LSU, 28S-like) rRNA sequences in the libraries confirmed that the M. cavernosa fragments hosted Symbiodinium in clade C (data not shown). Removal of rRNA is a known challenge in microbial metatranscriptomes (Frias-Lopez et al., 2008; Hewson et al., 2009; Poretsky et al., 2009). For example, bacterial rRNA represented 74 to 83% of the total reads in recently published microbial metatranscriptomes (Stewart et al., 2010). Our analysis suggests that host rRNA removal from viral metatranscriptomes also is difficult, but to a lesser degree. Future experiments may further benefit from the use of (1) oligo-dt primers for amplification, and/or (2) alternative approaches common in microbial metatranscriptomics (Stewart et al., 2010). Nevertheless the approach presented here significantly reduced host and bacterial RNA within the prepared cDNA, yet maintained sequence length and permitted the identification of both RNA viral genomes and DNA viral transcripts.
Evidence for an ssRNA virus associated with Symbiodinium
A majority of our cDNA reads had no known similarity to any viral protein or nucleotides, yet ∼1500 eukaryotic virus-like sequences were identified from this data set. Excitingly, some of these sequences constitute the first evidence for an RNA virus associated with the coral meta-organism. The stressed M. cavernosa virome contained unique similarities to Heterocapsa circularisquama RNA virus (HcRNAV, family Alvernaviridae, Figure 1a). HcRNAV is a 30 nm, icosahedral, +ssRNA dinornavirus (Tomaru et al., 2004; Nagasaki et al., 2005) that infects free-living dinoflagellate algae often responsible for toxic blooms (Nagasaki, 2008). Interestingly, VLPs of this description have previously been reported from stony coral tissues, including the gastrodermal layer where Symbiodinium reside (see Figure 2c in Vega Thurber and Correa (2011)). Two of the HcRNAV-like M. cavernosa virome sequences were not homologous to HcRNAV ORFs (for example, putative polyprotein ‘ORF-1' in Nagasaki et al., 2005), but were homologous to intergenic regions identified within the HcRNAV genome. The other three sequences were similar to the major capsid protein (mcp, ‘ORF-2' in Nagasaki et al., 2005), but these HcRNAV-like sequences clearly differed from published HcRNAV mcp sequences (Figure 2). Our alignment and the high divergence exhibited by corresponding HcRNAV-like nucleotide reads thus suggest that the HcRNAV-like sequences presented here represent novel undescribed members of the Alvernaviridae.
Although two of our HcRNAV-like mcp reads overlapped, they were significantly divergent from one another (Figure 2). Nagasaki et al. (2005) similarly documented divergence between and within HcRNAV ecotypes UA and CY, although this was largely restricted to the hypervariable regions of the mcp. Divergence between the M. cavernosa virome HcRNAV-like mcp proteins could thus represent different viral strains within a larger population. HcRNAV-like strains could target distinct Symbiodinium sub-genera (clades), which can reside in the same M. cavernosa coral colony (e.g., Correa et al., 2009b). HcRNAV homologs have not been detected within the two Bayer et al. Symbiodinium EST draft libraries nor any published EST library from corals (data not shown, Bayer et al. (2012)). If population-level variation exists within coral-associated dinornaviruses, this may explain why homologs to our annotated HcRNAV-like mcp sequences were not detected in these other libraries. Additional sequencing and microscopy efforts, as well as infection experiments, will clarify and expand our knowledge of these first dinornaviruses identified from coral colonies.
Evidence for other RNA viruses in M. cavernosa
Similarities to RNA viruses comprised only ∼10% of the eukaryotic viral similarities detected within viromes (Table 2). Furthermore, the majority (∼92%) of RNA viral similarities in this experiment were to retroviruses (Table 2, Figure 1a), yet many of these sequences were indicative of both retroviruses and retro-elements. Gene similarities to most retroviral genomes were based on fewer than five reads each (Figure 1b), obscuring the origin of these genes.
An additional PCR-based search for RNA viruses using degenerate primers (Culley and Steward, 2007) to the RNA-dependent RNA polymerase (RdRp, an RNA virus-specific gene) also failed to amplify RNA virus-specific sequences (data not shown). Several issues could have contributed to low RNA virus sequence recovery. First, long VLPs may have unintentionally been removed before ultracentrifugation. This could explain why no similarities to filamentous, +ssRNA closteroviruses were detected in the M. cavernosa viromes, despite a previous report of such VLPs by Lohr et al. (2007). Second, fewer marine RNA virus sequences are present in databases owing to the relatively low research effort that has been expended historically on environmental marine eukaryotic viruses.
Transcripts from dsDNA viruses suggest possible NCLDV infection of the coral meta-organism
The most abundant eukaryotic viral similarities detected in this study were to dsDNA viruses. Since a DNase step was used to digest DNA viral genomes during the extraction process, sequence similarities to DNA viruses in the M. cavernosa viromes are most parsimoniously described as the products of DNA viral genes actively being transcribed at the time of sampling. This interpretation is supported by the (1) relatively short lengths (for example, 321–620 bp in Table 4) of sequence similarities to DNA viruses observed, (2) overall inability to assemble longer contigs from M. cavernosa virome reads, (3) detection of numerous and strong similarities to DNA viral proteins (and lack of similarities to intergenic regions, Table 4), as well as the (5) detection of abundant similarities to our annotated NCLDV-like sequences (Table 5) in Symbiodinium transcriptomes produced independently by other laboratories (Bayer et al., 2012).
Although among previous coral virus studies, the relative abundance of similarities to many of these dsDNA viral Families (for example, Asfarviridae, Baculoviridae, Herpesviridae, Parvoviridae and Poxviridae) has varied, herpesvirus-like sequences have generally dominated (Wegley et al., 2007; Marhaver et al., 2008; Vega Thurber et al., 2008). In this study, however, many of the transcripts coding for structural components (that is, capsid proteins) and enzymes (for example, DNA topisomerases) were indicative of NCLDVs. Over 800 transcripts similar to NCLDVs were observed in our cDNA libraries, and these sequences were among the strongest (10−79 ⩽ e-values ⩽10−7, Table 4) and most abundant (14–18%, Table 3, Figure 1c) similarities identified in each virome. Many of these transcripts were similar to the early genes in another virus (Family: Phycodnaviridae) that infects an algal symbiont (Kang et al., 2005), indicating that our libraries were constructed near the onset of active NCLDV infection. Importantly, the boutique viral database used to identify virus-like sequences in our M. cavernosa viromes (Supplementary Figure 1) contained similar numbers of annotated herpesvirus genomes (N=47) and NCLDV genomes (N=45). Therefore, the observed dominance of phycodnavirus- and mimivirus-like transcripts most likely represents a biological signal, rather than a mathematical artifact of searching a biased database.
Multiple lines of genomic evidence indicate Symbiodinium are targeted by an ancient NCLDV
Viral metagenomes and viral metatranscriptomes can provide abundant and high-resolution sequence data regarding the viral consortia associated with environmental samples. Such data can rapidly provide insights into novel viral groups that are likely infecting different hosts. Sequence similarities in this study could theoretically represent viruses that target coral tissues, coral symbionts, or other reef-associated organisms (for example, corallivores). As the processed material contained millions of coral and symbiont cells, but few cells from other organisms, it is reasonable to assume that abundant cDNA similarities in the data set represent viruses that target coral or symbiont cells. Thus, given that some NCLDVs (Mimiviridae and Phycodnaviridae) commonly infect eukaryotic microalgae (Monier et al., 2008; Wilson et al., 2009), it is highly plausible that the NCLDVs reported here are associated with Symbiodinium. This hypothesis is supported by comparisons between our NCLDV-like sequences and two Symbiodinium draft transcriptomes (Bayer et al., 2012), which revealed that ∼79% of similarities in the transcriptomes were to putative NCLDVs (Table 5). In contrast, few to no similarities to retrovirus and ssDNA virus-like query sequences were detected in the Symbiodinium transcriptomes. Despite differences in read length among the M. cavernosa viromes and Symbiodinium transcriptomes (Tables 1 and 6), there was strong agreement between the gene annotations for each M. cavernosa virome query sequence and its best tBLASTx hit within a given Symbiodinium EST library. These data suggest that low-level persistent viral infections may have been present within the cultures as previously suggested by Wilson (2011). Aliquots of the Symbiodinium cultures were exposed to thermal (heat or cold) or irradiance (high light or dark) stressors before cDNA library generation, which may have triggered the transcription of viral genes (Bayer et al., 2012).
It is notable that the Symbiodinium cultures used for the transcriptomes were isolated from taxonomically disparate hosts (Bayer et al., 2012). Specifically, the more ancestral clade A culture was isolated from the upside-down jellyfish, Cassiopeia sp., whereas the more-derived clade B culture came from the coral, Montastraea faveolata (a relative of M. cavernosa). Therefore, the roughly equal numbers of unique NCLDV similarities within the two Symbiodinium transcriptomes (52 and 48 in clade A and B, respectively) suggest that these putative infections likely are of ancient origin, given that their dinoflagellate host sub-genera diverged approximately 35 million years ago (Pochon et al., 2006). Alternatively, if NCLDVs infect Symbiodinium, perhaps they gained the ability to do so more than once. If Symbiodinium-associated NCLDVs have codiverged with their dinoflagellate hosts, then distinct viral populations whose relationships track those of their hosts should be evident in future Symbiodinium EST libraries generated from sub-genera (clades) C through I.
Conclusions
This study constitutes the first genomic report of viruses associated with symbiotic dinoflagellates. We have identified the first RNA virus from a coral colony and also present metagenomic evidence that nuclear cytoplasmic large DNA viruses associate with the algal symbionts of corals in hospite, as well as in their free-living state (that is, in culture). Symbiodinium is a highly diverse genus containing nine clades or sub-genera, some of which are as genetically divergent from each other as are orders of free-living dinoflagellates. Future attempts to isolate phycodna-like and HcRNA-like viruses from Symbiodinium should therefore explore the diversity of this dinoflagellate genus more comprehensively, and also target Symbiodinium within non-scleractinian coral hosts (for example, soft corals, octocorals, foraminiferans, giant clams, ciliates), as well as free-living algal individuals. Such endeavors will allow us to better characterize these viruses and provide the direct evidence necessary to confirm that they infect a broad diversity of Symbiodinium. If these viruses induce Symbiodinium mortality, alter algal physiology (for example, reduce photosynthetic efficiency), and/or disrupt host–Symbiodinium partnerships, then such infections have significant implications for coral reef health and survival.
Acknowledgments
We thank Dana Willner for bioinformatic assistance, Jesse Zaneveld for helpful discussions regarding the construction of phylogenetic trees, Monica Medina for access to the Symbiodinium draft EST libraries before their publication, and three anonymous reviewers for their helpful comments on earlier versions of this manuscript. We are grateful to the Florida Keys National Marine Sanctuary for supplying the coral fragments (permit no. FKNMS-2009-062) and to Alex Culley for providing advice and the positive control for the RdRp PCR. The Gordon and Betty Moore Marine Virus Sequencing Project supported pyrosequencing for this study, and research was supported by a National Science Foundation grant (OCE-0960937) to RLVT.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ainsworth T, Fine M, Roff G, Hoegh-Guldberg O. Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica. ISME J. 2008;2:67–73. doi: 10.1038/ismej.2007.88. [DOI] [PubMed] [Google Scholar]
- Baker AC. The Symbiosis Ecology of Reef-Building Corals. University of Miami: Coral Gables, FL; 1999. [Google Scholar]
- Baker AC. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Ann Rev Ecol Evol Syst. 2003;34:661–689. [Google Scholar]
- Baker AC, Glynn PW, Riegl B. Climate change and coral reef bleaching: an ecological assessment of long-term impacts, recovery trends and future outlook. Estuar Coast Shelf Sci. 2008;80:435–471. [Google Scholar]
- Baker AC, Starger CJ, McClanahan TR, Glynn PW. Corals' adaptive response to climate change. Nature. 2004;430:741. doi: 10.1038/430741a. [DOI] [PubMed] [Google Scholar]
- Bayer T, Aranda M, Sunagawa S, Yum LK, DeSalvo M, Lindquist E, et al. Symbiodinium Transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One. 2012;7:e35269. doi: 10.1371/journal.pone.0035269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim Y, Banim E, Kushmaro A, Loya Y, Rosenberg E. Inhibition of photosynthesis and bleaching of zooxanthellae by the coral pathogen Vibrio shiloi. Environ Microbiol. 1999;1:223–229. doi: 10.1046/j.1462-2920.1999.00027.x. [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, et al. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol. 2003;53:309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- Berkelmans R, van Oppen MJH. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope' for coral reefs in an era of climate change. Proc R Soc B. 2006;273:2305–2312. doi: 10.1098/rspb.2006.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin NE, Van Oppen MJH, Willis BL, Mieog JC, Negri AP. Juvenile corals can acquire more carbon from high-performance algal symbionts. Coral Reefs. 2009;28:405–414. [Google Scholar]
- Claverie JM, Abergel C, Ogata H.2009aMimivirusIn: VanEtten JL, (ed).Lesser Known Large dsDNA Viruses Springer-Verlag: Berlin Heidelberg; 89–121. [Google Scholar]
- Claverie JM, Grzela R, Lartigue A, Bernadac A, Nitsche S, Vacelet J, et al. Mimivirus and Mimiviridae: giant viruses with an increasing number of potential hosts, including corals and sponges. J Invertebr Pathol. 2009b;101:172–180. doi: 10.1016/j.jip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Correa AMS, Brandt ME, Smith TB, Thornhill DJ, Baker AC. Symbiodinium associations with diseased and healthy scleractinian corals. Coral Reefs. 2009a;28:437–448. [Google Scholar]
- Correa AMS, McDonald MD, Baker AC. Development of clade-specific Symbiodinium primers for quantitative PCR (qPCR) and their application to detecting clade D symbionts in Caribbean corals. Mar Biol. 2009b;156:2403–2411. [Google Scholar]
- Culley AI, Steward GF. New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl Environ Microbiol. 2007;73:5937–5944. doi: 10.1128/AEM.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy SK, Burchett SG, Dale AL, Davies P, Davy JE, Muncke C, et al. Viruses: agents of coral disease. Dis Aquat Organ. 2006;69:101–110. doi: 10.3354/dao069101. [DOI] [PubMed] [Google Scholar]
- Downs CA, Fauth JE, Halas JC, Dustan P, Bemiss J, Woodley CM. Oxidative stress and seasonal coral bleaching. Free Radical Biol Med. 2002;33:533–543. doi: 10.1016/s0891-5849(02)00907-3. [DOI] [PubMed] [Google Scholar]
- Dunigan DD, Fitzgerald LA, Van Etten JL. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Fine M, Loya Y. Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc R Soc B. 2002;269:1205–1210. doi: 10.1098/rspb.2002.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin DJ, Hoegh-Guldberg P, Jones RJ, Berges JA. Cell death and degeneration in the symbiotic dinoflagellates of the coral Stylophora pistillata during bleaching. Mar Ecol Prog Ser. 2004;272:117–130. [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates RD, Baghdasarian G, Muscatine L. Temperature stress causes host cell detachment in symbiotic cnidarians: Implications for coral bleaching. Biol Bull. 1992;182:324–332. doi: 10.2307/1542252. [DOI] [PubMed] [Google Scholar]
- Glynn PW. Coral reef bleaching: facts, hypotheses and implications. Glob Change Biol. 1996;2:495–509. [Google Scholar]
- Goodbody-Gringley G, Woollacott RM, Giribet G. Population structure and connectivity in the Atlantic scleractinian coral Montastraea cavernosa (Linnaeus, 1767) Mar Ecol. 2012;33:32–48. [Google Scholar]
- Hall BG.2011Phylogenetic Trees Made Easy: A How-to Manual4th edn. Sinauer Associates, Inc.: Sunderland.
- Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ, et al. Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 2009;3:1286–1300. doi: 10.1038/ismej.2009.75. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Prieto R, Beltran VH, LaJeunesse TC, Reyes-Bonilla H, Thome PE. Different algal symbionts explain the vertical distribution of dominant reef corals in the eastern Pacific. Proc R Soc B. 2004;271:1757–1763. doi: 10.1098/rspb.2004.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LA, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Jones AM, Berkelmans R, van Oppen MJH, Mieog JC, Sinclair W. A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc R Soc B. 2008;275:1359–1365. doi: 10.1098/rspb.2008.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Dunigan DD, Van Etten JL. Chlorovirus: a genus of Phycodnaviridae that infects certain chlorella-like green algae. Mol Plant Path. 2005;6:213–224. doi: 10.1111/j.1364-3703.2005.00281.x. [DOI] [PubMed] [Google Scholar]
- Knowlton N, Rohwer F. Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am Nat. 2003;162:S51–S62. doi: 10.1086/378684. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. Vibrio shiloi sp nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol. 2001;51:1383–1388. doi: 10.1099/00207713-51-4-1383. [DOI] [PubMed] [Google Scholar]
- Kushmaro A, Loya Y, Fine M, Rosenberg E. Bacterial infection and coral bleaching. Nature. 1996;380:396. [Google Scholar]
- LaJeunesse TC, Smith RT, Finney J, Oxenford H. Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching' event. Proc R Soc B. 2009;276:4139–4148. doi: 10.1098/rspb.2009.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser MP. Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in symbiotic dinoflagellates. Limnol Oceanogr. 1996;41:271–283. [Google Scholar]
- Lesser MP. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs. 1997;16:187–192. [Google Scholar]
- Little AF, van Oppen MJH, Willis BL. Flexibility in algal endosymbioses shapes growth in reef corals. Science. 2004;304:1492–1494. doi: 10.1126/science.1095733. [DOI] [PubMed] [Google Scholar]
- Littman RA, Willis BL, Bourne DG. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ Microbiol Rep. 2011;3:651–660. doi: 10.1111/j.1758-2229.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- Lohr J, Munn CB, Wilson WH. Characterization of a latent virus-like infection of symbiotic zooxanthellae. Appl Environ Microbiol. 2007;73:2976–2981. doi: 10.1128/AEM.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10:2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The Metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieog JC, Van Oppen MJH, Cantin NC, Stam WT, Olsen JL. Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs. 2007;26:449–457. [Google Scholar]
- Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H. Marine mimivirus relatives are probably large algal viruses. Virol J. 2008;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K. Dinoflagellates, diatoms, and their viruses. J Microbiol. 2008;46:235–243. doi: 10.1007/s12275-008-0098-y. [DOI] [PubMed] [Google Scholar]
- Nagasaki K, Shirai Y, Takao Y, Mizumoto H, Nishida K, Tomaru Y. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl Environ Microbiol. 2005;71:8888–8894. doi: 10.1128/AEM.71.12.8888-8894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- Ogata H, Ray J, Toyoda K, Sandaa RA, Nagasaki K, Bratbak G, et al. Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment. ISME J. 2011;5:1143–1151. doi: 10.1038/ismej.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Toyoda K, Tomaru Y, Nakayama N, Shirai Y, Claverie J-M, et al. Remarkable sequence similarity between the dinoflagellate-infecting marine girus and the terrestrial pathogen African swine fever virus. Virol J. 2009;6:178. doi: 10.1186/1743-422X-6-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochon X, Montoya-Burgos JI, Stadelmann B, Pawlowski J. Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus. Symbiodinium Mol Phylogenet Evol. 2006;38:20–30. doi: 10.1016/j.ympev.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun SL, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nature Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Rowan R, Knowlton N, Baker AC, Jara J. Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature. 1997;388:265–269. doi: 10.1038/40843. [DOI] [PubMed] [Google Scholar]
- Silverstein RN, Correa AMS, Baker AC. Specificity is rarely absolute in coral-algal symbiosis: implications for coral response to climate change. Proc R Soc B. 2012;279:2609–2618. doi: 10.1098/rspb.2012.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altinatas I, Lin A, Peltier S, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KP, Porter JW, Torres C. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar Ecol Prog Ser. 2004;266:273–302. [Google Scholar]
- Tarutani K, Nagasaki K, Itakura S, Yamaguchi M. Isolation of a virus infecting the novel shellfish-killing dinoflagellate Heterocapsa circularisquama. Aquat Microb Ecol. 2001;23:103–111. [Google Scholar]
- Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, et al. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat Microb Ecol. 2004;34:207–218. [Google Scholar]
- Vega Thurber R, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Vega Thurber RL, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber RL, Correa AMS. Viruses of reef-building scleractinian corals. J Exp Mar Biol Ecol. 2011;408:102–113. [Google Scholar]
- Wegley L, Edwards R, Beltran R-B, Hong L, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Wilcox TP. The Evolution of Algal-Invertebrate Symbioses. University of Houston: Houston, TX; 1997. [Google Scholar]
- Wilson WH.2011Coral virusesIn: Hurst CJ, (ed).Studies in Viral Ecology: Animal Host Systemsvol 2. John Wiley & Sons, Inc.: Hoboken, NJ, USA; 141–149. [Google Scholar]
- Wilson WH, Dale AL, Davy JE, Davy SK. An enemy within? Observations of virus-like particles in reef corals. Coral Reefs. 2005;24:145–148. [Google Scholar]
- Wilson WH, Van Etten JL, Allen MJ. The phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol. 2009;328:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.