Abstract
Recent work has shown that dispersal has an important role in shaping microbial communities. However, little is known about how dispersed bacteria cope with new environmental conditions and how they compete with local resident communities. To test this, we implemented two full-factorial transplant experiments with bacterial communities originating from two sources (freshwater or saline water), which were incubated, separately or in mixes, under both environmental conditions. Thus, we were able to separately test for the effects of the new environment with and without interactions with local communities. We determined community composition using 454-pyrosequencing of bacterial 16S rRNA to specifically target the active fraction of the communities, and measured several functional parameters. In absence of a local resident community, the net functional response was mainly affected by the environmental conditions, suggesting successful functional adaptation to the new environmental conditions. Community composition was influenced both by the source and the incubation environment, suggesting simultaneous effects of species sorting and functional plasticity. In presence of a local resident community, functional parameters were higher compared with those expected from proportional mixes of the unmixed communities in three out of four cases. This was accompanied by an increase in the relative abundance of generalists, suggesting that competitive interactions among local and immigrant taxa could explain the observed ‘functional overachievement'. In summary, our results suggest that environmental filtering, functional plasticity and competition are all important mechanisms influencing the fate of dispersed communities.
Keywords: dispersal, competition, metacommunity, species sorting, neutral model, salinity
Introduction
During the last decade, there has been a growing number of studies focusing on the role of dispersal for microbial diversity (Whitaker et al., 2003; Logue and Lindström, 2008; Jones and McMahon, 2009; Lindström and Östman, 2011; Martiny et al., 2011), which was, to a great extent, triggered by the introduction of metacommunity concept (Leibold et al., 2004) to microbial ecology. However, the actual mechanisms by which dispersed bacteria can survive, establish and influence the local communities they arrive into have not been addressed. In general, the fate of dispersed organisms depends on how they cope with two main selective forces: the filtering effect of environmental changes constituted by the new habitat and the interactions with local communities (Gómez et al., 2010).
With regard to the filtering effect, adaptation to the new environment is a crucial requirement for immigrants to ensure their survival and establishment in a new habitat. According to Comte and del Giorgio (2011), functional adaptations occur primarily at the single cell level, but also determine the physiological structure of the community and ultimately its overall metabolic performance. The same authors generated a conceptual framework regarding the adaptation of bacterioplankton to environmental gradients and put forward two possible scenarios. First, the ‘replacement scenario' in which communities have a low functional plasticity, so that compositional changes are required for functional adaptation; and second, the ‘adjustment scenario' in which communities are dominated by generalists, which have a high functional plasticity, so that compositional changes are not anticipated for functional adaptation.
The ‘replacement scenario' is similar to the species-sorting concept of the metacommunity framework (Leibold et al., 2004), which assumes that microbial communities have the potential to adapt rapidly to new environmental conditions by adjusting their composition. Species sorting has shown to be an important process influencing the bacterial community composition (Langenheder and Ragnarsson, 2007; Van der Gucht et al., 2007; Logue and Lindström, 2010; Hovatter et al., 2011). Species sorting also suggests that there are habitat-specific communities with a pre-defined composition, so that compositional changes are required for successful adaptation. In other words, species sorting presumes that microbial communities have a relatively low functional plasticity.
On the other hand, it has been shown that bacterial community assembly is often, to a considerable extent, influenced by regional processes (for example, dispersal) and is conform to predictions made by the neutral model (Sloan et al., 2006; Östman et al., 2010). As the main assumption made by the neutral model is the functional equivalence of taxa (Hubbell, 2001), no compositional changes, but rapid functional adaptations of dispersed populations, should occur when they arrive in a new habitat, as functional plasticity is expected to be high. This corresponds to the adjustment scenario of Comte and del Giorgio (2011) and is supported by studies that have shown that microbial communities can functionally adapt to different environments even without drastic changes in composition, and that microbial communities of different composition can perform equally owing to their functional redundancy (Wohl et al., 2004; Wertz et al., 2007; Comte and del Giorgio, 2010; Werner et al., 2011).
The second major challenge dispersed bacteria have to face is competition for resources and interaction with the local resident communities. According to the metacommunity framework mentioned above, the following scenarios are possible: first, if communities are neutrally assembled, interactions between immigrants and local residents should be insignificant and communities will resemble a proportional mix of the immigrant and local communities. Second, in case of species sorting, the presumably better-adapted taxa of the local community will, in the simplest scenario, outcompete and overgrow the immigrating ones. Third, there could also be a more complex interaction-ruled scenario in which bacteria are selected, irrespective of their origin, according to their competitive abilities, which should have the consequence that generalist taxa that have a high competitive ability because of their wide habitat tolerance and good exploitation ability (Dall and Cuthill, 1997) will become dominant.
Here we present an experimental study, which had the major aim to disentangle the filtering effect of environmental changes and the interactions with local communities that bacteria have to face when immigrating into new habitats. We collected water from a freshwater and a saline rock pool, and implemented a full-factorial transplant experiment in which bacterial communities from either pool were, separately and in mixes, incubated under both environmental conditions. Thus, we were able to separately test for the effects of the new habitat with and without interactions with local communities. We determined community composition using 454-pyrosequencing of bacterial 16S rRNA to specifically target the active fraction of the communities, and measured biomass and specific bacterial production (SBP) to be able to address the degree of functional plasticity of the communities when exposed to environmental changes. To evaluate the interaction outcome in case of the mixed communities, we compared the measured values for relative abundances of taxa and functional parameters with those expected from proportional mixes of the unmixed communities. If the measured values showed values equal to those theoretically expected, this would support neutral processes, whereas deviations would indicate that species sorting was important. Furthermore, we classified all detected taxa according to their habitat specificity, and tested whether specialists and generalists showed differences in relative abundance (1) under local or new environmental conditions in case of the unmixed communities, and (2) compared with the expected ratios in case of the mixed communities.
We implemented two experiments, which differed with regard to the potential of competitive ability of the immigrating, that is, transplanted, community. In the first experiment, we used the original communities in which the competitive ability of the immigrants was hypothesized to be low. In the second experiment, we used communities that were pre-adapted (PA) to the new environment, and we hypothesize that this would lead to a pre-selection of more competitive immigrants, that is, generalists, compared with the original communities, and ultimately affect the outcome of the interaction with the local community.
Materials and methods
Experimental design
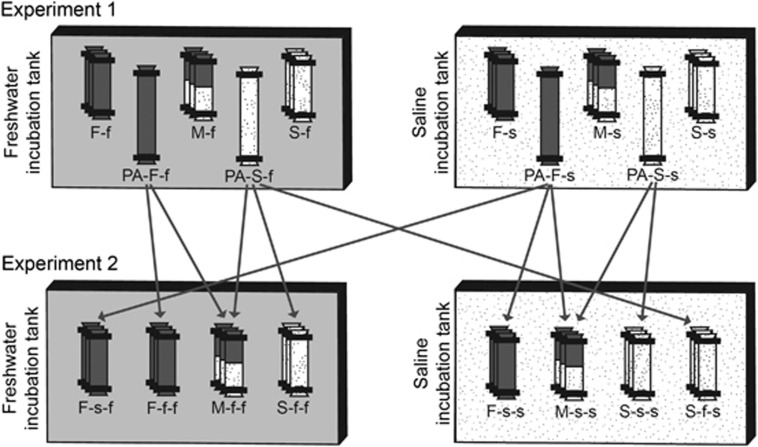
Water samples were collected from two rock pools from Uggelhällorna peninsula (N 60°39.895′, E 18°25.836′) located on the Gräsö island along the Swedish Baltic Sea Coast on 7 August 2009. The pools were chosen to differ notably in salinity, but not so much in other basic parameters (Supplementary Table S1). Chemical properties of the samples were analyzed as described in Langenheder and Ragnarsson (2007). Before the experiment, water was filtered through 1.2 μm, 142 mm Type A/E glass filter (Pall Corporation, Port Washington, NY, USA) to remove phytoplankton. Two 40 l tanks, kept in a 20 °C dark constant-temperature room, were filled with the filtered water to act as saline (s) and freshwater (f) incubation media, respectively. Throughout the manuscript, capital letters refer to the source of the community and small letters to the incubation environment.
To remove bacterivorous protozoans, water was further filtered through 0.7 μm, 47 mm glass microfiber filter (Whatman, Maidstone, UK) before each transplant experiment. Pre-treated (de-ionized water rinsed and autoclaved) dialysis bags (diameter 45 mm, molecular weight cut-off 12–14 kDa, Zellu Trans, Carl Roth, Karlsruhe, Germany) were filled with 70 ml of bacterivore-free saline (S), freshwater (F) or 1:1 volume ratio mix of both (M). Bags (S, F and M) were placed, in triplicates, into each tank (s and f), and after 3 days of incubation, all bags were sampled in a destructive way (Figure 1). This set-up allowed us to measure (1) the effect of new environmental conditions on transplanted unmixed communities (S-f and F-s); (2) the composition and functional performance of the local communities (S-s and F-f); and (3) the fate of the mixed communities (M-s and M-f). In parallel, to pre-adapt them for the second experiment (experiment 2), two additional bags for each kind of water (S and F) were filled with 350 ml of water and incubated in both tanks (PA-S-s, PA-S-f, PA-F-f and PA-F-s).
Figure 1.
Experimental setup. In both experiments, dialysis bags containing freshwater (F), saline water (S) or a 1:1 volume mix of both (M) were incubated in freshwater (f) or saline (s) incubation tanks for 3 days. In experiment 1, additional pre-adaptation bags (PA) were also incubated to serve as source of experiment 2. The lower-case ‘f' and ‘s' in the name of the bags stand for the applied incubation environment in experiment 1 and experiment 2, respectively.
Directly after termination of experiment 1, we implemented experiment 2 using the PA waters, and otherwise identical preparation and set-up procedures as in the first experiment. Water samples previously incubated under saline conditions (PA-S-s and PA-F-s) were used to fill the bags that were going to be incubated in the saline tank (saline (S-s-s), freshwater (F-s-s) and 1:1 volume ratio mix of both (M-s-s)), whereas for incubation in the freshwater tank, water samples adapted to freshwater conditions (PA-S-f and PA-F-f) were used (saline (S-f-f), freshwater (F-f-f) and 1:1 volume ratio mix of both (M-f-f); Figure 1). In addition, as we presumed that acclimatization to new incubation conditions might have caused differences even among communities originating from the same source (that is, PA-S-s and PA-S-f; PA-F-f and PA-F-s), bags filled with water previously incubated under new conditions (PA-S-f and PA-F-s) were placed back to their source environment (S-f-s and F-s-f). The latter samples served as a measure of functional and compositional performance of local communities, and by comparing them with the transplanted communities (S-f-f and F-s-s), we were able to assess the effect of pre-adaptation on the environmental adjustment capacities of the communities.
Osmotic equalization of salinity between the bags and the tanks was monitored with separate transplant bags every 30 min until reaching equal values (4 h), whereas microbial impermeability of the dialysis bags was tested with bags filled with respective sterile-filtered water (0.2 μm, 47 mm Supor-200 membrane filters, Pall Corporation) that were incubated in the same way as the experimental bags. The relatively short incubation time (3 days) assured that the permeability of the bags was not constrained due to the possible formation of biofilms on their surfaces.
Microbial abundance, cell size and biovolume
Immediately after sampling, 5 ml of the samples were fixed with sterile-filtered formaldehyde at a final concentration of 4% and stored at 4 °C. Bacterial abundance (BA) was determined according to del Giorgio et al., (1996). Briefly, samples were mixed with 1.25 μℳ final concentration of SYTO13 nucleic acid stain (Invitrogen, Eugene, OR, USA) and counted using a flow cytometer (CyFlow space, Partec, Münster, Germany). Microbial cells were identified according to their forward scatter and green fluorescence patterns. Average relative cell size was estimated using the mean forward scatter value and referred as individual cell size (ICS). Biovolume (BV) of the samples was calculated by multiplying ICS with respective BA (Hammes and Egli, 2010). Throughout the manuscript, we use BV as an estimator of the standing stock of bacterial biomass in the different treatments (Gasol and del Giorgio, 2000). Owing to the full-factorial nature of our experimental set-up, we expect BV to allow us to compare the stress effect of new environmental conditions with competition between bacterial communities. More specifically, if no stress effect is present, the BV of communities should depend only on the carrying capacity of the incubation environment, but not on their source.
Bacterial production
Heterotrophic bacterial production (BP) was measured using the radiolabeled leucine incorporation technique (Smith and Azam, 1992). Duplicate aliquots of each sample and trichloroacetic acid killed controls (5% final concentration) were incubated with 100 nℳℒ-leucine (15% ℒ-[4,5-3H] leucine, TRK510, 139 Ci mmol–1 (Amersham, Buckinghamshire, UK), and 85% cold substrate). Reactions, radioactivity measurement and bacterial carbon incorporation calculations were performed as described previously (Langenheder et al., 2006). BP is strongly correlated to the active part of bacterial biomass (del Giorgio et al., 1997). Therefore, we calculated SBP by dividing leucine incorporation with BV. This way the data was controlled for biomass and could be used as an estimator of heterotrophic bacterial activity in the experiments.
Active bacterial community composition
Active bacterial community composition (ABCC) was determined by 454-pyrosequencing of the reverse transcripted 16S rRNA to exclude inactive or dormant bacteria (Nikolausz et al., 2004). Hence, our analysis focuses specifically on the active taxa that are likely to influence the dynamics and function of a community. First, 50 ml of water from the dialysis bags were filtered onto 0.2 μm, 47 mm Supor-200 filters (Pall Corporation) and stored at −80 °C until processing. Nucleic acids were extracted from the filters using the Easy DNA kit (Invitrogen) following protocol number 3 for small amounts of cells, but included an extra bead-beating step using 0.2 ml of 0.1 mm silica beads. Then DNA was eliminated from the aliquots using DNAse I (Invitrogen) according to the manufacturer's protocol and complete removal of DNA was checked by doing PCRs on the untranscribed RNA samples using the same amplification conditions as for preparing the amplicons for 454-pyrosequencing. cDNA was synthesized from RNA using random hexamer primers and SuperScript II first-strand synthesis kit (Invitrogen).
The amplicons for 454-pyrosequencing were prepared, sequenced and the sequences were processed (quality checked, aligned, clustered, identified and normalized) as described before (Langenheder and Székely, 2011). Accordingly, singletons and operational taxonomic units (OTUs) with low abundance (<0.24% of total abundance) were not considered. Clustering was done into 3% dissimilarity OTUs, and only sequences with at least 95% similarity to their closest Ribosomal Database Project (rdp.cme.msu.edu) sequence match were included in the analyses. All sequences used in this study have been deposited to the NCBI sequence read archive under accession number SRP010301.
To follow the fate of the different OTUs, they were grouped into four environmental affinity categories based on their detection in the unmixed samples: (1) freshwater OTUs—OTUs detected only in samples originating from the freshwater source (F); (2) saline OTUs—OTUs detected only in saline samples (S); (3) common OTUs—OTUs detected in both freshwater and saline samples; and (4) undetermined OTUs—OTUs for which origin could not be determined, as they were not detected in any of the unmixed samples but only in mixed ones (M). Accordingly, OTUs corresponding to freshwater and saline environmental affinity groups are defined as habitat specialist (either freshwater or saline) and common OTUs as habitat generalists throughout the manuscript. The suitability of this approach was confirmed by analyzing the environmental distribution of the closest relatives of our OTUs in envDB, a database in which 16S rRNA sequences are environmentally characterized on the basis of their origin (Pignatelli et al., 2009; Supplementary Information S1).
Statistical analysis
In case of the unmixed communities (experiment 1: F-f, S-f, F-s and S-s; experiment 2: F-s-f, S-f-f, F-s-s and S-f-s; Figure 1), two-way analysis of variance (ANOVA) was used to test how the source and incubation environment influenced BA, ICS, BV, BP and SBP. Tukey's honestly significant difference test was applied for post-hoc pairwise comparison of the levels for single factors and Z-scores of the skew (zSkew) were analyzed to check data suitability for ANOVA. All these analyses were conducted using the ezANOVA software (http://www.cabiatl.com/mricro/ezanova/). For ABCC, the effect of source and incubation environment was tested by permutational multivariate ANOVA (PERMANOVA) using Bray–Curtis similarities based on relative abundances of the OTUs. The multivariate version of the t-statistic was used for pairwise a posteriori comparisons. Both analyses were conducted using PERMANOVA (Anderson, 2001; McArdle and Anderson, 2001). The ratio of the different OTU categories among treatments with the same source but different incubation environments (for example, S-s and S-f, or F-s-f and F-s-s) were compared by one-tailed Student's t-test.
The expected value of each analyzed parameter for the mixed communities (M) was calculated by summing the values measured for the unmixed communities (experiment 1: F-f and S-f for M-f, and F-s and S-s for M-s; experiment 2: F-f-f and S-f-f for M-f-f, and F-s-s and S-s-s for M-s-s) in a ratio equivalent to their abundance ratio in the corresponding mixes. The expected relative abundance of each OTU in mixed communities was calculated in the same way and was compared with the observed ABCC of the mixes by PERMANOVA (McArdle and Anderson, 2001; Anderson, 2006). Relative abundances of each OTU category (freshwater, saline, common and undetermined) were summed both for measured data and for expected values. The expected values of the functional parameters and the relative abundances of the OTU categories were compared with the measured values using one-tailed Student's t-test.
Results
Functional parameters
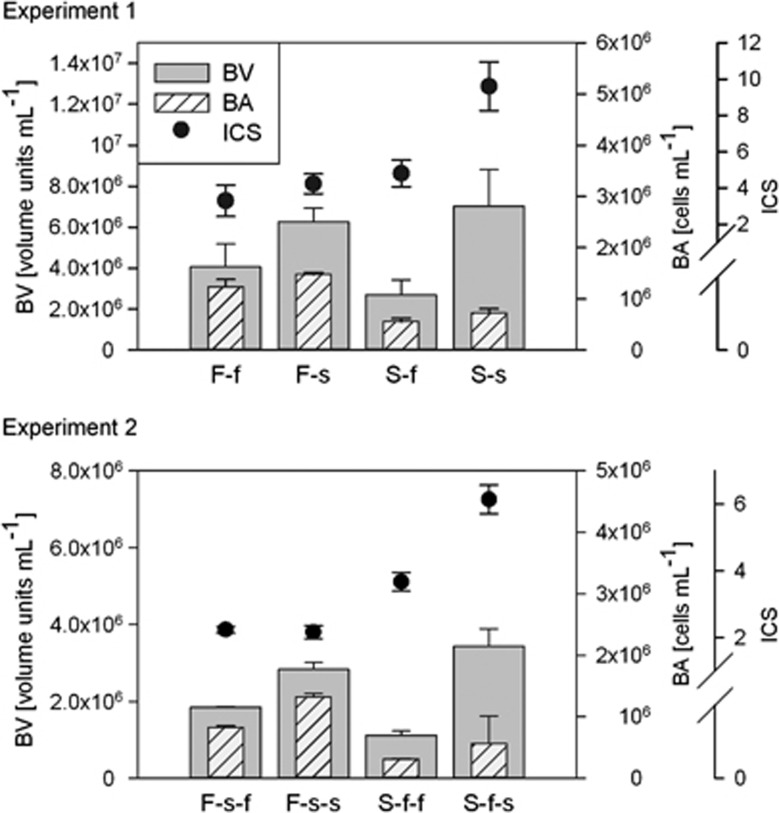
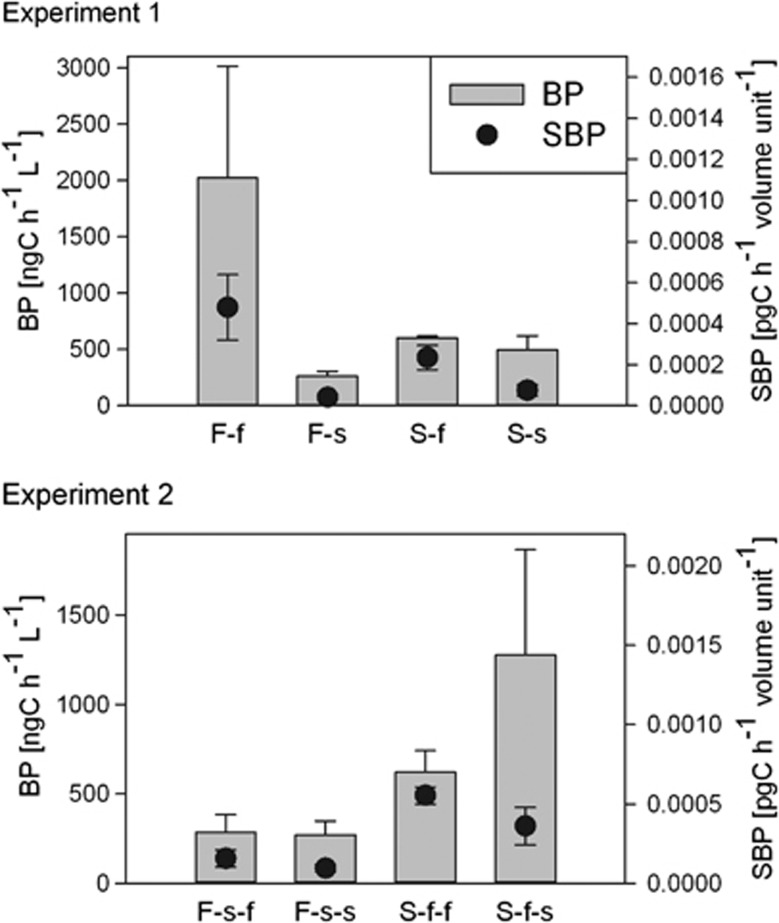
In case of unmixed communities, BV was primarily determined by the environment with higher values detected in the saline incubation, whereas the origin of the community was less important (Figure 2, Table 1, Supplementary Table S2). On the other hand, SBP values were always higher under freshwater conditions when comparing samples originating from the same source (for example, F-f and F-s, or S-f-f and S-f-s; Figure 3, Supplementary Table S2). The source effect on SBP was either direct (experiment 2) or interacted with the incubation conditions (experiment 1, Table 1).
Figure 2.
Biovolume (BV), abundance (BA) and average cell size (ICS) of unmixed samples incubated under local or new environmental conditions. Bars represent mean values of BV and BA, and dots represent mean values of ICS in experiment 1 and experiment 2. Error bars indicate s.d. ‘F' stands for freshwater source of the samples and ‘S' for saline, lower-case ‘f' and ‘s' stand for incubation environments in experiment 1 and experiment 2, respectively.
Table 1. Effects of source and incubation environment on functional parameters (two-way ANOVA) and ABCC (two-way PERMANOVA) of unmixed communities.
|
Source |
Environment |
Source × environment |
||||
|---|---|---|---|---|---|---|
| F | P-values | F | P-values | F | P-values | |
| Experiment 1 | ||||||
| BA | 188 | <0.001 | 15.5 | 0.004 | 0.46 | 0.517 |
| ICS | 41.8 | <0.001 | 29.1 | <0.001 | 13.1 | 0.007 |
| BV | 0.192 | 0.673 | 23.6 | <0.001 | 2.53 | 0.15 |
| BP | 4.23 | 0.074 | 10.5 | 0.012 | 8.18 | 0.021 |
| SBP | 4.49 | 0.067 | 35.9 | <0.001 | 7.74 | 0.024 |
| ABCC | 43.6 | <0.001 | 6.73 | 0.006 | 2.48 | 0.087 |
| Experiment 2 | ||||||
| BA | 760 | <0.001 | 258 | <0.001 | 25.9 | <0.001 |
| ICS | 287 | <0.001 | 56 | <0.001 | 63.4 | <0.001 |
| BV | 0.171 | 0.69 | 132 | <0.001 | 21.3 | 0.002 |
| BP | 14.3 | 0.005 | 3.27 | 0.108 | 3.57 | 0.096 |
| SBP | 65.4 | <0.001 | 9.51 | 0.015 | 2.56 | 0.148 |
| ABCC | 8.66 | <0.001 | 3.44 | 0.017 | 1.67 | 0.145 |
Abbreviations: ABCC, active bacterial community composition; ANOVA, analysis of variance; BA, bacterial abundance; BP, bacterial production; BV, biovolume; ICS; Individual cell size; PERMANOVA, permutational multivariate ANOVA; SBP specific BP.
Significant values (P<0.05) are in bold.
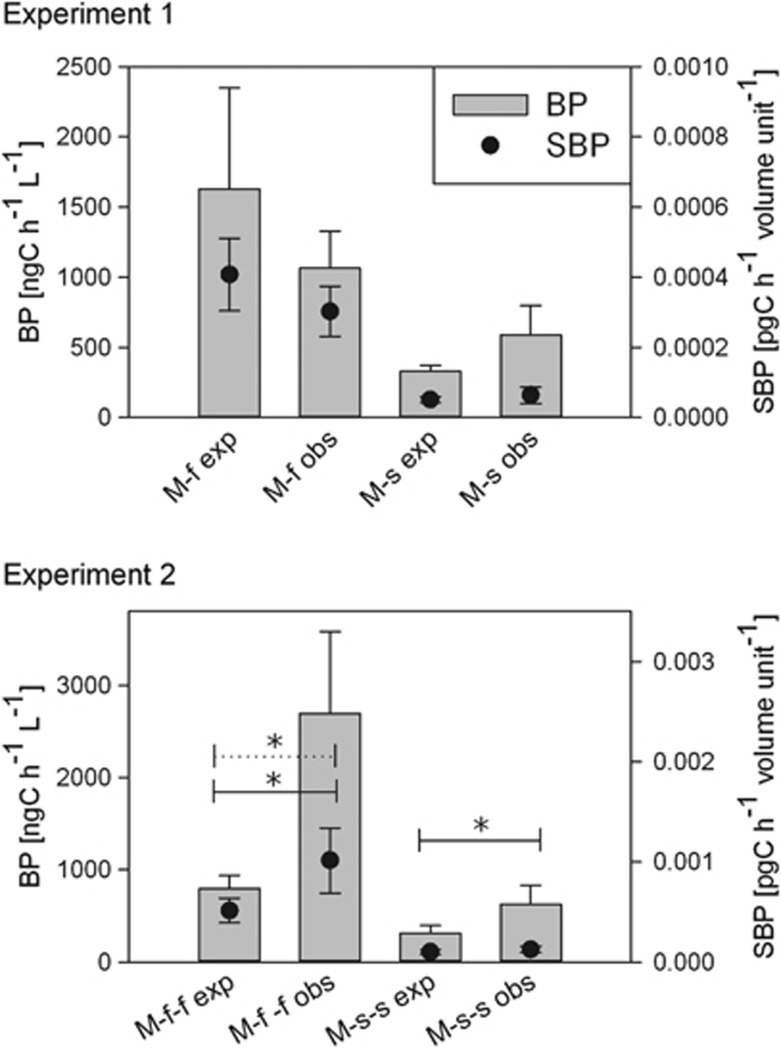
Figure 3.
Bacterial production (BP) and specific BP (SBP) of unmixed samples incubated under local or new environmental conditions. Bars represent mean values of BP and dots represent mean values of SBP in experiment 1 and experiment 2. Error bars indicate s.d. ‘F' stands for freshwater source of the samples and ‘S' for saline, lower-case ‘f' and ‘s' stand for incubation environments in experiment 1 and experiment 2, respectively.
Even though F and S communities showed similar BV when incubated in the same environment (for example, F-s and S-s, or F-s-s and S-f-s; Figure 2), they differed in the way they reached it. Under saline conditions, BA and ICS showed higher values in most of the cases, but microbial communities originating from freshwater had always significantly higher BA compared with communities originating from saline water, whereas the opposite was the case for ICS (Figure 2, Table 1, Supplementary Table S2). Moreover, saline communities showed a strong response to the incubation environment by producing significantly larger cells under saline conditions, whereas cell sizes of freshwater communities were not affected by the incubation environment (Figure 2, Supplementary Table S2).
For SBP, the source-dependent differences of the unmixed samples resulted mainly from the differences in BP, that is, in experiment 1, the effect of the incubation environment was strong in case of the freshwater samples and weaker in case of the saline samples, whereas in experiment 2, the situation was the opposite (Figure 3).
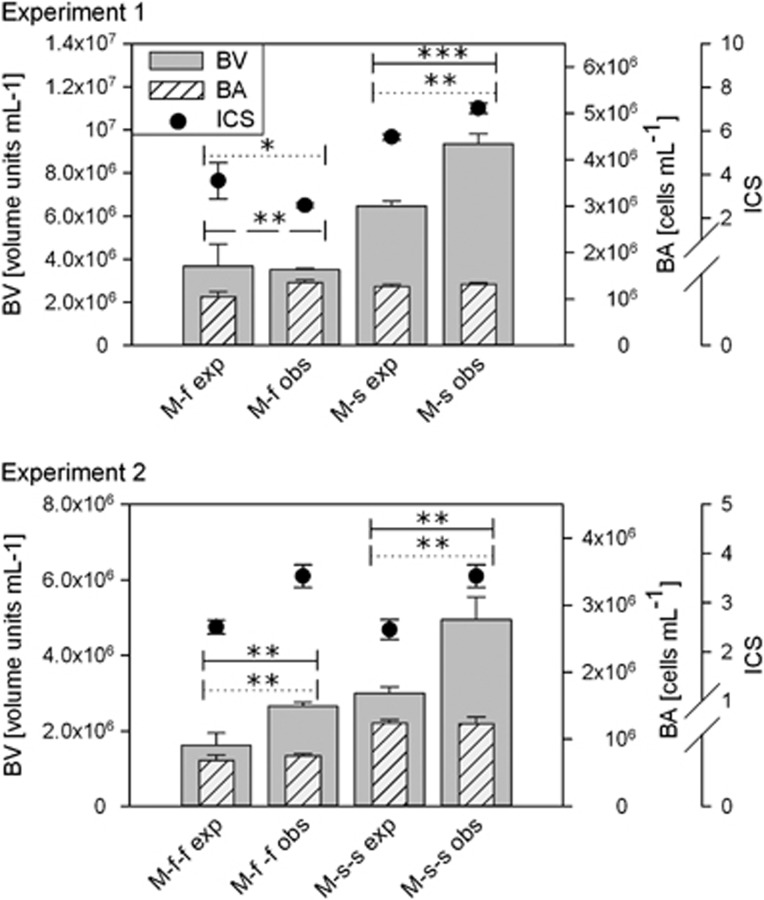
For the mixed communities, BV values were significantly higher than expected from BV values of the unmixed communities in three cases (experiment 1: M-s; experiment 2: M-f-f, M-s-s), and only in case of the freshwater incubation in the first experiment (M-f), measured BV was as expected (Figure 4). For the three mixes with higher BV values than expected, BA was not significantly different than expected, whereas ICS was significantly higher (Figure 4). For the M-f mix, BA was significantly higher compared with what we expected from unmixed samples, whereas ICS was significantly lower leading to no difference in BV (Figure 4). For BP and SBP, M-f communities showed lower measured values and the other three mixes showed higher measured values than expected. However, these differences were only significant in experiment 2, in case of BP for both M-f-f and M-s-s, and for SBP in case of the freshwater incubation (M-f-f; Figure 5).
Figure 4.
Expected and observed values of Biovolume (BV), abundance (BA) and average cell size (ICS) of mixed communities. Bars represent mean values of BV and BA, and dots represent mean values of ICS in experiment 1 and experiment 2. Error bars indicate s.d. ‘M' stands for mixed community, ‘f' and ‘s' stand for incubation environment in experiment 1 and experiment 2, respectively; ‘exp' stands for expected values and ‘obs' for observed values. Horizontal lines represent significant differences between expected and observed values. Solid lines stand for BV, dashed for BA and dotted for ICS. The asterisks represent the level of significance of the corresponding t-test in the following way: *P<0.05; **P<0.01 and ***P<0.001.
Figure 5.
Expected and observed values of bacterial production (BP) and specific BP (SBP) of mixed communities. Bars represent mean values of BP and dots represent mean values of SBP in experiment 1 and experiment 2. ‘M' stands for mixed community, ‘f' and ‘s' stand for incubation environment in experiment 1 and experiment 2, respectively; ‘exp' stands for expected values and ‘obs' for observed values. Error bars indicate s.d. Horizontal lines represent significant differences between expected and observed values. Solid lines stand for BP and dotted for SBP. The asterisks represent the level of significance of the corresponding t-test: *P<0.05.
Active bacterial community composition
Out of 52 683 good-quality sequences, 51 132 could be assigned with at least 95% similarity to known Ribosomal Database Project sequences. The 42 samples analyzed had 1381 identified sequences in average (minimum: 437; maximum: 3275). After normalization and singelton exclusion, sequences were grouped into 152 OTUs with an average of 38 OTUs per sample (minimum: 23; maximum: 53).
In case of the unmixed communities (F and S), composition was significantly influenced by both the origin of the communities and the incubation environment in both experiments (Table 1). To some extent, effects of both the source and the incubation environment were mitigated by adaptation as reflected by the decrease of the F-values in the PERMANOVA tests (Table 1) and the pairwise a posteriori comparisons (Supplementary Table S2). Regarding the relative abundance of habitat specialists and generalists, no apparent differences could be detected for the different incubation environments (Supplementary Table S3).
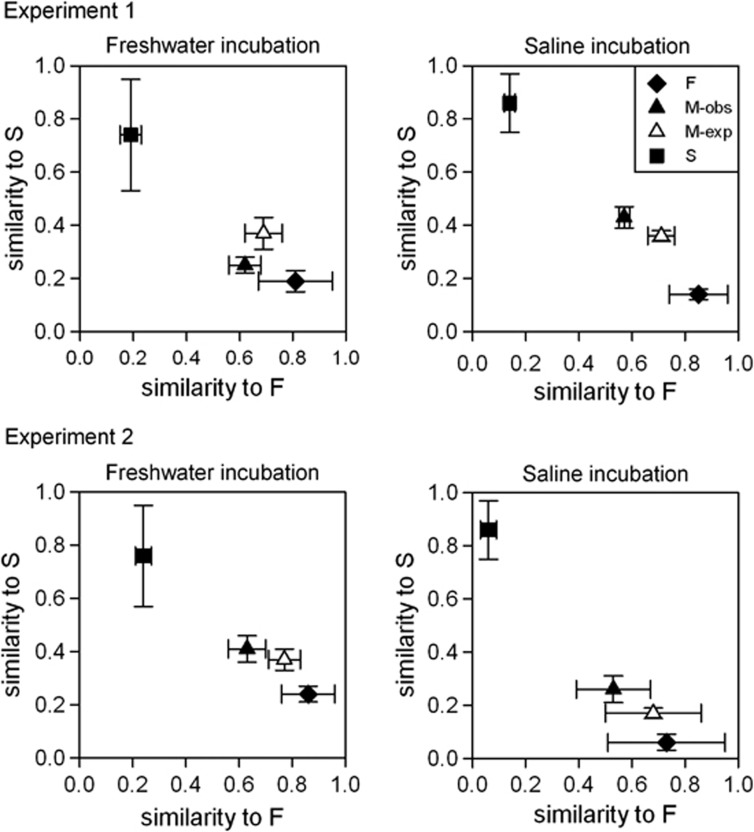
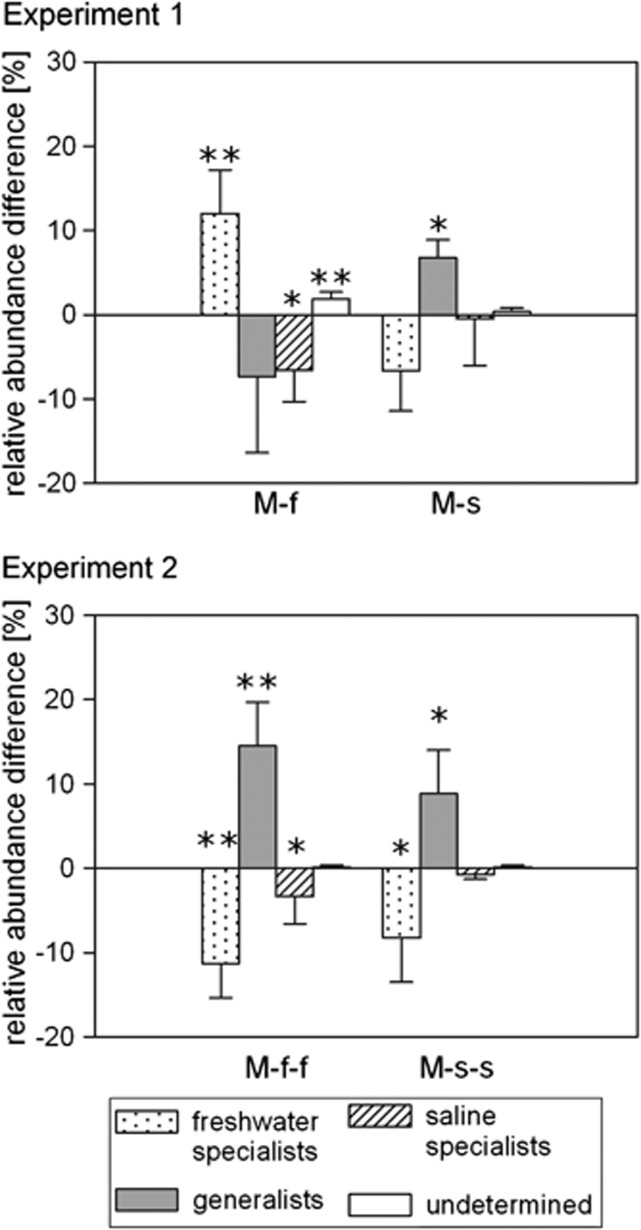
The actual composition of the mixed communities differed only to some extent from what we expected (PERMANOVA: Monte Carlo P-value: 0.029–0.070; Figure 6). In freshwater incubations of experiment 1, ABCC of the mixed samples was more dissimilar from the unmixed saline communities (S) than expected (Figure 6), whereas in the other three cases, the mixed communities were slightly closer to the unmixed saline communities (S) and further from the freshwater communities (F) than expected (Figure 6). However, when comparing expected and actual abundances of habitat specialists and generalists, there were several significant differences (Figure 7). In the freshwater incubation of experiment 1, the relative abundance of freshwater specialists was significangly higher than expected whereas the relative abundance of saline specialists and generalists was lower. In the saline incubation of experiment 1, and in both incubations in experiment 2, generalists were significantly more abundant, whereas freshwater specialists were less abundant. Saline specialists were less abundant than expected in all of the mixes. However, these differences were significant only in case of the communities incubated under freshwater conditions, whereas undetermined OTUs emerged in significant numbers only in case of the mixed communities incubated in freshwater in experiment 1 (M-f).
Figure 6.
Comparison of bacterial community composition based on Bray–Curtis similarities. Mean values of the Bray–Curtis similarities to the mean unmixed freshwater and saline samples are plotted under freshwater and saline conditions in experiment 1 and in experiment 2. ‘F' stands for freshwater source of the sample, ‘S' for saline, ‘M-obs' for observed mixed communities and ‘M-exp' for composition expected for mixed communities based on the unmixed samples; ‘f' and ‘s' stand for incubation environment in experiment 1 and experiment 2, respectively. Error bars indicate s.d. Along the horizontal axes, similarity to freshwater samples (F) is shown, whereas the vertical axes indicate similarity to saline samples (S). Deviation from one in case of the similarity of the unmixed samples (F and S) to their corresponding axes is due to the differences among replicates.
Figure 7.
Differences between observed and expected relative abundances of OTU categories of mixed bacterial communities. Bars represent mean values of the differences between expected and measured relative abundance of habitat specialists (freshwater or saline) or habitat generalists in the mixed samples in experiment 1 and in experiment 2. ‘M' stands for mixed communities, ‘f' and ‘s' stand for incubation environment in experiment 1 and experiment 2, respectively. Error bars indicate s.d. The asterisks represent the level of significance of the corresponding t-test in the following way: *P<0.05 and **P<0.01.
Discussion
The major aim of this study was to experimentally disentangle the functional and compositional consequences of the two major challenges that bacteria face when immigrating to new habitats, namely the change in abiotic conditions, as well as competition with the local community. Our results show that immigrating communities are able to adjust to new environmental conditions by different mechanisms, and are further influenced by interactions with the local community.
Abiotic differences between the two rock pools that we used to set up the experiments were mainly due to salinity and, to a lesser extent, also phosphorous concentrations (Supplementary Table S1). External osmolarity is the major factor shaping microbial community composition (Lozupone and Knight, 2007; Tamames et al., 2010), as both hyper- and hypo-osmotic environmental changes constitute a major stress for single cell organisms (Morbach and Krämer, 2002). Moreover, primary productivity has also been shown to influence the diversity and composition of bacterial communities (Horner-Devine et al., 2003; Smith, 2007). In our experiments, both freshwater (F) and saline (S) communities coped functionally fairly well with environmental changes when incubated under new conditions, and were similar to the respective ‘in situ' communities. Biomass depended only on the incubation conditions, and the trend of heterotrophic bacterial activity was also primarily determined by differences in the incubation environments (Figures 2 and 3, Table 1). Our results do, however, highlight that functional adjustments to new environments can follow different pathways (Comte and del Giorgio, 2011), as communities of freshwater origin adapted to changes in environmental conditions by increasing or decreasing their cell numbers, whereas saline ones modified their cell size (Figure 2). Morphological modifications have been known for a long time as cellular osmoregulation mechanism (Zahran 1997; Kültz 2001) and survival strategies (Justice et al., 2008). Thus, the observed higher morphological flexibility of the saline communities might suggest that these bacteria possess wider environmental potentials or at least have different adaptive strategies towards salinity changes. Possible reasons for the observed overall functional equivalence of both communities in both incubation environments, respectively, could be, for example, that the salinity differences between the samples were not high enough to significantly stress the communities (0 vs 4.5 psu). However, it has previously been shown that similar ‘mild' salinity differences can affect microbial communities both in terms of function and composition (for example, Langenheder et al., 2003). Another possibility could therefore be that the rock pool communities were already ‘pre-conditioned' to environmental fluctuations, as they regularly experience quite drastic differences in environmental conditions (Jocque et al., 2010).
Community composition of the unmixed saline and freshwater samples was determined by both the source of the samples and the incubation environment (Table 1). The fact that the communities maintained strong source-dependent compositions shows that environmental adaptation did not require conversion to a pre-defined habitat-specific composition and indicates that many taxa possess a high degree of functional plasticity. Hence, our results do not support predictions made by the replacement or species-sorting scenarios (Leibold et al., 2004; Comte and del Giorgio, 2011). On the other hand, the effect of the incubation environment conflicts with predictions made by the ‘adjustment scenario' or neutral model (Hubbell, 2001; Comte and del Giorgio, 2011). Thus, our study shows that several mechanisms determine community composition and functioning at the same time, and is in congruence with previous studies showing that species sorting and neutral processes have simultaneous roles during community assembly (Ofiteru et al., 2010; Langenheder and Székely, 2011). Moreover, environmental changes are prone to increase the abundance of generalists (Clavel et al., 2011), and also the adjustment scenario (Comte and del Giorgio, 2011) predicts a conversion towards generalists upon exposure to new environmental conditions. The fact that this was not observed here (Supplementary Table S3) might indicate that complex interactions within communities are more important in defining environmental affinities of taxa than abiotic conditions. In particular, it has been shown that many intrinsic properties of microbial communities, such as the complexity of microbial food-webs, the prevalence of syntrophy, the wide range of multicellular behaviors (West et al., 2006) and/or the close associations among bacterioplankton (Malfatti and Azam, 2009) may have important roles.
Functional parameters of mixed communities showed significant differences to those expected from the performance of the unmixed communities (Figures 4 and 5), pointing out that the mixed communities were not simply proportional mixes of the unmixed communities as predicted by the neutral model. When mixed communities were incubated under freshwater conditions in experiment 1 (M-f treatment), a functional adjustment strategy implying conversion towards unmixed freshwater communities, that is, an increase in abundance coupled with cell size decrease, was found (Figure 4). Furthermore, freshwater specialists had significantly higher relative abundance than expected, whereas generalists and saline specialists were underrepresented in the mixed community (Figure 7). This suggests that the communities were, to a considerable extent, structured by species sorting that led primarily to the emergence of a community more similar to the corresponding unmixed freshwater community than expected (Figure 6). On the contrary, the other three mixed communities (M-s, M-f-f and M-s-s) showed higher relative abundance of habitat generalists than expected (Figure 7). Interestingly, freshwater specialists had lower relative abundances than expected even in the freshwater incubation (M-f-f). Saline specialists, on the other hand, were neither favored nor disfavored under saline conditions (M-s and M-s-s), but had lower relative abundances under freshwater conditions as expected by the species-sorting scenario. Hence, under saline incubation conditions, mixed communities were structured as predicted by the interaction ruled scenario, whereas in the freshwater incubations, both species sorting (experiment 1), and the combination of species sorting and interaction-ruled scenario (experiment 2) had a role shaping the composition of the mixed community. The latter result also shows that pre-adaptation influenced the outcome of the encounter between the local and immigrant communities in the freshwater incubations.
As predicted by the interaction-ruled scenario, the observed increase of generalists is most likely a consequence of their higher competitive ability (Dall and Cuthill, 1997). However, even though protozoans, the most important bacterial grazers (Sherr and Sherr, 2002), have been removed before the experiments, we could not exclude the effect of viruses and prokaryotic bacterivores such as the Bdellovibrio species, which may also influence the outcome of the encounter between different communities, and against which generalists are supposed to be more resistant to.
A possible explanation for the differences observed for freshwater and saline environments regarding the assembly mechanisms of the mixed communities composition and the importance of freshwater specialists versus generalists could be that saline conditions are more ancestral (Logares et al., 2009), and that moderately saline conditions, like the ones in the salinity incubations of our experiment, are closer to the ionic conditions within cells (Kültz 2001; Cossins et al., 2011). Low-conductivity environments, such as the freshwater incubations, represent hypo-osmotic conditions that require a greater extent of specific adaptations from bacteria. For example, this could be achieved through increasing cell surface to cell volume ratios by producing smaller cells (Figure 2) to facilitate membrane and periplasma-associated osmoregulation (Kültz, 2001). Thus, freshwater bacteria may, to a greater extent, be habitat specialists, whereas saline bacteria may have wider range of environmental optima as indicated also by the higher morphological flexibility observed in case of the unmixed saline communities (Figure 2). Similar speculations may also explain the differences between the two consecutive freshwater incubations of the mixed communities. In the first experiment, the ratio of freshwater specialists increased compared with what was expected, whereas generalists that had higher relative abundance than expected in the second experiment required a period of pre-adapation before they could ‘take advantage' of the freshwater specialists (Figure 7).
In case of the mixed communities that were assembled according to the interaction-ruled scenario, the measured functional parameters were in all cases significantly higher than expected (Figures 4 and 5). Taking into count the parallel increase in abundance of common OTUs (Figure 7), this seems to contradict the ‘jack of all trades is master of none' theory (MacArthur, 1972), according to which generalists should be functionally inferior to specialists. However, the functional parameters that we measured in this study, biomass and BP derived from a labile and generally available compound (leucine), comprise rather general functions that are unlikely to require specific specialization of bacterial cells, and different results might have been obtained if we had measured more specific functions (Peter et al., 2011). Hence, the generalists that occurred in the experiments were taxa that could cope well with both salinities and performed generally well, resulting in higher biomass and production compared with the unmixed communities.
In summary, we could show that the fate of dispersed bacteria is affected by both, the environmental changes due to the exposure to the new habitat and competition with local communities. In the absence of competition from a local community, immigrant communities were able to adapt to the new environments with equivalent functional performances compared with local communities. When a local community was present in most of the cases, this resulted in a remarkable functional ‘overachievement' of communities, at least partly caused by the increased representation of generalists and points out the importance of competitive interactions for community performance, and ultimately ecosystem functioning. Even though our study was done over relatively short time periods and with only one environmental gradient, it provides some first insights that clearly show that environmental filtering, functional plasticity and competition are all important mechanisms influencing the fate of dispersed communities. Future studies should investigate how the relative importance of these mechanisms changes when communities are recurrently sampled over longer time periods, and when other and stronger environmental gradients as well as different levels of rates of dispersal are considered.
Acknowledgments
We thank Hannes Peter, Jan Johanson and Zoltán Török for technical assistance, advice and help during fieldwork, and the anonymous reviewers for their valuable comments on the manuscript. This study was funded by grants from the Swedish Research Council Formas, the Carl Tryggers foundation and the Centre for Metagenomic Sequence analysis at Uppsala University.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Clavel J, Julliard R, Devictor V. Worldwide decline of specialist species: toward a global functional homogenization. Front Ecol Environ. 2011;9:222–228. [Google Scholar]
- Comte J, del Giorgio PA. Linking the patterns of change in composition and function in bacterioplankton successions along environmental gradients. Ecology. 2010;91:1466–1476. doi: 10.1890/09-0848.1. [DOI] [PubMed] [Google Scholar]
- Comte J, del Giorgio PA. Composition influences the pathway but not the outcome of the metabolic response of bacterioplankton to resource shifts. PLoS One. 2011;6:e25266. doi: 10.1371/journal.pone.0025266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins BP, Jacobson MP, Guallar V. A new view of the bacterial cytosol environment. PLoS Comput Biol. 2011;7:e1002066. doi: 10.1371/journal.pcbi.1002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall SRX, Cuthill IC. The information costs of generalism. Oikos. 1997;80:197–202. [Google Scholar]
- del Giorgio PA, Bird DF, Prairie YT, Planas D. Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain SYTO 13. Limnol Oceanogr. 1996;41:783–789. [Google Scholar]
- del Giorgio PA, Prairie YT, Bird DF. Coupling between rates of bacterial production and the abundance of metabolically active bacteria in lakes, enumerated using CTC reduction and flow cytometry. Microb Ecol. 1997;34:144–154. doi: 10.1007/s002489900044. [DOI] [PubMed] [Google Scholar]
- Gasol JM, del Giorgio PA. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Method Cell Biol. 2000;64:197–224. [Google Scholar]
- Gómez JP, Bravo GA, Brumfield RT, Tello JG, Cadena CD. A phylogenetic approach to disentangling the role of competition and habitat filtering in community assembly of Neotropical forest birds. J Anim Ecol. 2010;79:1181–1192. doi: 10.1111/j.1365-2656.2010.01725.x. [DOI] [PubMed] [Google Scholar]
- Hammes F, Egli T. Cytometric methods for measuring bacteria in water: advantages, pitfalls and applications. Anal Bioanal Chem. 2010;397:1083–1095. doi: 10.1007/s00216-010-3646-3. [DOI] [PubMed] [Google Scholar]
- Horner-Devine CM, Leibold MA, Smith VH, Bohannan BJM. Bacterial diversity patterns along a gradient of primary productivity. Ecol Lett. 2003;6:613–622. [Google Scholar]
- Hovatter SR, Dejelo C, Case AL, Blackwood CB. Metacommunity organization of soil microorganisms depends on habitat defined by presence of Lobelia siphilitica plants. Ecology. 2011;92:57–65. doi: 10.1890/10-0332.1. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press: Princeton; 2001. [Google Scholar]
- Jocque M, Vanschoenwinkel B, Brendonck L. Freshwater rock pools: a review of habitat characteristics, faunal diversity and conservation value. Freshwater Biol. 2010;55:1587–1602. [Google Scholar]
- Jones SE, McMahon KD. Species-sorting may explain an apparent minimal effect of immigration on freshwater bacterial community dynamics. Environ Microbiol. 2009;11:905–913. doi: 10.1111/j.1462-2920.2008.01814.x. [DOI] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol. 2008;6:162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- Kültz D. Cellular osmoregulation: beyond ion transport and cell volume. Zoology. 2001;104:198–208. doi: 10.1078/0944-2006-00025. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Kisand V, Wikner J, Tranvik LJ. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol Ecol. 2003;45:189–202. doi: 10.1016/S0168-6496(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Sobek S, Tranvik LJ. Changes in bacterial community composition along a solar radiation gradient in humic waters. Aquat Sci. 2006;68:415–424. [Google Scholar]
- Langenheder S, Ragnarsson H. The role of environmental and spatial factors for the composition of aquatic bacterial communities. Ecology. 2007;88:2154–2161. doi: 10.1890/06-2098.1. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Székely AJ. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 2011;5:1086–1094. doi: 10.1038/ismej.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett. 2004;7:601–613. [Google Scholar]
- Lindström ES, Östman Ö. The importance of dispersal for bacterial community composition and functioning. PLoS One. 2011;6:e25883. doi: 10.1371/journal.pone.0025883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Logue JB, Lindström ES. Biogeography of bacterioplankton in inland waters. Freshw Rev. 2008;1:99–114. [Google Scholar]
- Logue JB, Lindström ES. Species sorting affects bacterioplankton community composition as determined by 16S rDNA and 16S rRNA fingerprints. ISME J. 2010;4:729–738. doi: 10.1038/ismej.2009.156. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH. Geographical Ecology: Patterns in the Distribution of Species. Harper and Row: New York; 1972. [Google Scholar]
- Malfatti F, Azam F. Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquat Microb Ecol. 2009;58:1–14. [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-devine MC. Drivers of bacterial β-diversity depend on spatial scale. Proc Natl Acad Sci USA. 2011;108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- Morbach S, Krämer R. Body shaping under water stress: osmosensing and osmoregulation of solute transport in bacteria. Chembiochem. 2002;3:384–397. doi: 10.1002/1439-7633(20020503)3:5<384::AID-CBIC384>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Nikolausz M, Márialigeti K, Kovács G. Comparison of RNA- and DNA-based species diversity investigations in rhizoplane bacteriology with respect to chloroplast sequence exclusion. J Microbiol Meth. 2004;56:365–373. doi: 10.1016/j.mimet.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östman Ö, Drakare S, Kritzberg ES, Langenheder S, Logue JB, Lindström ES. Regional invariance among microbial communities. Ecol Lett. 2010;13:118–127. doi: 10.1111/j.1461-0248.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- Peter H, Ylla I, Gudasz C, Romaní AM, Sabater S, Tranvik LJ. Multifunctionality and diversity in bacterial biofilms. PLoS One. 2011;6:e23225. doi: 10.1371/journal.pone.0023225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M, Moya A, Tamames J. EnvDB, a database for describing the environmental distribution of prokaryotic taxa. Environ Microbiol Rep. 2009;1:191–197. doi: 10.1111/j.1758-2229.2009.00030.x. [DOI] [PubMed] [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Antonie Leeuwenhoek. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine 1. Mar Biol Res. 1992;6:107–114. [Google Scholar]
- Smith VH. Microbial diversity-productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol. 2007;62:181–186. doi: 10.1111/j.1574-6941.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- Tamames J, Abellán JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10:85. doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, Declerck S, et al. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA. 2007;104:20404–20409. doi: 10.1073/pnas.0707200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci USA. 2011;108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Guillaumaud N, et al. Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ Microbiol. 2007;9:2211–2219. doi: 10.1111/j.1462-2920.2007.01335.x. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Wohl DL, Arora S, Gladstone JR. Functional redundancy supports biodiversity and ecosystem function in a closed and constant environment. Ecology. 2004;85:1534–1540. [Google Scholar]
- Zahran HH. Diversity, adaptation and activity of the bacterial flora in saline environments. Biol Fertil Soils. 1997;25:211–223. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.