Abstract
Microcystins (MCs) are widespread cyanobacterial toxins in freshwater systems, and have been linked to both acute and chronic health effects. A growing number of studies suggest that MC can bioaccumulate in food webs. Although, several methods (i.e. ELISA, LC-MS) have been developed for analysis of MC in water, extraction (for subsequent analysis) of the toxin from biological matrices (i.e. animal tissues) is impeded owing to covalent binding of toxins and active sites of their cellular targets, i.e. protein phosphatases. As an alternative approach, chromatographic methods for analysis of a unique marker, 2-methyl-3-methoxy-4-phenylbutanoic acid (MMPB), the product of the Lemieux oxidation of MCs, have been previously developed, and shown to measure total (bound and unbound) MC. Application, however, has been limited by poor recovery of the analyte. An improved recovery method is proposed – specifically the use of solidphase microextraction (SPME). The MMPB analogue, 4-phenylbutanoic acid (4PB), and oxidized MC, were used to develop methods, and we specifically investigated several SPME fibres, and post-oxidation steps. Specifically, a method employing post-oxidation methyl esterification, followed by headspace SPME recovery of MMPB, was developed, and subsequently applied to analysis of environmental samples (i.e. fish tissues) previously shown to contain MCs. The method shows high linearity for both water and tissues spiked with MC, and an improved limit of quantitation of approximately 140 ng g−1. Evaluation of field samples by SPME-GC/MS detected considerably higher levels of MC, than detected by conventional methods (i.e. ELISA), and it is proposed that this technique reveals MC (particularly in the bound form) that is not detected by these methods. These results indicate that the developed method provides improved detection capability for MC in biological matrices, and will enhance our ability to understand bioaccumulation in freshwater food webs, as well as monitor exposure.
Keywords: solid-phase microextraction (SPME), microcystin, 2-methyl-3-methoxy-4-phenylbutanoic acid (MMPB), Lemieux oxidation, cyanobacteria, bioaccumulation
1. Introduction
Cyanobacteria (‘blue-green algae’) are recognized to produce a diversity of toxic or otherwise bioactive metabolites. In freshwater systems, in particular, several of these toxins have been associated with human and environmental health concerns [1–3]. Specifically, it is well established that cyanobacterial toxins pose potential health concerns via direct exposure to drinking or recreational waters [1–3]. Although not as well investigated, recent studies have suggested that bioaccumulation, or even bio-magnification, of cyanobacterial toxin in food webs, including higher trophic levels, such as fish or other animals consumed by humans, may also occur [4–15], representing an additional route of exposure to these toxins. In order to better understand the bioaccumulation of such cyanotoxins, and the health threats posed by this route of exposure, as well as to more generally improve our ability to detect exposure, development of analytical methods for measurement of cyanobacterial toxin content in biological matrices, such as animal tissue, will be critical.
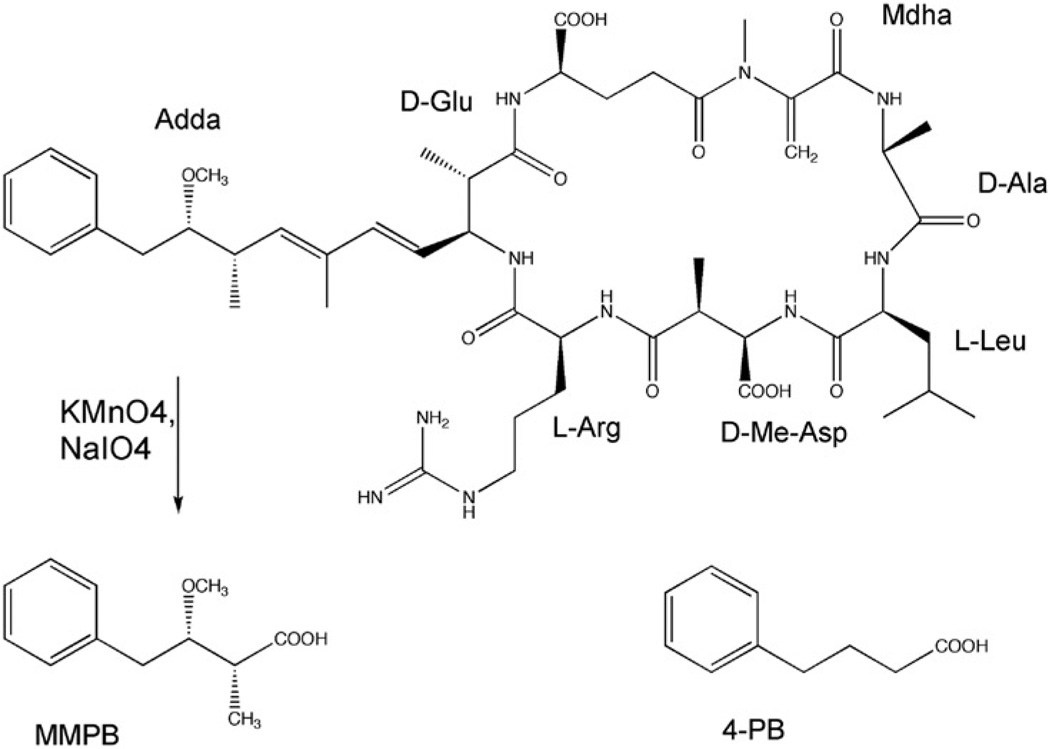
Microcystins (MCs) (Figure 1) are perhaps the most widespread cyanobacterial toxins, and are found worldwide in marine and freshwater environments [16]. MCs are a secondary metabolite of several cyanobacterial genera, including Microcystis, Anabaena, Oscillatoria and Nodularia, among others [16]. They are hepatotoxic cyclic heptapeptides with the general structure of cyclo(D-Ala-X-D-MeAsp-Y-Adda-D-Glu-Mdha), as shown in Figure 1, where ‘X’ and ‘Z’ are variable amino acids. Microcystin-LR (MC-LR), in which X and Z are leucine (L) and arginine (R), respectively, is among the most common of the MC family that consists of over 70 other variants [16].
Figure 1.
Structure of microcystin-LR (MC-LR) and the product, 2-methyl-3-methoxy-4-phenylbutanoic acid (MMPB), of the Lemieux oxidation of this compound, as well as of the MMPB analogue, 4-phenylbutanoic acid (4-PB).
Toxicologically, MCs are classified as ‘hepatotoxins’ as they accumulate primarily in the hepatocytes where they are acutely toxic, leading to necrosis and hemorrhaging of liver cells and associated tissues [16]. This condition manifests itself in severe gastroenteritis and even death [16]. In addition, MCs have been shown to promote tumors at nanomolar concentrations, and chronic exposures have been specifically linked to increased rates of primary liver cancer [3, 17–19]. The recognized target of MCs is specific inhibition of type 1 and 2A serine/threonine protein phosphatases (PPase) in a two-phase interaction [20–22]. The first phase is a rapid reversible binding that is responsible for the inhibition of the catalytic activity within the PPases. The second phase is the formation of a covalent and irreversible bond to cysteine in PPases via Michael-type addition to the Mdha of the MC molecule [23]. Mdha is not present in all MC variants, and is specifically replaced by dehydrobutyrine (Dhb), for example, in a few such variants [24], and would not, in these cases, be expected to contribute to the tissue-bound MCs. However, these generally do not represent common or particularly abundant variants. Accordingly, difficulties in the analysis of MC concentrations in biological matrices include the presence of two forms of the molecule – the free and covalently bound MCs.
Current methods commonly employed for detection of MCs are PPase inhibition assays, enzyme-linked immunosorbent assays (ELISA) and liquid chromatography/mass spectrometry (LC-MS) analysis [25]. Although these methods are generally effective for measuring the dissolved toxin in water, application of all of these methods to analysis of MC in tissues requires prior solvent extraction. Given the recognized covalent binding of MCs to PPase, as discussed above, it has become increasingly clear that detection of MCs in biological matrices following solvent extraction is, therefore, limited to only the free (i.e. non-covalently bound) toxin, and that quantitative analysis based on these techniques may significantly underestimate total toxin content [26, 27]. For example, in prior experimental studies, Williams et al. [28] found that methods based on solvent extraction detect only approximately 24% of total MC exposure dose, specifically based on intraperitoneal injection of salmon. Moreover, based on subsequently developed methods (see below), it was suggested [28] that conventional techniques (e.g. PPase inhibition) that rely on solvent extraction might underestimate total MC burden by as much as 104-fold.
In order to address this analytical limitation, a detection method, based on the unique nature of the pendant Adda moiety of MC (Figure 1), has been previously developed [26, 27]. Specifically, treatment of MC with potassium permanganate under carefully controlled conditions cleaves via Lemieux oxidation (Figure 1) the Adda moiety to produce 2-methyl-3-methoxy-4-phenylbutanoic acid (MMPB) that can be readily detected by either GC-MS or LC-MS, as well as GC-FID and HPLC-FL [27–33]. Since there is sufficient evidence that the Mdha portion of the MC molecule (Figure 1) is the site for the covalent bond to PPases, MMPB produced by Lemieux oxidation of the Adda will be identical for both free and covalently bound MCs. Moreover, MMPB represents a unique marker for the presence of MC, as it has no naturally occurring counterpart, and is expected to provide a highly specific method of detecting free and bound MCs [27, 28]. It has also been shown that the Lemieux oxidation of MCs provides a high yield of the butanoic acid that is detectable in low concentrations by GC/MS and LC/MS techniques [27–33].
Although the Lemieux oxidation reaction method described above has been successfully employed for the determination of MC content in various tissues [27–30], it requires extraction followed by ‘clean-up’ and other various post-oxidation procedures. Extraction methods typically employed are liquid-liquid extraction (LLE) or solid-phase extraction (SPE). These different extraction methods have been tried with varying degrees of success [27–30], but recovery of the MMPB analyte has been generally low in biological matrices (i.e. tissues) using these techniques, and this step has generally represented the most limiting aspect of the MMPB method. Specifically, recovery by LLE and SPE has limited detection of MC in relevant tissues (i.e. liver) to approximately 15 µg g−1 [28] and 5 µg g−1 [29], respectively. Here, we explore of the use of solid-phase microextraction (SPME) as an improved technique for the recovery of MMPB prior to analysis by GC-MS.
SPME, which has not been previously applied to the quantitative analysis of MMPB, utilizes a fused polymer-based fibre as an adsorption medium for the analyte [34]. Different fibres can be used for various analytes depending on the chemical nature of the analyte, including molecular size and functionalities present in the molecule. Samples can be analyzed for recovery by two methods: direct sampling (i.e. the SPME fibre is emerged in the liquid reaction solution) or headspace sampling. Fibres used with direct sampling have a lifetime of a few dozen runs whereas fibres used in headspace sampling have a lifetime of a few hundred runs. SPME fibres are desorbed for analysis by injecting fibres straight into the inject port/valve of the appropriate instrument which includes GC-MS, LC-MS and HPLC-UV [34].
Here we investigated the use of SPME, coupled to GC-MS, for recovery and analysis of MMPB, as a means to measure total MC content, using oxidized samples of MC-LR and the MMPB analogue, 4-phenylbutanoic acid (4-PB). In addition to the general assessment of this proposed technique, the study specifically investigated (1) several, appropriate SPME fibre types; (2) comparison of direct and headspace sampling; and (3) consequent development of a post-oxidation methyl esterification as a means to increase headspace recovery. Subsequently, the developed method was applied to the analysis of several environmental (i.e. fish tissue) samples previously identified by conventional methods (i.e. ELISA) to contain MCs as a means to compare the effectiveness of the developed technique.
2. Experimental
2.1 Materials
4-Phenylbutanoic acid (4PB) was purchased from Sigma Aldrich, Inc. Purified MC-LR was obtained from the NIH-NIEHS ARCH Toxic Algae Culture Core Facility of FIU. Chemicals used in the proposed methodologies include the following: diethyl ether (Fisher Scientific, USA), hydrochloric acid (Fisher Scientific, USA), potassium carbonate (Fisher Scientific, USA), potassium permanganate (EMD Chemicals, USA), silver carbonate (Sigma Aldrich, USA), sodium disulfite (EMD Chemicals, USA) and sodium metaperiodate (Alfa Aesar Chemicals, USA). SPME fibres and holders were purchased from Supelco, Inc.; selected fibres specifically included polydimethylsiloxane/divinylbenzene (PDMS-DVB), divinylbenzene/Carboxen™/polydimethylsiloxane (DVB-CAR-PDMS) and polyacrylamide (PA).
A standard of 2-methyl-3-methoxy-4-phenylbutyric acid (MMPB) as sodium salts were purchased from Wako Chemicals (Richmond, VA). To prepare the methyl ester of MMPB (meMMPB), for confirmation of the analyte in headspace SPME, aliquots of the MMPB standard (100 µg each) were taken to dryness in vacuo, and methyl esterified with both 14% trifluoroborate in methanol (Supelco, Bellafonte, PA), as per Kaya and Sano [33], and 5% HCl in methanol [32] as described below (see Section 2.5).
Fish tissue samples, including liver and muscle from Rainbow Trout, Kokanee and Yellow Perch from lakes in western Washington State were kindly provided by the Washington State Department of Ecology. Sample tissues from Goodea sp. were collected from Lago Patzcuaro (Michoacán, Mexico).
2.2 Lemieux oxidation procedure
Lemieux oxidation reactions were done using a method modified from Williams et al. [27, 28] with varying amounts of 4PB (2–14 µg) and MC-LR. Stock oxidant solution was prepared by dissolving potassium bicarbonate (30.0 mg), potassium permanganate (9.0 mg) and sodium metaperiodate (330.0 mg) in deionized water (0.016 L). Lemieux oxidation reactions were prepared by dissolving 4-PB or MC-LR (at varying analyte concentrations) in stock oxidant solution (2.7 mL) and deionized water (10.0 mL). Excess potassium bicarbonate was added periodically to samples in order to adjust the pH to ~9 (and maintain the solution’s purple colour). The oxidation was carried out for 3 hours at room temperature. The reaction was terminated by adding sodium disulfite and adjusting the pH to ~2 with 10% sulfuric acid (1.0 mL) until the reaction solution became clear [27, 28].
2.3 Preparation of Lemieux oxidation reactions with liver tissue background
To assess the effects of biological matrix on Lemieux oxidation, fish liver slurry was prepared by homogenizing (Fisher Scientific, Power Gen 125) Rainbow Trout liver (~7 g) in deionized water (5 mL). Rainbow trout were specifically collected from Alder Lake (Washington State), and selected as ‘background’ tissue due to the lack of apparent cyanobacterial blooms in the lake. Lemieux oxidation reactions with liver tissue were prepared by dissolving 4-PB or MC-LR in deionized water (9.0 mL) and fish liver slurry (1.0 mL), and subsequently stock oxidant solution (2.7 mL), at the previously prepared analyte concentrations. Samples where then treated according to the previously described Lemieux oxidation protocol. After termination, samples were centrifuged at 3000 rpm for 5 minutes to remove tissue debris (Beckman Coulter, Allegra X-15R Centrifuge).
2.4 Evaluation of direct SPME for recovery of 4PB and MMPB
To evaluate SPME as a means to recover the analyte, MMPB, and its analogue, 4-PB, Lemeiux oxidation reactions were extracted, by direct sampling using several SPME fibres (PDMS-DVB, PA and DVB-CAR-PDMS), for 1 hour following termination of the reactions. Presence of the analytes was evaluated by GC-MS analysis (see below).
2.5 Methyl esterification of 4PB and MMPB followed by SPME recovery
In order to assess the possibility of methyl esterification, as a means to volatilize analyte for subsequent headspace SPME sampling, we adapted a method based on that of Harada et al. [32]. For initial studies, 4PB (102.6 mg) was dissolved directly in 5% HCl methanol solution (2 mL) and heated for an hour at 70°C. In subsequent studies, aliquots (100 µL) of the oxidation products of 4PB and MC-LR, spiked into both water and liver tissue, were dried down (Speed Vac SVC100, SAVANT) and dissolved in 5% HCl methanol solution (1 mL), followed by heating for 1 h at 70°C. In each case, silver carbonate was subsequently added to neutralize the solutions as per Harada et al. [32]. The reaction mixtures were sampled by SPME (headspace, for 1 hour exposure), specifically using the PDMS-DVB and DVB-CAR-PDMS fibres, and analyzed by GC-MS (see below), for the presence of me4PB or meMMPB.
2.6 Lemieux oxidation and measurement of MCs in environmental samples
Fish tissue samples (~2 g) were homogenized and dissolved in deionized water (5 mL), and oxidized as described above (for standards). After termination of reactions, samples were centrifuged at 3000 rpm for 5 minutes (Beckman Coulter, Allegra X-15R Centrifuge) to remove tissue. Aliquots (100 µL) of oxidation products in the supernatant were dried down (SpeedVacSVC100,SAVANT)and dissolved in5%HClmethanol solution (1 mL), followed by heating for 1 h at 70°C, and neutralization with silver carbonate, for methyl esterification. Total MC content was calculated specifically based on the GC-MS peak area of meMMPB recovered from headspace by PDMS-DVB SPME (discussed below). Calculated amounts of MC were converted to MC content (µg) per gram of fish tissue (wet weight).
2.7 GC-MS analysis
Samples were analyzed by GC-MS under the following conditions using an Agilent Technologies-6890N Network GC System with DB5 column (30m × 0.25mm ID, phase thickness 0.25 mm) with He carrier gas (splitless): initial temperature (80°C) for 1 minute, followed by ramp (8°C min−1 to 280°C). Analytes were detected using Agilent Technologies-5973 Network Mass Selective Detector. Analytes, 4PB and MMPB, were detected using SIM mode. The ions 176, 146, 104, 91 m/z were selected for 4PB; 178, 146, 104, 91 m/z for me4PB; and 190, 135, 131, 91 and 75 m/z for the meMMPB [31, 32]. ChemStation and RTE integration software was used for the quantitative analysis of these analytes.
2.8 ELISA of MCs in environmental samples
To compare analyses of MCs by the SPME-GC-MS method to conventional detection techniques, we evaluated solvent extracts of fish tissues, previously shown [35, 36] to contain MCs, by ELISA. Fish tissue samples were extracted with a two-fold solvent extraction method adapted from the Wilson et al. [37]. Homogenized tissue was first extracted in 75% methanol (20 mL), followed by 75% methanol with 0.5% acetic acid (20 mL). Pooled extracts were dried down and retaken into phosphate buffered saline (PBS). An ELISA kit from Abraxis, Inc. (Warminster, PA), specific for the Adda of MCs and nodularin, was used to analyze the samples as per the manufacturer’s instructions.
2.9 Calculation of LOD/LOQ
Limit of detection (LOD) and limit of quantitation (LOQ) were determined using standard methods [38]. The signal (i.e. peak area) for replicate samples (n=7) of both blank (0 µg) and 2.2 µg, of MC-LR were measured. The LOD/LOQ was calculated using the slope of the calibration curve (m=1389.2 signal/µg MC-LR) and the standard deviation (3 s and 10 s, respectively, for LOD and LOQ) of the blank replicates. Calibration curve of meMMPB in liver tissue was corrected for the baseline signal, observed in the blank, when MC content in fish tissues was determined.
3. Results and discussion
3.1 Detection of 4PB and MMPB in Lemieux oxidation reactions by SPME-GC-MS
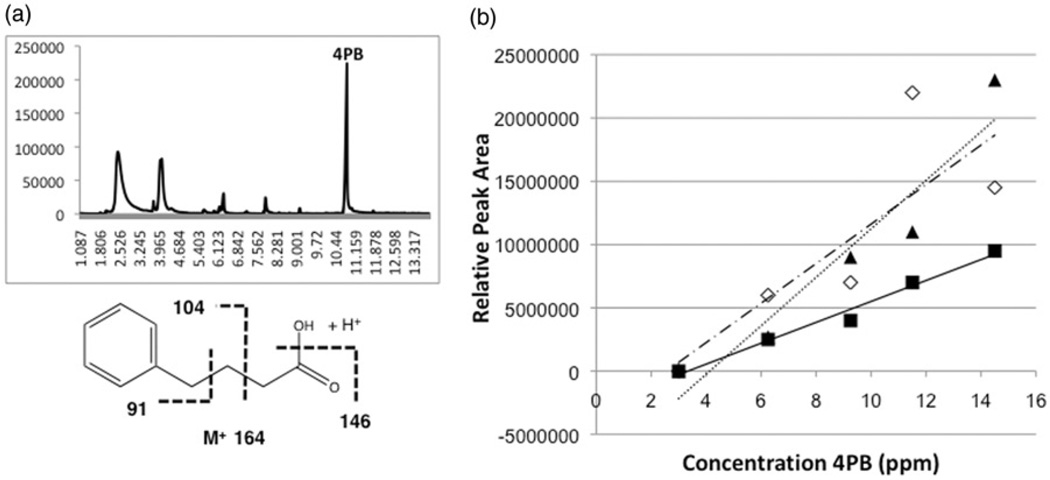
In the current studies, as in prior studies [29, 32], 4PB was used as an analogue due to chromatographic and structural similarities to the MMPB analyte (Figure 1). Initial experiments determined that 4PB can be detected by direct sampling of pure 4PB in water (data not shown) and Lemieux oxidation reactions (Figure 2a) with retention time of ~10.9 minutes using the proposed SPME-GC-MS method. Observed mass spectra for 4PB include molecular ion M+ at 164, 146 (M+ −18 [H2O]), 104 and 91 m/z (Figure 2a). This peak was not present in negative controls (i.e. no 4PB). This experiment showed that the 4PB can be recovered using the SPME fibres, and detected with the previously discussed GC-MS method; however, when direct sampling of the 4PB was performed in Lemieux oxidation solution, SPME fibres quickly deteriorated. Moreover, direct sampling by SPME surprisingly did not seem to recover MMPB (as evaluated by GC-MS), although concurrent analyses using LLE for recovery do, indeed, confirm (data not shown) the presence of the analyte in oxidation reaction mixtures. Neither 4PB nor MMPB – even following attempts at, for example, warming of the reaction mixture to volatilize the analytes post-oxidation – were detected by headspace sampling with SPME.
Figure 2.
Total ion chromatogram (top) showing peak (10.9 min), and mass spectrum and expected fragmentation pattern (bottom), of 4PB in Lemieux oxidation reactions sampled by SPME (a), as well as linearity of response for peak area versus 4-PB concentration for SPME fibres (▲/……: PDMS-DVB, ■/ —: DVB-CAR-PDMS and ◊/ – –: PA) evaluated in the study.
SPME fibres, PDMS-DVB, DVB-CAR-PDMS and PA, were chosen based on generic and semi-volatile detection characteristics. Based on the data collected it was observed that the relationship between amount of 4PB in solution and peak area was linear (Figure 2b) for both PDMS-DVB (R2=0.9224) and DVB-CAR-PDMS (R2=0.9779) fibres over the range of 4PB evaluated (0.15–1.0 µgmL−1). The PA fibres were found to be the least linear (R2=0.6646) for this method when sampling the Lemieux oxidation solution over the range of 4PB analyte tested. Therefore, for the remainder of this study, only the PDMSDVB and DVB-CAR-PDMS fibres were used.
3.2 Detection of methyl ester 4PB (me4PB) by SPME-GC-MS
In preliminary analysis of the Lemieux oxidation product of MCs, it was noted (as mentioned above) that the SPME fibres could not detect the MMPB analyte by either headspace or direct sampling of the Lemieux oxidation reaction, although other methods, including LLE and SPE were able (data not shown) to recover the analyte as per previous studies [27–30]. Moreover, although analysis of 4PB in Lemieux oxidation reaction with SPME fibres did, indeed, confirm this analyte could be recovered and detected by direct sampling, the harsh conditions of the reaction solutions, which led to rapid deterioration of SPME fibres, was not acceptable for repetitive use of these fibres for routine analyses. Accordingly we investigated methyl esterification as a means to volatize the analyte for headspace sampling. This would additionally allow for a reduction in the damage caused to the SPME fibres in direct sampling.
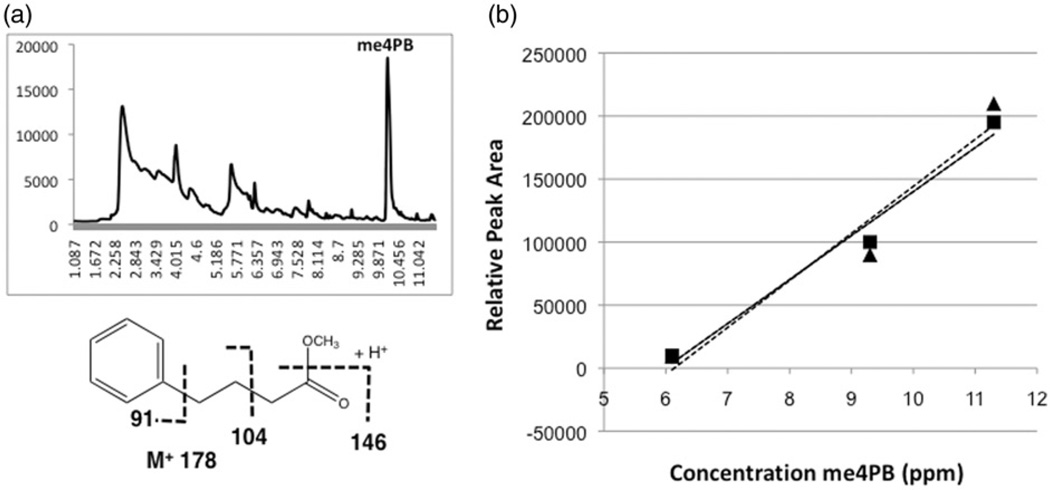
Initial analysis of pure 4PB with this method showed that me4PB can be detected at a retention time of ~10.1 minutes with the previously discussed GC-MS method (Figure 3a). Full scan and SIM of the me4PB by SPME-GC-MS identified me4PB fragments within a mass spectrum that confirms that the peak being observed is the analyte (Figure 3a). This peak was not observed in negative controls (i.e. no 4PB). Observed mass spectra for the me4PB specifically included the molecular ion M+ at 178, 146 (M+_32 [CH3OH]), 104 and 91 m/z (Figure 3a).
Figure 3.
Total ion chromatogram (top) showing peak (10.1 min), and mass spectrum and expected fragmentation pattern (bottom), of me4PB in Lemieux oxidation reactions sampled by SPME (a), as well as linearity of response for peak area versus 4-PB concentration for SPME fibres (▲/ ……: PDMS-DVB, ■/ —: DVB-CAR-PDMS) evaluated in the study.
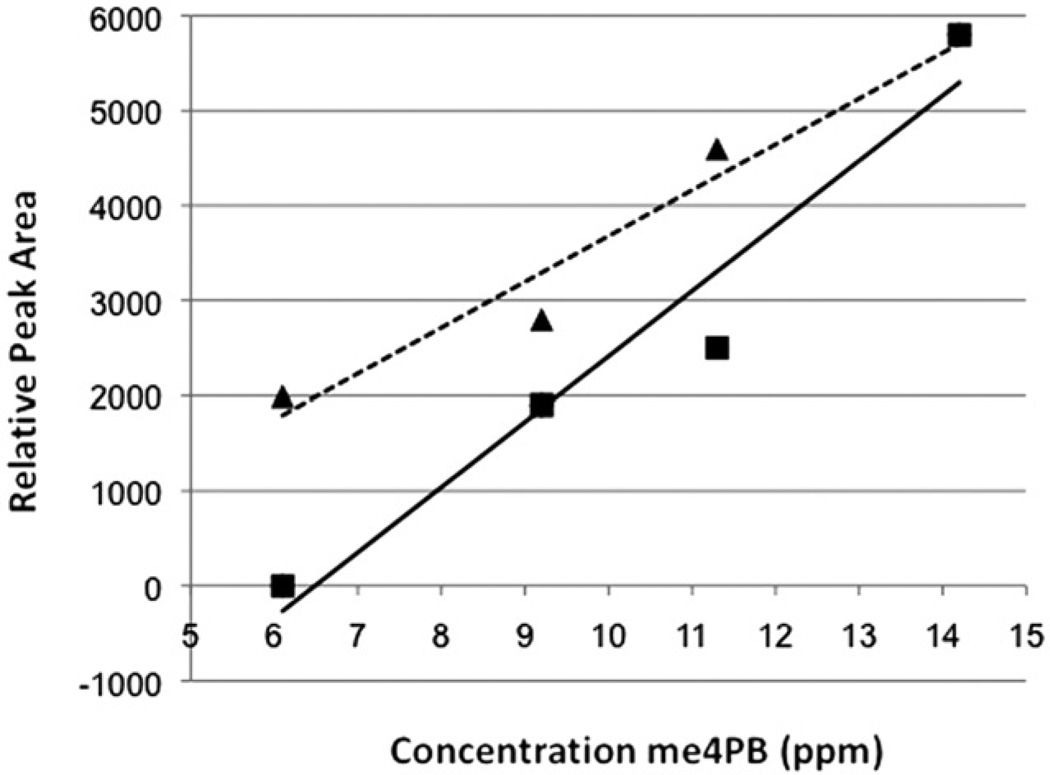
It was subsequently determined that the SPME-GC-MS method detected the me4PB in the Lemieux oxidation reaction with a linear relationship over the concentration range of 4PB tested (6–11 µg). The experiment was, therefore, repeated with liver tissue as a background, to determine the effects of biological matrixes on the proposed method, and SPME-GC-MS detected the methyl ester in the same manner with and without liver tissue (Figure 3b and 4). As with the previous 4PB experiment, both fibres were shown to give largely similar results, including a slightly higher linearity for the DVB-CAR-PDMS fibre. Linearity without liver (PDMS-DVB: R2=0.9437, DVB-CAR-PDMS: R2=0.9839) and with liver tissue (PDMS-DVB: R2=0.8899, DVB-CAR-PDMS: R2=0.9497) was observed, although it was noted that there was a slight decrease in linearity with the addition of the liver tissue for both fibres (Figure 4).
Figure 4.
Linearity of response for me4PB peak area versus 4PB concentration in liver tissue for SPME fibres (▲/……: PDMS-DVB, ■/ —: DVB-CAR-PDMS) evaluated in the study.
3.3 Detection of methyl ester MMPB (meMMPB) by SPME-GC-MS
The methyl esterification method developed for 4PB was developed in order to utilize the technique of SPME recovery of MMPB. As initial tests with SPME fibres showed, the MMPB molecule did not absorb onto the fibres and therefore could not be analyzed quantitatively. After the development of the me4PB protocol, a similar method for analysis (i.e. methyl esterification and SPME recovery) of MMPB produced from Lemieux oxidation of a sample (50 µg) of pure MC-LR was evaluated.
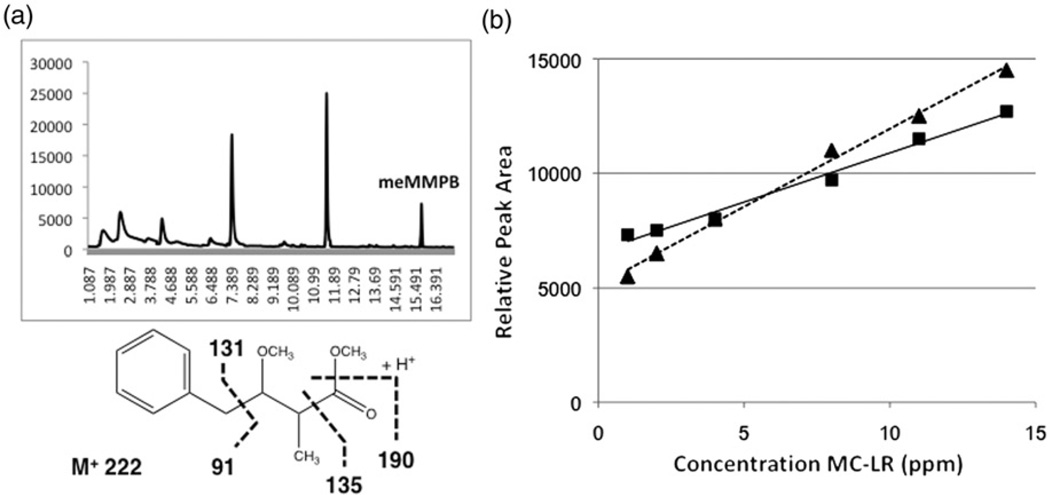
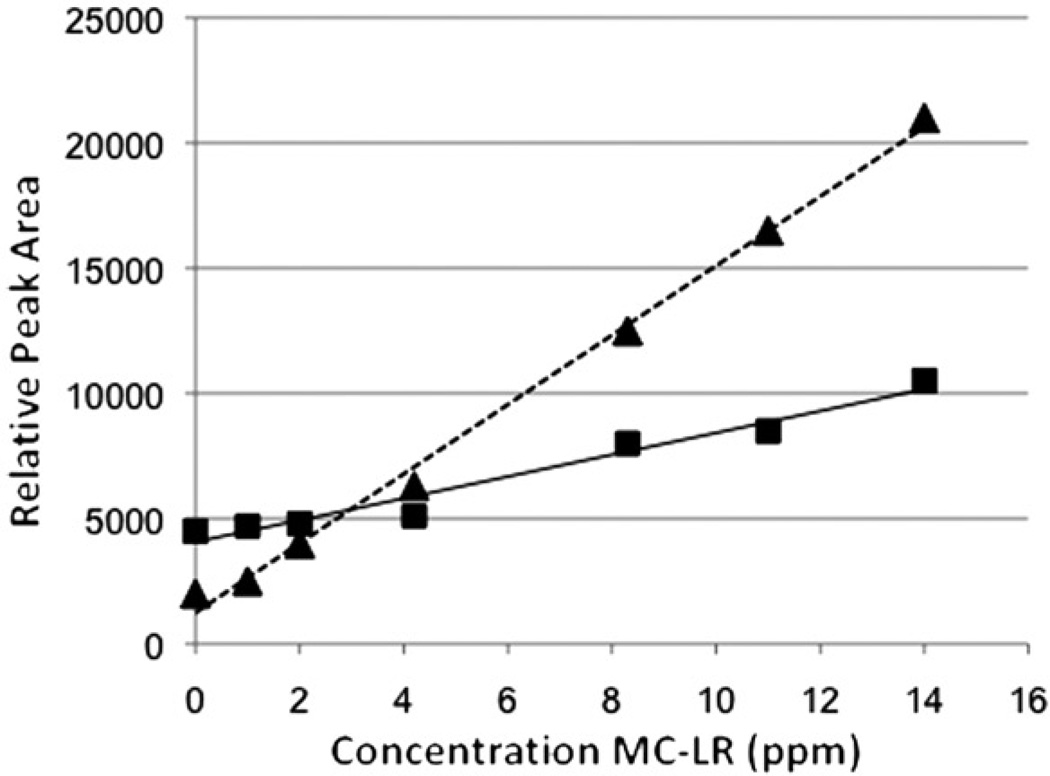
As in prior studies [31–33], fragmentation of the meMMPB with EI-MS spectrum does not present a molecular ion peak (M+ 222), but the fragments at m/z 190 (M+_32 [CH3OH]), 135, 131, 91 and 75 (17) were observed (Figure 5a). In SIM mode, however, GC-MS detected three significant peaks at 7.3, 11.5 and 15.7 minutes with these expected fragments (Figure 5a). These peaks were evaluated for correlation between peak area with increasing MC-LR concentration, and it was found that the 15.7 peak was the only peak that had a high linearity (Table 1 and Figure 5). Indeed, peak area of the presumptive meMMPB was found to be highly linear (R2=0.9909–0.9986; Table 1) relative to concentration of MC-LR, particularly when compared to the peaks at 11.5 and 7.3 minutes (R2=0.00821–0.62638; Table 1). In fact, previous studies from Kaya and Sano [33] using LLE recovery, likewise, determined that the meMMPB analyte had a retention time ~15.8 minutes using similar post-oxidation procedures and GC-MS methodology, further supporting the identity of this peak as the expected analyte. Furthermore, it was found that the fibres detected the same putative meMMPB in the presence and absence of liver tissue with consistently linear trends (Figure 5b and 6), and that a more or less equally high linearity was found for both fibrefibres (Table 1 and Figures 5b and 6). The identity of the analyte was subsequently confirmed by recovery of meMMPB prepared from MMPB standard (data not shown). Although peak area versus concentration of MC was highly linear for both fibres, linearity for PDMS-DVB was slightly higher, and subsequently used to develop a working standard curve for analysis of environmental samples (see below).
Figure 5.
Selected ion monitoring (SIM) chromatogram showing putative peak (15.7 min), and mass spectrum and expected fragmentation (inset), of meMMPB in Lemieux oxidation reactions sampled by SPME (a), as well as linearity of response for peak area versus microcystin (MC-LR) concentration for SPME fibres (▲/……: PDMS-DVB, ■/ —: DVB-CAR-PDMS) evaluated in the study.
Table 1.
Correlation coefficients (R2) for peak area versus MC concentration for peaks observed by selected ion monitoring (SIM) for meMMPB following Lemieux oxidation of MC-LR, and subsequent methyl esterification.
| Peak Retention Time (minutes) |
||||||||
|---|---|---|---|---|---|---|---|---|
| 15.7 |
11.5 |
9.6 |
7.3 |
|||||
| Fiber | LmOx | Liver | LmOx | Liver | LmOx | Liver | LmOx | Liver |
| PDMS-DVB | 0.9986 | 0.9985 | 0.0232 | 0.6263 | 0.2357 | 0.5426 | 0.0082 | 0.5426 |
| DVB-CAR-PDMS | 0.9909 | 0.9913 | 0.0272 | 0.5884 | 0.0050 | 0.0023 | 0.0462 | 0.0023 |
“LmOx” is Lemieux oxidation in water; “Liver” is Lemieux oxidation with added liver tissue.
Figure 6.
Linearity of response for putative meMMPB peak area versus microcystin (MC-LR) concentration in liver tissue for SPME fibres (▲/……: PDMS-DVB, ■/ —: DVB-CAR-PDMS) evaluated in the study.
Percent recovery of the oxidation product was not determined here. However, very recent studies [39] investigated the recovery of MMPB following oxidation of spiked samples of fetal bovine serum, and found recovery rates were approximately 22–38%, suggesting that estimates based on the method may, in fact, considerably underestimate the total MCs. Future studies on the recovery of the oxidation products by the current method (and in fish tissues) will be required.
Based on the meMMPB peak, LOD and LOQ for the method (with liver tissue as the matrix, and using the PDMS-DVB fibre) were calculated to be 0.04 µg g−1 and 0.14 µg g−1 of tissue, respectively. This is a considerable (10- to 100-fold) improvement in the limit of detection of the MMPB method as previously developed [27–30]. Specifically, use of LLE [28] and SPE [29] for recovery of the analyte limited detection to 15 µg g−1 and 5 µg g−1respectively, in prior studies.
3.4 SPME-GC/MS for detection of MCs in environmental samples
In order to further assess the method for quantitative analysis of MCs, we evaluated several environmental (i.e. fish tissue) samples previously shown by ELISA to contain bioaccumulated levels of (solvent extractable) MCs. Specifically, MC content was calculated based on peak area of meMMPB measured for samples using a standard curve of oxidized, MC-spiked liver tissue (as sample matrix) with recovery by a PDMSDVB fibrefibre. Samples analyzed included tissue (muscle and/or liver) from fish collected from three separate locations, namely Kokanee from American Lake (Washington State, USA), Yellow Perch from Steilacoom Lake (Washington State, USA) and Goodea sp. from Lago Patzcuaro (Michoacán, Mexico). Total MC content for environmental tissues, analyzed by both methods, is summarized in Table 2.
Table 2.
MC content (ng/g) measured for samples of fish tissue evaluated by solvent extraction/ELISA and SPME-GC-MS method (described here).
| Species | Tissue(s) | Sample Weight (g) | SPME-GC-MS Method MC (µg g−1) |
ELISA Method MC (µg g−1) |
|---|---|---|---|---|
| Goodea sp. | Viscera | 1.3046 | 4.69 (±0.12) | 0.867 (±0.315) |
| Muscle | 2.1529 | 1.97 (±0.06) | 0.157 (±0.062) | |
| Kokanee | Liver | 2.5873 | 1.09 (±0.05) | 0.084 (±0.038) |
| Muscle | 2.2999 | 1.07 (±0.06) | 0.041 (±0.020) | |
| Yellow Perch | Liver | 1.9587 | 1.43 (±0.07) | 0.268 (±0.372) |
| Muscle | 2.2327 | 0.84 (±0.06) | 0.018 (±0.017) |
In all three of the fish samples tested, the MC content determined by the meMMPB method was considerably greater than measured by the ELISA method (Table 2). This is consistent with previous findings [26–28] that suggest that methods based on solvent extraction of free MC significantly underestimate total content. Williams et al. [27, 28], for example, compared the use of PPase inhibition assay and the MMPB method (with LLE recovery) for measure of MC in wild-caught crab larvae and saltwater mussels, and found the latter method detects approximately 104 and 103 times more toxin, respectively, than the former. It was, thereby, specifically suggested that the vast majority of MC in these tissues was present in the covalently bound form [27, 28]. In addition, to inaccessibility of bound MC, it has been pointed-out that, due to chemical variability, including lipophilicity, and modified or lack of Adda, not all variants of MC react with antibodies used in the ELISA. As such, many of the more than 70 variants cannot be detected by ELISA, and underestimation observed here may be additionally related to this inherent limitation. On the other hand, a recent study [39], as discussed above, suggests that percent recovery of the oxidation products may be rather low (e.g. 22–38% in foetal bovine serum), and as such it likely that the MC concentrations measured here by the SPME-GC/ MS may, in contrast, further underestimate actual values; recovery rates for the current method, however, remain to be determined.
Although toxicity of MC covalently bound to PPases remains to be clarified, at least one study [40] to-date has suggested that tissue-bound MC may be bioavailable in a potentially toxic form. Specifically, Smith et al. [40] generated MC-conjugated peptide fragments, representative of those that would be expected following enzymatic digestion of PPase with pepsin, trypsin and chymotrypsin, and found that all MC-peptides were at least partially (~58%) active in the inhibition of PP1 (as a relevant Ser/Thr protein phosphatase).
Previous studies, based on analysis of free, solvent extractable MCs, have generally shown that liver tissue has a higher content of accumulated toxin than muscle tissues, presumably due to accumulation of the toxin, via active transport by organic anion transporter proteins, in hepatocytes [41]. Surprisingly, the meMMPB method has shown that this is not consistently the case with the fish tissues examined in this study (Table 2). In both the Kokanee and perch tissues, in particular, the MC content of liver tissue is comparable to that of muscle tissue. We propose that this may reflect the measurement of covalently bound MCs in muscle tissue that may consequently accumulate over time in these tissues, but not be otherwise detectable by conventional methods of solvent extraction (for detection). In other words, although the largest pool of unbound MC may occur (due to active transport) in the liver, recurring exposure of both muscle and liver to MC may lead to an equivalent accumulation of the covalently bound form in both tissues over time. Alternatively, it may be attributable to the use of an improper matrix for the muscle tissues (i.e. liver tissue was used as a matrix for the standard curve). Both possibilities, however, remain to be clarified by future research.
4. Conclusions
The method developed in this study utilizes SPME as a unique and novel application to the post-oxidation protocols. Specifically, this method employs methyl esterification and subsequent headspace SPME sampling as a means to recover the analyte, meMMPB, as a seemingly sensitive and accurate surrogate for total MC content. This method was developed using fish tissue spiked with MC-LR, and/or the analogue, 4PB, and subsequently evaluated with appropriate environmental samples (i.e. fish tissues). In agreement with previous studies [27–28, 30], MC concentration in fish tissues measured by the SPME-GC-MS were considerably higher than those measured by solvent extraction and subsequent ELISA analysis.
These findings indicate that the method developed may represent an effective and efficient means of measuring total MC content of biological matrices (e.g. animal tissues), and therefore a powerful tool for understanding the bioaccumulation of this widespread cyanobacterial toxin, including its potential implications for human health, and for assessment of human exposure to the toxin. To be sure, at present, SPME is not as widely available to, or used in, as many labs when compared to other techniques (e.g. SPE, LLE), and further studies, including evaluation of recovery efficiency and possible adaptation to other analytical techniques (e.g. HPLC-MS), are needed before the proposed method might replace currently routine analytical methods (e.g. solvent extraction of unbound MC coupled to ELISA or LC-MS, LLE/SPE-coupled LC- and GC-MS analysis of MMPB). However, these findings minimally suggest that SPME represents an effective procedural modification for recovery meMMPB, and that the SPME-GC-MS or possibly related (e.g. SPME-LC-MS) techniques – owing to both a calculated improvement with regards to sensitivity (i.e. decreased LOD/LOQ) and inherent advantages (e.g. speed and ease of SPME recovery compared to LLE and SPE) – may, in the future, provide a methodological option to MC analysis. Moreover, as a growing technology, advancements in SPME [34] – particularly, for example, available SPME autosamplers – may make SPME-based techniques, such as that developed here, increasingly efficient and high-throughput analytical tools for investigation of this cyanobacterial toxin.
Acknowledgements
This work was partially supported by a grant from the Great Lakes Fishery Commission. The authors would particularly like to thank Dr Kenneth Furton and his laboratory, and particularly Dr Maiko Kusano, Howard Holness and Katylynn Beltz, at Florida International University (Department of Chemistry and Biochemistry) for use of their GC-MS and general guidance in developing the SPME method. We would also like to the Washington State Department of Ecology, including Art Johnson, and WA State Department of Fish and Wildlife, including Adam Couto and Richard Eltrich, for providing fish tissue from Western Washington Lakes, and Dr Fernando Bernal-Brooks of the Universidad Michoacana de San Nicolás de Hidalgo for assistance in collection of fish from Lago Patzcuaro.
References
- 1.Funari E, Testai E. Crit. Rev. Toxicol. 2008;38:97. doi: 10.1080/10408440701749454. [DOI] [PubMed] [Google Scholar]
- 2.Codd GA, Morrison LF, Metcalf JS. Toxicol. Appl. Pharmacol. 2005;203:264. doi: 10.1016/j.taap.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Rao PV, Gupta N, Bhaskar AS, Jayaraj R. J. Environ. Biol. 2002;23:215. [PubMed] [Google Scholar]
- 4.Berry JP, Lind O. Toxicon. 2010;55:930. doi: 10.1016/j.toxicon.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 5.Ibelings BW, Chorus I. Environ. Pollut. 2007;150:177. doi: 10.1016/j.envpol.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhang D, Xie P, Liu Y, Chen J, Liang G. Environ. Toxicol. Chem. 2007;26:171. doi: 10.1897/06-222r.1. [DOI] [PubMed] [Google Scholar]
- 7.White SH, Dulvenvoorden LJ, Fabbro LD, Eaglesham GK. Environ. Pollut. 2007;147:158. doi: 10.1016/j.envpol.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Lance E, Brient L, Bormans M, Gerard C. Aquat. Toxicol. 2006;79:140. doi: 10.1016/j.aquatox.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Cazenave J, Wunderlin DA, de Los Angeles Bistoni M, Amé MV, Krause E, Pflugmacher S, Wiegand C. Aquat. Toxicol. 2005;75:178. doi: 10.1016/j.aquatox.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Xie L, Xie P, Guo L, Li L, Miyabara Y, Park HD. Environ. Toxicol. Chem. 2005;20:293. doi: 10.1002/tox.20120. [DOI] [PubMed] [Google Scholar]
- 11.Kankaanpää HT, Turunen AK, Karlsson K, Bylund G, Meriluoto J, Sipiä V. Chemosphere. 2005;59:1091. doi: 10.1016/j.chemosphere.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Pereira P, Dias E, Franca S, Pereira F, Carolino M, Vasconcelos V. Aquat. Toxicol. 2004;68:339. doi: 10.1016/j.aquatox.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Cox PA, Banack SA, Murch SJ. Proc. Natl. Acad. Sci. 2003;100:13380. doi: 10.1073/pnas.2235808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magalhä es VF, Marinho MM, Domingos P, Oliveira AC, Costa SM, Azevedo LO, Azevedo SM. Toxicon. 2003;42:289. doi: 10.1016/s0041-0101(03)00144-2. [DOI] [PubMed] [Google Scholar]
- 15.Sipiä VO, Kankaanpä ä HT, Pflugmacher S, Flinkman J, Furey A, James KJ. Ecotoxicol. Environ. Saf. 2002;53:305. doi: 10.1006/eesa.2002.2222. [DOI] [PubMed] [Google Scholar]
- 16.De Figueiredo DR, Azeiteiro UM, Esteves SM, Goncalves FJ, Pereira MJ. Environ. Saf. 2004;59:151. doi: 10.1016/j.ecoenv.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Svircev Z, Krstic S, Miladinov-Mikov M, Baltic V, Vidovic M. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2009;27:36. doi: 10.1080/10590500802668016. [DOI] [PubMed] [Google Scholar]
- 18.Yu S, Zhao N, Zi X. Zhonghua Zhong Li Za Zhi. 2001;23:96. [PubMed] [Google Scholar]
- 19.Ueno Y, Nagata S, Tsutsumi T, Hasegawa A, Watanabe MF, Park HD, Chen GC, Chen G, Yu SZ. Carcinogenesis. 1996;17:1317. doi: 10.1093/carcin/17.6.1317. [DOI] [PubMed] [Google Scholar]
- 20.Maynes JT, Luu HA, Cherney MM, Andersen RJ, Williams D, Holmes CF, James MN. J. Mol. Biol. 2006;356:111. doi: 10.1016/j.jmb.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Bagu JR, Sönnichesen FD, Williams DE, Andersen RJ, Sykes BD, Holmes CFB. Nature Struct. Biol. 1995;2:114. doi: 10.1038/nsb0295-114. [DOI] [PubMed] [Google Scholar]
- 22.Craig M, Luu HA, McCready T, Williams DE, Andersen RJ, Holmes CFB. Biochem. Cell Biol. 1996;74:569. doi: 10.1139/o96-061. [DOI] [PubMed] [Google Scholar]
- 23.MacKintosh RW, Dalby KN, Campbell DG, Cohen PTW, Cohen P, MacKintosh C. Fed. Eur. Biochem. Soc. Lett. 1995;371:236. doi: 10.1016/0014-5793(95)00888-g. [DOI] [PubMed] [Google Scholar]
- 24.Sano T, Takagi H, Kaya K. Phytochem. 2004;65:2159. doi: 10.1016/j.phytochem.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 25.McElhiney J, Lawton LA. Toxicol. Appl. Pharmacol. 2005;203:219. doi: 10.1016/j.taap.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Williams DE, Craig M, Dawe SC, Kent ML, Andersen RJ, Holmes CF. Toxicon. 1997;35:985. doi: 10.1016/s0041-0101(96)00196-1. [DOI] [PubMed] [Google Scholar]
- 27.Williams DE, Dawe SC, Kent ML, Andersen RJ, Craig M, Holmes CF. Toxicon. 1997;35:1617. doi: 10.1016/s0041-0101(97)00039-1. [DOI] [PubMed] [Google Scholar]
- 28.Williams DE, Craig M, Dawe SC, Kent ML, Holmes CFB, Andersen RJ. Chem. Res. Toxicol. 1997;10:463. doi: 10.1021/tx9601519. [DOI] [PubMed] [Google Scholar]
- 29.Ott JL, Carmichael WW. Toxicon. 2006;47:734. doi: 10.1016/j.toxicon.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Yuan M, Carmichael WW, Hilborn ED. Toxicon. 2006;48:627. doi: 10.1016/j.toxicon.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 31.Sano T, Nohara K, Shiraishi F, Kaya K. Intern. J. Environ. Anal. Chem. 1992;49:163. [Google Scholar]
- 32.Harada K-I, Murata H, Qiang Z, Suzuki M, Kondo F. Toxicon. 1996;34:701. doi: 10.1016/0041-0101(95)00163-8. [DOI] [PubMed] [Google Scholar]
- 33.Kaya K, Sano T. Anal. Chim. Acta. 1999;386:107. [Google Scholar]
- 34.Risticevic S, Niri VH, Vuckovic D, Pawliszyn J. Anal. Bioanal. Chem. 2009;393:781. doi: 10.1007/s00216-008-2375-3. [DOI] [PubMed] [Google Scholar]
- 35.Berry JP, Lee E, Walton K, Wilson AE, Bernal-Brooks F. Environ. Chem. Toxicol. doi: 10.1002/etc.548. in press. [DOI] [PubMed] [Google Scholar]
- 36.Johnson A. Washington State Department of Ecology Pub. No. 10-03-011. 2010 [Google Scholar]
- 37.Wilson AE, Gossiaux DC, Höök TO, Berry JP, Landrum PF, Dyble J, Guildford SJ. Can. J. Fish Aquat. Sci. 2008;65:1487. [Google Scholar]
- 38.Harris DC. Quantitative Chemical Analysis. 7th ed. W. H. Freeman; New York: 2007. [Google Scholar]
- 39.Neffling M-R, Lance E, Meriluoto J. Environ. Pollut. 2010;158:948. doi: 10.1016/j.envpol.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 40.Smith JL, Schulz KL, Zimba PV, Boyer GL. Ecotoxicol. Environ. Saf. 2010;73:757. doi: 10.1016/j.ecoenv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Fischer WJ, Altheimer S, Cattori V, Meier PJ, Dietrich DR, Hagenbuch B. Toxicol. Appl. Pharmacol. 2005;203:257. doi: 10.1016/j.taap.2004.08.012. [DOI] [PubMed] [Google Scholar]