Abstract
In eukaryotes, Rad51 protein is responsible for the recombinational repair of double-strand DNA breaks. Rad51 monomers cooperatively assemble on exonuclease-processed broken ends forming helical nucleo-protein filaments that can pair with homologous regions of sister chromatids. Homologous pairing allows the broken ends to be reunited in a complex but error-free repair process. Rad51 protein has ATPase activity but its role is poorly understood, as homologous pairing is independent of adenosine triphosphate (ATP) hydrolysis. Here we use magnetic tweezers and electron microscopy to investigate how changes of DNA twist affect the structure of Rad51-DNA complexes and how ATP hydrolysis participates in this process. We show that Rad51 protein can bind to double-stranded DNA in two different modes depending on the enforced DNA twist. The stretching mode is observed when DNA is unwound towards a helical repeat of 18.6 bp/turn, whereas a non-stretching mode is observed when DNA molecules are not permitted to change their native helical repeat. We also show that the two forms of complexes are interconvertible and that by enforcing changes of DNA twist one can induce transitions between the two forms. Our observations permit a better understanding of the role of ATP hydrolysis in Rad51-mediated homologous pairing and strand exchange.
INTRODUCTION
Although many molecular details of Rad51-mediated homologous pairing are unknown, it is known that the active role in this process is played by helical filaments formed by Rad51 protein polymerized on single-stranded DNA (ssDNA) or partially single–stranded DNA molecules (1,2). These helical filaments have then the potential to interact with protein-free duplexes in a process that allows homologous DNA sequences to pair and then start strand exchange (3). It is also known that helical Rad51-DNA filaments can assemble in different forms depending on the bound cofactors such as adenosine triphosphate (ATP) or adenosine diphosphate (ADP) (4). Rad51 belongs to an intensively studied group of RecA-like proteins including RecA, RadA, UvsX and Dmc1 (5). Many earlier studies revealed that these proteins can form two principal types of helical complexes: stretched and non-stretched (6). The stretched filaments form in the presence of ATP or its non-hydrolyzable analogs on ssDNA or double-stranded DNA (dsDNA). The DNA in these complexes adopts an unusual structure with ∼19 bases or base pairs per ∼95 Å pitch (7,8). The stretched filaments were shown to constitute the active state in the process of pairing and strand exchange (9–12). Non-stretched filaments have a less well-defined structure and it is likely that there are several distinct forms of them (6). Structural studies of one of the forms of non-stretched filaments indicated that it has different protein:DNA stoichiometry than stretched filaments (13). This finding decreased the interest in non-stretched filaments because it seemed to exclude their participation in the catalytic cycle of DNA pairing and strand exchange. However, relatively recent studies using total internal reflection fluorescence microscopy (TIRFM) revealed that human Rad51 filaments formed on linear dsDNA molecules can reversibly transit several times between stretched and non-stretched forms depending on whether ATP was added or removed from the reaction buffer (14). Because there was no free Rad51 in the flow cells used for the experiments, these reversible transitions strongly indicated that stretched and non-stretched helical filaments constitute simply different allosteric forms of Rad51-DNA complexes where the transition between the two forms is regulated by ATP binding and hydrolysis (14). This finding reinitiated interest in non-stretched forms of the Rad51-DNA filaments and in the mechanism by which helical Rad51-DNA complexes can switch between stretched and non-stretched forms.
MATERIALS AND METHODS
Production and purification of Rad51 protein
Human Rad51 gene was inserted at the NdeI site of the pET15b expression vector (Novagen) and expressed in the Escherichia coli JM109 (DE3) strain that also carried an expression vector for the minor transfer RNAs [Codon(+)RIL®, Novagen]. The protein was purified on Nickel-nitrilotriacetic acid agarose (Invitrogen, France). The hexahistidine tag was then removed from human Rad51 protein sequence by incubation with 1.5 units of thrombin protease (Amersham Biosciences) per mg of Rad51 during 18 h. The tag-free protein was further purified by chromatography on a MonoQ column (Amersham Biosciences). The Rad51-containing fractions are dialysed against storage buffer (20 mM Tris–HCl, pH 8, 0.25 mM ethylenediaminetetraacetic acid (EDTA), 20% glycerol, 5 mM dithiothreitol (DTT) and 200 mM KCl) and kept at −80°C. Protein concentrations were determined using the Bio-Rad protein assay kit with bovine serum albumin (Pierce) as a standard.
Magnetic tweezers
DNA construction.
DNA molecules held in the magnetic tweezers were composed of 14 438 bp fragment, ligated at one end to a multidigoxigenin-labelled DNA fragment of 672 bp and at the other end to a multibiotin-labelled fragment of 834 bp. All the DNA fragments were obtained by polymerase chain reaction from phage λ DNA as previously described (15). The biotin-labeled ends of DNA molecules were then bound to streptavidin-coated 2.8 µm magnetic beads (Dynabeads® M-280 Streptavidin) in a binding buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl) by interaction of the biotin label with the streptavidin. The DNA-bound bead suspension was then introduced at a controlled flow rate into a polydimethylsiloxane (PDMS) microchannel. After 30 min of incubation, most of the unbound beads were washed out of the channel with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5). Flow chamber, a PDMS microchannel 2 cm × 2 mm × 110 μm (total volume: 4 μl), was prepared according to Fulconis et al. (15) and placed on a glass coverslip of 24 × 40 mm (Erie Scientific Company, France) treated with Sigmacote® (Sigma-Aldrich) followed by anti-digoxigenin (Roche, France) for subsequent binding of digoxigenin-labelled DNA molecules. Before first use of the channel, Pluronic F-127® (Sigma-Aldrich) was injected into it and incubated overnight at 4°C to minimize adsorption of Rad51 onto the glass surface and onto the PDMS walls.
Experimental protocol.
The length of the DNA molecule was recorded by 3D tracking of the magnetic bead, as described (16). Rad51 at 200 nM concentration was bound to dsDNA in a buffer containing 15 mM Tris–HCl, pH 7.5, 25 mM NaCl, 1 mM ATP, 1 mM DTT, 0.05% Tween 20. The buffer was supplemented either with 2 mM CaCl2 or with 2 mM MgCl2. Buffers with and without Rad51 were injected in the flow chamber with a constant flow between 200 and 500 μl/h for more than 3 min (the total volume of fluid exceeds by 10 times the chamber capacity). During buffer injection and buffer exchange, the molecule length cannot be recorded accurately because of the viscous drag. In all experiments, there was a large excess of Rad51 during the initial incubation to assure formation of saturated complexes. The incubations and measurements were performed at a temperature of 25°C ± 2°C. We systematically verified that only one DNA molecule was bound to the observed bead.
Electron microscopy
Human Rad51 protein was bound to commercial pUC18 preparations (Fermentas) that contained ∼5% of nicked DNA circles. Reactions were performed in the same buffer as used for magnetic tweezers experiments presented in Figure 1, but after 30 min of incubation at 20°C, the formed complexes were stabilized with 1 mM ATPγS for 1 min. Afterwards the samples were diluted 10 times in 4 mM magnesium acetate and were adsorbed to glow-discharged carbon grids. Specimens were negatively stained with 2% uranyl acetate solution as previously described (17).
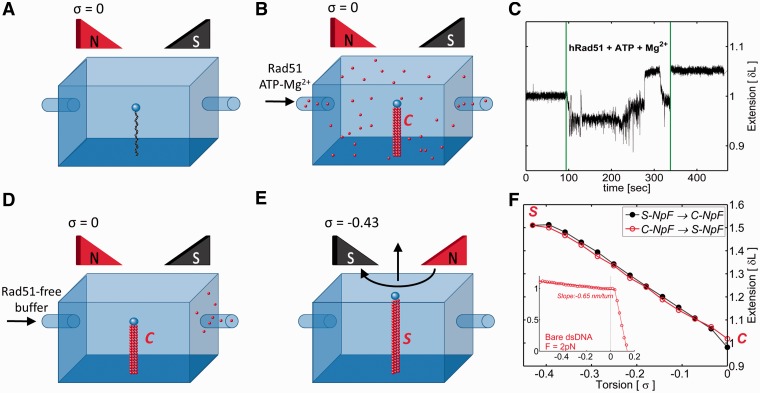
Figure 1.
DNA twist change induces interconversion of preformed Rad51-dsDNA complexes between stretched and non-stretched forms. (A and B) Rad51 in a buffer containing ATP and Mg2+ is added to the DNA that is prevented from rotation by rotationally locked magnetic bead. Such a DNA maintains its original twist with ca. 10.5 bp/turn and thus has the specific linking difference, σ = 0. (C) The tracing of magnetic bead position shows that the extension of Rad51-covered DNA increases by ∼5% as compared with protein-free DNA (strong perturbation of bead position lasting for ∼200 sec are induced by the buffer exchange in the flow cell). (D and E) After elimination of unbound Rad51, the DNA in the preformed complex is progressively unwound towards the specific linking difference σ = −0.43, which results in a helical repeat of ∼18.6 bp/turn when the DNA is prevented from writhing. (F) Fluctuation-corrected extension measurements show that Rad51-dsDNA complex progressively increases its length as the filament is unwound towards the helicity of 18.6 bp/turn and then shrinks back to the non-stretched form when the DNA is wound back towards the helical repeat of 10.5 bp/turn. The inset shows the extension profile of protein-free DNA at the pulling force of 2 pN. In contrast to Rad51-dsDNA filaments, it hardly increases its length on DNA unwinding. S-NpF and C-NpF denote stretched and compact nucleo-protein filaments, respectively.
RESULTS
Robertson et al. (14) showed that stretched filaments shrink when ATP is removed from the reaction buffer. However, using TIRFM it was not possible to find out whether the observed change of the length was associated with a change of DNA twist in the filaments. Magnetic tweezers constitute the method of choice to investigate the coupling between the helical repeat and length of helical filaments (15,18–22), and therefore, we decided to use this method to investigate this structural aspect of Rad51-dsDNA complexes. In addition to controlling DNA rotation, the magnetic tweezers allow us also to control the stretching force exerted on the DNA molecule (16). Knowing from earlier electron microscopy studies that Rad51 can completely cover circular dsDNA molecules even if these are prevented from unwinding by covalent closure (23), we decided to investigate the consequences of Rad51 binding to linear dsDNA molecules that were maintained in their natural twist by a magnetic bead that was not permitted to rotate (see Figure 1). The applied pulling force of 2 pN kept the DNA close to its crystallographic length. As shown in Figure 1A and B, adding Rad51, ATP and MgCl2 resulted in a very small increase of DNA length of 4 ± 2% (25 measured molecules). This small length increase was consistent with formation of non-extended Rad51-dsDNA complexes that by their increased stiffness, as compared with protein-free DNA, undergo smaller thermally induced undulations and thus show a slightly larger end-to-end distance at this relatively weak pulling force. We subsequently tested how formed complexes react to DNA unwinding towards the helicity of ∼18.6 bp/turn [the helicity of the DNA in the stretched filaments (7,8)], which corresponds to supercoiling density or torsion σ = −0.43. To avoid the possibility that free Rad51 in the surrounding solution could bind to the DNA during unwinding, we removed unbound Rad51 by flushing the flow cell using protein-free buffer with original concentrations of ATP and MgCl2. As shown in Figure 1D, E and F, the unwinding process caused the formed Rad51-DNA complexes to increase their length by 49 ± 5% (27 measured molecules), as would be expected if non-stretched filaments were progressively converted to stretched filaments. It is important to realize that under the same conditions, protein-free DNA increases its length by <10% on unwinding (16) (see the inset in Figure 1F). The pulling force of 2 pN is sufficient to prevent unwound DNA from underwinding-induced DNA writhing (24,25). Under such conditions, strongly unwound protein-free DNA minimizes its energy by accumulating stretches of L-DNA that mainly consist of DNA portions adopting left-handed Z-type DNA structure (24,25). In Z-type DNA structure, base pairs maintain stacking interactions (16) and therefore entire molecules maintain practically the same length as in torsionally unconstrained B-DNA.
We subsequently checked whether by rewinding the DNA back to a form with B-DNA helicity we could again induce formation of a non-stretched form. Figure 1F shows that this is indeed the case.
In principle, the perceived extension and shrinking as reported in Figure 1F could be also explained by plectonemic coiling of entire Rad51-DNA filaments. Because these filaments reach torsionally relaxed state at σ of ∼−0.43 they would not form plectonemes at this σ, but as the DNA is wound towards σ = 0 the entire filaments could fold on themselves and form plectonemes. However, such a plectonemic coiling of the entire filaments is expected to give a much higher slope of perceived length change than this is the case of protein-free DNA. This expectation results from the fact that the stiffness of Rad51-dsDNA filaments is ca. 10 times higher than that of protein-free DNA and the physical diameter of filaments is ca. 5 times bigger than that of protein-free DNA (5). The observed slope of the extension change is, however, less steep than that of overwound DNA forming plectonemes, where a much smaller number of DNA rotations resulted in bringing together two ends of plectonemically coiled DNA molecules (see inset in Figure 1F). In addition, electron micrographs of Rad51-dsDNA complexes formed under comparable conditions on torsionally constrained plasmids (see Figure 3) revealed them to be essentially free from plectonemes, thus suggesting that the torque induced by Rad51 binding to DNA is weak.
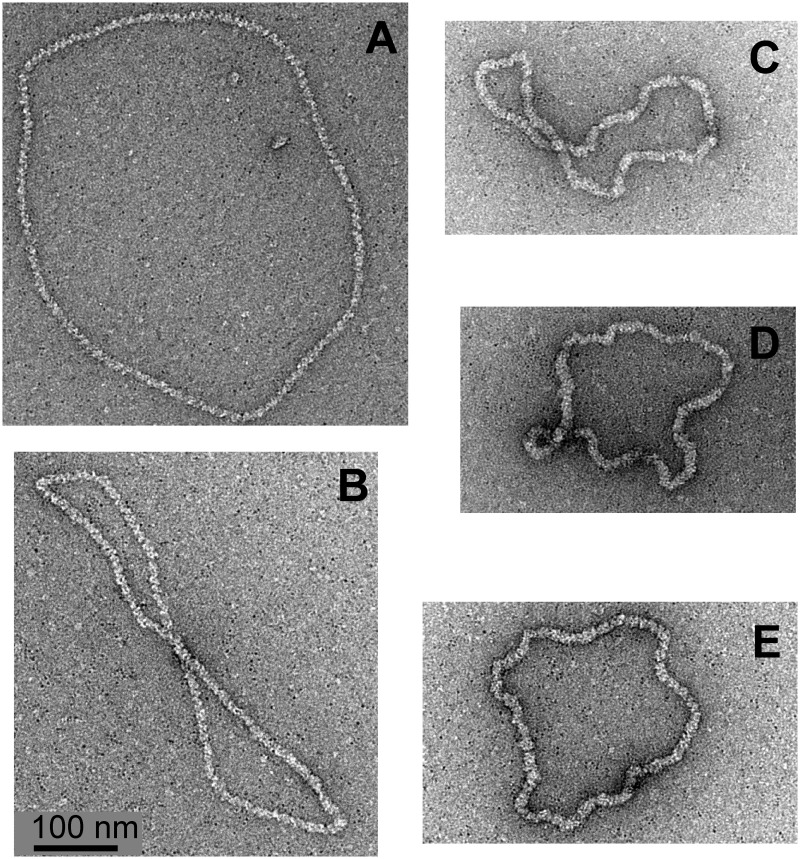
Figure 3.
Electron micrographs of Rad51-dsDNA complexes in stretched (A, B) and non-stretched form (C–E).
It is important to note that Rad51-dsDNA filaments maintained in the absence of free Rad51 in the surrounding buffer reacted to changes of DNA twist in essentially the same way as filaments kept in the presence of saturating amounts of Rad51 in solution (Supplementary Figure S1).
In a separate experiment, we investigated the behaviour of Rad51-dsDNA complexes over a broader range of enforced DNA helicity values than those spanning the interval between 10.4 and 18.6 bp per turn. We observed that in the absence of Rad51 in solution, the Rad51-dsDNA complexes formed in the presence of ATP reached their maximum extension of enforced DNA helicity corresponding to ∼18.6 bp/turn (Supplementary Figure S2). When the DNA was overwound beyond the natural helicity of the DNA, the filaments continued to shrink but the shrinking rate was decreasing. Because we observed protein dissociation when the DNA was overwound beyond its natural helicity (data not shown), the further shortening of the filaments may be explained by progressive dissociation of Rad51 and formation of localized plectonemes (See Supplementary Figure S2).
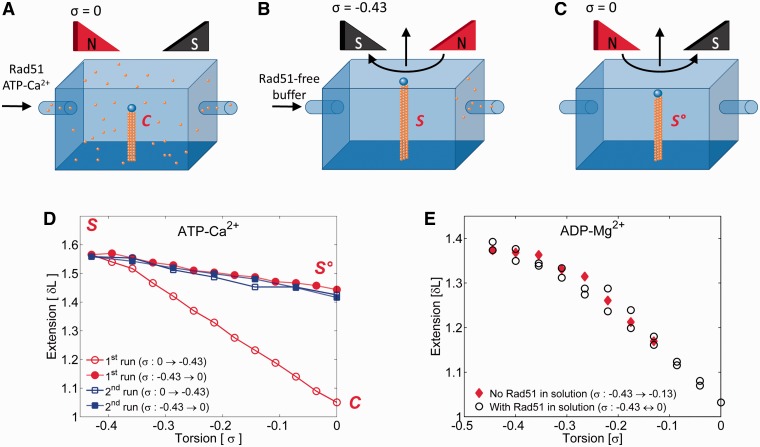
Robertson et al. (14) reported that an inhibition of ATP hydrolysis traps the stretched Rad51-dsDNA filaments and prevents their transition to a non-stretched form. Because ATPase activity of Rad51 is strongly inhibited when Mg2+ is replaced by Ca2+ (26), we repeated an experiment similar to the one shown in Figure 1 but incubations of Rad51 with DNA were performed in a buffer containing 2 mM CaCl2 instead of 2 mM MgCl2. Complexes formed with dsDNA that was kept at its natural helicity stretched upon unwinding in essentially the same way as complexes formed in buffers containing MgCl2 (Figure 2A, B and D). However, once the filament reached its stretched form, it was locked in that structure and could not return to the non-stretched form on DNA rewinding towards natural B-DNA helicity (Figure 2C and D). The filament only shrank a bit on rewinding, which may be explained by a partial Rad51 detachment or by an elastic deformation of a helical filament that slightly decreased its pitch on rewinding. We favour the second interpretation because the filament seems to maintain all bound Rad51, as can be concluded from the fact that upon several rounds of underwinding and rewinding, the stretched filaments maintained practically the same extension (see Figure 2D).
Figure 2.
Ca2+-stabilized Rad51-dsDNA-ATP complexes get locked in the stretched form, whereas Rad51-dsDNA-ADP filaments can freely pass between non-stretched and stretched forms of the filament. (A) Rad51 in a buffer containing ATP and Ca2+ is added to the DNA that is prevented from rotation and thus maintains its natural twist. (B and D; the lowest profile) After elimination of unbound Rad51, the preformed complex progressively increases its length on DNA unwinding towards a structure with ca 18.6 bp/turn. (C and D) Rewinding of stretched complexes does not convert them to a non-stretched form, although some shortening is observed. (E) Extension profiles of Rad51-dsDNA-ADP filaments formed in the presence of Mg2+ as the helicity of DNA is changed from 10.5 bp/turn (σ = 0) to 18.6 bp/turn (σ = −0.43).
To shed more light on the role of ATP hydrolysis in the conversion between stretched and non-stretched forms of Rad51-dsDNA filaments, we investigated the behaviour of Rad51-dsDNA filaments formed in the presence of ADP and MgCl2. Interestingly, these filaments behaved similarly to those formed in the presence of ATP under conditions where ATP can be hydrolysed. Irrespective of whether free Rad51 was present in the solution or not, the filaments formed in the presence of ADP extended by nearly 50% (43 ± 6% for 10 molecules measured) as the imposed DNA twist of bound DNA changed stepwise from 10.5 to 18.6 bp/turn (see Figure 2E). We presume that the equilibrium twist of DNA in filaments formed in the presence of ADP is close to that of free B-DNA, whereas in filaments where most of bound ATP is still not hydrolysed the DNA equilibrium twist is likely to be ∼18.6 bp/turn. Such an interpretation agrees with DNA curtain experiments where the length of Rad51-dsDNA filaments decreased as ATP was hydrolysed and then increased again when fresh ATP was added (14).
To complement magnetic tweezers studies of the coupling between the length and twist of individual Rad51-DNA complexes, we wanted to directly observe how the structure of Rad51-DNA complexes changes when the DNA is permitted or prohibited from freely changing its twist. With this aim, we studied by electron microscopy Rad51-dsDNA complexes formed on circular DNA molecules that were either covalently closed and thus restricted in their change of twist or were nicked and thus could freely change their twist.
Because calcium salts are not suited for the magnesium spreading method (17), we decided to inhibit ATP hydrolysis by adding ATPγS to the complexes initially formed in the presence of ATP and Mg2+. Figure 2E–G shows images obtained in the reactions using preparations of supercoiled pUC18 DNA molecules that contain ∼5% of nicked DNA molecules. We observed that all DNA molecules formed complexes with Rad51 but that there were two types of complexes: stretched with a contour length of ∼1300 nm (1314 ± 115, n = 3) (Figure 3A and B) and non-stretched (Figure 3C–E) with a contour length of ∼900 nm (890 ± 26, n = 11). Non-stretched complexes constituted the great majority, and their length was close to the expected length of protein-free pUC18 DNA (913 nm). The stretched complexes constituted ∼5% of observed molecules and clearly showed their helical structure typical, for example, of RecA-DNA complexes formed in the presence of ATPγS (9,27,28). Non-stretched complexes had a much smoother and denser structure than stretched complexes, suggesting a slinky-like transition between stretched and non-stretched complexes. From the relative frequency of observed stretched and non-stretched complexes, we concluded that DNA molecules that were nicked and thus could freely change their twist were free to form stretched Rad51-DNA filaments, whereas covalently closed DNA molecules that were unable to freely change their twist formed non-stretched Rad51-DNA filaments. Electron microscopy observations corroborated, therefore, magnetic tweezers experiments, in which DNA molecules that were not permitted to change their twist formed complexes that did not stretch the DNA, whereas a subsequent unwinding of DNA permitted these complexes to adopt a stretched form. To conserve the stoichiometry, while permitting the DNA to change from an extended helix with ∼18.6 bp per 95Å pitch to a B-DNA–like helix with ∼10.5 bp per 35Å pitch, the filaments would need to change from a structure with ∼6.2 Rad51 monomers per 95Å pitch to a structure with ∼3.5 Rad51 monomers per 35Å pitch. Such a change would require ∼30% increase in the linear density of filaments, which could easily be achieved by closing the prominent groove in the stretched filaments, consistent with the observed structure of two forms of complexes (see Figure 3). It is important to mention that the negative staining technique applied here allows us to unambiguously visualize protein-free DNA (23). Therefore, it is unlikely that there are substantial uncovered portions of DNA molecules, in contrast to the situation where RecA protein interacts with covalently closed DNA molecules and leaves a substantial portion of uncovered DNA (23).
DISCUSSION
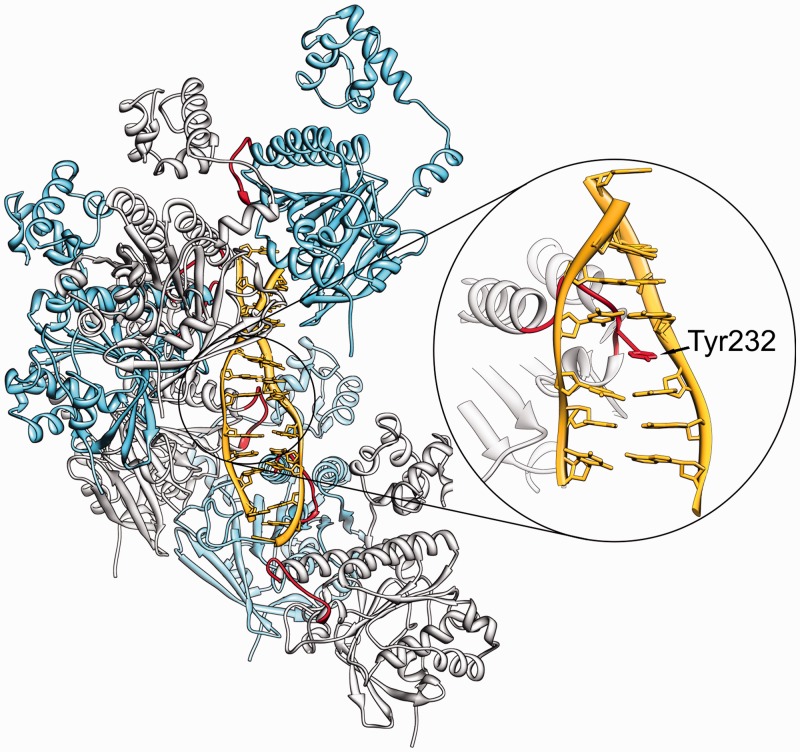
Based on magnetic tweezers experiments and electron microscopy observations presented here and earlier TIRFM results (14), we conclude that human Rad51 protein shows two interconvertible binding modes to dsDNA: a stretching and a non-stretching mode. To reach the stretching mode in living cells, two necessary requirements have to be met: the DNA needs to be free to undergo axial rotation, which allows the change in twist needed to reach the stretched form, and in addition, Rad51 monomers have to be in the ATP bound form. If the bound DNA is not free to rotate or if ATP is hydrolysed, the non-stretched form is observed. The crystal structure of stretched RecA-DNA complexes reveals that the unwinding and extension of the DNA is due to the stacking disruptions occurring every 3 bp, where the space between unstacked base pairs is filled by suitably placed hydrophobic amino acids from so-called L1 loop (8). Although the atomic structure of stretched Rad51-DNA complexes is not yet determined, all available data suggest that a similar amino acid insertion occurs in these complexes (29,30) (see Figure 4). It is known that ATP binding pockets in RecA and Rad51 are placed in the interface zone between subsequent monomers in the complex (31–33). Therefore, ATP binding can naturally lead to changes of filament architecture resulting, for example, in the correct positioning of Tyr232 from L1 loop for the intercalation-like insertion into the DNA (29,30). The critical role of Tyr232 in Rad51-DNA binding was previously shown in studies of Rad51 mutant proteins, in which Tyr232 was replaced by various amino acids (34).
Figure 4.
A model of Rad51-dsDNA filaments explaining how the intercalation of Tyr232 into the DNA can critically regulate the structure and stability of Rad51-DNA complexes. Earlier studies provided the evidence that Tyr232 from L1 loop of Rad51 intercalates into the bound DNA and this process is implicated in DNA stretching (29,30). Because DNA intercalation necessitates DNA unwinding, the impediment of DNA rotation by magnetic beads or by covalent closure permits only formation of non-stretched filaments. If the DNA is free to rotate and the L1 loop is in a favourable orientation as in Rad51 in the ATP-bound state, the complexes reach the stretched form of high stability. Once the Tyr232 is intercalated, it can be retracted from the DNA on ATP hydrolysis or on rewinding of the DNA. However, in the presence of Ca2+, the intercalation is so stable that DNA rewinding to natural helicity is not able to expel the intercalated Tyr232 from the DNA.
Two earlier studies showed that stretched Rad51 filaments shrink when ATP is hydrolysed, however the helicity of the non-stretched filaments was not determined in these studies (4,14). We have shown here that transitions between stretched and non-stretched forms of Rad51-DNA filaments are coupled to changes of DNA twist. We believe that the intrinsic helicity of compact filaments such as those formed in the presence of ADP is close to that of B-DNA, as non-intercalating binding of Rad51 would not be expected to change the DNA twist in a significant way. However, further studies using a different form of magnetic tweezers (20) will be needed to determine the intrinsic helicity of compact Rad51 filaments.
Although DNA pairing and strand exchange do not require ATP hydrolysis and can be catalysed by Rad51 or RecA helical filaments stabilized in the stretched form (9,10,12), the complete reaction requires ATP hydrolysis to release the DNA from complexes with recombination promoting proteins (35). It is likely that ATP hydrolysis induces such changes in the filament structure that may cause the inserted amino acids to move out from unstacked dinucleotide steps. This permits then the DNA to return to a non-stretched form together with the bound Rad51 and eventually leads to dissociation of Rad51 from the bound DNA. In fact, recent studies using direct imaging of Rad51 dynamics revealed that shrinking of complexes precedes their dissociation (4). Because the removal of any intercalating agent from the DNA is in part driven by the natural tendency of unstacked base pairs to ‘snap back’, it is natural to expect that if the DNA is maintained under a significant pulling force then even after ATP hydrolysis there will be no tendency of inserted amino acids to move out of the DNA and, therefore, Rad51 or RecA will not dissociate from the DNA. This is exactly what was observed in recent single molecule studies (21,36). The remaining question is why in the presence of Ca2+ the filaments are locked in the extended form and do not shrink on rewinding, whereas such shrinking is observed in the presence of Mg2+. Recent linear dichroism study combined with molecular modelling indicated that in the presence of Ca2+, the L1 loop of Rad51 changes its configuration in such a way that it enforces intercalation of its residues into the DNA (30). This would then lock the filaments in the stretched form (see Figure 3). In the presence of Mg2+ the L1 loop is more flexible, which permits easier insertion of its hydrophobic amino acids between unstacked bases when the DNA is free to rotate, but also facilitates removal of the loop from the DNA when the DNA is rewound and exerts a pressure on the inserted residues. Our magnetic tweezers experiments have been performed with dsDNA, as this permits to induce torsional tension in the bound DNA. However, we believe that the mechanism of Ca2+-induced stabilization of stretched Rad51 filaments applies also to Rad51-ssDNA complexes. In the known structure of RecA-ssDNA complexes, their stretched structure is maintained by insertion of amino acids into unstacked steps separating stacked triplets of bases (8).
Here, changes in DNA twist were induced by changing DNA torsional stress with the primary objective of better understanding the molecular mechanism of Rad51-DNA interaction. It is an open question whether changes in the torsional tension of bound DNA can induce interconversions between stretched and non-stretched forms of the formed complexes during Rad51-mediated homologous pairing and strand exchange, which is needed for double-strand break repair occurring in vivo. Such a possibility may seem unlikely because free ends created as a result of double-strand breaks are free to rotate and this provides an easy possibility to dissipate the torsional stress resulting from changes of DNA twist. However, the situation changes when Rad51 filaments formed on single-stranded terminal regions, resulting from the processing of double-stranded ends (1,2), start to interact with a homologous DNA region in the sister chromatid. Although the precise structure of the formed joints involving ssDNA from processed broken ends and double-stranded homologous regions in sister duplexes is still unknown, most plausible scenarios restrict the freedom of rotation (37–39). Therefore, the torsional tension resulting from the change in DNA twist in the homologously paired duplex DNA (40,41) may regulate transitions between stretched and non-stretched forms of Rad51-DNA and affect one or more stages of Rad51-mediated repair of double-strand breaks in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Agence nationale de la recherche [ANR-10-BLAN-1013 1 – ‘DynRec’ to G.C., A.R.C. and M.T.]; Region Ile-de-France in the framework of DIM Centre de compétences en nanosciences en Ile-de-France (‘RecombinaTarget’) (to G.C.); Swiss National Science Foundation [31003A-138367 to A.S.]. Funding for open access charge: Agence nationale de la recherche (France).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anna Reymer for preparing Figure 4. We warmly thank Jacques Prost for his essential help to identify the signature of a phase transition of the NpF (stretched-collapsed) in the Magnetic Tweezers experiments and Julie Plastino for having kindly read and corrected the manuscript.
REFERENCES
- 1.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 2.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ristic D, Kanaar R, Wyman C. Visualizing RAD51-mediated joint molecules: implications for recombination mechanism and the effect of sequence heterology. Nucleic Acids Res. 2011;39:155–167. doi: 10.1093/nar/gkq766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl Acad. Sci. USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheridan SD, Yu X, Roth R, Heuser JE, Sehorn MG, Sung P, Egelman EH, Bishop DK. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 2008;36:4057–4066. doi: 10.1093/nar/gkn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egelman EH, Stasiak A. Electron microscopy of RecA-DNA complexes: two different states, their functional significance and relation to the solved crystal structure. Micron. 1993;24:309–324. [Google Scholar]
- 7.Stasiak A, Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 9.Honigberg SM, Gonda DK, Flory J, Radding CM. The pairing activity of stable nucleoprotein filaments made from recA protein, single-stranded DNA, and adenosine 5'-(gamma-thio)triphosphate. J. Biol. Chem. 1985;260:11845–11851. [PubMed] [Google Scholar]
- 10.Menetski JP, Bear DG, Kowalczykowski SC. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl Acad. Sci. USA. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy G, Burnett B, Radding CM. Uptake and processing of duplex DNA by RecA nucleoprotein filaments: insights provided by a mixed population of dynamic and static intermediates. Biochemistry. 1995;34:10194–10204. doi: 10.1021/bi00032a013. [DOI] [PubMed] [Google Scholar]
- 12.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J. Mol. Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 14.Robertson RB, Moses DN, Kwon Y, Chan P, Chi P, Klein H, Sung P, Greene EC. Structural transitions within human Rad51 nucleoprotein filaments. Proc. Natl Acad. Sci. USA. 2009;106:12688–12693. doi: 10.1073/pnas.0811465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulconis R, Bancaud A, Allemand JF, Croquette V, Dutreix M, Viovy JL. Twisting and untwisting a single DNA molecule covered by RecA protein. Biophys. J. 2004;87:2552–2563. doi: 10.1529/biophysj.104.043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 17.Stasiak A, Di Capua E, Koller T. Elongation of duplex DNA by recA protein. J. Mol. Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 18.Mine J, Disseau L, Takahashi M, Cappello G, Dutreix M, Viovy JL. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arata H, Dupont A, Mine-Hattab J, Disseau L, Renodon-Corniere A, Takahashi M, Viovy JL, Cappello G. Direct observation of twisting steps during Rad51 polymerization on DNA. Proc. Natl Acad. Sci. USA. 2009;106:19239–19244. doi: 10.1073/pnas.0902234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipfert J, Kerssemakers JW, Jager T, Dekker NH. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 23.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 25.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 26.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 28.Heuser J, Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J. Mol. Biol. 1989;210:473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 29.Reymer A, Frykholm K, Morimatsu K, Takahashi M, Norden B. Structure of human Rad51 protein filament from molecular modeling and site-specific linear dichroism spectroscopy. Proc. Natl Acad. Sci. USA. 2009;106:13248–13253. doi: 10.1073/pnas.0902723106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fornander LH, Frykholm K, Reymer A, Renodon-Corniere A, Takahashi M, Norden B. Ca2+ improves organization of single-stranded DNA bases in human Rad51 filament, explaining stimulatory effect on gene recombination. Nucleic Acids Res. 2012;40:4904–4913. doi: 10.1093/nar/gks140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 32.Grigorescu AA, Vissers JH, Ristic D, Pigli YZ, Lynch TW, Wyman C, Rice PA. Inter-subunit interactions that coordinate Rad51's activities. Nucleic Acids Res. 2009;37:557–567. doi: 10.1093/nar/gkn973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Villanueva N, Rould MA, Morrical SW. Insights into the mechanism of Rad51 recombinase from the structure and properties of a filament interface mutant. Nucleic Acids Res. 2010;38:4889–4906. doi: 10.1093/nar/gkq209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuo Y, Sakane I, Takizawa Y, Takahashi M, Kurumizaka H. Roles of the human Rad51 L1 and L2 loops in DNA binding. FEBS J. 2006;273:3148–3159. doi: 10.1111/j.1742-4658.2006.05323.x. [DOI] [PubMed] [Google Scholar]
- 35.Rosselli W, Stasiak A. Energetics of RecA-mediated recombination reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J. Mol. Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 36.Conover AJ, Danilowicz C, Gunaratne R, Coljee VW, Kleckner N, Prentiss M. Changes in the tension in dsDNA alter the conformation of RecA bound to dsDNA-RecA filaments. Nucleic Acids Res. 2011;39:8833–8843. doi: 10.1093/nar/gkr561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radding CM. Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. Biol. Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 38.Stasiak A. Three-stranded DNA structure; is this the secret of DNA homologous recognition? Mol. Microbiol. 1992;6:3267–3276. doi: 10.1111/j.1365-2958.1992.tb02194.x. [DOI] [PubMed] [Google Scholar]
- 39.Prevost C, Takahashi M. Geometry of the DNA strands within the RecA nucleofilament: role in homologous recombination. Q. Rev. Biophys. 2003;36:429–453. doi: 10.1017/s0033583504003956. [DOI] [PubMed] [Google Scholar]
- 40.Kiianitsa K, Stasiak A. Helical repeat of DNA in the region of homologous pairing. Proc. Natl Acad. Sci. USA. 1997;94:7837–7840. doi: 10.1073/pnas.94.15.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voloshin ON, Camerini-Otero RD. The duplex DNA is very underwound in the three-stranded RecA protein-mediated synaptic complex. Genes Cells. 1997;2:303–314. doi: 10.1046/j.1365-2443.1997.1240322.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.