Abstract
Telomere position effect (TPE) is the influence of telomeres on subtelomeric epigenetic marks and gene expression. Previous studies suggested that TPE depends on genetic background. As these analyses were performed on different chromosomes, cell types and species, it remains unclear whether TPE represents a chromosome—rather than genetic background-specific regulation. We describe the development of a Linear Human Artificial Chromosome (L-HAC) as a new tool for telomere studies. The L-HAC was generated through the Cre-loxP-mediated addition of telomere ends to an existing circular HAC (C-HAC). As it can be transferred to genetically distinct cell lines and animal models the L-HAC enables the study of TPE in an unprecedented manner. The HAC was relocated to four telomerase-positive cell lines via microcell-mediated chromosome transfer and subsequently to mice via blastocyst injection of L-HAC+-ES-cells. We could show consistent genetic background-dependent adaptation of telomere length and telomere-associated de novo subtelomeric DNA methylation in mouse ES-R1 cells as well as in mice. Expression of the subtelomeric neomycin gene was inversely correlated with telomere length and subtelomeric methylation. We thus provide a new tool for functional telomere studies and provide strong evidence that telomere length, subtelomeric chromatin marks and expression of subtelomeric genes are genetic background dependent.
INTRODUCTION
Human artificial chromosomes combine unique properties for transgenic (1) and putative gene therapeutic (2) applications and are essential to study the function and architecture of chromosome structural elements (3–6)—mainly centromeres and telomeres.
Telomeres are specialized nucleoprotein complexes that protect chromosomal ends against degradation, fusion and recombination (7,8). In addition they promote specific chromosomal movements (9,10), have a role in holding sister chromatids together before resolution at mitotic anaphase (11) and can influence the expression of subtelomeric genes in a process known as telomere position effect (TPE) (12).
Vertebrate telomeres have a tandemly repeated (TTAGGG) DNA-unit of which the number of repeats (total length) can differ between species, individuals, cell types, homologous chromosomes and even sister chromatids (13–15). In humans and likely in all mammals, the telomeric repeat ends in a single-stranded G-rich 3′ overhang that loops back into part of the double-stranded (TTAGGG)n repeats by homologous strand invasion. Capped telomeres, generated via the T-loop model, most likely protect chromosomal ends in vivo (16,17). A complex of six telomere-specific proteins (TRF1, TRF2, POT1, TIN2, RAP1 and TPP1) (8,18) is proposed to control the structure and modification of the telomeric DNA, thus ensuring the integrity of the chromosomal end. Dysfunctional telomeres result from either (sub-) telomeric DNA or protein alterations causing the destruction of the protective end-cap (19).
Telomerase is a ribonucleoprotein complex that can lengthen telomeres by using its reverse transcriptase (TERT) activity and its RNA subunit (TERC) as template (20,21). Aside from germ cells, stem cells and most cancerous cells, adult human cells lack telomerase activity and thus telomeres will shorten with each cell cycle due to the end-replication problem. In addition telomeres can be lengthened by alternative recombination-based mechanisms. Some human cancers and immortalized cell lines make use of these alternative lengthening of telomere (ALT)-pathway(s) (22,23).
It was recently shown that telomere length affects the epigenetic status of mouse telomeres and subtelomeres. Benetti et al. (2007) (24) showed that lack of telomerase results in a decrease in trimethylation of H3K9 and H4K20, less CBX3-binding, increased levels of H3K9 acetylation in the telomeric as well as in the subtelomeric chromatin, and decreased DNA methylation of the subtelomeric chromatin. These data suggest that telomere shortening results in the loss of (sub-) telomeric heterochromatin features leading to a more ‘open’ chromatin state. Additionally, it was demonstrated that mouse cells that are knocked-out for specific chromatin building bricks undergo abnormal telomere elongation (25–27). Taken together, these results revealed important links between (sub-) telomeric chromatin conformation and telomere length homeostasis.
Because telomeres affect the architecture of nearby chromatin, they have the ability to influence the expression of subtelomeric genes via a process known as TPE. Although a molecular understanding of the TPE-mechanism is currently lacking, its effect appears to be dependent on telomere length (28), local chromatin structure (29) and the distance relative to the telomere (30). Furthermore, the analysis of TPE in cells with different genetic backgrounds suggested that the subtelomeric chromatin and thus also the molecular mechanisms that underpin TPE may differ between different vertebrate genetic backgrounds (mouse ES-cells versus human tumor cell lines versus avian cell lines) (12,28,31). However, these studies were performed on chromosomes from different species obscuring whether the TPE-differences are chromosome rather than genetic background-specific effects.
In this study, we linearize a circular human artificial chromosome (C-HAC) using artificial telomeres. The C-HAC was derived from a naturally occurring small accessory chromosome consisting of a 4 Mb region of the short arm of human chromosome 1 (1p21.3-22.1) and alphoid sequences of human chromosome 20, previously called chromosomal vector (CV) (32). This C-HAC/CV has previously been applied for transgenesis in cell lines and in vivo in mice (32,33) and for studying meiotic chromosome behavior and checkpoints (6). We apply the linearized human artificial chromosome (L-HAC) for analysing the influence of genetic background switching on (sub-) telomere processing and TPE. Our data provide substantial evidence that the structure, DNA methylation status and TPE of a specific chromosomal end alter in a cell-specific manner.
MATERIALS AND METHODS
The isolation of DNA and RNA from cells and tissues, Southern blot analysis, and DNA sequencing were performed according to standard procedures.
Cell cultures
Hamster E10B1 hprt−/− cells, CHL hprt−/− cells and mouse BALB/c 3T3 cells were grown in DMEM/F12 (Life Technologies, NY) containing L-glutamine, pyridoxine HCl and 10% FBS (Thermo Fisher Scientific, MA). Chicken DT40-CRE cells (34) were grown as described on the DT40 website (http://pheasant.gsf.de/DEPARTMENT/dt40.html). ES-cells (R1, 129/SVx129/SV-CP) (35) were grown on mitomycinC-treated G418-resistant mouse embryonic fibroblast feeder cells (Thromb-X, Leuven, Belgium) in TX-WES medium (Thromb-X) supplemented with L-glutamine (Life Technologies).
Plasmids pSP73-TEL08-5′HPRT1-loxP and pSP73-TEL08-BLAS-loxP
Plasmid pSP73-TEL08-5′HPRT1-loxP contains 800 bp of cloned telomeric (TTAGGG)n repeats, followed by the 5′ end of the human HPRT1 minigene (controlled by a SV40 promoter) and a loxP sequence. It was created by ligating a 1254 bp BamHI restriction fragment carrying the ‘SV40-5′HPRT1-loxP’ DNA cassette from plasmid pBS-Hyg/SV40 5′Hprt/loxP (60) into the unique BamHI site of plasmid pSP73.Sty11. Plasmid pSP73.Sty11 contains 800 bp of cloned TTAGGG repeats and was a gift from Dr de Lange, T. (The Rockefeller University, NY). Two plasmids, pSP73-TEL08-5′HPRT1-loxP and pSP73-TEL08-loxP-5′HPRT1, were obtained depending on the orientation of the ‘SV40-5′HPRT1-loxP’ cassette. Plasmid pSP73-TEL08-5′HPRT1-loxP that has a ‘(CCCTAA)n-SV40 promoter-5′HPRT1-loxP’ cassette was used for linearization of the circular HAC. Plasmid pSP73-TEL08-loxP-5′HPRT1 was further modified into plasmid pSP73-TEL08-BLAS-loxP. To generate plasmid pSP73-TEL08-BLAS-loxP, the SV40 promoter and 5′ end of the HPRT1 minigene of plasmid pSP73-TEL08-loxP-5′HPRT1 were deleted by PvuII digestion, followed by purification and self-ligation of the remaining 3845 bp DNA fragment. Subsequently, a 2824 bp BamHI/XhoI fragment from plasmid MS2bsr (a gift of Dr Hiroshi Arakawa (Institute for Molecular Radiobiology, Munich, Germany)) containing a blasticidin-resistance gene driven by a chicken beta-actin promoter was blunted and ligated into the SmaI site of pSP73-TEL08-loxP, resulting in plasmid pSP73-TEL08-BLAS-loxP.
Transfections
Plasmids pOG231 (expressing CRE) (18.75 µg), pSP73-TEL08-5′HPRT1-loxP (25 µg) and pSP73-TEL08-BLAS-loxP (25 µg) were co-transfected into 12.5 × 106 E10B1 cells (resuspended in 0.5 ml ice-cold DMEM/10% FCS) by electroporation at 230 V and 960 µF using the Bio-Rad Gene-Pulser (Bio-Rad, Hercules, CA). Twenty four hours later, selective HAT—(Life Technologies) and blasticidin—(Life Technologies) containing medium was added. After 8 days resistant colonies became visible and were picked. Transfections of plasmid pBS-TEL-aSAT-HYG into the chicken DT40-CRE cells that contain the L-HAC were performed as described on the DT40 website (http://pheasant.gsf.de/DEPARTMENT/dt40.html). DT40-CRE clones were selected in medium containing 1.5 mg/ml hygromycin (Life Technologies).
Microcell-mediated chromosome transfer
All microcell-mediated chromosome transfers (MMCTs) were performed according to the following basic protocol. To induce micronucleation, colcemid was added to a final concentration of 0.5 µg/ml to 3–4 × 107 donor cells in exponential growth. After 36 h, cells were trypsinized and pelleted by centrifugation at 800 rpm for 10 min (4°C). The pellet was resuspended in 4 ml serum-free medium (SFM) at room temperature (RT) and mixed with two pre-warmed (37°C) 40 ml 1:1 v/v Percoll (Amersham Biosciences, Uppsala, Sweden)/DMEM gradients containing 20 µg/ml cytochalasin B (Sigma, St. Louis, MO) and 0.1 µg/ml colcemid (Life Technologies). The mixture was centrifuged in a pre-warmed centrifuge at 43 000g (19 000 rpm) for 70 min at 37°C. Microcells (15 ml gradient aliquots) were washed with 35 ml SFM, pelleted by centrifugation at 1500 rpm for 10 min (4°C), resuspended and pooled in 5 ml medium. Microcell formation was checked under the microscope. The microcells were pelleted, resuspended in 1 ml SFM, and added to a 25 cm2 culture flask containing 90% confluent adherent acceptor cells (in the presence of 100 µg (Sigma)) or mixed with ±3 × 107 log phase acceptor cells in suspension and were allowed to adhere to the acceptor cells by incubation at 37°C for 10 min. The medium was removed and 1 ml pre-warmed (37°C) 50% w/v polyethylene glycol 1500 (Roche, Basel, Switzerland) was added for 1 min. Subsequently, the acceptor cells were washed 3 times with medium and were finally plated out in medium containing penicillin/streptomycin (P/S) (Life Technologies). After 24 h, selective medium was added. To allow counter-selection of contaminating donor cells, BALB/c 3T3 acceptor cells were modified to be puromycin resistant by transfection of plasmid pPNT-PURO/TK. Upon MMCT (Figure 2) the BALB/c cells were selected in medium supplemented with 10 µg/ml puromycin (BD Biosciences, Franklin Lakes, NJ) and either 4 µg/ml blasticidin or 400 µg/ml G418 (Life Technologies). DT40-CRE cells receiving a L-HAC from the L-HAC+ BALB/c 3T3 cells were selected in medium containing 6 µg/ml puromycin and 30 µg/ml blasticidin. The DT40-CRE acceptor cells contained plasmid BML4-PURO (a gift of Dr Arakawa H., Institute for Molecular Radiobiology, Munich, Germany) and were thus puromycin resistant. The BALB/c 3T3 L-HAC+ cells were applied as donor cells to provide the CHL hprt−/− cells with the L-HAC. The latter were selected in 30 µM 6-Thioguanine (Sigma-Aldrich, Saint Louis, MO) and 3 µg/ml blasticidin. CHL hrpt−/− cells receiving the size-reduced L-HACs from the HAC+ DT40-CRE donor cells were selected in 30 µM 6-Thioguanine and 700 µg/ml hygromycin. ES-R1 clones with a L-HAC from CHL hrpt−/− cells were grown on mitomycinC-treated neoR-MEFs in TX-WES medium supplemented with HAT and either 300 or 500 µg/ml G418.
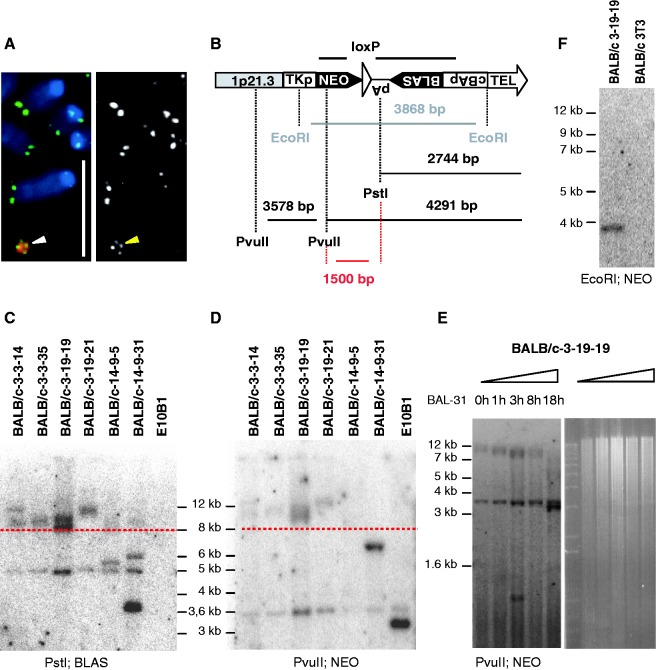
Figure 2.
Proof of HAC-linearization in mouse BALB/c 3T3 cells. (A) FISH with alphoid-20 DNA (red) and PNA telomeric (green) probes. Arrowheads point toward the HAC. Grayscale representation of the telomeric signal is shown on the right. Bar, 10 μm. (B) Schematic representation of TRF restriction digests and expected fragment lengths. Bold bars represent probes for neomycin (NEO) and blasticidin (BLAS). (C and D) Comparative TRF analyses of genomic DNA cut with PstI (C) or PvuII (D) that was size-separated by agarose gel electrophoresis and probed with the BLAS and NEO probes, respectively. Except for BALB/c 14-9-5 and 14-9-31 the BALB/c clones display an identical fragment pattern in the 8–12 kb region (above red-dashed line) for both restriction digests, with a 1.5 kb length difference. Internal control fragments of 5 kb (shown in C) and 3.6 kb (shown in D) were detected. (E) BAL-31 experiment on clone 3-19-19. Increasing the BAL-31 incubation-time preceding the TRF analysis resulted in decreasing fragment lengths. TRFs were monitored after 1, 3, 8 and 18 h of BAL-31 incubation. The 3.6 kb control fragment (D) remained unaltered. Right panel shows the EtBr-stained agarose gel. (F) Southern blot analysis (left panel) of EcoRI digested genomic DNA from clone BALB/c 3-19-19 probed with NEO resulted in the expected 3.9 kb fragment.
BAL-31 exonuclease treatment
We mixed 360 µg genomic DNA (prepared by phenol-chloroform extraction without vortexing) with 5U BAL-31 exonuclease (New England Biolabs, Ipswich, MA) in a 800 µl reaction and incubated it at 30°C. At time points 0, 8 and 20 h, a portion of the mixture was removed and the BAL-31 exonuclease was inhibited by adding 80 µl of 0.5 M EDTA pH 8.0. DNA was extracted by phenol-chloroform and 10 µg was used for restriction digestion and Southern blot analysis.
Fluorescent in situ hybridization
Hamster Cot1 DNA was obtained from Life Technologies. Chicken Cot1 DNA was prepared from sonicated DT40 genomic DNA. The DNA fragments (ranging between 200 bp and 800 bp) were denatured (5 min at 95°C in 0.3 M NaCl), repetitive sequences were renatured at 65°C for t (=1/Co) seconds (Co = start concentration of the DNA in mol/l) and finally, the remaining ssDNA-fragments were hydrolyzed (30 min at 37°C) with Nuclease S1 (Roche). The α-satellite DNA probe specific for the HAC was generated by low stringency PCR using primers 5′-AGTAAGTTCT TTGTGTTGCC TC and 5′-CAGAGTGTTT CCAAACTACT CTATG and E10B1 DNA as template (32). P1-derived artificial chromosomes (RPCI-5 837O21 and RPCI-4 573H3) were used as probes for detecting human chromosome 1p21-22 sequences. Probes were labeled with either digoxygenin-11-dUTP or biotin-16-dUTP using, respectively, the DIG-Nick or the BIOTIN-Nick Translation Mix (Roche). Denaturation of the metaphase slides and probes, hybridization and subsequent detection of the fluorescent signals were performed as described previously (32). Fluorescent in situ hybridization (FISH) with a FITC-(C3TA2)3 peptide nucleic acid (PNA) probe to detect telomere sequences was performed as described (36). Chromosomes were counterstained with DAPI and the slides were mounted in Vectashield mounting medium (Vector Laboratories Inc., Burlingame, CA). The signal was visualized by digital imaging microscopy using a cooled charge-coupled device camera (Photometrics Ltd, Tucson, AZ) coupled to the QuipsFISH system (Abott laboratories, Illinois).
Pulsed-field gel electrophoresis
Cells were cultured, trypsinized (if needed), washed two times with 1 × PBS (Life Technologies) and diluted to 3 × 107 cells/ml in 1 × PBS. An equal volume of 1.5% low-melting point agarose (Life Technologies) was added. The mixture was divided into block molds and allowed to set overnight at 4°C. Upon addition of 20 volumes of lysis buffer (100 mM EDTA, pH 8.0; 10 mM Tris–HCl, pH 8.0; 1% (w/v) Sarkosyl; 100 μg/ml proteinase K) the blocks were incubated overnight at 50°C. After repeating the lysis step at 37°C, the blocks were washed 3 times for 1 h with 10 volumes of storage buffer (10 mM Tris–HCl, pH 8.0; 10 mM EDTA, pH 8.0) supplemented with PMSF at RT. Subsequently, the blocks were washed 3 times 30 min at RT with 10 volumes of Tris–HCl (pH 8.0). The DNA fragments were size separated using chromosomal grade agarose (Bio-Rad Laboratories) on a CHEF mapper XA system (Bio-Rad Laboratories). The standard was a Schizosaccharomyces Pombe size standard from Bio-Rad Laboratories.
Quantitative PCR
Quantitative real-time PCR (qPCR) was carried out on a LC480 apparatus (Roche). Reactions were performed using the ‘LightCycler® 480 SYBR Green I master’ mix (Roche) and 10 ng genomic DNA or 50 ng of cDNA. Primers were designed with the LightCycler Probe Design2 software (Roche) and are given in Supplementary Table S1 together with the PCR primers. All experiments were run in duplicate and each time a non-template control was included. Data were analysed with the instrument software and further dissected in Excel (Microsoft) via the comparative ddCt method (Sequence Detection System, bulletin no. 2; Life Technologies). Neomycin expression levels were normalized against two individual murine housekeeping genes (Hprt, Actb,) to correct for potential variation in housekeeping gene expression levels between the ES-R1 and BALB/c genetic backgrounds. In addition, NEO expression data were corrected for the number of HACs per cell. For that, genomic DNA was collected for each sample and qPCR was performed using primers for the HAC (NEO and PAC 837O21) and for the mouse Rer1 locus. Each dataset represents the pooled data per genetic background (ES-R1 circular: ES-g and ES-h, ES-linear: ES-R1-3-19-19-33-47 and ES-R1-3-19-19-32-27 and BALB/c: BALB/c-E and BALB/c-L which represent BALB/c-3-19-19 cells from early (E) and late (L) passage) with the standard deviation (SD) reflecting the variation between both samples. BALB/c cells were grown in the absence of blasticidin; ES-cells were cultured without feeder cells in G418 medium.
Telomeric repeat amplification protocol
TRAP-assays were performed with the TeloTAGGG Telomerase PCR ELISAPLUS kit from Roche according to the manufacturer’s instructions.
Bisulfite sequencing
Bisulfite treatment of 1 µg genomic DNA was performed with the EZ DNA Methylation Kit™ from Zymo Research (Irvine CA) according to the manufacturer’s instructions. Bisulfite sequencing primers were designed using the Methyl Primer Express® software (v1.0) from Life Technologies. The primers that were used are: subTEL forward 5′-GTTTGGTGGTTTTTAAGAGG and subTEL reverse 5′-ACAATCCATCTTATTCAATAACC. PCR reactions were performed in 50 µl reactions (16.6 mM (NH4)2SO4; 67 mM Tris, pH 8.8; 6.7 mM MgCl2.6H2O; 10 mM β-mercapto-ethanol; 0.2 mM dNTP) with 10 pmol of each primer using 2 µl (±200 ng) of bisulfite-treated genomic DNA and 1 U JumpStart™ REDTaq® DNA Polymerase (Sigma). PCR products were directly sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies) and samples were run on a 3130x Genetic Analyzer (Life Technologies). Data analysis was based on the chromatograms obtained from the Genetic Analyzer. For the in vivo data signal intensities of the C’s and T’s were compared between the C-HACs and the L-HACs. For the in vitro data we additionally calculated the percentage of methylation for each of the 18 CpGs surrounding the EcoRI(2) site. To this extent the initial PCR products were subcloned into the pGEM-T easy vector and for each PCR product 12 clones were sequenced. The percentages that are depicted in Figure 4 represent the percentage of clones that have the C-nucleotide methylated at that specific CpG.
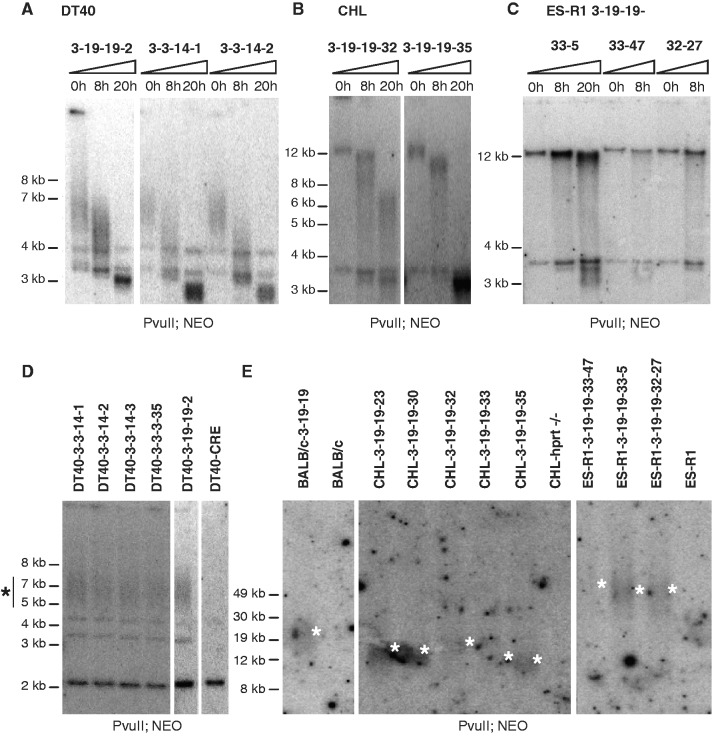
Figure 4.
TRF analyses of the L-HACs in the different genetic backgrounds. (A–C) Genomic DNA incubated with the BAL-31 exonuclease for 0, 8 or 20 h was digested with the PvuII restriction enzyme, blotted and probed for neomycin sequences. (A) The TRFs of the L-HAC present in the DT40 clones 3-19-19-2, 3-14-14-1 and 3-14-14-2 migrate between 5 kb and 8 kb (0 h). Increasing the BAL-31 incubation-time results in a decrease in TRF length. Typical BAL-31 degradation products are observed together with fragments inherent to the DT40 background. (B) A similar TRF pattern is observed upon BAL-31 incubation for CHL-clones 3-19-19-32 and 3-19-19-35. (C) Although in the ES-R1 background a decrease in TRF length is less pronounced, the typical degradation products for BAL-31 treatment are clearly present. (D–E) TRF analysis of genomic DNA, cut with PvuII and detected with the NEO probe. (D) TRF analysis of 5 DT40 clones (DT40-3-3-14-1, DT40-3-3-14-2, DT40-3-3-14-3, DT40-3-3-3-35, DT40-3-19-19-2) and the WT DT40-CRE line. TRFs consistently locate between 5 kb and 8 kb in all the tested clones. (E) PFGE of the TRFs of the linear HAC in the BALB/c, CHL and ES-R1 cells lines. TRFs for the BALB/c, CHL and ES-R1 cell lines were about 12.5–25 kb, 12–20 kb and 50 kb in size, respectively.
RESULTS
Generation and structural characterization of the L-HAC
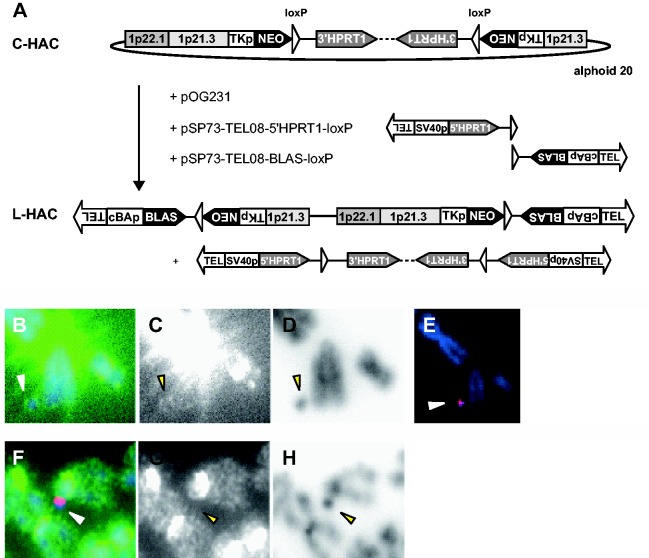
The circular HAC (32), was capped with artificial telomeres in the hprt−/− telomerase-positive hamster CHL cell line (E10B1) by CRE-loxP-mediated recombination. Two linear DNA-constructs carrying 800 bp of cloned (TTAGGG)n repeats as well as a positive selectable marker gene (BLAS or 5′HPRT1) and a loxP site were co-transfected with a transient CRE-expression plasmid pOG231 to the E10B1 hamster cell line that carries the circular HAC (Figure 1A). 123 HAT- and blasticidin-resistant clones were picked and analysed further with PCR using primer sets spanning the loxP recombination sites as well as sequencing of the amplification products (data not shown). The expected recombination had occurred in 90% of the clones. Subsequently, 15 clones were subjected to telomere FISH analysis (Figure 1B to D). Six clones displayed the expected four telomeric signals on the HAC upon hybridization of a PNA telomeric probe (Figure 1B to D) and rehybridization of the metaphases with an alphoid-20 DNA probe recognizing the HAC specifically (Figure 1E). To ensure that no hamster sequences had integrated into the HAC that could explain its stable maintenance as an independent chromosome hamster Cot-1 and alphoid-20 probes were co-hybridized to the metaphases. In five of the clones no hamster Cot-1 signals were detected on the L-HAC (Figure 1F and H). Finally, the structure of the five potentially linear HACs was further analysed via Southern blot. Although correct CRE/loxP recombinations were detected between the C-HAC and the linear targeting constructs, other recombination events, putatively not on the HAC were detected as well (data not shown). Hence, to separate the HAC from these background signals it was transferred to a BALB/c background by MMCT.
Figure 1.
Linearization of the C-HAC. (A) Plasmids pSP73-TEL08-5′HPRT1-loxP, pSP73-TEL08-BLAS-loxP and the CRE-expression plasmid pOG231 were co-transfected into the hprt−/− E10B1 hamster cell line carrying the C-HAC. CRE/loxP-induced recombination resulted in a linearized HAC with a blasticidin selection marker and reconstituted the HPRT1 minigene, which was excised from the HAC. The orientation of the loxP sites in the constructs pSP73-TEL08-5′HPRT1-loxP and pSP73-TEL08-BLAS-loxP is designed in such a way that it does not allow any other recombination. (B–D) FISH analysis on E10B1 metaphase spreads with a PNA telomeric probe (green) counterstained with DAPI (blue). HACs displayed the expected four telomeric signals. Strong telomere signals were present on the hamster chromosomes due to the presence of interstitial telomeric repeats in the CHL cell line. (E) Rehybridization of the metaphases with an alphoid-20 DNA probe (red). (F–H) FISH with hamster Cot1 DNA (green) and alphoid-20 DNA probes (red). (C and G) Detailed image showing the PNA telomeric signal/hamster Cot1 signal in grayscale. (D and H) Corresponding color-inverted, grayscale DAPI images. Arrowheads point to the HACs.
The mouse BALB/c 3T3 acceptor cells were telomerase positive by TRAP analysis and displayed telomere lengths ranging from 10 kb to 100 kb by terminal restriction fragment (TRF) analysis. These cells moreover do not carry long interstitial DNA-(TTAGGG)n arrays in their genome as hamster cells do thus simplifying the cytogenetic analysis of telomeres on the transferred chromosome. More than 200 BLAS resistant BALB/c clones grew out per MMCT experiment. To ensure clonality for downstream analyses six BALB/c subclones (3-3-14, 3-3-35, 3-19-19, 3-19-21, 14-9-5 and 14-9-31) were selected by FISH analysis (Figure 2A). TRF analyses were performed on these six subclones. For this, genomic DNA was digested with either PstI or PvuII, size separated by agarose gel electrophoresis, Southern blotted and probed for BLAS and NEO sequences, respectively (Figure 2B to D). Clones carrying a linear HAC were expected to display similar band patterns for both TRF analyses but with a band shift of 1.5 kb between the PstI and PvuII HAC-telomere restriction fragments. This was indeed the case for the band pattern detected above 8 kb in four (3-3-14, 3-3-35, 3-19-19 and 3-19-21) of the six BALB/c subclones analysed, suggesting that these contain a linear HAC with de novo seeded telomeres of at least 5.4 kb (Figure 2B to D). Further proof for HAC linearization was obtained by incubating the genomic DNA with a BAL-31 exonuclease before HAC-specific TRF analysis was performed. Decreased TRFs with increasing BAL-31 incubation-time were detected in the four BALB/c subclones demonstrating that the BLAS and NEO were indeed located at a chromosomal end (Figure 2E). Bands that represented interstitial DNA sequences served as internal controls for BAL-31 treatment. The BLAS probe labels an aspecific 5 kb PstI fragment of the host genetic background (Figure 2C), whereas the NEO probe detects a 3.6 kb PvuII fragment of the HAC adjacent to the TRF (Figure 2B and D). Neither of those bands reduced in size or intensity immediately following BAL-31 treatment (Figure 2E).
The anticipated subtelomeric structure of the L-HACs was further confirmed by Southern blot using EcoRI and the NEO probe (Figure 2F) as well as by PCR analyses (data not shown). A more profound structural analysis demonstrated that the linear HAC was capped with identical (sub-) telomeres (Figure 1A, Supplementary Methods S1, Supplementary Figure S1). Furthermore, the human 1p21.3 and 1p22 sequences that mapped to the circular HAC were also found on the L-HAC as tested by STS-PCR analysis suggesting that the genomic content had not altered upon linearization (Supplementary Figure S2).
Telomere length is genetic background dependent
To study the function of the artificial telomeres as well as the epigenetic alteration of its (sub-) telomere in different genetic backgrounds, we transferred the L-HAC from the mouse BALB/c genetic background to a chicken DT40-CRE cell line, hamster hprt−/− CHL cells and the mouse embryonic stem cell line ES-R1 by MMCT according to the scheme presented in Figure 3.
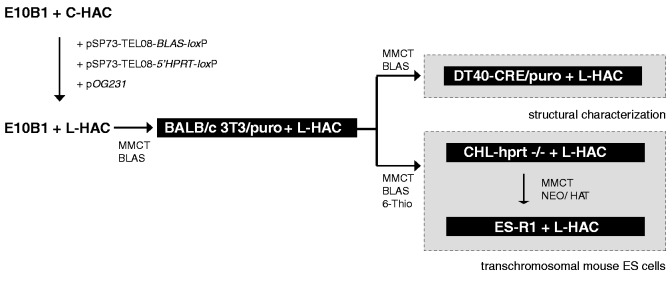
Figure 3.
Overview of the transfer of the L-HAC to four different genetic backgrounds. The circular HAC originally resided in the E10B1 cells. Linearization was acquired via the co-transfection of two linear constructs pSP73-TEL08-BLAS-loxP and pSP73-TEL08-5′HPRT1-loxP with the CRE-expression plasmid pOG231. Putative L-HACs were transferred to a BALB/c 3T3/puro cell line by MMCT and subsequently to the DT40-CRE cell line, where they were structurally characterized (Supplementary Data). In addition they were relocated from the BALB/c cells to an hprt−/− CHL cell line and next to the ES-R1 cell line. The latter was applied to generate transchromosomal mice. As such the linear HAC is present in four unique genetic backgrounds (gray rectangles) that can be used to study TPE and (sub-) telomeric alteration at a single telomeric end. Selection criteria are displayed.
TRF analysis preceded by time-lapsed BAL-31 incubation displayed the expected decrease in TRF length, confirming that the linear structure of the L-HAC was maintained in all 5 DT40 (Figure 4A), 6 CHL (Figure 4B) and 4 ES-R1 (Figure 4C) clones tested.
Pulsed-field gel electrophoresis (PFGE) of HAC-TRFs revealed a background-dependent variability in telomere length while HAC-TRFs of similar lengths were detected among different clones from the same genetic background. The BALB/c donor cells contained HACs with telomere lengths varying from 9 kb to 21.5 kb (Figure 4E). Upon shuttling to the chicken DT40-CRE acceptor cells HAC-telomeres had shortened significantly, despite high telomerase activity. PvuII-digested DT40-CRE genomic DNA yielded HAC-TRFs between 5 kb and 8 kb in length (Figure 4D), indicative of telomere lengths between 1.5 kb and 4.5 kb. The original telomere length of 9–21.5 kb on the shuttled chromosomes was reduced to equal telomere lengths in all five independent DT40 clones. This suggests that telomere length homeostasis is strictly regulated in the chicken DT40-CRE cells. The latter is reinforced by the fact that the five DT40-CRE clones received their L-HAC from three different donor BALB/c subclones and via three separate MMCT experiments (3-3-14, 3-3-35 and 3-19-19). In contrast, L-HAC telomeres measured 12–20 kb in the CHL hamster cells and ∼50 kb in the ES-R1 cells. As such, an increase in telomere length of at least 35 kb was induced upon transfer of the L-HACs from their respective donor CHL cell line to the ES-R1 acceptors (Figure 4E). Taken together, these results indicate that telomere length of the same chromosome is adjusted in a genetic background-specific manner rather than it is maintained in the recipient telomerase-positive cell.
Subtelomeric chromatin of the L-HAC alters in a genetic background-dependent manner
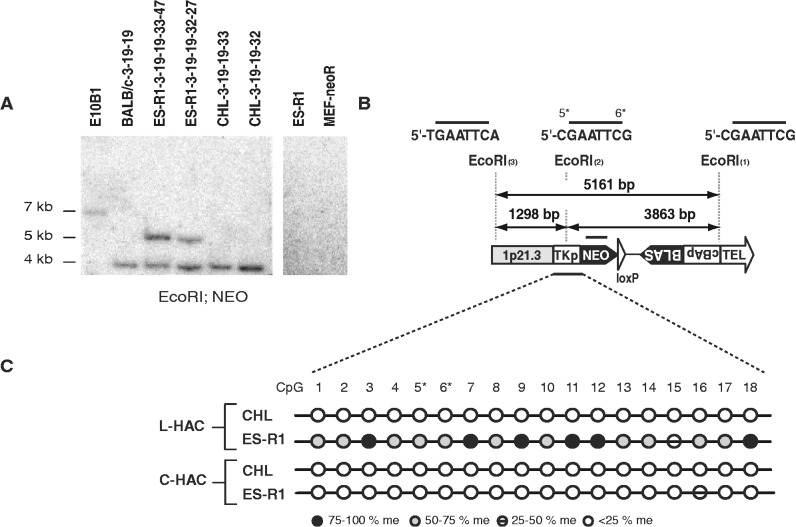
Besides telomere length, the subtelomeric chromatin state was altered by the genetic background as well. L-HAC subtelomeric DNA was analysed by Southern blot of EcoRI-digested DNA and NEO sequences as a probe. Whereas the L-HAC present in the BALB/c and CHL clones resulted in the expected 3.9 kb subtelomeric fragment, all analysed ES-R1 clones revealed an aberrant band pattern that was not due to the ES-R1 of MEF-neoR genetic background (Figure 5A and B). The EcoRI site that was located 3870 bp upstream of the telomeric repeat (EcoRI(2), 5′-CGAATTCG) (Figure 5B) contained 2 CpGs (5* and 6*) that when methylated could explain the unexpected Southern blot band pattern shown in Figure 5A. Bisulfite sequencing of 2 ES-R1 clones (ES-R1-3-19-19-33-47 and ES-R1-3-19-19-32-27) as well as their respective donor cells (CHL-3-19-19-33 and CHL-3-19-19-32) confirmed this hypothesis. To quantify the level of methylation per CpG-site, DNA-amplicons obtained after bisulfite DNA-treatment were first cloned in the pGEM-T cloning vector and subsequently, for each PCR product 12 unique clones were sequenced. In the ES-cells, 17 of the 18 subtelomeric CpGs were found to be methylated in 50–75% of the cells, explaining the occurrence of the 5.1 kb fragment. In the CHL cells on the other hand the equivalent CpGs were almost never methylated (<25% of the cells) (Figure 5C), hence the CHL clones always displayed the expected 3.9 kb EcoRI fragment. Bisulfite sequencing of the same methylation-sensitive EcoRI-locus on the C-HAC in two independent mouse ES-R1 acceptor clones as well as in the hamster E10B1 donor cell line indicated that this site remained hypo-methylated on the C-HAC (Figure 5C). This strongly suggests that the de novo methylation of the subtelomeric CpG dinucleotides on the L-HAC is directly linked to the neighboring telomere rather than a trait inherent to the genetic background of the ES-R1 cells. In addition, we analysed via bisulfite sequencing the subtelomeric region of the L-HAC in the BALB/c and DT40 cells. The level of L-HAC subtelomeric CpG methylation, deduced from the chromatograms of the Genetic Analyzer, was similar in these genetic backgrounds when compared to the L-HAC in the CHL background (Supplementary Figure S3).
Figure 5.
Analysis of the L-HAC subtelomeric region. (A) Southern blot analysis of genomic DNA derived from E10B1 cells with the C-HAC and BALBc, ES-R1 and CHL cells carrying the L-HAC, digested with EcoRI and probed for NEO. In addition to the 3.9 kb expected fragment an aberrant band of 5.1 kb was observed in clones ES-R1-3-19-19-32-27 and ES-R1-3-19-19-33-47. The 5.1 kb fragment was never detected in the wild-type ES-R1 cells or in the MEF-neoR cells. (B) Schematic overview of the subtelomeric region on the L-HAC. The latter encloses three EcoRI sites [(1), (2) and (3)] spaced 1298 bp [(2) and (3)] and 3863 bp [(1) and (2)] apart. EcoRI(2) is prone to CpG methylation, both at the C-nucleotide 5′ from its recognition site as on its terminal C-nucleotide (5* and 6*). EcoRI(3) contains no CpGs. Depending on the methylation status of EcoRI(2), Southern blot analysis with the NEO or BLAS probes will result in a fragment of either 3.9 kb (EcoRI(2) unmethylated) or 5.1 kb (EcoRI(2) methylated). The black bar above the NEO cassette represents the recognition site for the NEO probe. (C) Bisulfite treatment of an identical subtelomeric region from both the CHL and ES-R1 cells and for both the circular and the linear HAC. The selected region contains both the TK promotor and part of the Neomycin resistance gene and is represented by a black bar at the bottom of (B). The area contains 18 CpGs among which those of EcoRI(2) (5* and 6*). The methylation status of each of the CpGs is depicted by a colored circle according to the legend. Black circle: CpG methylated in >75% of the cells; dark gray circle: CpG is methylated in 50–75% of the cells; strikethrough circle: CpG methylated in 25–50% of the cells; white circle: CpG methylated in <25% of the cells.
We conclude that both telomere length and subtelomeric chromatin, measured by DNA CpG methylation, are altered in a genetic background-specific manner.
TPE is genetic background dependent
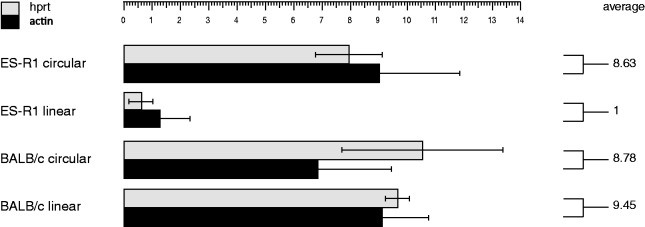
The influence of the telomere on the expression of the subtelomeric neomycin resistance gene was investigated. The expression of NEO present on the L-HAC in the ES-R1 cells was downregulated 9-fold compared to its circular counterpart in the same background and 9.5-fold compared to the expression levels from the L-HAC in the BALB/c background (Figure 6). NEO expression levels did not differ between BALB/c cells carrying the circular or the linear HAC.
Figure 6.
Relative expression levels of the neomycin resistance gene from the circular and linear HAC in the BALB/c and ES-R1 cells. NEO expression levels are normalized to the expression of housekeeping genes Hprt and Actb and are calculated as the relative expression level per L-HAC. The average expression level per background is depicted. Data are presented such that the average expression level of the L-HAC in the ES-R1 cells is 1. SDs from three independent clones for each background are shown.
These data strongly suggest that TPE acting on the same chromosome is genetic background dependent. The drop in NEO expression levels is consistent with the de novo methylation of the subtelomeric region of the L-HAC in the mouse ES-R1 cells.
Linear and circular artificial chromosomes as tools for in vivo functional studies in mice
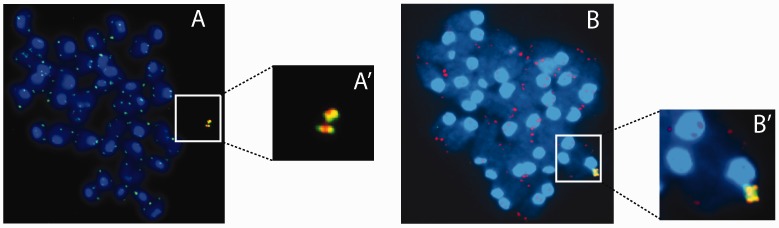
To analyse telomere function in vivo, transchromosomal mice carrying the L-HAC were generated. ES-cells from clone 3-19-19-33-47 were injected into Swiss blastocysts. This ES-clone displayed the highest mitotic stability (0.045% loss rate) upon 40 population doublings in absence of any selective pressure (data not shown). FISH analysis with the alphoid-20 probe together with a telomere probe revealed the expected 4 telomeric signals at the L-HAC and demonstrated that it was maintained as an individual chromosome (Figure 7A and A′). A single male chimera with 10% chimerism was obtained. Backcrossing this mouse to a WT Swiss female enabled the transfer of the L-HAC to 6% of the first generation (F1) offspring. Similar to the transgenic mice carrying the C-HAC (32) the obtained F1-L-HAC+-mice were backcrossed to both the NMRI and C57Bl/6 mouse genetic backgrounds.
Figure 7.
Telomere co-FISH on metaphase spreads derived from L-HAC+ ES-cells from clone 3-19-19-33-47 (A) and from mouse tail fibroblasts (F1-2 mice) (B). The HAC was identified via the alphoid-20 probe, labeled either in red (A) or green (B) whereas a telomeric probe, labeled in the complementary color, was used to screen for the presence of telomeric sequences. A′ and B′ display a magnification of the regions indicated in A and B, which display the complete metaphase. In panels A and A′ sister chromatids already separated whereas in panels B and B′ they are still connected.
FISH analysis of tail fibroblast metaphases derived from F1-L-HAC+-mice using a combination of the alphoid-20 and telomere probes confirmed that also in vivo the L-HAC was maintained as an independent chromosome with the expected telomeric signals (Figure 7B and B′). Crossing six F2 males with WT NMRI females for a period of 6 months demonstrated a male germline transmission efficiency of 34% (Supplementary Table S2).
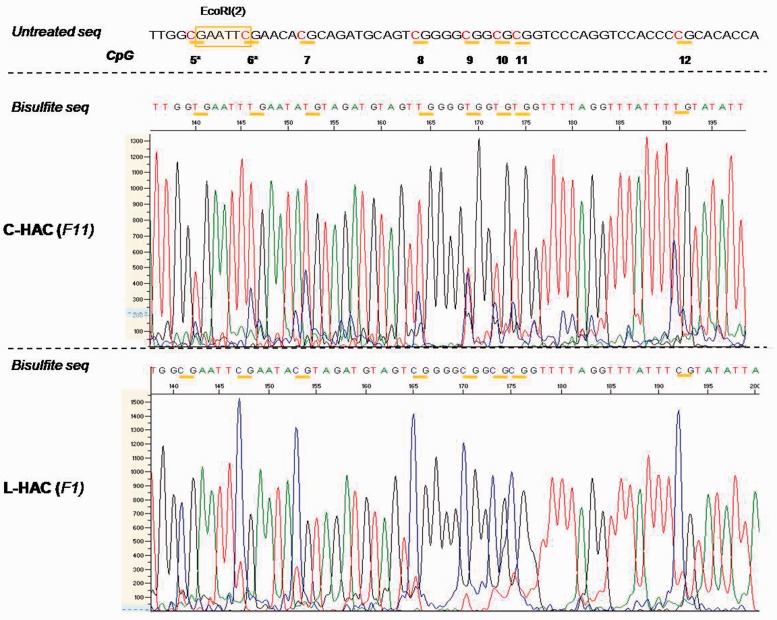
To study the artificial telomeres in vivo we performed TRF analysis on genomic DNA isolated from mouse lung tissue using PstI and the BLAS probe. Four littermates displayed telomeres of similar length between 30 kb and 70 kb (Supplementary Figure S4). Bisulfite sequencing of tail fibroblast DNA derived from two F1-L-HAC+-mice as well as from three mice carrying the C-HAC (backcrossed to the NMRI and 129/Sv backgrounds for 15 and 11 generations, respectively) was performed to check the methylation status of the HAC-subtelomere. All analysed CpG sites were fully methylated in mice carrying the L-HAC whereas in mice carrying the C-HAC the methylation status at each CpG was between 25% and 50% at most (Figure 8).
Figure 8.
In vivo bisulfite sequencing data for the C- and L-HAC. Analogous to the in vitro analysis, a region of 60 bp surrounding EcoRI(2) was analysed. All the CpGs are marked by horizontal yellow bars. They are termed 5–12 and correspond to CpGs 5–12 in Figure 5. All C nucleotides that precede a G nucleotide are highlighted in red. Genomic DNA of the mice containing the C-HAC was extracted from mouse tail fibroblast derived from mice backcrossed to the NMRI background for 11 generations. For the L-HAC, solely mice from generation 1 (F1) were available. The graphs represent screenshots from the chromatogram obtained from the 3130x Genetic Analyzer (Life Technologies). The scale on the left (0–1300 and 0–1500) reflects the peak intensity for each nucleotide readout. The CpG′s in the subtelomeric region of the L-HAC are clearly hypermethylated compared to their counterparts on the circular chromosome. Untreated seq: the untreated wild-type sequence at that specific locus; bisulfite seq: the sequence that was obtained upon bisulfite treatment and sequencing PCR; F11: the 11th generation of offspring; F1: the 1st generation of offspring.
DISCUSSION
We developed a tool to study (sub-) telomere architecture and TPE on an identical chromosome in different genetic backgrounds. For the first time, firm evidence is provided that telomere length, subtelomeric chromatin and TPE are mainly driven by features inherent to the genetic background and can vary extensively in different telomerase-positive genetic contexts. Furthermore, this tool allows the monitoring of the modulation of (sub-) telomeres upon transfer of a chromosome between different genetic backgrounds.
A L-HAC was engineered from a C-HAC (32) by CRE-loxP-mediated linearization and de novo telomere seeding in a telomerase-positive cell line (E10B1). Both the L-HAC and the C-HAC can be shuttled between different cell lines of different species by MMCT and both have been demonstrated to efficiently migrate through the male and the female mouse germline enabling in vivo studies as well.
Upon transfer of the L-HAC from the BALB/c 3T3 cell line to the chicken DT40-CRE line, the majority of HAC-telomeric lengths decreased from 9–21.5 kb to 1.5–4.5 kb despite comparable levels of telomerase activity in both cell lines. Although the DT40 clones were created from three different BALB/c donor clones HAC-telomeres were reduced to a similar length in all five DT40 clones, strongly suggesting that telomere length adaptation is a recipient-cell-specific process independent from the MMCT technique. In contrast, HAC-telomeres were significantly elongated in the mouse ES-R1 genetic context when shuttled from the hamster CHL cells. HAC-TRFs measured ∼50 kb in the mouse ES-R1 cells whereas they were 12–20 kb in length in the donor CHL cells. Genetic-dependent adaptation of telomere length has been observed before on mouse chromosomes (37). Together these results prove that telomere length is not chromosome-specific per se, but that it most likely is determined by the genetic context. Furthermore, they demonstrate that the L-HAC is a valid tool for studying cell-specific telomere structure on an identical chromosomal context.
A number of studies in yeast as well as in mammalian cells have shown that telomeres have the ability to silence nearby genes. Since TPE is dependent on telomere length (28) and on the distance of a particular gene relative to a telomere (30), TPE has been suggested to play a role in a variety of human syndromes through the silencing of genes relocated to telomeres. In addition, TPE might be implicated in a mechanism to control subtelomeric gene expression throughout the replicative life-span of cells (38) as well as in embryo development (12). However, the exact function of TPE remains elusive as its molecular mechanism. TPE involves changes in subtelomeric chromatin conformation but it is hypothesized that different genetic backgrounds have different TPE molecular mechanisms (30). For instance, TPE in normal mouse cells is associated with DNA-methylation and complete gene silencing (12), which has not been observed for TPE in human tumor cells (28,31). Similarly, TPE in mouse ES-cells could not be reversed by trichostatin A treatment as was the case in human tumor cell lines (12). Caveat for the hypothesis however, is that these studies were performed in different species and on different chromosomes with potentially different local chromatin structures. The latter has been shown to influence TPE (29,39). By analysing TPE from an identical chromosome in different genetic contexts, we provided in this study significant evidence that telomere length and subtelomeric DNA methylation, are indeed genetic context dependent.
In the mouse ES-R1 cells, the expression levels of the aminoglycoside phosphotransferase gene, conferring resistance to G418, dropped by 90% when expressed subtelomerically from the L-HAC compared to its expression levels from a C-HAC. This observation confirmed that the developed L-HAC allowed the study of TPE. Furthermore, our observations were in line with the data of Pedram et al. (2006), who showed that transgenes, integrated in close proximity to different telomeres, were expressed at very low levels compared to the same transgenes integrated at interstitial sites (12). Subsequent analyses of the expression level of the neomycin resistance gene from the L-HAC in other cell lines are fully in line with the hypothesis that TPE and subtelomeric chromatin are genetic background dependent. Upon transfer of the L-HAC to ES-R1 cells and in vivo in mice, the HAC-subtelomeric DNA was de novo methylated. Cytosine methylation was not observed for the same locus on the C-HAC in the same ES-R1 background confirming that the de novo subtelomere DNA methylation was instigated by the HAC-telomere. However, the HAC-telomere did not invoke de novo cytosine methylation in the genetic context of other mouse (BALB-c) or chicken (DT40) cell lines. Although we did not detect extensive subtelomere DNA-methylation in chicken DT40-CRE cell lines, Rincon-Arano et al. (2007) showed that subtelomeric DNA methylation is an important player in TPE in chicken HD3 cells (39). This again emphasizes that important mechanistic differences in TPE exist. By analysing TPE and subtelomeric chromatin of the same chromosomal locus in different genetic contexts we provide firm evidence that not only telomere length, but also TPE and subtelomere chromatin are dictated by the genetic context for a specific chromosomal end-cap. Our conclusion is in line with the report of Gao et al. (2008) who noticed that telomeric transgenes were differentially expressed in mouse embryonic stem cells when compared to other tissues of the transgenic mice (40).
As the subtelomeric constructs for NEO and BLAS resistance gene expression are tail-to-tail on the L-HAC we cannot rule out that the quantitation of NEO mRNA expression is confounded by potential read-through transcripts of the neighboring BLASR gene. Such hypothetical read-through transcripts could have an inhibitory role on NEO transcription due to mRNA binding, or could artificially augment the amounts of NEO cDNA produced by reverse transcription. However, if this would be the case, read-through transcription from the BLASR expression construct should be differentially regulated in mouse ES and BALB/c cells. Although rather unlikely we cannot neglect this hypothesis for which however, no hard experimental data are yet available.
The artificial chromosome we developed allows to further dissect the molecular mechanisms driving telomere length homeostasis in an unprecedented manner. Appropriate telomere metabolism appears to be linked to an epigenetic imprint present on the telomeric as well as on the subtelomeric chromatin. Mouse embryonic stem cells and mouse embryonic fibroblasts null for Suv39h1 and Suv39h2 histone methyltransferases have elongated telomeres on random sets of chromosomes, but maintained telomeric protective end function (25). Similarly, mouse embryonic fibroblasts deficient for Rb1 (retinoblastoma 1), Rbl1 (retinoblastoma-like 1) and Rbl2 (retinoblastoma-like 2)—also known as TKO cells—have elongated telomeres that retain their end-capping function (26). Additionally, subtelomeric DNA methylation also seems to have an important role. Mouse embryonic stem cells deficient for DNMT1, or both DNMT3a and DNMT3b showed elongated telomeres but increased recombinational processing (27). However, caveat in all of these studies is that the mutation does not specifically alter the (sub-) telomere chromatin conformation but affects the entire epigenetic profile of the genome. Hence, the telomere aberrations seen in those mutated cells can also be explained by the deregulated expression of, e.g., proteins involved in telomere length processing. By inserting the lac I operon sequences in the subtelomeric region of the L-HAC and using chromatin remodelers fused to the lac I repressor, which specifically recognizes the lac I operon sequences, we aim to site-specifically alter the HAC-subtelomeric chromatin conformation and dissect the effect on HAC-telomere length homeostasis as well as its protective end function in different unique genetic backgrounds.
Our data clearly demonstrate that artificial chromosomes are not only useful transgene vehicles, but are also important tools for the study of chromosome structure and function. The L-HAC will give us the unique opportunity to further clarify telomere biology. Our data furthermore indicate that one has to be cautious with the use of HACs as CVs given the potential silencing of subtelomeric genes. This is the first report that describes the targeted linearization of a C-HAC and that reports on the processing of de novo telomeres in different genetic backgrounds after MMCT. We found two processes that are active on telomeres of MMCT-transferred chromosomes: (i) telomere length adaptation (reduction or elongation); and (ii) de novo subtelomeric DNA methylation together with TPE alteration.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4, Supplementary Method 1 and Supplementary Reference [43].
FUNDING
Innovation by Science and Technology (IWT) fellowship (to T.V.); ‘Vlaams Instituut voor Biotechnologie’ (VIB) [young investigator award to T.V.]; Research Foundation Flanders (FWO) [fellowship to A.W.]. Funding for open access charge: KU Leuven.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol. Ther. 2011;19:1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Luz S, az-Nido J. Prospects for the use of artificial chromosomes and minichromosome-like episomes in gene therapy. J. Biomed. Biotechnol. 2010;2010:1–16. doi: 10.1155/2010/642804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimes BR, Monaco ZL. Artificial and engineered chromosomes: developments and prospects for gene therapy. Chromosoma. 2005;114:230–241. doi: 10.1007/s00412-005-0017-5. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl Acad. Sci. USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voet T, Liebe B, Labaere C, Marynen P, Scherthan H. Telomere-independent homologue pairing and checkpoint escape of accessory ring chromosomes in male mouse meiosis. J. Cell Biol. 2003;162:795–807. doi: 10.1083/jcb.200305065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm W, de LT. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 9.Scherthan H. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 10.Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol. Life Sci. 2007;64:117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 12.Pedram M, Sprung CN, Gao Q, Lo AW, Reynolds GE, Murnane JP. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol. Cell Biol. 2006;26:1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bekaert S, Derradji H, Baatout S. Telomere biology in mammalian germ cells and during development. Dev. Biol. 2004;274:15–30. doi: 10.1016/j.ydbio.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 15.Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, Baird DM. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- 16.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 17.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 18.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 19.Martinez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 20.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 22.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 2005;14:R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 23.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 24.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Cao M, Gonzalo S, Dean D, Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat. Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 28.Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 29.Ning Y, Xu JF, Li Y, Chavez L, Riethman HC, Lansdorp PM, Weng NP. Telomere length and the expression of natural telomeric genes in human fibroblasts. Hum. Mol. Genet. 2003;12:1329–1336. doi: 10.1093/hmg/ddg139. [DOI] [PubMed] [Google Scholar]
- 30.Kulkarni A, Zschenker O, Reynolds G, Miller D, Murnane JP. Effect of telomere proximity on telomere position effect, chromosome healing, and sensitivity to DNA double-strand breaks in a human tumor cell line. Mol. Cell Biol. 2010;30:578–589. doi: 10.1128/MCB.01137-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voet T, Vermeesch J, Carens A, Durr J, Labaere C, Duhamel H, David G, Marynen P. Efficient male and female germline transmission of a human chromosomal vector in mice. Genome Res. 2001;11:124–136. doi: 10.1101/gr.159901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voet T, Schoenmakers E, Carpentier S, Labaere C, Marynen P. Controlled transgene dosage and PAC-mediated transgenesis in mice using a chromosomal vector. Genomics. 2003;82:596–605. doi: 10.1016/s0888-7543(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 34.Arakawa H, Lodygin D, Buerstedde JM. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeesch JR, Falzetti D, Van Buggenhout G, Fryns JP, Marynen P. Chromosome healing of constitutional chromosome deletions studied by microdissection. Cytogenet. Cell Genet. 1998;81:68–72. doi: 10.1159/000014991. [DOI] [PubMed] [Google Scholar]
- 37.Zhu LX, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl Acad. Sci. USA. 1998;95:8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright WE, Shay JW. Telomere positional effects and the regulation of cellular senescence. Trends Genet. 1992;8:193–197. doi: 10.1016/0168-9525(92)90232-s. [DOI] [PubMed] [Google Scholar]
- 39.Rincon-Arano H, Furlan-Magaril M, Recillas-Targa F. Protection against telomeric position effects by the chicken cHS4 beta-globin insulator. Proc. Natl Acad. Sci. USA. 2007;104:14044–14049. doi: 10.1073/pnas.0704999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Q, Reynolds GE, Wilcox A, Miller D, Cheung P, Artandi SE, Murnane JP. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair (Amst) 2008;7:1233–1249. doi: 10.1016/j.dnarep.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.