Abstract
SUMO-targeted ubiquitin ligases (STUbLs) recognize sumoylated proteins as substrates for ubiquitylation and have been implicated in several aspects of DNA repair and the damage response. However, few physiological STUbL substrates have been identified, and the relative importance of SUMO binding versus direct interactions with the substrate remains a matter of debate. We now present evidence that the ubiquitin ligase Rad18 from Saccharomyces cerevisiae, which monoubiquitylates the sliding clamp protein proliferating cell nuclear antigen (PCNA) in response to DNA damage, exhibits the hallmarks of a STUbL. Although not completely dependent on sumoylation, Rad18’s activity towards PCNA is strongly enhanced by the presence of SUMO on the clamp. The stimulation is brought about by a SUMO-interacting motif in Rad18, which also mediates sumoylation of Rad18 itself. Our results imply that sumoylated PCNA is the physiological ubiquitylation target of budding yeast Rad18 and suggest a new mechanism by which the transition from S phase-associated sumoylation to damage-induced ubiquitylation of PCNA is accomplished.
INTRODUCTION
Ubiquitin and the small ubiquitin-related modifier (SUMO) are small post-translational protein modifiers that alter the properties of their targets by affecting their activities, interactions, stabilities or intracellular localization (1). Despite non-overlapping conjugation machineries, there is extensive cross-talk between the two modification systems (2). A growing number of proteins are being identified as targets of both ubiquitin and SUMO. One example is IκBα, an inhibitor of the transcriptional activator NF-κB. IκBα is either ubiquitylated or sumoylated on the same lysine, but with opposite consequences for protein stability (3). A rather different relationship applies to a class of ubiquitin ligases (E3s) that recognize sumoylated targets as substrates for ubiquitylation and have been implicated in various aspects of DNA repair and genome maintenance (4–9). These SUMO-targeted ubiquitin ligases (STUbLs) are RING finger E3s harbouring conserved SUMO interaction motifs (SIMs), which consist of a hydrophobic core flanked by several acidic residues (10). By interacting predominantly with poly-SUMO chains, they mediate ubiquitylation of the SUMO moieties themselves, as well as the proteins to which these are attached. Hence, sumoylation can serve as a signal for subsequent ubiquitylation, often followed by proteasome-mediated degradation. Despite a profound influence on the homeostasis of SUMO conjugates in the cell, few physiological STUbL substrates have been identified to date.
Post-translational modifications of the budding yeast sliding clamp protein, proliferating cell nuclear antigen (PCNA), present a unique example of how ubiquitin and SUMO cooperate in the context of DNA replication and repair (11). In response to replication-stalling DNA damage, PCNA is monoubiquitylated at a highly conserved lysine, K164, by the E2–E3 complex Rad6–Rad18 (12). This promotes the recruitment of a class of specialized polymerases capable of using damaged DNA as a template for translesion synthesis (13–15). Extension to a polyubiquitin chain activates an error-free pathway of damage bypass that likely involves template switching (12). In contrast to ubiquitylation of PCNA, which is common to all eukaryotes, modification by SUMO appears to be less prevalent. In Saccharomyces cerevisiae, the SUMO E3 Siz1 promotes attachment of the SUMO homologue Smt3 mainly to K164 (12). K127 is modified to a lesser degree in a Siz1-independent manner (16). Sumoylation of budding yeast PCNA during S phase prevents unscheduled recombination events by enhancing the binding of an anti-recombinogenic helicase, Srs2 (17,18). Hence, the modification enables ubiquitin-dependent damage bypass by blocking alternative processing pathways.
The cooperation between SUMO and ubiquitin in orchestrating lesion bypass raises the question of how the transition from the S phase-associated sumoylated form of PCNA to the damage-induced ubiquitylated form is accomplished. The ubiquitin E3 Rad18, which is rate limiting for both mono- and polyubiquitylation of PCNA (19,20), is likely to play a critical role in this process. Intriguingly, a previous report suggested that Rad18 physically interacts with the SUMO E2 Ubc9 (12). We have now identified a SIM in Rad18 that strongly stimulates its ubiquitin ligase activity towards the sumoylated form of PCNA. We propose that budding yeast Rad18 is adapted to act primarily on sumoylated PCNA and discuss the implications for the switch between the two modifications in response to DNA damage.
MATERIALS AND METHODS
Yeast strains
All experiments involved the use of isogenic strains. HisPOL30, siz1Δ, rad18Δ and the lysine mutants of POL30 have been described previously (15,19). The 9myc epitopes were appended by a polymerase chain reaction strategy to RAD18 wildtype (WT) or SIM* alleles that had been inserted into the URA3 locus of rad18Δ on integrative vectors derived from YIplac211, bearing the RAD18 promoter.
Proteins
Recombinant budding yeast HisPCNA, HisUb-PCNA, Replication Factor C (RFC), Rad6, HisAos1-Uba2His, Ubc9His, HisSmt3 and HisSiz1(1-508) and human Rad6–HisRad18 complex were produced in Escherichia coli and purified as previously described (16,20–25). The yeast Rad6–HisRad18 complex was produced by over-expression in S. cerevisiae and purified as described previously (20). The Rad18 SIM* mutation (L139A, I141A and V142A) was introduced by polymerase chain reaction. HisRad18(1-255) was produced in E. coli strain BL21-Rosetta(DE3) (Stratagene) and purified by Ni–NTA affinity, anion exchange and gel filtration chromatography. Human HisUba1 was purchased from BioMol and ubiquitin was from Sigma.
The N-terminal fusion of Smt3 to yeast PCNA (HisSmt3-PCNA) was constructed by combining Smt3 lacking the C-terminal GG motif in frame with the PCNA open reading frame in the vector pQE-POL30 (Qiagen). The protein was purified as HisPCNA. Human HisPCNA was produced in E. coli from plasmid pRSF-PCNA (a gift from S. Petersen-Mahrt) and purified by Ni–NTA affinity, anion exchange and gel filtration chromatography. A linear fusion of SUMO-2 to this construct was generated by inserting the sequence of SUMO-2 lacking the C-terminal GG motif between the His6 tag and the PCNA sequence within the same vector. The protein was purified as human HisPCNA. Glutathione S transferase (GST) fusion proteins used for interaction assays (GST, GSTSmt3, GSTPCNA and GSTUbc9) were expressed from pGEX-4T-1 or pGEX-2TK (GE Healthcare) and purified by glutathione affinity chromatography.
In vitro protein–protein interaction assays
In vitro interaction assays were performed in phosphate-buffered saline with 0.05% Triton X-100 by immobilizing the relevant GST-tagged protein on glutathione Sepharose for 1 h at 4°C. After washing the beads, the respective binding partner was added for a further incubation of 90 min at 4°C. The beads were then washed three times with buffer, and bound material was eluted by boiling in sodium dodecyl sulphate (SDS) sample buffer before analysing the samples by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting using an anti-His antibody (Sigma).
In vitro protein modification assays
All protein names are given without specifying His6 tags. Unless otherwise noted, 10 µl reactions were set up in a buffer containing 40 mM HEPES, pH 7.4; 50 mM NaCl; 8 mM magnesium acetate and 1 mM adenosine triphosphate, incubated at 30°C for 1 h, terminated by addition of SDS sample buffer and denatured at 95°C for 3 min. In vitro sumoylation of Rad18 was analysed in reactions containing 200 nM Aos1-Uba2, 50 nM Ubc9, 20 nM Siz1(1-508), 8 µM Smt3, 2.5 nM nicked plasmid DNA and 500 nM Rad6–Rad18 complex, unless otherwise noted. Products were analysed by 8% SDS-PAGE and western blotting with polyclonal anti-Rad18 antibody. Modification of Rad6 in the same reactions was analysed by 12% SDS-PAGE and western blotting with polyclonal anti-Rad6 antibody. PCNA monoubiquitylation reactions were set up with 50 nM Uba1, 200 nM Rad6–Rad18 complex, 1 µM ubiquitin, 20 nM RFC, 3 nM nicked plasmid DNA and 50 nM PCNA trimer as described previously (20). Products were analysed by 10% SDS-PAGE and western blotting with polyclonal anti-PCNA antibody. The same conditions were applied to modification of human PCNA or SUMO-PCNA on DNA. In the absence of DNA, human PCNA was used at 500 nM. Peptide inhibition assays were performed in the presence of 30, 60 and 600 µM peptide as indicated. Peptide sequences (26) were KVDVIDLTIE (WT), KVDVADLTIE (* = mutated) and VKDVLTDEIE (sc = scrambled). Where sumoylation and ubiquitylation of yeast PCNA were analysed simultaneously, reactions additionally contained 50 nM Aos1-Uba2, 50 nM Ubc9, 8 µM Smt3 and variable amounts of Siz1(1-508) as indicated. For quantification, chemiluminescence signals were recorded on a Fuji LAS-3000 imager, and averages and standard deviations were determined from two to three independent experiments.
Two-hybrid analysis
Two-hybrid assays for detection of in vivo protein–protein interactions were performed in the reporter strain PJ69-4 A as described previously (27), using the Gal4 system. Constructs for Rad18, PCNA, PCNA* and Smt3-PCNA* have been described previously (27–29), and those for Ubc9 and Smt3 were constructed analogously by insertion of the open reading frame. Protein levels were analysed in total extracts by western blotting, using antibodies against the Gal4 activation and DNA-binding domains (Clontech).
In vivo analysis of yeast proteins and damage sensitivities
Total cell lysates were prepared under denaturing conditions as described previously (12,15). Rad189myc (WT or SIM*) was detected by western blotting using a monoclonal anti-myc antibody (Santa Cruz Biotechnology). For detection of PCNA ubiquitylation, cultures of appropriate HisPOL30 (WT or K127R) strains were treated with 0.02% methyl methanesulfonate (MMS) for 90 min where indicated, or a time course analysis was performed after addition of 0.025% MMS. Cells were lysed under denaturing conditions, HisPCNA was isolated by Ni–NTA affinity chromatography and conjugates were detected by anti-ubiquitin western blotting. Sensitivities towards MMS and ultraviolet (UV) (254 nm) were determined as previously described (15).
RESULTS
A SIM in Rad18 promotes covalent and non-covalent interactions with SUMO
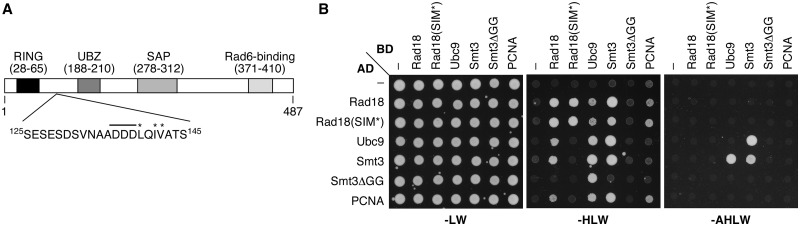
We noticed a sequence within budding yeast Rad18 that conforms to the consensus of a SIM, spanning amino acids 136–142 (Figure 1A). To determine whether Rad18 indeed binds to Smt3, we analysed the interaction in the two-hybrid system (Figure 1B). The assay confirmed previously reported interactions of Rad18 with itself, with Ubc9 and with PCNA (12,27). In addition, we found an association of Rad18 with full-length Smt3. Mutation of the hydrophobic core residues within the putative SIM to alanine (L139A, I141A and V142A, named SIM*) resulted in a loss of the interaction in one of the two orientations. Interactions with Ubc9 and with PCNA were affected in a similar manner. Surprisingly, truncation of the C-terminal diglycine (GG) motif in Smt3 abolished most interactions involving Smt3, suggesting that covalent rather than non-covalent interactions gave rise to these signals.
Figure 1.
Identification of a SUMO-interacting motif in Rad18. (A) Domain structure of Rad18 from S. cerevisiae. The acidic region and the hydrophobic core of the SIM are highlighted by a bar and asterisks, respectively. (B) Yeast two-hybrid assay showing interactions between Rad18, Ubc9, Smt3 and PCNA. Reporter constructs, based on Gal4 activation (AD) and DNA-binding (BD) domains, confer growth on selective medium (-LW). Positive interactions were scored by growth on medium lacking histidine (-HLW) or, for stronger interactions, histidine and adenine (-AHLW). SIM*: L139A, I141A and V142A.
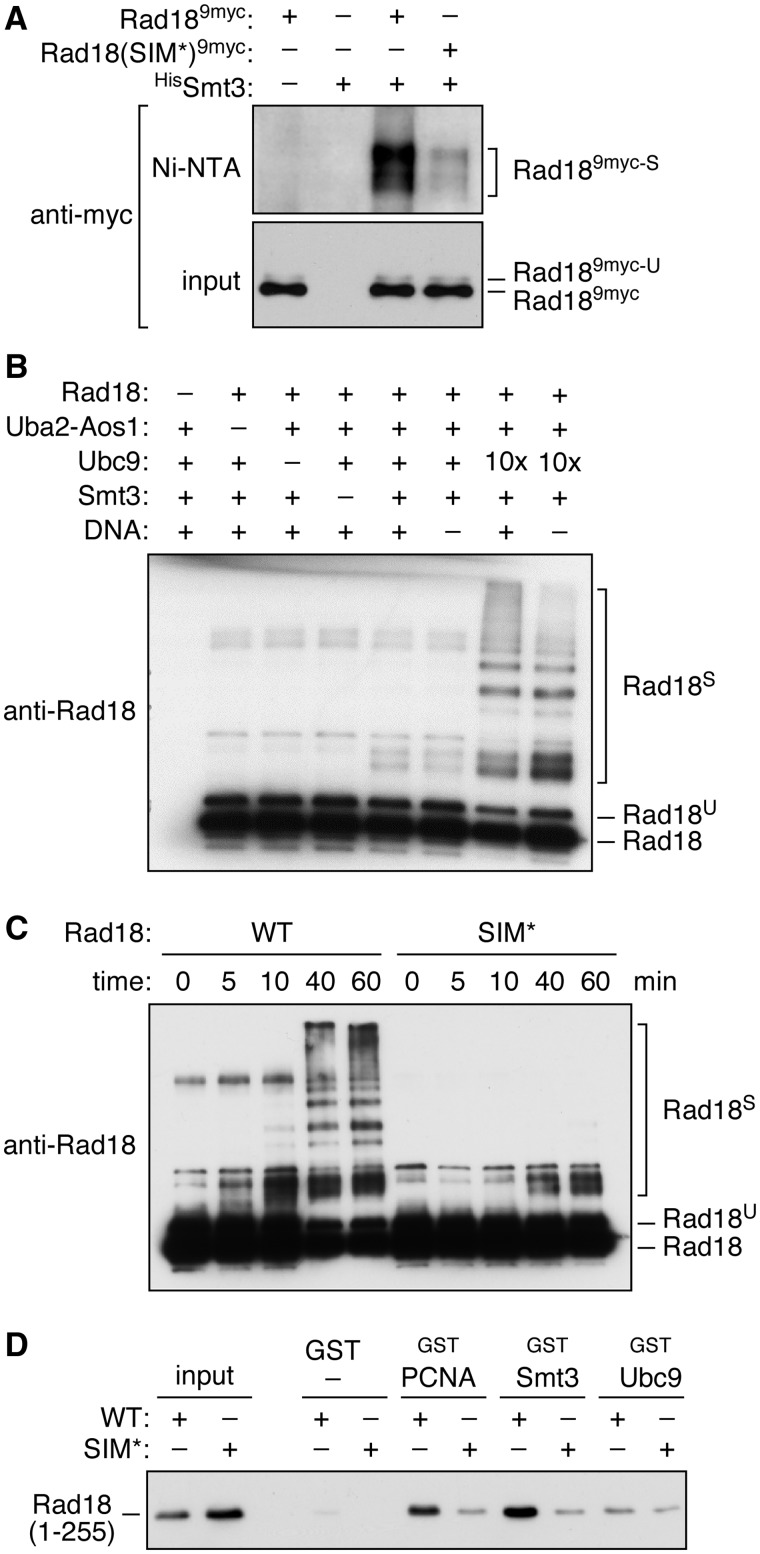
We therefore expressed a His7-tagged form of Smt3 in a strain bearing a 9myc-tagged allele of RAD18 to isolate total Smt3 conjugates under fully denaturing conditions. High-molecular-weight forms of Rad189myc were detectable in the isolated material, indicating that Rad18 is indeed covalently sumoylated in vivo (Figure 2A). Modification of the mutant Rad18(SIM*) protein was much reduced, although not completely abolished. Similar results were obtained in vitro, where addition of SUMO-specific E1 and E2 (Aos1-Uba2 and Ubc9) promoted sumoylation of purified Rad18 (Figure 2B). The reaction was independent of the presence of DNA, which is known to be required for Rad18’s ubiquitin ligase activity towards PCNA (30). Rad18’s cognate E2, Rad6, was not modified, and the SUMO ligase Siz1 moderately enhanced the reaction (Supplementary Figure S1). Importantly, the SIM* mutant was sumoylated much less efficiently (Figure 2C).
Figure 2.
Covalent and non-covalent interactions of Rad18 with Smt3. (A) Rad189myc is sumoylated in vivo in a SIM-dependent manner. Total Smt3 conjugates were isolated by Ni–NTA pull-down from a strain expressing HisSmt3, and sumoylated Rad189myc (marked ‘S’) was detected in the isolated material by anti-myc western blot. Endogenous ubiquitylated Rad18 (‘U’) is also detectable. (B) Rad18 is sumoylated in vitro. Sumoylation assays were set up under standard conditions with the indicated components, and modified Rad18 was detected by western blot. The ‘10×’ indicates a 10-fold higher concentration of Ubc9. The presence of ubiquitylated Rad18 is a consequence of producing the protein in yeast. (C) Rad18 sumoylation is enhanced by the SIM. Time course analysis of in vitro sumoylation was performed with WT Rad18 and the SIM* mutant protein. (D) Rad18 interacts non-covalently with Smt3 in a SIM-dependent manner. GSTPCNA, GSTSmt3 or GSTUbc9 was immobilized on glutathione Sepharose, and retention of HisRad18(1-255), either WT or SIM*, was analysed by anti-His western blot. GST was used as a negative control.
Taken together, these findings suggest that the interaction between Rad18 and Smt3 observed in the two-hybrid system was mainly caused by the sumoylation of Rad18 itself. However, as the modification was strongly dependent on the putative SIM, it is likely that this motif was responsible for directing Ubc9-bound Smt3 towards Rad18, which would imply a non-covalent association after all. We therefore investigated the interaction between Rad18 and Smt3 with recombinant proteins in vitro. A truncated construct that includes the putative SIM, Rad18(1-255), was efficiently retained by immobilized GSTSmt3 in a SIM-dependent manner (Figure 2D). This indicates that the motif indeed mediates non-covalent interaction of Rad18 with Smt3. The weak interaction observed with the SIM* mutant might be because of either a residual affinity of the mutated SIM or a second unidentified binding site in the construct. As observed in the two-hybrid system, the interaction between Rad18 and PCNA was also affected by mutation of the SIM, whereas recombinant Ubc9 bound to Rad18 weakly, but in a SIM-independent manner. Overall, our data show that Rad18 interacts non-covalently with Smt3 via a canonical SIM, which also promotes the covalent sumoylation of Rad18 in vivo and in vitro.
The SIM directs Rad18’s ubiquitin ligase activity towards sumoylated PCNA
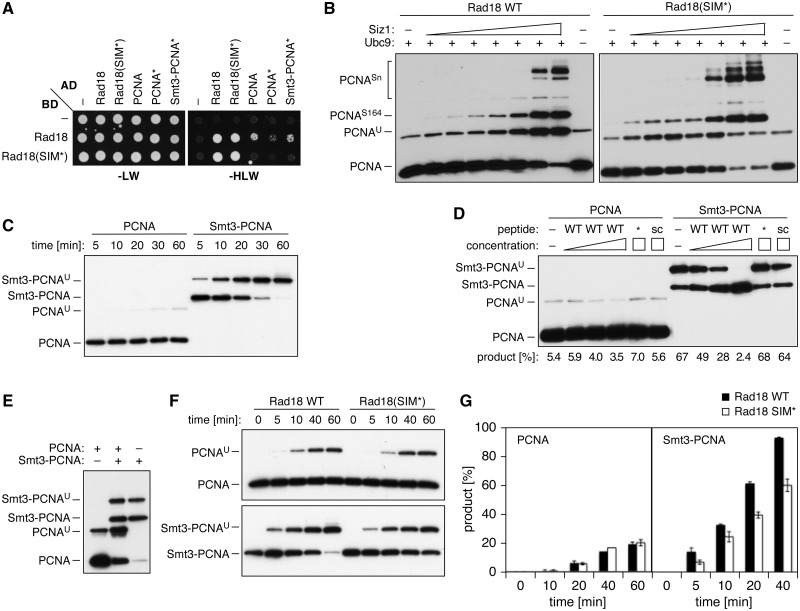
The notion that a significant portion of PCNA is sumoylated during S phase (24) raised the question of whether the SIM would mediate a preferential interaction of Rad18 with the sumoylated form of PCNA. We addressed this in the two-hybrid system. When compared with wild-type PCNA, a mutant that cannot be modified by ubiquitin or SUMO, PCNA(K127/164R), designated as PCNA*, gave a reduced interaction signal with Rad18, suggesting that sumoylation of PCNA within the two-hybrid construct might be important for the association (Figure 3A, Supplementary Figure S2). Consistent with this notion, covalent fusion of Smt3 to the N-terminus of PCNA* restored the interaction. This result prompted us to compare the catalytic activity of Rad18 as a ubiquitin ligase towards PCNA in the presence or absence of Smt3 in vitro. We first followed the extent of PCNA ubiquitylation in vitro under conditions where the clamp would also be sumoylated. In the presence of all the required ubiquitylation components and the SUMO-specific E1 and E2 enzymes, increasing amounts of the SUMO E3, Siz1, resulted in an enhancement of PCNA ubiquitylation, roughly parallel to the amount of sumoylated PCNA that was produced (Figure 3B). Use of the Rad18(SIM*) mutant abolished the enhancement of PCNA ubiquitylation by Siz1 without decreasing the efficiency of sumoylation. To verify that this effect was attributable to the conjugation of SUMO to PCNA and not to any other reaction component, such as Rad18 itself, we examined in vitro ubiquitylation of a linear fusion of Smt3 to the N-terminus of PCNA in the absence of any sumoylation enzymes. Compared with native PCNA, the Smt3 fusion was ubiquitylated with strongly enhanced efficiency, as evident from time course experiments (Figure 3C), as well as titrations of Rad18 (Supplementary Figure S3A). The stimulating effect of the Smt3 moiety was reversed by the addition of an excess of a canonical SIM peptide derived from the PIAS2 protein (26), but not a mutated or scrambled version, indicating that it is indeed caused by a genuine SIM–SUMO interaction (Figure 3D). When a mixture of native PCNA and Smt3-PCNA was used as a substrate, ubiquitylation was stimulated not only on the fusion protein but also on native PCNA (Figure 3E). Considering that the two species can form mixed trimers, this result suggests that the position of Smt3 within the PCNA ring is not important for its stimulatory effect.
Figure 3.
Interactions and activities of Rad18 towards sumoylated PCNA. (A) Fusion of Smt3 to PCNA enhances interaction with Rad18. Two-hybrid analysis was performed as in Figure 1B. The mutant PCNA(K127/164R) is labelled PCNA*. Expression levels of the constructs are shown in Figure S2. (B) Ongoing sumoylation enhances the ubiquitin ligase activity of Rad18, but not Rad18(SIM*), towards PCNA. In vitro modification reactions were set up with all factors required for PCNA ubiquitylation, as well as Aos1-Uba2 and Smt3. Siz1 concentrations were varied from 0.8 to 50 nM. Products were detected by anti-PCNA western blot. (C–F) Standard in vitro ubiquitylation reactions, analysed as earlier in text. (C) Time course analysis of in vitro ubiquitylation of PCNA and Smt3-PCNA. (D) Reversal of the stimulatory effect of Smt3 by a canonical SIM peptide (WT), but not a mutated (*) or scrambled (sc) version. Peptide concentrations ranged from 30 to 600 µM, and the amount of product formed is indicated below the blot. (E) Presence of Smt3-PCNA enhances ubiquitylation of native PCNA. (F) The enhanced activity of Rad18 towards Smt3-PCNA relies on an intact SIM. Time course analysis was performed with PCNA (upper panel) and Smt3-PCNA (lower panel), using Rad18 WT or the SIM* mutant protein. (G) Quantification of the blots shown in panel F.
A number of controls demonstrated that the Smt3-stimulated activity of Rad18 closely reflects its natural behaviour towards PCNA: Rad6 in the absence of Rad18 was unable to modify PCNA or Smt3-PCNA (Supplementary Figure S3B), the reaction required RFC-dependent loading of the substrate onto DNA (Supplementary Figure S3C), and ubiquitylation required the physiological site, K164 (Supplementary Figure S3D). Fusion of ubiquitin instead of Smt3 to the N-terminus of PCNA afforded comparatively minor stimulation (Supplementary Figure S3E). Importantly, although WT and SIM* proteins exhibited similar activities towards native PCNA, fusion of Smt3 to PCNA resulted in a much larger enhancement of the reaction for WT Rad18 compared with the mutant (Figure 3F and G and Supplementary S3F). Again, a residual affinity for Smt3 (Figures 1B and 2D) was likely responsible for the moderate effect of the Smt3 moiety on the activity of Rad18(SIM*). Taken together, these results imply that Rad18’s SIM is capable of promoting a productive interaction with an Smt3 moiety on the PCNA trimer, resulting in a strongly enhanced ubiquitin ligase activity.
The SIM–SUMO interaction contributes to Rad18 function in vivo
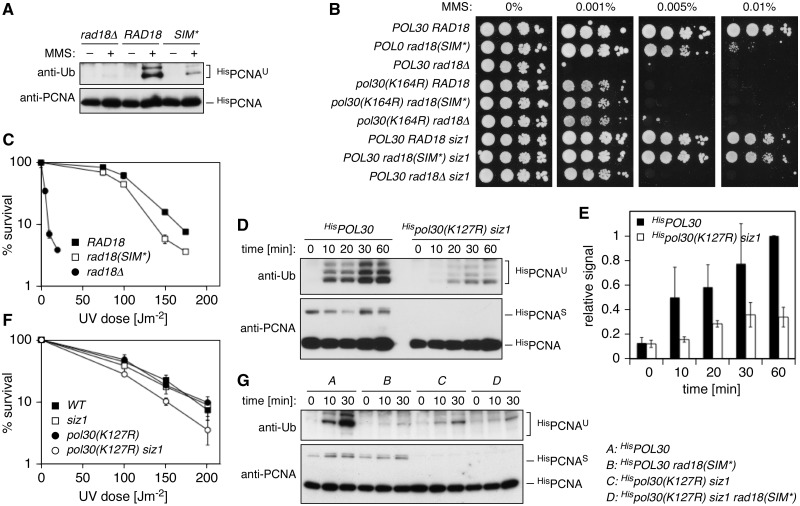
Consistent with a relevance of the SIM for Rad18 function in vivo, damage-induced PCNA ubiquitylation was reduced in the mutant (Figure 4A), and the cells exhibited moderate sensitivity towards the alkylating agent MMS and UV radiation (Figure 4B and C). Mutation of K164 of PCNA abolished this effect, confirming that the damage sensitivity is specific to a defect in PCNA ubiquitylation (Figure 4B). Deletion of SIZ1 suppressed the phenotype of rad18(SIM*), as expected from the notion that a failure to recruit Srs2 allows damage processing by homologous recombination. The partial loss of function was not likely caused by a destabilization of the protein, as equal levels of wild-type and mutant Rad189myc were detectable in total cell extracts (Figure 2A). Yet, despite a WT-like activity towards native PCNA in vitro (Figure 3F), we had observed a reduced interaction of the mutated protein with PCNA (Figures 1B and 2D). To rule out that this reduction in PCNA binding was causing the phenotype of the mutant in vivo, we examined a strain bearing WT RAD18, but devoid of PCNA sumoylation, pol30(K127R) siz1. In this strain, PCNA ubiquitylation was also reduced >3-fold (Figure 4D and E), and cells again exhibited UV sensitivity (Figure 4F). Combination of these mutations with rad18(SIM*) did not lead to a loss of PCNA ubiquitylation beyond the level of the rad18(SIM*) single mutant, demonstrating an epistatic relationship between the two defects (Figure 4G). Hence, a functional SIM–SUMO interaction contributes to full activity of Rad18 towards PCNA, and its loss gives rise to a phenotype comparable with that of mutants in other components of the pathway, such as rev3 or rad30 (15).
Figure 4.
Relevance of the SIM–SUMO interaction for Rad18 function in vivo. (A) Damage-induced PCNA ubiquitylation is reduced in the SIM* mutant. HisPCNA was isolated by Ni–NTA pull-down under denaturing conditions from extracts of MMS-treated cells, and ubiquitin conjugates were detected by anti-ubiquitin western blot. (B) Mutation of the SIM confers enhanced sensitivity to MMS in a manner dependent on K164 of PCNA, but the phenotype is suppressed by deletion of SIZ1. Deletion mutants of rad18 were complemented with RAD18 (WT or SIM*) for spot assays. An empty vector served as control. (C) Mutation of the SIM confers enhanced UV sensitivity. (D) Damage-induced PCNA ubiquitylation is reduced in a mutant deficient in PCNA sumoylation, pol30(K127R) siz1. A modification time course was analysed as in panel A. (E) Quantification of the blots shown in panel D, relative to the WT signal at 60 min. (F) The pol30(K127R) siz1 mutant displays enhanced UV sensitivity. (G) The PCNA ubiquitylation defects of rad18(SIM*) and pol30(K127R) siz1 show an epistatic relationship. Modifications were analysed as in D.
Preference for sumoylated PCNA is not a conserved feature of Rad18
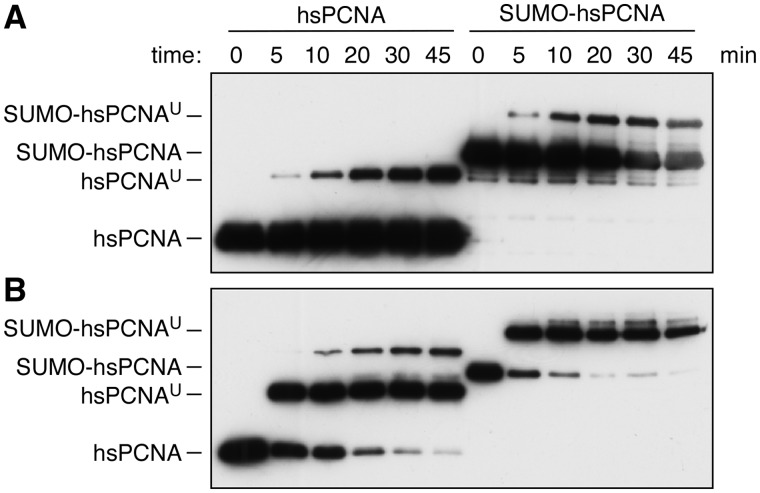
We asked whether the stimulation of Rad18 activity by the presence of a SUMO moiety on PCNA was a conserved phenomenon. SUMO-2 was therefore fused to the N-terminus of human PCNA, and the construct was used as a substrate for the human Rad6–Rad18 complex, either free in solution (Figure 5A) or loaded onto DNA (Figure 5B). As in the yeast system, RFC and DNA strongly stimulated the overall efficiency of the reaction. However, the presence of SUMO on PCNA did not enhance Rad18’s activity, consistent with the lack of an obvious SIM in the human Rad18 sequence. Although sumoylated PCNA has recently been observed in human cells, its abundance is low, and its regulation may well differ from the situation in yeast (31). Hence, the observed phenomenon does not seem to be a universal property of Rad18. Rather, it appears that the SIM in Rad18 from S. cerevisiae is a feature that enables the ligase to specifically react to this abundant modification of chromatin-bound PCNA and facilitates the transition to the damage-associated state.
Figure 5.
Human Rad18 is unaffected by fusion of SUMO to PCNA. In vitro ubiquitylation reactions were set up with human Rad6–Rad18 complex and human PCNA or a linear fusion with SUMO-2. Products were detected by western blot with an antibody against human PCNA. (A) Reactions set up without DNA and RFC. (B) Reactions set up in the presence of DNA and RFC.
DISCUSSION
Implications of SUMO binding for Rad18 function in DNA damage bypass
Based on these observations, we propose that a DNA-bound PCNA trimer bearing at least one SUMO moiety is the physiological substrate of Rad18 in S. cerevisiae. As we previously showed, a significant portion of the total cellular PCNA pool is sumoylated during DNA replication, and—similar to ubiquitylation—its residence on DNA is a prerequisite for sumoylation (24). Hence, our model implies a direct transition from sumoylation to ubiquitylation on replication problems or DNA damage, without an intervening desumoylation step. Isolation of a species bearing SUMO on K127 and ubiquitin on K164 within the same subunit indicates that the two modifiers can coexist on a single clamp in vivo (16).
This model stipulates that neither sumoylation nor ubiquitylation affects all three subunit of the PCNA trimer in concert. Although initial observations had suggested that ubiquitin can be conjugated to all three subunits of the clamp in damage-treated human cells (14), indirect evidence from yeast and mouse PCNA lends support to our model: we have shown that co-expression of a linear ubiquitin–PCNA fusion and a native non-modifiable PCNA allele supports translesion synthesis, despite the low abundance of fully ‘ubiquitylated’ trimers expected from a stochastic arrangement (28). Along the same lines, normal activation of the damage-tolerant DNA polymerase η in heterozygous PCNAK164R murine B cells suggests that modification of all three subunits within a trimer is not required for ubiquitin-dependent translesion synthesis (32).
Thus, Rad18 activity towards PCNA appears to be controlled on two levels: on the one hand, we previously showed that the replication protein A complex recruits the E3 to sites where replication problems have caused the accumulation of single-stranded DNA (19); on the other hand, the SIM–SUMO interaction described here directly enhances the recognition of PCNA as a relevant substrate. In this way, the properties of budding yeast Rad18 not only ensure an ordered transition from sumoylated to ubiquitylated PCNA but they may also limit inappropriate PCNA modification under conditions where the clamp is not engaged in replication.
Relevance of SIM-dependent Rad18 sumoylation
The SIM not only stimulates ubiquitin ligase activity of Rad18 towards sumoylated PCNA but also enhances the sumoylation of Rad18 itself. The reaction is reminiscent of a phenomenon called coupled monoubiquitylation, which is based on the interaction of a ubiquitylation target with a ubiquitin-charged E2 or a ubiquitylated E3, mediated by a ubiquitin-binding domain within the substrate (33,34). Whether Rad18’s SIM recognizes the thioester-bound modifier or a SUMO moiety attached to a lysine on Ubc9 (35) is unknown, but the reaction does not require a SUMO ligase. In the two-hybrid system, mutation of the Rad18 SIM results in a partial loss of interaction with Ubc9 (Figure 1B), and in pull-down assays the SIM-independent affinity of Rad18 for unmodified Ubc9 is low (Figure 2D), indicating that the contact between Rad18 and Ubc9 is largely mediated by SUMO. Given that SUMO conjugates of Rad18 are much less abundant in vivo than the ubiquitylated forms of the E3 (Figure 2A), sumoylation might be an inevitable consequence of SUMO binding and of little physiological relevance for Rad18 function. This, however, will need to be examined in a future study.
Substrate recognition by Rad18 and other STUbLs
Based on the properties of its SIM, Rad18 exhibits the hallmarks of a STUbL. Although our data show that interactions with both SUMO and PCNA contribute to the efficient recognition of sumoylated PCNA by Rad18, the situation is not always that clear for other STUbLs. Most notably, few physiological substrates have so far been identified. Prominent examples are the sumoylated forms of human promyelocytic leukaemia protein PML or the PML–RARα fusion, which are ubiquitylated by RNF4; however, the E3 predominantly recognizes these substrates via their poly-SUMO chains (7). In vitro, STUbLs often act relatively unspecifically ubiquitylating artificial test substrates such as GST-SUMO, sumoylated Siz2 or free poly-SUMO chains (6,36). There are a few notable exceptions to this rule: budding yeast Slx5–Slx8 complex mediates the ubiquitylation and subsequent degradation of the transcription factor Matα2 (37), and the Drosophila STUbL, Degringolade, ubiquitylates the transcriptional repressor Hairy (38). Intriguingly, sumoylation was found to be dispensable for ubiquitylation in both cases, suggesting that STUbLs can also function in a SIM-independent manner. In terms of substrate properties, the transcription factor Mot1 from S. cerevisiae may resemble Rad18 most closely, as the Slx5–Slx8 complex recognizes this substrate through SUMO, as well as epitopes on the protein itself (39). However, ubiquitylation apparently correlated with misfolding in this case, suggesting a possible role of the STUbL in general protein quality control rather than the targeting of specific substrates. Clearly, much remains to be learned about the mechanisms by which STUbLs select their targets. Our identification of budding yeast Rad18 as a substrate-specific member of this enzyme family has given insight into a new aspect of STUbL function by showing how the enzyme can coordinate an ordered progression from one modification to another.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Funding for open access charge: Cancer Research UK [A7123].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Kay Hofmann for valuable discussions, Titia Sixma for providing the expression vector for human Rad6–Rad18 complex, Svend Petersen-Mahrt for pRSF-PCNA and Adelina Davies and Jonathan Lowther for technical assistence.
REFERENCES
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15:525–532. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 4.Hu XV, Rodrigues TM, Tao H, Baker RK, Miraglia L, Orth AP, Lyons GE, Schultz PG, Wu X. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc. Natl Acad. Sci. USA. 2010;107:15087–15092. doi: 10.1073/pnas.1009025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 8.Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 9.Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3-Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 10.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair. 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 13.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 14.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 15.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 16.Windecker H, Ulrich HD. Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J. Mol. Biol. 2008;376:221–231. doi: 10.1016/j.jmb.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 19.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 22.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 2007;35:5819–5830. doi: 10.1093/nar/gkm615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker JL, Bucceri A, Davies AA, Heidrich K, Windecker H, Ulrich HD. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008;27:2422–2431. doi: 10.1038/emboj.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulrich HD. Protein-protein interactions within an E2-RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J. Biol. Chem. 2003;278:7051–7058. doi: 10.1074/jbc.M212195200. [DOI] [PubMed] [Google Scholar]
- 26.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 27.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saugar I, Parker JL, Zhao S, Ulrich HD. The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res. 2012;40:245–257. doi: 10.1093/nar/gkr738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J. Exp. Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 2006;8: 1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 35.Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell. 2008;31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Mullen JR, Brill SJ. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 2008;283:19912–19921. doi: 10.1074/jbc.M802690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, Rubenstein EM, Matt T, Hochstrasser M. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 2010;24:893–903. doi: 10.1101/gad.1906510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abed M, Barry KC, Kenyagin D, Koltun B, Phippen TM, Delrow JJ, Parkhurst SM, Orian A. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011;30:1289–1301. doi: 10.1038/emboj.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell. Biol. 2009;29:1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.