Abstract
Hepatitis C virus (HCV) translation initiation is directed by an internal ribosome entry site (IRES) and regulated by distant regions at the 3′-end of the viral genome. Through a combination of improved RNA chemical probing methods, SHAPE structural analysis and screening of RNA accessibility using antisense oligonucleotide microarrays, here, we show that HCV IRES folding is fine-tuned by the genomic 3′-end. The essential IRES subdomains IIIb and IIId, and domain IV, adopted a different conformation in the presence of the cis-acting replication element and/or the 3′-untranslatable region compared to that taken up in their absence. Importantly, many of the observed changes involved significant decreases in the dimethyl sulfate or N-methyl-isatoic anhydride reactivity profiles at subdomains IIIb and IIId, while domain IV appeared as a more flexible element. These observations were additionally confirmed in a replication-competent RNA molecule. Significantly, protein factors are not required for these conformational differences to be made manifest. Our results suggest that a complex, direct and long-distance RNA–RNA interaction network plays an important role in the regulation of HCV translation and replication, as well as in the switching between different steps of the viral cycle.
INTRODUCTION
The hepatitis C virus (HCV) genome is a 9.6-kb-long, positive, single-stranded RNA molecule (1,2). It consists of a single open reading frame (ORF) flanked by untranslatable regions (UTRs) that contain structural elements essential to the execution of the viral cycle (3–9). Viral translation initiation is mediated by a highly structured element mainly located at the 5′-UTR, which acts as an internal ribosome entry site (IRES) (3,4) (Figure 1A). IRES-dependent translation differs greatly from the mechanism used by most cellular mRNAs, operating by the direct recruitment of the 40S ribosomal subunit (10,11), which promotes the formation of the 48S particle via the further recruitment of eIF3 and the eIF2/GTP/Met–tRNA ternary complex. The configuration of the 48S particle permits the appropriate positioning of the codon–anticodon pairs in the P site during protein synthesis initiation (12–14). This apparently simple process is guided by a subset of regulatory structural RNA elements included in the IRES element. Thus, the conformation of the IRES plays a crucial role in viral protein synthesis (14–18).
Figure 1.
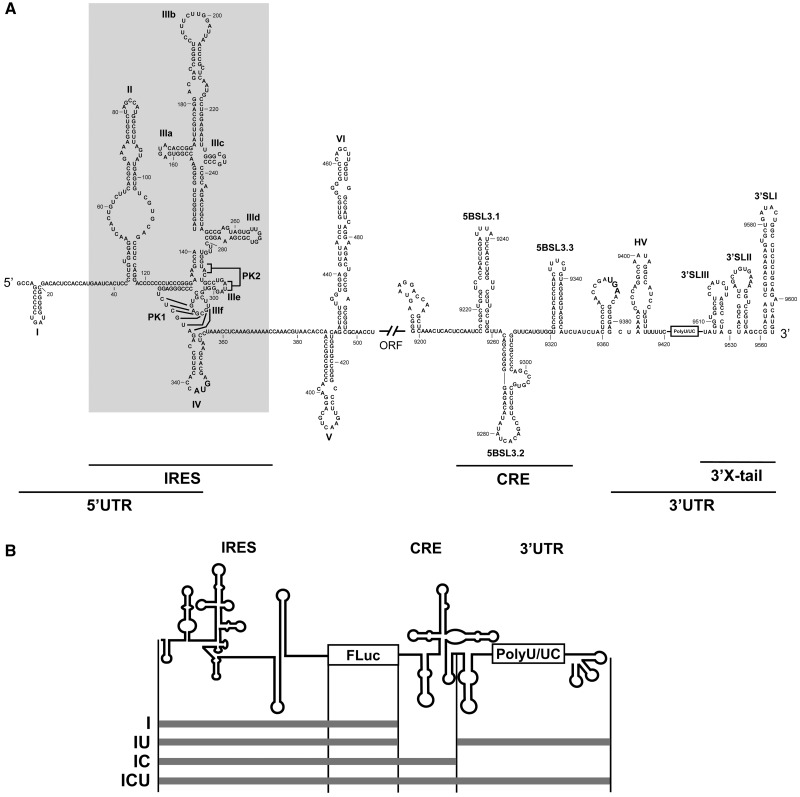
Secondary structure of the 5′- and 3′-ends of the HCV genome. (A) Sequence and secondary structure of the 5′- and 3′-regions of the HCV genome. The 5′-UTR plus domains V and VI located inside the HCV coding sequence are included. The minimum region for IRES activity is boxed. The 3′-end of the viral genomic RNA is organized into two structural elements: the CRE region and the 3′ X-tail, separated by a hypervariable sequence (HV) and a polyU/UC stretch (also considered inside the 3′-UTR). Numbers refer to the nucleotide positions of the HCV Con1 isolate (GenBank accession number AJ238799). Start and stop translation codons placed at positions 342 and 9371, respectively, are shown in bold. Pseudoknot elements are marked as PK1 and PK2. (B) Diagram of the HCV transcripts harboring the IRES–FLuc fusion attached to different 3′ viral genomic domains.
Under physiological conditions, the minimum HCV IRES element required for translation initiation folds into a well-defined structure composed of two major domains (19) (II and III) and a short stem–loop (domain IV) containing the initiation codon (20) (Figure 1A). Domains II and III are aligned at both sides of a complex double pseudoknot structure allowing the correct positioning of the start codon (14). Importantly, many of the crucial structural regions required for IRES-dependent translation are located in the highly branched domain III (21). The latter is organized into three- and four-way junctions defined by stem–loop motifs (designated subdomains IIIa to IIIf) that mark the anchoring sites for protein factors. Thus, the IIIabc junction directs the interaction with eIF3 (13,22), whereas subdomains IIIc to IIIf cooperate in the recruitment of the ribosomal subunit 40S (13,23). Importantly, subdomain IIId has been described as the core 40S binding center (24–26). Its structure is that of a dynamic stem–loop with an internal E loop motif and an apical loop with typical U-turn geometry (15,27). The conservation of all these structural motifs in the IRES element is critical for its proper function (21).
The presence of structural elements at the 3′-end of the viral genome may also modulate the initiation and elongation steps involved in HCV translation (28–33). This might be mediated via the HCV genomic RNA, assuming a circular topology resembling the closed-loop structure adopted by cellular cap-mRNAs. Such circular topologies were initially thought to be strictly dependent on the recruitment of protein factors able to simultaneously bind to the 5′- and 3′-UTRs of the genomic HCV RNA (28–31,34–36). Importantly, the existence of a direct, long-range RNA–RNA interaction has recently been proposed. This involves subdomain IIId of the IRES element and the essential stem–loop 5BSL3.2 of the CRE (cis-acting replication element) region at the 3′-end of the viral protein coding sequence (37) (Figure 1A). This circularization mechanism resembles that evolved by other closely related RNA viruses of the family Flaviviridae (38). End-to-end communication mediated by direct RNA–RNA interactions would likely alter the conformation of the HCV IRES, allowing a regulatory mechanism for viral protein synthesis dependent on RNA high-order structure. The effect of the 3′ genomic elements on IRES folding is required to gain deeper insights into the mechanism of HCV translation and its regulation.
The present work provides evidence that IRES conformation is fine-tuned by the 3′-end of the HCV genome. 3′-End-mediated IRES structural changes are mainly detected in the essential subdomains IIIb and IIId, and in domain IV, suggesting this interaction to be important in the recruitment of the 40S particle and eIF3. These results are in good agreement with our previous data demonstrating long-range contacts between the apical loop of subdomain IIId and the internal loop of 5BSL3.2 of CRE region, and highlighting the relevance of both the CRE and the 3′-UTR for the proper IRES activity (33). To our knowledge, this is the first report describing the requirement of two 3′ genomic elements, CRE and 3′-UTR, for the fine-tuning of the structural and functional organization of the HCV IRES element. Further, this interaction occurs in the absence of protein factors. The obtained results point to local conformational rearrangements as key events in the regulation of viral translation and replication, as well as in switching between different steps of the HCV life cycle.
MATERIALS AND METHODS
DNA templates and RNA synthesis
DNA templates for the transcription of RNA molecules I, IU, IC and ICU were obtained by amplification as previously described (33) to get the precise 3′-termini. Briefly, molecules T7pIU and T7pICU encoding the variants IU and ICU were amplified from their respective plasmid constructs pGL-IU and pGL-ICU, using the primers 5′T7pHCV and 3′HCV. DNA templates containing either the I or the IC constructs (T7pI and T7pIC) were PCR amplified from the plasmid pGL-ICU: primers 5′T7pHCV and asFLuc (5′-TCTAGAATTACACGGCGATCTTTCCGC-3′) were used for the amplification of the variant T7pI, whereas T7pIC was synthesized by using the oligonucleotides 5′T7pHCV and asHCV-9321 (33).
The plasmid encoding the 5′HCV-691 transcript has been previously described (39). DNA templates coding for constructs I + U, I + C and I + CU were generated by site-directed mutagenesis using the Phusion site-directed mutagenesis kit (Finnzymes) from the template vector pGL-ICU (33), employing the following primers: HCV-9181 (5′-GGGCAGTAAGGACCAAGCTCAAA-3′) and asHCV-698 + HindIII for pGLI + C and pGLI + CU plasmids, and HCV-9383 and asHCV-698 + HindIII for pGLI + U construct. These vectors were used as DNA templates for PCR amplification of the molecules T7pI + U, T7pI + C and T7pI + CU from which the respective RNA variants (I + C, I + U and I + CU) were in vitro synthesized (see below). Primers 5′T7pHCV and 3′HCV were used for the amplification of T7pI + U and T7pI + CU, whereas 5′T7pHCV and asHCV-9321 allowed the production of T7pI + C. The integrity of each construction was checked by DNA sequencing.
The DNA vector containing the replication-competent construct, pFK-I389FLucNS3-3′ET (40), was kindly provided by Dr R. Bartenschlager. The DNA template for the generation of the replicative RNA transcript was obtained as previously described (40).
All RNAs were synthesized using the RiboMAX™-T7 large scale RNA production system (Promega), following the manufacturer’s instructions. The resulting transcripts were purified by phenol extraction and non-incorporated nucleotides were removed by two consecutive steps of size exclusion chromatography (Sephadex G-25, GE Healthcare). The amount of RNA was determined by UV spectrophotometry (A260) and the extent of protein and carbohydrate/phenolic contaminations was assessed by A260/A280 and A260/A230 ratios, respectively. The integrity of the RNA molecules was determined by denaturing agarose-formaldehyde gel electrophoresis.
Microarray design, RNA hybridization and data analysis
Antisense DNA oligonucleotides complementary to IRES domains III and IV of HCV (genotype 1b) were designed to specifically hybridize with sequences of 14 consecutive nucleotides of the viral genome, overlapping by seven nucleobases with the adjacent oligos. Each HPLC-purified oligonucleotide (IBA GmbH) was composed of a C6-amino linker motif at its 5′-end, followed by either (TCC)5 or CCCTCCCTCCCTCCC tracks (designed to avoid self-annealing within the oligo) as spacer regions, and the specific antisense sequence at the 3′-side of the oligo (Supplementary Table S1). Their names correspond to the nucleotide on the IRES sequence complementary to the 3′-end of the oligo. Two different oligonucleotides with unrelated sequences (belonging to either the 5′-genomic region of foot-and-mouth disease virus or the pol gene of human immunodeficiency virus type 1) were included as negative controls. The 35 oligonucleotides were diluted in 1× spotting solution (Telechem-Arrayit) and spotted onto epoxi-modified glass slides (Telechem-Arrayit). Two concentrations (5 and 20 µM) of each oligonucleotide probe were printed in triplicate 150 µm Ø spots, with a distance of 300 µm between groups of triplicate spots. Microarrays containing two duplicated grids of 210 spots were printed using a GSM 417 DNA arrayer (Affymetrix).
The HCV RNAs were fluorescently labeled with Alexa 647 (Invitrogen) as previously described (41). Briefly, 1 µg of RNA was incubated in 30 µl of labeling buffer for 15 min at 4°C, prior to the addition of 2 µl of the fluorescent dye, and the final mixture was incubated for 10 min at 90°C. The labeling reaction was stopped by transferring the sample to ice for 10 min and purified by Microcon (Millipore). The final volume was adjusted to 5–10 µl in a speed vacuum.
The printed microarrays were pre-treated with 2× SSC and 0.1% N-lauril sarcosine for 2 min at room temperature (RT). The unbound DNA oligos and the excess of spotting solution were washed by an additional incubation in 2× SSC for 2 min. Then, printed oligos were denatured by baking the microarrays for 2 min in boiling milli-Q water, cooling for 10 s at RT and immediately fixed in ice-cold 100% ethanol as described (42). The microarrays were prehybridized in 6× SSC, 0.5% SDS and 10 µg/µl BSA for 45 min at 42°C. A total of 300 ng of the fluorescent transcripts were denatured by heating at 95°C for 3 min and subsequent cooling at 4°C prior to incubation with the slides. Hybridization was performed under non-denaturing conditions (100 mM HEPES pH 8.0, 100 mM NaCl, 1 mM MgCl2) for 2 h at 37°C. Slides were washed twice in 1× washing buffer (20 mM HEPES pH 8.0, 50 mM NaCl, 1 mM MgCl2) in the presence of 0.2% SDS for 15 min at RT and then once again in washing buffer alone for 15 min at RT. After a final rinse with water, the slides were dried by centrifugation for 1 min at 500 g and immediately scanned using a Genepix 4100 scanner. Data were retrieved using Genepix Pro 6.0 software.
The differential capacity of each RNA transcript for hybridizing to the microarrays was measured in three independent experiments. In all cases, the raw fluorescence signal was corrected by subtraction of the local background. To compare the signal among the three independent hybridization assays, the mean fluorescence signal yielded by the 20 µM spots was normalized against the mean fluorescence of oligonucleotides 113 and 176 (Supplementary Table S1). The signal range that defines the accessibility level was defined as follows: the interquartile range of the hybridization values was calculated to identify the outliers, i.e. the positions showing the highest relative accessibility. The lowest of these outliers was established as the upper limit of the range. Quartile 3 defined the lower limit of the range.
Dimethyl sulfate probing
A total of 1 pmol of purified HCV RNA (∼1 µg) was denatured for 3 min at 95°C in folding buffer (50 mM HEPES pH 8.0, 100 mM NaCl,1 mM MgCl2) before transferring to ice for 15 min. The RNA was then incubated for 5 min at 37°C. Reactions were initiated by the addition of 1 µl of freshly diluted dimethyl sulfate (DMS) in ethanol (1:5). Chemical probing proceeded for 1 min at 37°C in the presence of tRNA (1 µg) and was stopped by the addition of 300 mM sodium acetate pH 5.2. Untreated DMS reactions were performed in parallel. The RNA was ethanol-precipitated, washed three times with 80% ethanol and subjected to primer extension for the detection of the modified A and C residues. Fluorescently labeled DNA oligonucleotides (Applied Biosystems) used for the determination of DMS and N-methyl-isatoic anhydride (NMIA) reactivity were purified using high-resolution denaturing polyacrylamide gels. Two 5′-end fluorescently labeled primers were used to map domains III and IV of the IRES element: asHCV-372 (5′-TTTTTCTTTGAGGTTTAGGATTCGTGCT-3′) and asHCV-539 (5′-GGGGATAGGTTGTCGCCTT-3′). Briefly, 2 pmol of gel-purified primer were hybridized with the total processed RNA by incubation for 2 min at 95°C, followed by slow cooling to 52°C. Extension reactions were performed for 30 min at 52°C in a 20-μl reaction volume with reverse transcriptase buffer, 0.5 mM dNTPs and 100 U SuperScript III RT (Invitrogen). NED fluorophore was used for mapping the untreated probes; VIC was employed to read the DMS assays. RNA sequencing reactions were performed under identical conditions with the FAM and PET fluorescently labeled primers. A total of 2 µl of the resulting cDNA samples were purified using the BigDye® XTerminator™ Purification kit (Applied Biosystems) and resolved in an Applied Biosystems 3130xl Genetic Analyzer capillary electrophoresis DNA sequencer, as previously described (43). Electropherograms were analyzed using ShapeFinder software (44). Normalized DMS reactivity values for each nucleotide position were obtained by dividing each value by the average intensity of the 10% most reactive residues, after excluding outliers calculated by box plot analysis (43). This novel protocol afforded a DMS probing method using the technology developed for SHAPE chemistry.
SHAPE analysis
Treatment with NMIA was performed essentially as previously reported (43). Briefly, 1 pmol of RNA was denatured as described earlier under the same ionic conditions. 2′-Acylation was initiated by the addition of 6 mM of NMIA in dimethyl-sulfoxide. Reactions proceeded for 3 min at 37°C. The formation of 2′-O-adducts was stopped by ethanol precipitation. The RNA was subsequently washed three times with 80% ethanol and subjected to primer extension as described earlier. Normalized NMIA reactivity values for each nucleotide position were calculated as indicated for DMS probing.
Statistical analysis
All accessibility and reactivity data are presented as mean ± SD. Data points were compared using the unpaired two-tailed Mann–Whitney test (45), as indicated in the figure legends. Significance was set at P ≤ 0.05. P-values are included in the corresponding Supplementary Tables and provided throughout the text when appropriate.
RESULTS
Microarray analysis detected RNA accessibility variations in the IRES element mediated by the 3′-end of the HCV genome
A direct RNA–RNA interaction involving subdomain IIId of the HCV IRES element and the stem–loop 5BSL3.2 located at the CRE region was initially demonstrated in trans (37) and its regulatory role in the HCV IRES function subsequently proven (33). Based on these observations, it was postulated that the IRES conformation might rely on long-distance RNA–RNA interaction in cis with the 3′-end of the viral genome. To test this hypothesis, IRES accessibility in the presence of different elements belonging to the 3′-end of the viral genome was tested by hybridizing (under non-denaturing conditions) each labeled transcript to a customized panel of overlapping 14-mer antisense DNA oligonucleotides (see ‘Materials and Methods’ section and Supplementary Table S1) complementary to domains III and IV of the IRES element. The use of DNA microarrays to analyze RNA structure is based on the strong correlation between the native structure of a target RNA and its ability to differentially hybridize with a panel of antisense oligonucleotides printed onto the microarray surface. This technology has been successfully used for detecting structural rearrangements affecting nucleotide stretches of target subgenomic RNA molecules from HCV (42), foot-and-mouth disease virus (41,46) and human immunodeficiency virus (47).
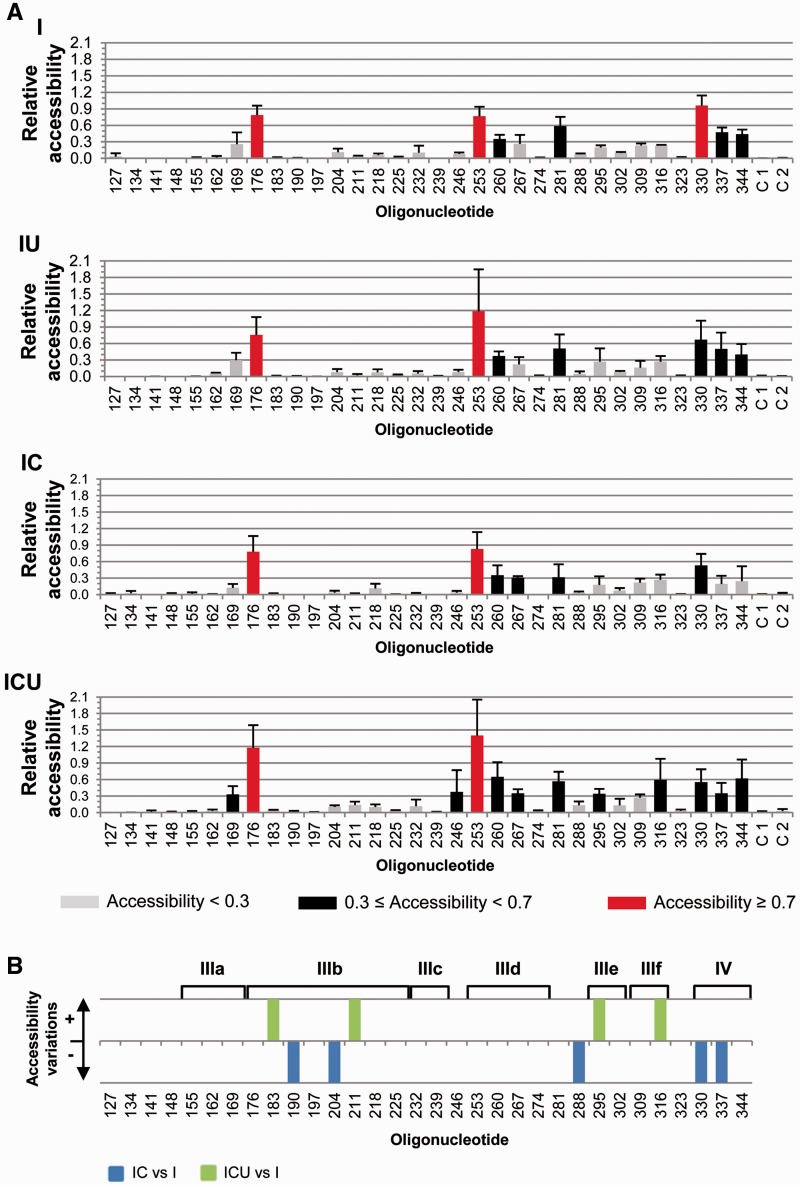
Microarray analyses were performed with previously described chimeric RNA molecules (33) encompassing the IRES element fused to the reporter gene fluc (termed molecule I) plus different 3′-end regions (Figure 1B). Thus, the variant IC carried the CRE element, the construct IU contained the 3′-UTR and the whole 3′-end of the HCV genome was included in the ICU molecule. The use of PCR-generated DNA templates assured the presence of the precise 3′-ends. The RNA constructs were subsequently transcribed and fluorescently labeled, renatured and hybridized on the microarray under native conditions (19) as previously described (41,42). The fluorescence signal for each oligonucleotide was normalized with respect to the average fluorescence detected for oligos 113 and 176 (see ‘Materials and Methods’ section). The mean and SD (from at least three independent assays) of the fluorescence signal was estimated for each oligonucleotide (Figure 2A).
Figure 2.
Comparative microarray analysis of the HCV IRES structure in the presence of different 3′ viral genomic domains. The fluorescently labeled RNA transcripts were hybridized under native conditions on microarrays containing 14-nt-long antisense DNA oligonucleotides (termed 127–344) complementary to the IRES domains III and IV, each of them overlapping by 7 nt with the adjacent oligos. (A) Normalized hybridization signal of transcripts I, IU, IC and ICU plotted against each oligonucleotide (mean fluorescence hybridization signal ± SD) averaged from, at least, three independent assays. Color code delimits the three accessibility levels, as described in ‘Materials and Methods’ section. Negative control oligonucleotides containing unrelated sequences were included as C 1 and C 2 (see Supplementary Table S1 for details). (B) Significant variations (P ≤ 0.05) in fluorescence intensity for constructs IC and ICU with respect to transcript I are represented qualitatively as gain (+) or loss (−) of accessibility. Functional IRES subdomains are shown. There were no significant differences in fluorescence intensity for construction IU with respect to transcript I.
An initial overview of the hybridization profile for the molecule I reported an expected low-accessibility pattern, many regions showing signals close to the background levels (Figure 2A). Nevertheless, well-defined nucleotide stretches were detected to display either medium or high accessibility. This suggested that the IRES folding is tight enough to preserve a unique, stable conformation. The addition of the whole 3′-end of the HCV genome (molecule ICU) revealed specific variations in the hybridization pattern (Figure 2A). These differences were analyzed by a statistical non-parametric test to further evaluate their significance. It should be noted that the low accessibility profile shown by the molecule I prompted us to consider even minor changes in the signal, as long as the statistical significance of the observation can be assessed. Moreover, since IRES folding is essentially autonomous (19), we paid special attention to those minor variations, which could reflect the conformational fine-tuning of functionally relevant domains. Thus, significant increases in the fluorescent signal of molecule ICU were detected at oligonucleotides 183 (P = 0.050) and 211 (P = 0.050), which mapped to subdomain IIIb (Figure 2B). Despite these relative increases, the apical region of subdomain IIIb remained fairly inaccessible (oligos 183–197) or only slightly accessible (204 and 211) in transcript ICU (Figure 2 and Supplementary Figure S1). Two other significant increases in molecule ICU with respect to I were found at oligonucleotides 295 (P = 0.050) and 316 (P = 0.050), which recognize the essential pseudoknots PK1 and PK2 (subdomains IIIef). This reflects the relative instability of these tertiary interactions in different structural environments when competing with longer antisense oligonucleotides. No additional changes in the hybridization profile induced by the CRE element or the 3′-UTR on their own (transcripts IC and IU, respectively) were preserved in the molecule ICU (Figure 2 and Supplementary Figure S1). Thus, the CRE region induced a significant reduction in the hybridization signal at oligos 190, 204, 288, 330 and 337, which was annulled in the presence of the 3′-UTR (Figure 2 and Supplementary Figure S1).
These data suggest that, independently of protein factors, the 3D folding adopted by the whole 3′-end of the HCV genome may be required for the acquisition of the functional conformation of the IRES subdomains IIIb and IIId, and domain IV, at its 5′-end.
DMS treatment showed the HCV IRES structure to be dependent on the 3′-end of the viral genome
The antisense oligonucleotide microarray informs about the accessibility of tracks of 14 nt of the HCV IRES element, thus providing a wide overview of the global conformation of the molecule. However, it cannot reveal individual nucleotides eventually involved in long-range RNA–RNA interactions. To solve such a limitation, DMS chemical probing was performed to map the precise IRES nucleotides affected by the presence of the CRE and 3′-UTR elements.
The RNA transcripts previously used in the microarray assays (I, IU, IC and ICU) were denatured and renatured under low-magnesium conditions to achieve a stabilized native structure (19) as described earlier and then subjected to chemical modification with DMS (see ‘Materials and Methods’ section). Reactive residues were detected by a high-throughput approach (43,48,49) in which the primer extension reactions were performed with two different sets of fluorescent primers (see ‘Materials and Methods’ section). The reverse transcription elongation products were subsequently resolved by capillary electrophoresis and analyzed using ShapeFinder software (43). To our knowledge, this is the first report describing the use of that technology, originally developed for SHAPE chemistry (43), for DMS chemical probing. Unpaired A and C residues present in domains III and IV of the IRES element were studied using this strategy. Occasionally, reverse transcriptase stop points were also detected 5′ of highly reactive G residues (50,51). The relative reactivity at each nucleotide position was normalized as previously described (43), dividing the absolute reactivity value by the average of the 10% most reactive residues after excluding outliers. Mean and SD values (derived from at least four independent experiments) were calculated and represented in a histogram to provide the DMS probing profile of domains III and IV (Supplementary Figure S2).
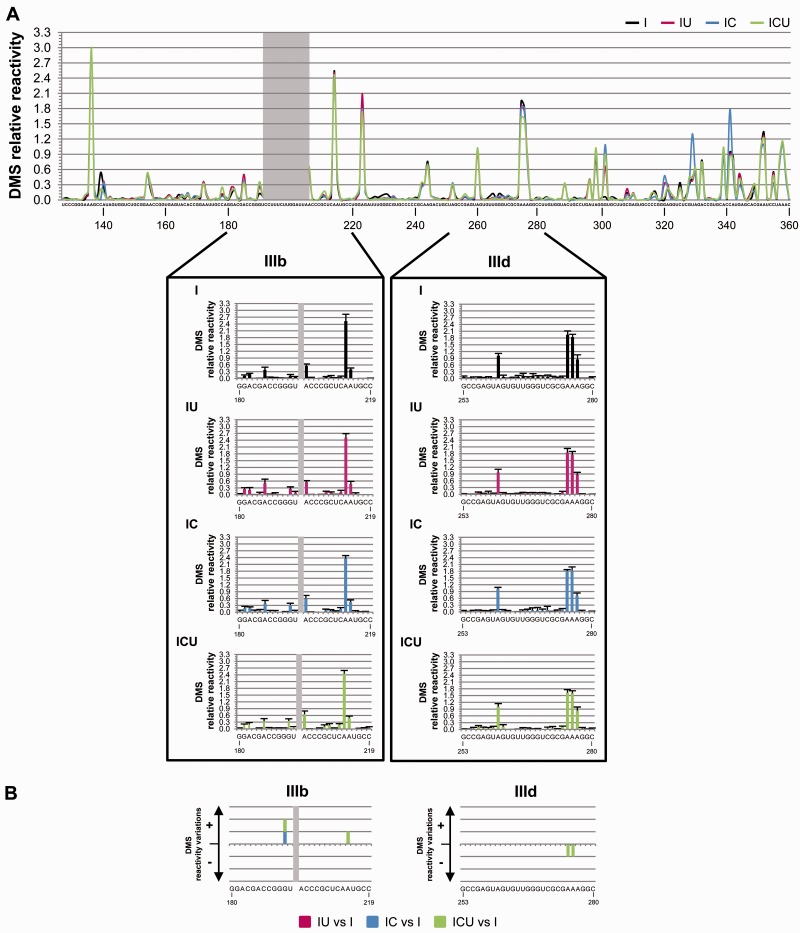
The results for construct I corresponded perfectly with the secondary structure proposed for domains III and IV of the IRES element (15,17–19,27,52) (Figure 3A and Supplementary Figure S2A). Similar analyses of the IU, IC and ICU transcripts allowed for evaluation of potential conformational changes, with respect to molecule I, mediated by the CRE and/or the 3′-UTR elements. The resulting reactivity patterns suggested the existence of differences in the DMS map (Figure 3A and Supplementary Figure S2A). Subsequent statistical analysis identified (independently of their degree of variation) the specific and significant variations in DMS reactivity (Figure 3B, Supplementary Figure S2B and Supplementary Table S2 for P values associated with each nucleotide position). Interestingly, the combined presence of the CRE and the 3′-UTR elements (molecule ICU) specifically induced significant reductions in DMS reactivity at nucleotides A275 (P = 0.020) and A276 (P = 0.020), located in the E loop motif of subdomain IIId (Figures 3 and 5; Supplementary Figure S2). This is in good agreement with previous results describing the existence of an interaction involving subdomain IIId of the IRES and the 3′-end of the HCV genome (37).
Figure 3.
Differential DMS sensitivity of the HCV IRES element induced by the CRE and the 3′-UTR motifs of the viral genome. (A) The graph shows a comparative analysis with the values of DMS reactivity at each nucleotide position for the four constructs under study (I, IU, IC and ICU). Noisy residues in the apical loop of subdomain IIIb are shadowed. Residues with DMS reactivity values <0.3 were classified as non-reactive positions. Lower panel: histogram representation showing a detailed view of the DMS reactivity pattern for subdomains IIIb (nucleotides 180–219) and IIId (nucleotides 253–280). Values are the mean of four independent experiments ± SD. (B) Significant variations (P ≤ 0.05) in DMS sensitivity at essential subdomains IIIb and IIId are represented qualitatively as gain (+) or loss (−) of reactivity for constructs IU, IC and ICU with respect to RNA I. Only reactive nucleotides (relative DMS reactivity ≥0.3) were considered in this analysis. Color code refers to the RNA construct: I, black; IU, magenta; IC, blue; ICU, green. Nucleotide positions are indicated as in Figure 1A.
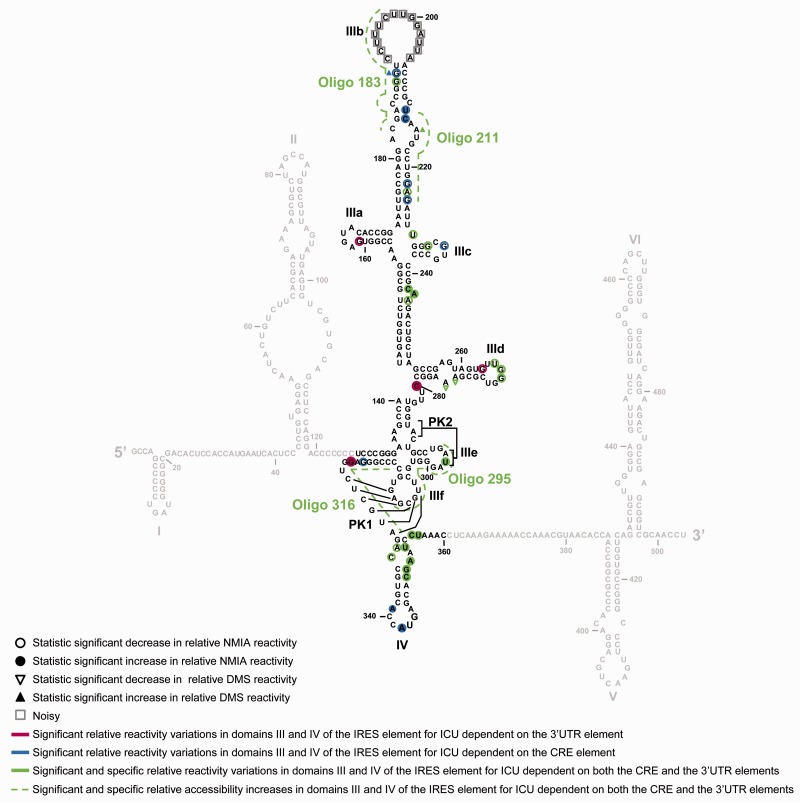
Figure 5.
IRES conformation is fine-tuned by the 3′-end of the HCV genome as detected by antisense DNA oligonucleotide microarrays, DMS chemical probing and SHAPE analysis. Comparison of the results obtained from three complementary methodologies for the study of the conformation of the IRES domains III and IV in the ICU molecule. Filled circles: significant increase (P ≤ 0.05) in NMIA reactivity in the transcript ICU with respect to molecule I; open circles: significant reduction (P ≤ 0.05) in NMIA reactivity for variant ICU with respect to construct I. Filled triangles: significant increase (P ≤ 0.05) in DMS reactivity in ICU with respect to construct I; open triangles: significant reduction (P ≤ 0.05) in DMS reactivity in ICU with respect to construct I. Additional color coding (also used in Figures 2–4) was included to indicate variations in specific reactivity derived from the presence of the CRE (blue), the 3′-UTR (magenta) or both elements (green), in the 3′-end of the viral genome. Gray squares: noisy residues.
In concordance with the microarray results, nucleotide A215 showed higher DMS reactivity in molecule ICU (P = 0.050), confirming the influence of the whole 3′-end in the folding of certain regions within the IRES element (Figure 5). The influence of either CRE or 3′-UTR elements on other positions of subdomain III (Figure 3 and Supplementary Figure S2B), was assessed by comparing the accessibility profiles for IC or IU molecules versus I. A significant increase in DMS sensitivity mediated by the CRE element was found at nucleotide G190 (P = 0.025), which was preserved in the ICU construct. Other changes in the DMS reactivity profile observed for the variants IC and IU were annulled when the whole 3′-end was used in the ICU construct (Figure 3 and Supplementary Figure S2). Importantly, most of the specific and significant reactivity changes detected were also found in the absence of the fluc gene (Supplementary Figure S3A and S3B; Supplementary Table S3), ruling out the possibility of an indirect effect of the luciferase coding sequence on IRES folding.
Together, these data show that the reactivity of certain specific residues in the HCV IRES element, including A275 and A276 of subdomain IIId, is affected by the presence of different 3′-tails, suggesting a direct influence of the CRE and 3′-UTR elements on IRES structure.
SHAPE analysis detected variations in HCV IRES conformation mediated by the 3′-end of the viral genome
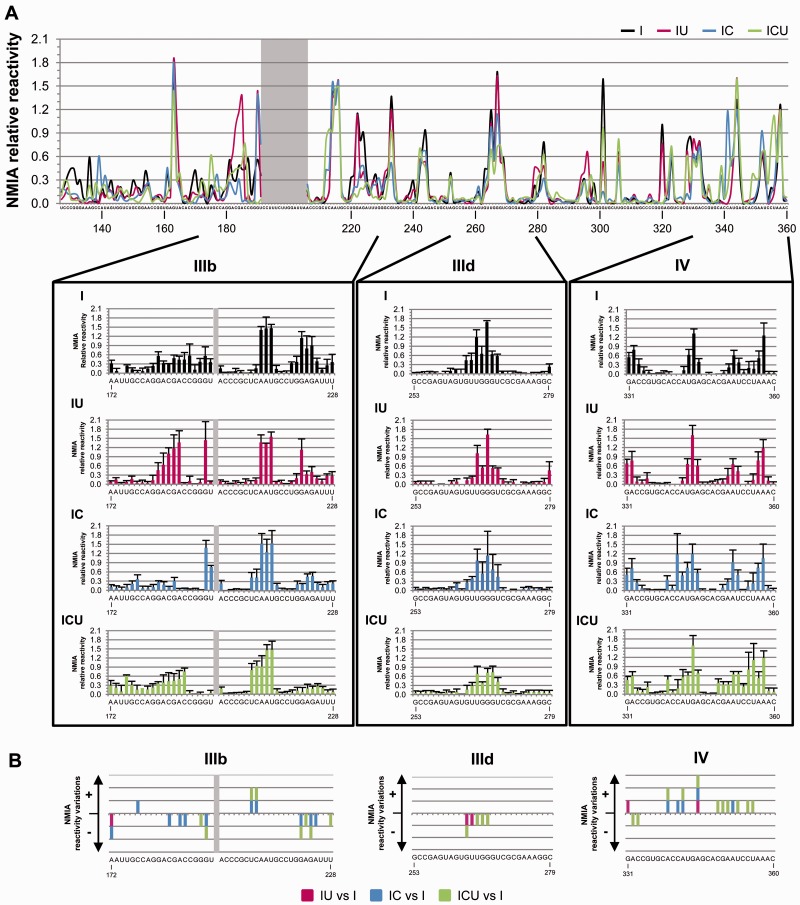
Antisense oligonucleotide microarray analysis and DMS probing can provide accurate information on the secondary and, eventually, higher-order structure of a target RNA molecule. However, in many cases, the folding features of a target molecule do not necessarily depend on base-pair interactions, but on the conformation of the ribose-phosphate backbone. This may ultimately define the flexibility and overall geometry of an RNA molecule. Selective 2′-hydroxyl acylation and primer extension (SHAPE) (53,54) analysis was therefore performed on transcripts I, IU, IC and ICU using NMIA as the acylating reagent. The formation of 2′-O-adducts was monitored by primer extension with two sets of fluorescently labeled DNA oligonucleotides and analyzed using ShapeFinder software as described earlier (see also ‘Materials and Methods’ section). The mean relative reactivity and SD (for at least five independent experiments) was calculated for each nucleotide position. This allowed the construction of the corresponding SHAPE profile of the studied region (Figure 4A and Supplementary Figure S4A).
Figure 4.
The SHAPE pattern of the HCV IRES is influenced by the 3′-end of the viral genome. (A) Mean NMIA reactivity values for IRES domains III and IV of the transcripts I, IU, IC and ICU calculated from, at least, five independent experiments and represented in a line graph according to color code indicated in Figure 3A. Nucleotides with mean reactivity values <0.3 were considered non-reactive. The gray box indicates noisy positions. Lower panel: detailed views of the SHAPE profile for subdomains IIIb (nucleotides 180–219) and IIId (nucleotides 253–280) and domain IV (nucleotides 331–360). (B) Graphs show significant variations (P ≤ 0.05) in the SHAPE pattern detected at subdomains IIIb, IIId and IV for molecules IU, IC and ICU with respect to RNA I at positions with reactivity values of ≥0.3. Reactivity changes are qualitatively indicated by (+) (increase) or (−) (decrease). Nucleotide numbering is indicated according to Figure 1A. Color code is indicated as in Figure 3.
The reactivity pattern obtained for transcript I showed no significant differences with respect to that previously reported (55) (Figure 4A). This reinforced the idea that the HCV IRES element adopts a preferential, stable conformation in native conditions. Interestingly, the comparative representation revealed remarkable differences in NMIA sensitivity for the transcripts encompassing different regions of the 3′-end with respect to molecule I (Figure 4A and Supplementary Figure S4A). Statistical analyses filtered the large number of nucleotides differentially modified to reveal specific and significant differences in the reactivity profile for the molecule ICU compared to variant I in precise nucleotide positions (Figure 4B and Supplementary Figure S4B; see also Supplementary Table S4 for a detailed summary of the P values at each reactive residue). As detected in DMS probing and microarray assays (Figures 2 and 3), modifications able to drastically influence the secondary structure were not expected. Hence, the study was focused on statistically significant reactivity variations affecting to nucleotides extended stretch at functionally relevant domains. Most of these differences affected structural elements essential for the IRES function (Figures 4 and 5). Interestingly, a significant reduction in NMIA sensitivity was observed for residues U265 (P = 0.034), G266 (P = 0.050) and G267 (P = 0.048), located in the apical loop of subdomain IIId. Again, this firmly supports the previous hypothesis that a long-range RNA–RNA interaction can be established between subdomains IIId and the CRE element (37). SHAPE analysis also revealed significant increases (P ≤ 0.050) in the flexibility of the residues flanking the translation initiation codon in domain IV (Figures 4 and 5; Supplementary Table S4). In addition, several nucleotides throughout the stem of subdomain IIIb showed differential NMIA sensitivity compared to transcript I, suggesting alterations in its local conformation. Deviations (mainly reductions) in the reactivity pattern related to single nucleotides were also significant in subdomains IIIa, IIIc and IIIe, suggesting the existence of sporadic folding reorganization processes upon the interaction of these subdomains with the 3′-end of the viral genome (Figures 4 and 5; Supplementary Table S4).
To analyze the individual role played by the CRE and 3′-UTR elements present in the ICU molecule, the IC and IU constructs were compared to molecule I using SHAPE chemistry. The presence of each of the 3′ elements also affected the reactivity pattern at single nucleotide positions, most of these modifications being preserved in the ICU molecule (Figures 4 and 5; Supplementary Figure S4). These results support the notion of a specific role for these 3′ elements in the direct regulation of IRES conformation. Thus, the presence of the CRE region promoted remarkable and significant variations in NMIA-reactivity at nucleotides located throughout the stem of subdomain IIIb and domain IV (Figures 4 and 5; Supplementary Table S4) in variants IC and ICU. Slight susceptibility to the presence of the independent 3′-UTR element (molecule IU) was also noted at single nucleotide positions throughout domains III and IV (Figures 4 and 5; Supplementary Table S4). All these reactivity modifications involving 1 nt are likely related to local conformational changes produced in the IRES element triggered by the presence of the different 3′ elements. Further modifications in the IRES SHAPE pattern promoted by the independent CRE and 3′-UTR elements were reversed when both domains were combined in the ICU transcript, as previously noted in the DMS analysis (Figure 4). Moreover, the reductions in reactivity detected for the variant ICU at subdomain IIId represent specific events mediated by the presence of the 3′-tail domains, as evidenced by the fact that most of the reactivity changes were preserved in the constructs lacking the fluc gene (Supplementary Figure S3C and Supplementary Table S4).
These results support the idea that conformational rearrangements affecting essential regions of the IRES element (such as subdomains IIIb and IIId, as well as domain IV) are strongly mediated by structural domains placed at the 3′-end of the HCV genome. Therefore, such long-range RNA–RNA interactions may contribute to the acquisition of a fully functional IRES structure.
IRES folding in a replication-competent viral RNA
The structural analyses described earlier were performed on IRES-dependent translationally active RNA constructs. DMS chemical probing and SHAPE structural analysis were then performed using a replication-competent viral RNA molecule. For this, a subgenomic HCV replicon RNA was obtained by in vitro transcription from the plasmid pFK-I389FLucNS3-3′ET (40). This construct (termed Rep) carries the HCV IRES derived from genotype 1b followed by the fluc gene, the IRES element belonging to the encephalomyocarditis virus (EMCV) and the coding sequence for the non-structural HCV proteins plus the HCV 3′-UTR (Figure 6A).
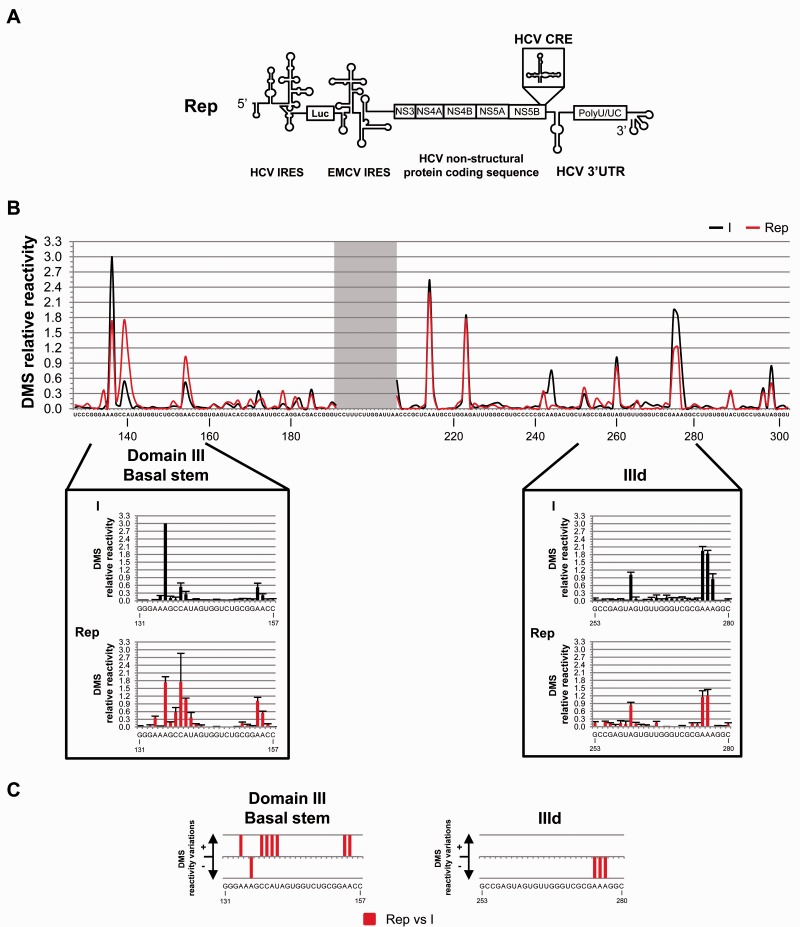
Figure 6.
DMS pattern for domain III of the HCV IRES within a replication-competent RNA molecule (Rep). An RNA transcript derived from the plasmid pFK-I389FLucNS3-3′ET (40) was subjected to DMS analysis. (A) Schematic representation of the replicative RNA construct Rep. (B) DMS reactivity profiles for transcripts I (black) and Rep (red). Reactivity values of <0.3 indicate non-reactive positions. Noisy nucleotides are shadowed. Lower panels: histograms of DMS reactivity for subdomain IIId and the short stem at the base of the domain III. Data are the mean of, at least, four independent assays ± SD. (C) Significant variations (P ≤ 0.05) in DMS sensitivity in the replicative RNA Rep with respect to construct I at reactive nucleotides of subdomain IIId and the basal stem of domain III. Qualitative increases (+) and reductions (−) in DMS reactivity are represented. Nucleotide positions (according to Figure 1A) are indicated.
Chemical probing with DMS was performed on Rep under the same experimental conditions as those described earlier. The mean of the relative reactivity values from four independent assays was estimated at each nucleotide position to evaluate the accessibility pattern (Figure 6B). Compared to construct I, notable differences were measured in the DMS reactivity profile affecting residues located in the E loop of subdomain IIId. Reduction in accessibility reached statistical significance at the trinucleotide A274-A276 of subdomain IIId (A274, P = 0.011; A275, P = 0.011; A276, P = 0.022; Figures 6B and 6C, as well as Supplementary Figure S5A and S5B; see Supplementary Table S2 for a complete list of the P values corresponding to those reactive positions). This is in good agreement with DMS probing data previously obtained for the ICU transcript. The DMS reactivity profile of the central stem of domain III was also affected in Rep (A243, P = 0.008; A244, P = 0.020; Figures 6B and 6C; Supplementary Table S2) compared to construct I. Accordingly, SHAPE analysis also revealed differential reactivity for these nucleotides in the ICU transcript (Figure 5). Compared to construct I; any other 1-nt modifications in the DMS sensitivity of Rep was hardly observed (Figure 6B and 6C; Supplementary Figure S5A and S5B).
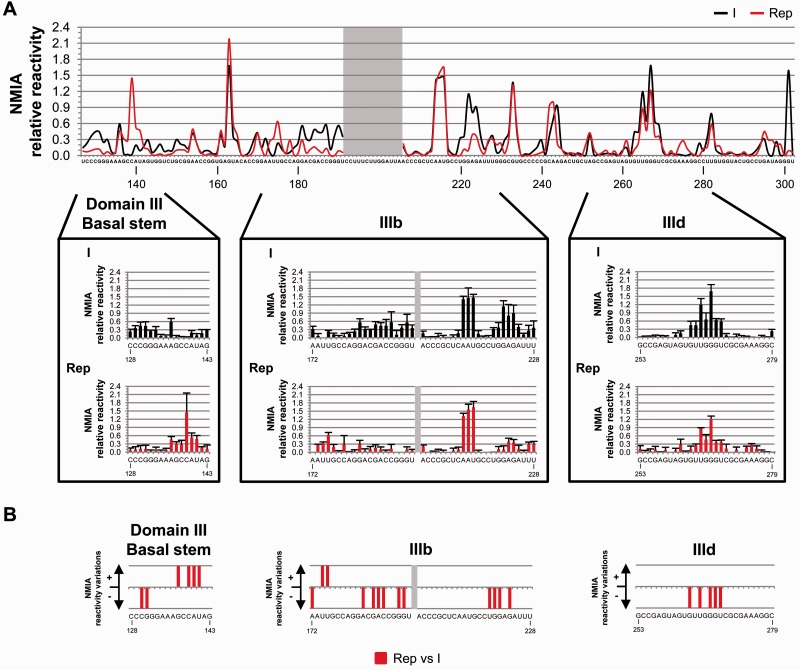
The study of the IRES conformation in the replication-competent RNA was complemented with SHAPE analyses. The comparison of the IRES reactivity profile in the construct Rep with that of variant I showed several changes analogous to those previously observed for the ICU transcript (Figure 7A and Supplementary Figure S5C), thus reinforcing the consistency of our findings. These changes consisted in significant reductions in NMIA reactivity in the apical loop of subdomain IIId (G263, P = 0.034; U265, P = 0.025; G267, P = 0.034; G268, P = 0.050 and U269, P = 0.034; Figures 7B and 8; Supplementary Figure S5C and S5D). This is a further proof supporting a long-range interaction between IRES subdomain IIId and the 3′-end of the HCV genome (37). Local rearrangements at the stem of subdomain IIIb were also inferred from the SHAPE reactivity pattern, in agreement with those detected for the ICU molecule (Figures 7 and 8; Supplementary Figure S5C and S5D; see also Supplementary Table S4 for P values at each reactive nucleotide). Additional variations in the SHAPE pattern were also detected at different nucleotides, suggesting that other RNA domains of the HCV genome play a role in the overall conformation of the IRES element. Among them, a relevant change in reactivity to chemical modification was detected at the short basal stem of domain III: compared to molecule I, the residues C139-U141 of Rep were more exposed to both NMIA and DMS probes (Figures 7 and 8).
Figure 7.
SHAPE analysis of domain III of the HCV IRES within a replication-competent transcript. (A) Comparative study of the mean NMIA reactivity values obtained from at least five independent experiments for RNA molecules I (black) and Rep (red). Mean values of <0.3 correspond to non-reactive positions. The gray box shows noisy residues. Lower panel: detailed views of the SHAPE reactivity profile for selected regions within domain III. Histograms of SHAPE data represent the mean NMIA relative reactivity value ± SD. (B) Significant variations (P ≤ 0.05) in the SHAPE pattern for the replicative RNA molecule Rep with respect to transcript I. A qualitative representation of the gain (+) or loss (−) in SHAPE reactivity at subdomains IIIb, IIId and the basal stem of domain III is shown. Nucleotide numbering is specified as described in Figure 1A.
Figure 8.
Impact of the non-structural proteins coding sequence on IRES folding. Summary of significant reactivity differences (P ≤ 0.05) observed in DMS and SHAPE profiles for the replication-competent RNA molecule (Rep) with respect to transcript I. Filled circles: significant increase (P ≤ 0.05) in NMIA reactivity in Rep with respect to molecule I; open circles: significant reduction (P ≤ 0.05) in NMIA reactivity for Rep with respect to construct I; filled triangles: significant increase (P ≤ 0.05) in DMS reactivity in Rep with respect to construct I; open triangles: significant reduction (P ≤ 0.05) in DMS reactivity in Rep with respect to construct I; grey squares: noisy residues.
Together, the present data support the existence of a complex, long-distance RNA–RNA interaction network in the HCV genome, which fine-tunes the 3D structure of the active IRES element.
DISCUSSION
HCV genome contains well-defined, phylogenetically conserved structural elements that mainly localize at its 5′- and 3′-termini and extend to the ORF (56–60). These regions are essential for the consecution of two key, closely related processes for the viral cycle: translation and replication. In particular, HCV translation initiation is dependent on an IRES element located at the 5′-end of the viral genome and influenced by the presence of RNA domains placed at the 3′-end of the genome. Both ends can participate in the acquisition of a circular conformation of the viral genome, which is typically associated to the interaction with cellular and viral protein factors (61). We recently reported that long-distant RNA–RNA interactions between both ends of the HCV genome may help form this circular topology, with important consequences for IRES function (37). The results of the present work show that the IRES conformation is fine-tuned by the presence of different RNA domains at the 3′-end, independently of protein factors. Our findings also suggest that the overall effect of the 3′-end on IRES folding reflects the combined contribution of CRE and 3′-UTR elements, since the presence of each of these 3′ regions induces conformational changes that map to different IRES nucleotides. These findings are reinforced by the fact that the structural rearrangements of the IRES induced by the 3′-end elements are functionally relevant in the context of translation and replication-active RNA transcripts. Therefore, though the IRES region acquires a functional conformation in the absence of additional RNA elements, as previously described (16,19), our results show that its structure can be further fine-tuned by the 3′-end of the HCV genome. This is in good agreement with previous observations demonstrating the functional role of the CRE and the 3′-UTR elements on IRES activity (33). To our knowledge, this is the first report describing complex, long-range RNA–RNA interactions between the 5′- and 3′-ends of HCV genome that influence the functional conformation of viral RNA elements.
Three experimental strategies were followed to study the structure of the IRES element in the presence of different 3′-tails: hybridization to antisense DNA oligonucleotide microarrays, DMS chemical probing and SHAPE structural analysis. These methodologies provide alternative and complementary information on the 3D structure of a target RNA molecule. Microarray analysis informs about the accessibility of consecutive, overlapping stretches of nucleotides, being the main limitation of the method that it cannot provide single-nucleotide resolution. In turn, classical chemical probing methods, such as DMS, inform about the solvent accessibility of the nucleobases at the residue level, although the data obtained are still limited since DMS does not recognize alternative conformational rearrangements that might affect the ribose-phosphate backbone without altering the base-pairing and stacking pattern. Finally, SHAPE structural analysis has minor but significant drawbacks that must be taken into account. It has been described that 2′-OH reactivity does not depend on solvent accessibility and may be slightly influenced by several factors including the pKa value of the surrounding nucleotides (62), the nucleobase identity (63) and, more importantly, the acquisition of alternative foldings in the RNA duplex distinct from the canonical A-form (64). This implies that all the conformationally dynamic residues can be detected by NMIA probing, although they remain solvent inaccessible. In this work, capillary electrophoresis followed by analysis using ShapeFinder software was used to detect not only NMIA modifications but also DMS signals. This experimental achievement facilitated comparisons between the results obtained using both types of probing methods.
Analysis of the IRES folding in the construct lacking any 3′-tail (molecule I) using these methodologies revealed that the global conformation of domains III and IV (Figures 2, 3 and 4) was consistent with the previously published results (14,17–20,55,65). Interestingly, structural features of functionally relevant regions of the IRES element were evidenced by this study. For example, the microarray analysis reported high fluorescence signal at the oligonucleotide 176, which recognizes the stem of subdomain IIIb (Figure 2 and Supplementary Figure S1). This observation was reinforced by the medium NMIA reactivity detected for this nucleotide stretch (Figure 4). In contrast, this region was barely susceptible to DMS modification (Figure 3 and Supplementary Figure S2). It should be noted that the subdomain IIIb of HCV IRES contains a GC-rich stem interrupted by a variable internal loop and a CC mismatch, followed by an apical hairpin loop (17). These features remarkably determine its geometry and suggest that certain flexibility might account at the backbone of this region while the base-pairing is preserved in the duplex, as detected by both previous NMR studies (17) and the DMS probing reported in this study (Figure 3). Therefore, our results evidence the usefulness of combining different methods to obtain a complete overview of the RNA folding in structurally complex regions.
Experimental mapping of HCV IRES structure was initiated with RNA transcripts bearing distinct 3′-tails that had been previously shown to exhibit differential translational efficiency (33). Significant changes in the NMIA and DMS reactivity patterns were detected in specific structural regions of the IRES domains III and IV, suggesting that local conformation rearrangements are tuned by the presence of CRE and/or 3′-UTR elements (Figures 2–5). Thus, in the presence of the whole 3′-end of the HCV genome (molecule ICU), a remarkable reduction in NMIA reactivity was detected at specific residues of the apical loop of subdomain IIId. Also, DMS reactivity was reduced at nucleotides belonging to the essential internal E loop of this subdomain (Figures 3 and 5). NMR and molecular modeling studies have previously shown that the formation of a typical U-turn motif (27) at the apical loop could be related to the proper recruitment of the ribosomal subunit 40S (11,16,19,25,26). Significantly, the subdomain IIId is also a primary determinant in the acquisition of the functional folding of surrounding structural elements (19). Hence, it seems likely that a decrease in the flexibility of this subdomain upon interaction with the whole 3′-end of the viral genome (Figures 4 and 5) would affect the local structural environment. This is in good agreement with the concomitant increase detected in the microarray hybridization signal for the molecule ICU at oligonucleotides 295 and 316, mapping at the subdomain IIIe and the PK1 element, respectively (Figures 2 and 5). This region has been recently related to the formation of a double pseudoknot structure that controls the orientation of the start codon at the ribosomal P site (14). Importantly, these findings were confirmed using a short RNA transcript lacking the fluc gene, what supports the specificity of the observed effect (Supplementary Figure S3 and Supplementary Table S5). These IRES rearrangements triggered by the interaction with the CRE and 3′-UTR elements may intervene in translation initiation by affecting to the 48S complex assembly, thus providing an explanation for the translational properties of the transcript ICU (33). The results obtained with this transcript showed that the 3′-UTR enhancer effect on HCV IRES function can be further regulated by the CRE element.
Important variations in the reactivity and hybridization profiles were also observed in the transcript ICU, compared to molecule I, in the eIF3 binding platform located at the major stem of domain III (Figures 2–5). It has been described that the binding of eIF3 is strictly dependent on the intact four-way helical junction defined by subdomains IIIabc (18). In addition, the typical S-turn geometry of the internal loop of the subdomain IIIb is also necessary for full IRES-dependent translational efficiency (17). The conformational variations throughout subdomain IIIb mediated by the 3′-end could influence its proper folding required for the interaction with eIF3 (17). This provides additional clues to understand the singular translational properties of the ICU construct (33).
Domain IV was also remarkably affected by the presence of the whole 3′-end of the viral genome, as derived from the NMIA reactivity pattern (Figures 4 and 5). It has been described that the interplay between domains II and IV of the HCV IRES and the ribosomal subunit 40S seems to be essential for the proper unwinding of the stem and the further recognition of the AUG codon (65). Therefore, it is assumed that the stability of domain IV is inversely correlated with the IRES-dependent translation efficiency (20). The significant gain observed in NMIA reactivity of the transcript ICU at numerous nucleotides of domain IV (Figures 4 and 5) points to an increase in the flexibility of this domain. Therefore, in a biological context, the presence of the whole 3′-end of the viral genome could promote a destabilization of domain IV additional to that exerted by domain II, thus contributing to the proper positioning of the start codon. A further investigation of this interaction would provide important insights into the efficiency and regulation of the IRES-dependent translation, as previously suggested (33).
The analysis performed on the synthetic construct ICU was extended to a replication-competent RNA transcript (Figures 6, 7 and 8). Significant reductions in DMS and NMIA reactivity were again measured at the apical and the internal loops of subdomain IIId. Variations in the reactivity profile at the major stems of domain III were also detected. These results confirm the robustness of the present findings and show that the conformation of important functional regions of the IRES element is modulated by the 3′-end of the viral genome. Further, in the context of the transcript Rep, the IRES element shows changes in reactivity that affect a larger number of nucleotides than those previously mapped in the transcript ICU. In particular, changes in DMS and NMIA reactivity involving the short basal stem of domain III were specifically detected in Rep (Figures 6 and 7). This suggests the existence of complex and still unexplored interaction networks that involve additional distant regions located throughout the non-structural proteins coding region of the HCV genome.
In summary, the present study shows that the 3D folding of domains III and IV of the IRES element is fine-tuned by RNA–RNA interactions involving the entire 3′-end of the HCV genome. Such long-range interactions occur in the absence of protein factors. This findings could have important consequences for a better understanding of IRES-dependent HCV translation, in agreement with previous observations demonstrating a regulatory role played by the entire viral 3′-end (33). Further studies will be required to clarify the mechanistic and functional details of the observed structural changes. Taken together, the reported observations, along with previous data, depict a more complex scenario for HCV IRES-dependent translation, in which both RNA–RNA and RNA–protein interactions cooperate to modulate viral protein synthesis. These findings might eventually be extended to other closely related, IRES-containing RNA viruses. Finally it is worth noting that our data exemplify that the structure of even well characterized RNA elements, such as the HCV IRES element, may be refined when they are analyzed in a genomic context that includes additional RNA regions. Thus, complex interaction networks determined by long-distance, direct RNA–RNA contacts, could play important roles in the regulation of essential steps in the HCV cycle.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–5.
FUNDING
Spanish Ministerio de Ciencia e Innovación, MICINN [BFU2009-08137 to A.B.-H., EUI2008-00158 and BIO2010-20696 to C.B.]; Junta de Andalucía [CTS-5077 to A.B.-H.]; University of Granada-GREIB Project [GREIB.PYR_2010_12 to C.R.-L.]; Spanish Council for Scientific Research [201120E004 to A.B.-H.]; FEDER funds from the EU [to A.B.-H. and C.B.]; Instituto de Salud Carlos III [to CIBERehd]. Funding for open access charge: Spanish Ministerio de Ciencia e Innovación, MICINN [BFU2009-08137 to A.B.-H.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Vicente Augustin and María Fernández Algar for excellent technical assistance. The authors acknowledge Jaime Iranzo for his advice and suggestions on the statistical analysis of experimental data.
REFERENCES
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001;75:12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 2002;76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi M, Lemon SM. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003;9:331–345. doi: 10.1261/rna.2144203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 16.Odreman-Macchioli FE, Tisminetzky SG, Zotti M, Baralle FE, Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875–885. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-Ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 18.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 19.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 20.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 21.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl Acad. Sci. USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 2000;74:10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–1151. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito T, Tahara SM, Lai MM. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- 30.Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bung C, Bochkaeva Z, Terenin I, Zinovkin R, Shatsky IN, Niepmann M. Influence of the hepatitis C virus 3′-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett. 2010;584:837–842. doi: 10.1016/j.febslet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Romero-López C, Berzal-Herranz A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 2012;69:103–113. doi: 10.1007/s00018-011-0729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito T, Lai MM. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 35.Fang JW, Moyer RW. The effects of the conserved extreme 3′ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J. Hepatol. 2000;33:632–639. doi: 10.1034/j.1600-0641.2000.033004632.x. [DOI] [PubMed] [Google Scholar]
- 36.Kong LK, Sarnow P. Cytoplasmic expression of mRNAs containing the internal ribosome entry site and 3′ noncoding region of hepatitis C virus: effects of the 3′ leader on mRNA translation and mRNA stability. J. Virol. 2002;76:12457–12462. doi: 10.1128/JVI.76.24.12457-12462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris E, Holden KL, Edgil D, Polacek C, Clyde K. Molecular biology of flaviviruses. Novartis Found. Symp. 2006;277:23–39. discussion 40, 71–23, 251–253. [PubMed] [Google Scholar]
- 39.Romero-López C, Barroso-delJesus A, Puerta-Fernández E, Berzal-Herranz A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005;386:183–190. doi: 10.1515/BC.2005.023. [DOI] [PubMed] [Google Scholar]
- 40.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernández N, García-Sacristán A, Ramajo J, Briones C, Martínez-Salas E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology. 2011;409:251–261. doi: 10.1016/j.virol.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Martell M, Briones C, de Vicente A, Piron M, Esteban JI, Esteban R, Guardia J, Gomez J. Structural analysis of hepatitis C RNA genome using DNA microarrays. Nucleic Acids Res. 2004;32:e90. doi: 10.1093/nar/gnh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann EL, D’Abrera HJM. Nonparametrics: Statistical Methods Based on Ranks. 3rd edn. New York: Springer-Verlag; 2006. [Google Scholar]
- 46.Fernández N, Fernández-Miragall O, Ramajo J, García-Sacristán A, Bellora N, Eyras E, Briones C, Martínez-Salas E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res. 2011;39:8572–8585. doi: 10.1093/nar/gkr560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooms M, Verhoef K, Southern E, Huthoff H, Berkhout B. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res. 2004;32:819–827. doi: 10.1093/nar/gkh206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 49.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005;6:361–366. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chamberlin SI, Weeks KM. Differential helix stabilities and sites pre-organized for tertiary interactions revealed by monitoring local nucleotide flexibility in the bI5 group I intron RNA. Biochemistry. 2003;42:901–909. doi: 10.1021/bi026817h. [DOI] [PubMed] [Google Scholar]
- 54.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 55.Pang PS, Elazar M, Pham EA, Glenn JS. Simplified RNA secondary structure mapping by automation of SHAPE data analysis. Nucleic Acids Res. 2011;39:e151. doi: 10.1093/nar/gkr773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA. 2002;8:824–841. doi: 10.1017/s1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 58.You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl Acad. Sci. USA. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You S, Rice CM. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J. Virol. 2008;82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgil D, Harris E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Velikyan I, Acharya S, Trifonova A, Foldesi A, Chattopadhyaya J. The pK(a)′s of 2′-hydroxyl group in nucleosides and nucleotides. J. Am. Chem. Soc. 2001;123:2893–2894. doi: 10.1021/ja0036312. [DOI] [PubMed] [Google Scholar]
- 63.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA. 2011;17:1258–1273. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.