Abstract
We present Quip, a lossless compression algorithm for next-generation sequencing data in the FASTQ and SAM/BAM formats. In addition to implementing reference-based compression, we have developed, to our knowledge, the first assembly-based compressor, using a novel de novo assembly algorithm. A probabilistic data structure is used to dramatically reduce the memory required by traditional de Bruijn graph assemblers, allowing millions of reads to be assembled very efficiently. Read sequences are then stored as positions within the assembled contigs. This is combined with statistical compression of read identifiers, quality scores, alignment information and sequences, effectively collapsing very large data sets to <15% of their original size with no loss of information. Availability: Quip is freely available under the 3-clause BSD license from http://cs.washington.edu/homes/dcjones/quip.
INTRODUCTION
With the development of next-generation sequencing (NGS) technology, researchers have had to adapt quickly to cope with the vast increase in raw data. Experiments that would previously have been conducted with microarrays and resulted in several megabytes of data, are now performed by sequencing, producing many gigabytes, and demanding a significant investment in computational infrastructure. While the cost of disk storage has also steadily decreased over time, it has not matched the dramatic change in the cost and volume of sequencing. A transformative breakthrough in storage technology may occur in the coming years, but the era of the $1000 genome is certain to arrive before that of the $100 petabyte hard disk.
As cloud computing and software as a service become increasingly relevant to molecular biology research, hours spent transferring NGS data sets to and from off-site servers for analysis will delay meaningful results. More often researchers will be forced to maximize bandwidth by physically transporting storage media (via the ‘sneakernet’), an expensive and logistically complicated option. These difficulties will only be amplified as exponentially more sequencing data are generated, implying that even moderate gains in domain-specific compression methods will translate into a significant reduction in the cost of managing these massive data sets over time.
Storage and analysis of NGS data centers primarily around two formats that have arisen recently as de facto standards: FASTQ and SAM. FASTQ stores, in addition to nucleotide sequences, a unique identifier for each read and quality scores, which encode estimates of the probability that each base is correctly called. For its simplicity, FASTQ is a surprisingly ill-defined format. The closest thing to an accepted specification is the description by Cock et al. (1), but the format arose ad hoc from multiple sources (primarily Sanger and Solexa/Illumina), so a number of variations exist, particularly in how quality scores are encoded. The SAM format is far more complex but also more tightly defined, and comes with a reference implementation in the form of SAMtools (2). It is able to store alignment information in addition to read identifiers, sequences and quality scores. SAM files, which are stored in plain text, can also be converted to the BAM format, a compressed binary version of SAM, which is far more compact and allows for relatively efficient random access.
Compression of nucleotide sequences has been the target of some interest, but compressing NGS data, made up of millions of short fragments of a greater whole, combined with metadata in the form of read identifiers and quality scores, presents a very different problem and demands new techniques. Splitting the data into separate contexts for read identifiers, sequences and quality scores, and compressing them with the Lempel-Zip algorithm and Huffman coding has been explored by Tembe et al. (3) and Deorowicz and Grabowski (4), who demonstrate the promise of domain-specific compression with significant gains over general-purpose programs like gzip and bzip2.
Kozanitis et al. (5) and Hsi-Yang Fritz et al. (6) proposed reference-based compression methods, exploiting the redundant nature of the data by aligning reads to a known reference genome sequence and storing genomic positions in place of nucleotide sequences. Decompression is then performed by copying the read sequences from the genome. Though any differences from the reference sequence must also be stored, referenced-based approaches can achieve much higher compression and they grow increasing efficient with longer read lengths, since storing a genomic position requires the same amount of space, regardless of the length of the read.
This idea is explored also in the Goby format (http://campagnelab.org/software/goby/), which has been proposed as an alternative to SAM/BAM, the primary functional difference being that sequences of aligned reads are not stored but looked up in a reference genome when needed (frequently they are not). For some applications, reference-based compression can be taken much further by storing only single nucleotide polymorphism (SNP) information, summarizing a sequencing experiment in several megabytes (7). However, even when SNP calls are all that is needed, discarding the raw reads would prevent any reanalysis of the data.
While a reference-based approach typically results in superior compression, it has a number of disadvantages. Most evidently, an appropriate reference sequence database is not always available, particularly in the case of metagenomic sequencing. One could be contrived by compiling a set of genomes from species expected to be represented in the sample. However, a high degree of expertise is required to curate and manage such a project-dependent database. Secondly, there is the practical concern that files compressed with a reference-based approach are not self-contained. Decompression requires precisely the same reference database used for compression, and if it is lost or forgotten, the compressed data becomes inaccessible.
Another recurring theme in the the growing literature on short read compression is lossy encoding of sequence quality scores. This follows naturally from the realization that quality scores are particularly difficult to compress. Unlike read identifiers, which are highly redundant, or nucleotide sequences, which contain some structure, quality scores are inconsistently encoded between protocols and computational pipelines and are often simply high-entropy. It is dissatisfying that metadata (quality scores) should consume more space than primary data (nucleotide sequences). Yet, also dissatisfying to many researchers is the thought of discarding information without a very good understanding of its effect on downstream analysis.
A number of lossy compression algorithms for quality scores have been proposed, including various binning schemes implemented in QScores-Archiver (8) and SCALCE (http://scalce.sourceforge.net), scaling to a reduced alphabet with randomized rounding in SlimGene (5), and discarding quality scores for bases which match a reference sequence in Cramtools (6). In SCALCE, SlimGene and Cramtools, quality scores may also be losslessly compressed. Kozanitis et al. (5) analyzed the effects of their algorithm on downstream analysis. Their results suggest that while some SNP calls are affected, they are primarily marginal, low-confidence calls between hetero- and homozygosity.
Decreasing the entropy of quality scores while retaining accuracy is an important goal, but successful lossy compression demands an understanding of what is lost. For example, lossy audio compression (e.g. MP3) is grounded in psychoacoustic principles, preferentially discarding the least perceptible sound. Conjuring a similarly principled method for NGS quality scores is difficult given that both the algorithms that generate them and the algorithms that are informed by them are moving targets. In the analysis by Kozanitis et al. (5), the authors are appropriately cautious in interpreting their results, pointing out that ‘there are dozens of downstream applications and much work needs to be done to ensure that coarsely quantized quality values will be acceptable for users.’
In the following sections, we describe and evaluate Quip, a new lossless compression algorithm which leverages a variety of techniques to achieve very high compression over sequencing data of many types, yet remains efficient and practical. We have implement this approach in an open-source tool that is capable of compressing both BAM/SAM and FASTQ files, retaining all information from the original file.
MATERIALS AND METHODS
Statistical compression
The basis of our approach is founded on statistical compression using arithmetic coding, a form of entropy coding which approaches optimality, but requires some care to implement efficiently [see Said (9) for an excellent review]. Arithmetic coding can be thought of as a refinement of Huffman coding, the major advantage being that it is able to assign codes of a non-integral number of bits. If a symbol appears with probability 0.1, it can be encoded near to its optimal code length of −log2(0.1) ≈ 3.3 bits. Despite its power, it has historically seen much less use than Huffman coding, due in large part to fear of infringing on a number of patents that have now expired.
Arithmetic coding is a particularly elegant means of compression in that it allows a complete separation between statistical modeling and encoding. In Quip, the same arithmetic coder is used to encode quality scores, read identifiers, nucleotide sequences and alignment information, but with very different statistical models for each, which gives it a tremendous advantage over general-purpose compression algorithms that lump everything into a single context. Furthermore, all parts of our algorithm use adaptive modeling: parameters are trained and updated as data are compressed, so that an increasingly tight fit and higher compression is achieved on large files.
Read identifiers
The only requirement of read identifiers is that they uniquely identify the read. A single integer would do, but typically each read comes with a complex string containing the instrument name, run identifier, flow cell identifier and tile coordinates. Much of this information is the same for every read and is simply repeated, inflating the file size.
To remove this redundancy, we use a form of delta encoding. A parser tokenizes the ID into separate fields which are then compared with the previous ID. Tokens that remain the same from read to read (e.g. instrument name) can be compressed to a negligible amount of space—arithmetic coding produces codes of less than 1 bit in such cases. Numerical tokens are recognized and stored efficiently, either directly or as an offset from the token in the same position in previous read. Otherwise, non-identical tokens are encoded by matching as much of the prefix as possible to the previous read's token before directly encoding the non-matching suffix.
The end result is that read IDs, which are often 50 bytes or longer, are typically stored in 2–4 bytes. Notably, in reads produced from Illumina instruments, most parts of the ID can be compressed to consume almost no space; the remaining few bytes are accounted for by tile coordinates. These coordinates are almost never needed in downstream analysis, so removing them as a preprocessing step would shrink file sizes even further. The parser used is suitably general so that no change to the compression algorithm would be needed.
Nucleotide sequences
To compress nucleotide sequences, we adopt a very simple model based on high-order Markov chains. The nucleotide at a given position in a read is predicted using the preceding 12 positions. This model uses more memory than traditional general-purpose compression algorithms (413 = 67 108 864 parameters are needed, each represented in 32 bits) but it is simple and extremely efficient (very little computation is required and run time is limited primarily by memory latency, as lookups in such a large table result in frequent cache misses).
An order-12 Markov chain also requires a very large amount of data to train, but there is no shortage with the data sets we wish to compress. Though less adept at compressing extremely short files, after compressing several million reads, the parameters are tightly fit to the nucleotide composition of the data set so that the remaining reads will be highly compressed. Compressing larger files only results in a tighter fit and higher compression.
Quality scores
It has been previously noted that the quality score at a given position is highly correlated with the score at the preceding position (5). This makes a Markov chain a natural model, but unlike nucleotides, quality scores are over a much larger alphabet (typically 41–46 distinct scores). This limits the order of the Markov chain: long chains will require a great deal of space and take a unrealistic amount of data to train.
To reduce the number of parameters, we use an order-3 Markov chain, but coarsely bin the distal two positions. In addition to the preceding three positions, we condition on the position within the read and a running count of the number large jumps in quality scores between adjacent positions (where a ‘large jump’ is defined as |qi − qi−1| > 1), which allows reads with highly variable quality scores to be encoded using separate models. Both of these variables are binned to control the number of parameters.
Reference-based compression
We have also implemented lossless reference-based compression. Given aligned reads in SAM or BAM format, and the reference sequence to which they are aligned (in FASTA format), the reads are compressed preserving all information in the SAM/BAM file, including the header, read IDs, alignment information and all optional fields allowed by the SAM format. Unaligned reads are retained and compressed using the Markov chain model.
Assembly-based compression
To complement the reference-based approach, we developed an assembly-based approach which offers some of the advantages of reference-based compression, but requires no external sequence database and produces files which are entirely self-contained. We use the first (by default) 2.5 million reads to assemble contigs which are then used in place of a reference sequence database to encode aligned reads compactly as positions.
Once contigs are assembled, read sequences are aligned using a simple ‘seed and extend’ method: 12-mer seeds are matched using a hash table, and candidate alignments are evaluated using Hamming distance. The best (lowest Hamming distance) alignment is chosen, assuming it falls below a given cutoff, and the read is encoded as a position within the contig set. This alignment method is simple and exceedingly fast, but it is most effective on platforms in which erroneous indels occur infrequently (e.g. Illumina).
Roughly, assembly-based compression can be thought of as a variation on the Lempel-Ziv algorithm: as sequences are read, they are matched to previously observed data, or in this case, contigs assembled from previously observed data. These contigs are not explicitly stored, but rather reassembled during decompression.
Traditionally, de novo assembly is extremely computationally intensive. The most commonly used technique involves constructing a de Bruijn graph, a directed graph in which each vertex represents a nucleotide k-mer present in the data for some fixed k (e.g. k = 25 is a common choice). A directed edge from a k-mer u to v occurs if and only if the (k − 1)-mer suffix of u is also the prefix of v. In principle, given such a graph, an assembly can be produced by finding an Eulerian path, i.e. a path that follows each edge in the graph exactly once (10). In practice, since NGS data have a non-negligible error rate, assemblers augment each vertex with the number of observed occurrences of the k-mer and leverage these counts using a variety of heuristics to filter out spurious paths.
A significant bottleneck of the de Bruijn graph approach is building an implicit representation of the graph by counting and storing k-mer occurrences in a hash table. The assembler implemented in Quip overcomes this bottleneck to a large extent by using a data structure based on the Bloom filter to count k-mers. The Bloom filter (11) is a probabilistic data structure that represents a set of elements extremely compactly, at the cost of elements occasionally colliding and incorrectly being reported as present in the set. It is probabilistic in the sense that these collisions occur pseudo-randomly, determined by the size of the table and the hash functions chosen, but generally with low probability.
The Bloom filter is generalized in the counting Bloom filter, in which an arbitrary count can be associated with each element (12). A very similar data structure, the CountMin Sketch, is described by Cormode and Muthukrishnan (13), which was arrived at independently of the prior Bloom filter research. Bonomi et al. (14) subsequently described the d-left counting Bloom filter (dlCBF), a refinement of the counting Bloom filter requiring significantly less space to achieve the same false positive rate.
We base our assembler on a realization of the dlCBF. Since we use a probabilistic data structure, k-mers are occasionally reported to have incorrect (inflated) counts. The assembly can be made less accurate by these incorrect counts, however a poor assembly only results in slightly increasing the compression ratio. Compression remains lossless regardless of the assembly quality, and in practice collisions in the dlCBF occur at a very low rate (this is explored in the ‘Results’ section).
Given a probabilistic de Bruijn graph, we assemble contigs using a very simple greedy approach. A read sequence is used as a seed and extended on both ends one nucleotide at a time by repeatedly finding the most abundant k-mer that overlaps the end of the contig by k − 1 bases. More sophisticated heuristics have been developed, but we choose to focus on efficiency, sacrificing a degree of accuracy.
Memory efficient assembly has been a goal of particular interest and is a topic of ongoing research. Conway and Bromage (15) have developed a more efficient means of representing de Bruijn graphs using sparse bitmaps compressed with Elias–Fano encoding. The String Graph Assembler described by Simpson and Durbin (16) takes an entirely different approach, relying on the FM-index (17) to build a compact representation of the set of short reads from which contigs are generated by searching for overlaps. Both of these algorithms sacrifice time-efficiency for their promising increases in space-efficiency, reporting significantly longer run times than traditional assemblers [e.g. both with compared with ABySS (18)].
Our work is a significant departure in that we abandon exact representations and rely on a probabilistic data structure, an idea that has only very recently been breached. Counting k-mers efficiently with the help of Bloom filters was previously explored by Melsted and Pritchard (19), who use it in addition, rather than in place, of a hash table. The Bloom filter is used as a ‘staging area’ to store k-mers occurring only once, reducing the memory required by the hash table.
Concurrently with our work, Pell et al. (20) explore the use of probabilistic data structures in assembly, implementing a probabilistic de Bruijn graph based on the Bloom filter. While we go somewhat further, developing a full assembler using the more versatile dlCBF data structure, we have limited our discussion to data compression. Pell et al. (20) on the other hand provide a compelling justification for the practicality of probabilistic de Bruijn graphs in the assembly problem, showing that the false-positive rate of the data structure is dwarfed by base-calling error in the data itself.
Metadata
In designing the file format used by Quip, we included several useful features to protect data integrity. First, output is divided into blocks of several megabytes each. In each block a separate 64-bit checksum is computed for read identifiers, nucleotide sequences and quality scores. When the archive is decompressed, these checksums are recomputed on the decompressed data and compared with the stored checksums, verifying the correctness of the output. The integrity of an archived data set can also be checked with the ‘quip-test’ command.
Apart from data corruption, reference-based compression creates the possibility of data loss if the reference used for compression is lost, or an incorrect reference is used. To protect against this, Quip files store a 64-bit hash of the reference sequence, ensuring that the same sequence is used for decompression. To assist in locating the correct reference, the file name, and the lengths and names of the sequences used in compression are also stored and accessible without decompression.
Additionally, block headers store the number of reads and bases compressed in the block, allowing summary statistics of a data set to be listed without decompression using the ‘quip-list’ command.
RESULTS
Compression of sequencing data
We compared Quip with three commonplace general-purpose compression algorithms: gzip, bzip2 and xz, as well as more recently developed domain-specific compression algorithms: DSRC (4) and Cramtools (6). Other methods have been proposed (3,5,21), but without publicly available software we were unable independently evaluate them. We have also restricted our focus to lossless compression, and have not evaluated a number of promising lossy methods (6,8), nor methods only capable of compressing nucleotide sequences (22). We invoked Quip in three ways: using only statistical compression (‘quip’), using reference-based compression (‘quip -r’), and finally with assembly-based compression (‘quip -a’). Table 1 gives an overview of the methods evaluated.
Table 1.
Methods evaluated in the ‘Results’ section
| Program | Input | Methods |
|---|---|---|
| gzip (1.4) | Any | Lempel–Ziv |
| bzip2 (1.0.6) | Any | Burrows–Wheeler and Huffman coding |
| xz (5.0.3) | Any | Lempel–Ziv with arithmetic coding |
| sra (2.1.10) | FASTQ | No published description |
| dsrc (0.3) | FASTQ | Lempel–Ziv and Huffman coding. |
| cramtools (0.85-b51) | BAM | Reference-based compression |
| quip (1.1.0) | FASTQ, SAM, BAM | Statistical modeling with arithmetic coding |
| quip -a (1.1.0) | FASTQ, SAM, BAM | Quip with assembly-based compression |
| quip -r (1.1.0) | SAM, BAM | Quip with reference-reference based compression |
All methods compared are lossless, with the exception of Cramtools which does not preserve the original read identifiers.
We acquired six data sets (Table 2) from the Sequence Read Archive (23), representing a broad sampling of recent applications of NGS. Genome and exome sequencing data were taken from 1000 Genomes Project (24), total and mRNA data from the Illumina BodyMap 2.0 data set (25), Runx2 ChIP-Seq performed in a prostate cancer cell line (26) and metagenomic DNA sequencing of biofilm found in the extremely acidic drainage from the Richmond Mine in California (27).
Table 2.
We evaluated compression on a single lane of sequencing data taken from a broad collection of seven studies
| Accession number | Description | Source | Read length | Size (GB) |
|---|---|---|---|---|
| SRR400039 | Whole genome | The 1000 Genomes Project (24) | 101 | 67.6 |
| SRR125858 | Exome | The 1000 Genomes Project (24) | 76 | 55.2 |
| SRR372816 | Runx2 ChIP-Seq | Little et al. (26) | 50 | 14.6 |
| ERR030867 | Total RNA | The BodyMap 2.0 Project, Asmann et al. (25) | 100 | 19.5 |
| ERR030894 | mRNA | The BodyMap 2.0 Project, Asmann et al. (25) | 75 | 16.4 |
| SRR359032 | Metagenomic DNA | Denef et al. (27) | 100 | 8.9 |
Except for the metagenomic data, each was generated from human samples. Uncompressed file sizes are shown in gigabytes.
The Sequence Read Archive provides data in its own SRA compression format which we also evaluate here. Programs for working with SRA files are provided in the SRA Toolkit, but a convenient means of converting from FASTQ to SRA is not, so we have not measured compression time and memory in this case.
In the case of reference-based compression implemented in Cramtools and Quip, we aligned reads from all but the metagenomic data to the GRCh37/hg19 assembly of the human genome using GSNAP (28) generating a BAM file. Splice-junction prediction was enabled for the two RNA-Seq data sets. In the case of multiple alignments, we removed all but the the primary (i.e. highest scoring). Quip is able to store secondary alignments, but the version of Cramtools we evaluated was not. When the purpose of alignment in simply compactly archiving the reads, secondary alignments have no purpose and merely inflate the file size, but in downstream analysis they are often extremely informative and retaining them may be desirable in some cases.
It is important to note pure lossless compression is not the intended goal of Cramtools. Though we invoked Cramtools with options to include all quality scores (––capture–all–quality–scores), optional tags (––capture–all–tags) and retain unaligned reads (––include–unmapped–reads), it is unable to store the original read IDs. Since all other programs evaluated were entirely lossless, measuring compressed file sizes is not an entirely fair comparison. Cramtools is put at some advantage simply by excluding identifiers. However, as the only other available reference-based compression method, it is a useful comparison.
With each method, we compressed then decompressed each data set on a server with a 2.8Ghz Intel Xeon processor and 64 GB of memory. In addition to recording the compressed file size (Figure 1), we measured wall clock run-time (Figure 3) and maximum memory usage (Table 3) using the Unix time command. Run time was normalized to the size of the original FASTQ file, measuring throughput in megabytes per second.
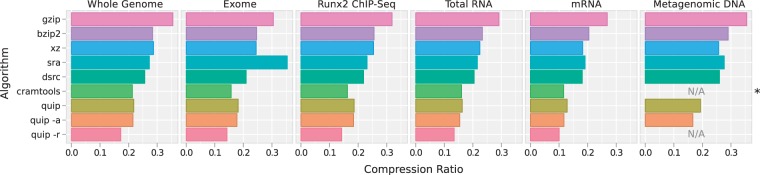
Figure 1.
One lane of sequencing data from each of six publicly available data sets (Table 2) was compressed using a variety of methods (Table 1). The size of the compressed data is plotted in proportion to the size of uncompressed data. Note that all methods evaluated are entirely lossless with the exception of Cramtools, marked in this plot with an asterisk, which does not preserve read IDs, giving it some advantage in these comparisons. Reference-based compression methods, in which an external sequence database is used, were not applied to the metagenomic data set for lack of an obvious reference. These plots are marked ‘N/A’.
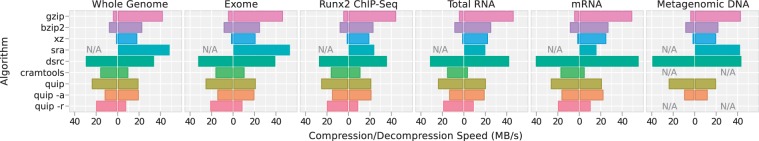
Figure 3.
The wall clock run-time of each evaluation was recorded using the Unix time command. Run-time is normalized by the size of the original FASTQ file to obtain a measure of compression and decompression throughput, plotted here in megabytes per second. Compression speed is plotted to the left of the zero axis, and decompression speed to the right. There is a not a convenient means of converting from FASTQ to SRA, so compression time is not measured for SRA. Additionally, reference-based methods are not applied to the metagenomic data. In the case of reference-based compression implemented in ‘cramtools’ and ‘quip -r’, input and output is in the BAM format, so the speeds plotted here include time needed to decompress and compress the BAM input and output, respectively.
Table 3.
Maximum memory usage was recorded for each program and data set
| Method | Average memory usage (MB) |
|
|---|---|---|
| Compression | Decompression | |
| gzip | 0.8 | 0.7 |
| bzip2 | 7.0 | 4.0 |
| xz | 96.2 | 9.0 |
| sra | NA | 46.9 |
| dsrc | 28.3 | 15.1 |
| cramtools | 6803.8 | 6749.3 |
| quip | 412.1 | 412.9 |
| quip -a | 823.0 | 794.1 |
| quip -r | 1710.0 | 1717.2 |
We list the average across data sets of these measurements. For most of the programs, memory usage varied by <10% across data sets and well summarized by the mean, with the exception of SRA. Decompression used <60 MB except in the human exome sequencing data set, in which >1.4 GB was used. We report the mean with this outlier removed.
For the reference-based compressors, time spent aligning reads to the genome was not included, though it required up to several hours. Furthermore, while Quip is able to compress aligned reads in an uncompressed SAM file and then decompress back to SAM (or FASTQ), the input and output of Cramtools is necessarily a BAM file, and so we also use BAM files as input and output in our benchmarks of ‘quip-r’. This conflates measurements of compression and decompression time: since BAM files are compressed with zlib, compression times for the reference-based compressors include time needed to decompress the BAM file, and decompression times include re-compressing the output to BAM. Consequently, decompression speeds in particular for both Cramtools and reference-based Quip (‘quip-r’) appear significantly lower than they otherwise might.
In the single-genome samples, we find that reference-based compression using Quip consistently results in the smallest file size (i.e. highest compression), yet even without a reference sequence, compression is quite high, even matching the reference-based approach used by Cramtools in two of the data sets. Assembly-based compression provides almost no advantage in the single-genome ChIP-Seq, Whole Genome and Exome data sets. Since we assembly only several million reads (2.5 million, by default), coverage is only enough to assemble representatives of certain families of repeats in these data sets.
In the RNA-Seq data sets, more is assembled (∼35 and 70% of the reads are aligned to assembled contigs in the total RNA-Seq and mRNA-Seq data sets, respectively), but this only provides a small increase in the compression overall. Mostly probably, the aligned reads in these data sets are relatively low-entropy, composed largely of protein-coding sequence, and would be encoded only moderately less compactly with the Markov chain method.
Nevertheless, assembly-based compression does provide a significant advantage in the metagenomic data set. Nearly 95% of reads align to assembled contigs, resulting in a large reduction in compressed file size. Metagenomic sequencing is performed on such a wide variety of samples, that these results can not be easily generalized to all metagenomic data sets. Since success in assembly relies on high coverage, a significant role is played by species diversity, which can vary widely. Yet these results do show that assembly-based compression is beneficial in certain cases. Though it comes at some cost, we are able to perform assembly with exceedingly high efficiently: memory usage is increased only by 400 MB and compression time is faster than all but DSRC and assembly-free Quip even while assembling 2.5 million reads and aligning millions more reads to the assembled contigs.
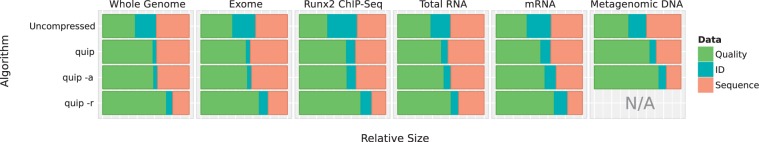
With all invocations of Quip, the size of the resulting file is dominated, sometimes overwhelmingly, by quality scores (Figure 2). There is not a direct way to measure this using the other methods evaluated, but this result suggests that increased compression of nucleotide sequences will result in diminishing returns.
Figure 2.
After compressing with Quip using the base algorithm (labeled ‘quip’), assembly-based compression (‘quip -a’) and reference-based compression (‘quip -r’), we measure the size of read identifiers, nucleotide sequences and quality scores in proportion to the total compressed file size. Reference-based compression (‘quip -r’) was not applied to the metagenomic data.
Characteristics of the dlCBF
Though our primary goal is efficient compression of sequencing data, the assembly algorithm we developed to achieve this is of independent interest. Only very recently has the idea of using probabilistic data structures in assembly been breached, and to our knowledge, we are the first to build a functioning assembler using any version of the counting Bloom filter. Data structures based on the Bloom filter make a trade-off between space and the probability of inserted elements colliding. Tables can be made arbitrarily small at the cost of increasing the number of collisions. To elucidate this trade-off, we performed a simple simulation comparing the dlCBF to a hash table.
Comparisons of data structures are notoriously sensitive to the specifics of the implementation. To perform a meaningful benchmark, we compared our dlCBF implementation with the sparsehash library (http://code.google.com/p/sparsehash/), an open-source hash table implementation with the expressed goal of maximizing space efficiency. Among many other uses, it is the core data structure in the ABySS (18) and PASHA (29) assemblers.
We randomly generated 10 million unique, uniformly distributed 25-mers and inserted them into a hash table, allocated in advance to be of minimal size while avoiding resizing and rehashing. We repeated this with dlCBF tables of increasing sizes. Upon insertion, a collision occurs when the hash functions computed on the inserted k-mer collide with a previously inserted k-mer. An insertion may also fail when a fixed size table is filled to capacity and no empty cells are available. For simplicity, we count these occurrences also as collisions. Wall clock run-time and maximum memory usage were both recorded using the Unix time command.
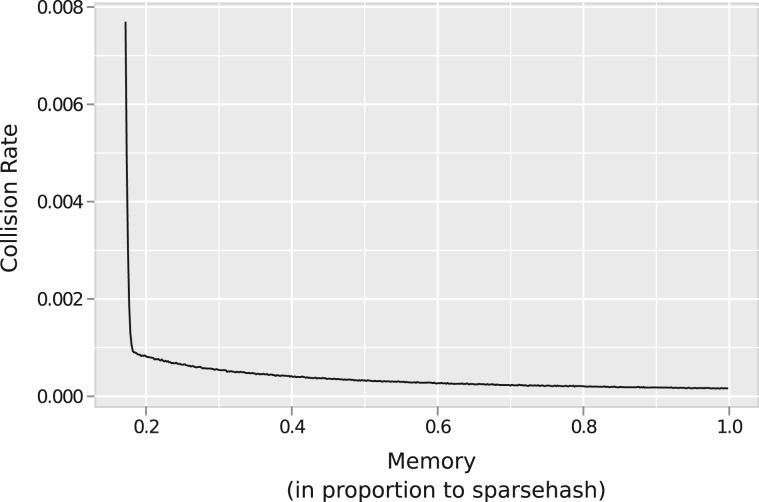
We find that with only 20% of the space of the hash table, the dlCBF accrues a collision rate of less than 0.001 (Figure 4). While the hash table performed the 10 million insertions in 7.34 s, it required only 4.48 s on average for the dlCBF to do the same, with both simulations run on a 2.8 Ghz Intel Xeon processor. Table size did not greatly affect the run time of the dlCBF. The assembler implemented in Quip uses a fixed table size, conservatively set to allow for 100 million unique k-mers with a collision rate of ∼0.08% in simulations.
Figure 4.
The trade-off between memory usage and false positive rate in the dlCBF is evaluated by inserting 10 million unique 25-mers into tables of increasing size. Memory usage is reported as the proportion of the memory used by a memory efficient hash table to do the same.
Though the authors of sparsehash claim only a 4-bit overhead for each entry, and have gone to considerably effort to achieve such efficiency, it still must store the k-mer itself, encoded in 64 bits. The dlCBF avoids this, storing instead a 14-bit ‘fingerprint’, or hash of a k-mer, resulting in the large savings we observe. Of course, a 25-mer cannot be uniquely identified with 14 bits. False positives are thus introduced, yet they are kept at a very low rate by the d-left hashing scheme. Multiple hash functions are used under this scheme, so that multiple hash function must collide to result in a collision, an infrequent event if reasonably high-quality hash functions are chosen.
A previous analysis of the dlCBF by Zhang et al. (30) compared it with two other variations of the counting Bloom filter and concluded that ‘the dlCBF outperforms the others remarkably, in terms of both space efficiency and accuracy.’ Overall, this data structure appears particularly adept for high efficiency de novo assembly.
DISCUSSION
Compared with the only other published and freely-available reference-based compressor, Cramtools, we see a significant reduction in compressed file size, despite read identifiers being discarded by Cramtools and retained by Quip. This is combined with dramatically reduced memory usage, comparable run-time (slightly faster compression paired with varying relative decompression speeds), and increased flexibility (e.g. Quip is able to store multiple alignments of the same read and does not require that the BAM file be sorted or indexed). Conversely, Cramtools implements some potentially very useful lossy methods not provided by Quip. For example, a number of options are available for selectively discarding quality scores that are deemed unnecessary, which can significantly reduce compressed file sizes.
We find that assembly-based compression offers only a small advantage in many data sets. Since we assemble a relatively small subset of the data, coverage is too low when sequencing a large portion of a eukaryotic genome. In principle, the assembler used could be scaled to handle exponentially more reads, but at the cost of being increasingly computationally intensive. In such cases, the reference-based approach may be more appropriate. Yet, in the metagenomic data set we evaluate, where there is no obvious reference sequence, nucleotide sequences are reduced in size by over 40% (Figure 2). While limited in scope, for certain projects assembly-based compression can prove to be invaluable, and the algorithm devised here makes it a tractable option.
The Lempel-Ziv algorithm, particularly as implemented in gzip/libz has become a reasonable default choice for compression. The zlib library has matured and stabilized over the course of two decades and is widely available. The BAM and Goby formats both use zlib for compression, and compressing FASTQ files with gzip is still common practice. Despite its ubiquity, our benchmarks show that it is remarkably poorly suited to NGS data. Both compression ratio and compression time were inferior to the other programs evaluated. For most purposes, the gains in decompression time do not make up for its shortcomings. With the more sophisticated variation implemented in xz, compression is improved but at the cost of tremendously slow compression.
Our use of high-order Markov chain models and de novo assembly results in a program that uses significantly more memory than the others tested, with the exception of Cramtools. Though limiting the memory used by a general purpose compression program enables it to be used on a wider variety of systems, this is less important in this domain-specific application. Common analysis of NGS data, whether it be alignment, assembly, isoform quantification, peak calling or SNP calling, all require significant computational resources. Targeting low-memory systems would not be of particular benefit: NGS precludes previous-generation hardware.
Though memory consumption is not a top priority, run time is important. Newer instruments like the HiSeq 2000 produce far more data per lane than previously possible. And, as the cost of sequencing continues to decrease, experiments will involve more conditions, replicates and timepoints. Quip is able to compress NGS data at three times the speed of gzip, while performing de novo assembly of millions of reads, and up to five times as fast without the assembly step. Only DSRC is faster, but with consistently lower compression. In addition, our reference-based compression algorithm matches the speed of Cramtools but with substantially better lossless compression.
As illustrated in Figure 2, the size of compressed NGS data is dominated by quality scores. More sophisticated compression of nucleotide sequences may be possible but will result in only minor reductions to the overall file size. The largest benefit would be seen by reducing quality scores. While it is easy to reduce the size of quality data by coarse binning or other lossy methods, it is very hard to determine the effect such transformations will have on downstream analysis. Future work should concentrate on studying lossy quality score compression, strictly guided by minimizing loss of accuracy in alignment, SNP calling and other applications. Quality scores are encoded in Quip using an algorithm that is suitably general so that lossy transformations (e.g. binning) can be automatically exploited, and can be treated as an optional preprocessing step.
Other aspects of the algorithm presented here can be improved, and will be with future versions of the software. A large body of work exists exploring heuristics to improve the the quality of de novo assembly, however the algorithm in Quip uses a very simple greedy approach. Assembly could likely be improved without greatly reducing efficiency by exploring more sophisticated methods. We currently perform no special handling of paired-end reads, so that mates are compressed independently of each other. In principle, some gains to compression could be made by exploiting pair information. We also intend to implement parallel compression and decompression. This is non-trivial, but quite possible and worthwhile given the abundance of data and ubiquity of multi-core processors.
Combining reference-based and assembly-based techniques, with carefully tuned statistical compression, the algorithm presented in Quip probes the limit to which NGS data can be compressed losslessly, yet remains efficient enough be a practical tool when coping with the deluge of data that biology research is now presented with.
FUNDING
Funding for open access charge: Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract [No. HHSN272200800060C]; Public Health Service [P51RR000166]; National Institutes of Health, in whole or in part.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to James Bonfield, Matt Mahoney, Seth Hillbrand and other participants in the Pistoia Alliance Sequence Squeeze competition, whose work inspired many improvements to our own. While preparing the manuscript, Richard Green provided valuable feedback and suggestions.
REFERENCES
- 1.Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–71. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tembe W, Lowey J, Suh E. G-SQZ: compact encoding of genomic sequence and quality data. Bioinformatics. 2010;26:2192–2194. doi: 10.1093/bioinformatics/btq346. [DOI] [PubMed] [Google Scholar]
- 4.Deorowicz S, Grabowski S. Compression of DNA sequence reads in FASTQ format. Bioinformatics. 2011;27:860–862. doi: 10.1093/bioinformatics/btr014. [DOI] [PubMed] [Google Scholar]
- 5.Kozanitis C, Saunders C, Kruglyak S, Bafna V, Varghese G. Compressing genomic sequence fragments using SlimGene. J. Comput. Biol. 2011;18:401–413. doi: 10.1089/cmb.2010.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz MH-Y, Leinonen R, Cochrane G, Birney E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christley S, Lu Y, Li C, Xie X. Human genomes as email attachments. Bioinformatics. 2009;25:274–275. doi: 10.1093/bioinformatics/btn582. [DOI] [PubMed] [Google Scholar]
- 8.Wan R, Anh VN, Asai K. Transformations for the compression of FASTQ quality scores of next generation sequencing data. Bioinformatics. 2011;28:628–635. doi: 10.1093/bioinformatics/btr689. [DOI] [PubMed] [Google Scholar]
- 9.Said A. Hewlett-Packard Laboratories Report, HPL-2004-76. Palo Alto, CA, USA: 2004. Introduction to arithmetic coding - theory and practice. [Google Scholar]
- 10.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloom BH. Space/time trade-offs in hash coding with allowable errors. Comm. ACM. 1970;13:422–426. [Google Scholar]
- 12.Fan L, Cao P, Almeida J, Broder AZ. Summary cache: a scalable wide-area Web cache sharing protocol. IEEE/ACM Trans. Netw. 2000;8:281–293. [Google Scholar]
- 13.Cormode G, Muthukrishnan S. An improved data stream summary: the count-min sketch and its applications. J. Algor. 2005;55:58–75. [Google Scholar]
- 14.Bonomi F, Mitzenmacher M, Panigrahy R. 14th Annual European Symposium on Algorithms, Vol. 4168. LNCS. Berlin/Heidelberg: Springer; 2006. An improved construction for counting Bloom filters; pp. 684–695. [Google Scholar]
- 15.Conway TC, Bromage AJ. Succinct data structures for assembling large genomes. Bioinformatics. 2011;27:479–486. doi: 10.1093/bioinformatics/btq697. [DOI] [PubMed] [Google Scholar]
- 16.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferragina P, Manzini G. In: Proceedings of the 41st Annual Symposium on Foundations of Computer Science. Washington, DC: IEEE Computer Society; 2000. Opportunistic data structures with applications; pp. 390–398. [Google Scholar]
- 18.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melsted P, Pritchard JK. Efficient counting of k-mers in DNA sequences using a Bloom Filter. BMC Bioinformatics. 2011;12:333. doi: 10.1186/1471-2105-12-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pell J, Hintze A, Canino-Koning R, Howe A, Tiedje JM, Brown CT. Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. 2011 doi: 10.1073/pnas.1121464109. Arxiv preprint arXiv, I, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhola V, Bopardikar AS, Narayanan R, Lee K, Ahn T. 2011 IEEE International Conference on Bioinformatics and Biomedicine. Washington, DC: IEEE Computer Society; 2011. No-reference compression of genomic data stored in FASTQ format; pp. 147–150. [Google Scholar]
- 22.Cox AJ, Bauer MJ, Jakobi T, Rosone G. Large-scale compression of genomic sequence databases with the Burrows-Wheeler transform. Bioinformatics. 2012;28:1415–1419. doi: 10.1093/bioinformatics/bts173. [DOI] [PubMed] [Google Scholar]
- 23.Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. The 1000 Genomes Consortium. (2010) A map of human genome variation from population-scale sequencing. Nature467, 1061–1073. [DOI] [PMC free article] [PubMed]
- 25.Asmann YW, Necela BM, Kalari KR, Hossain A, Baker TR, Carr JM, Davis C, Getz JE, Hostetter G, Li X, et al. Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer Res. 2012;72:1921–1928. doi: 10.1158/0008-5472.CAN-11-3142. [DOI] [PubMed] [Google Scholar]
- 26.Little GH, Noushmehr H, Baniwal SK, Berman BP, Coetzee GA, Frenkel B. Genome-wide Runx2 occupancy in prostate cancer cells suggests a role in regulating secretion. Nucleic Acids Res. 2011:1–10. doi: 10.1093/nar/gkr1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 28.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Schmidt B, Maskell DL. Parallelized short read assembly of large genomes using de Bruijn graphs. BMC Bioinformatics. 2011;12:354. doi: 10.1186/1471-2105-12-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Wu J, Lan J, Liu J. Performance evaluation and comparison of three counting Bloom filter schemes. J. Electrons. 2009;26:332–340. [Google Scholar]