Abstract
The ribonuclease Dicer plays a central role in the microRNA pathway by catalyzing the formation of microRNAs, which are known to regulate messenger RNA (mRNA) translation. In order to improve our understanding of the molecular context in which Dicer functions and how it is regulated in human cells, we sought to expand its protein interaction network by employing a yeast two-hybrid screening strategy. This approach led to the identification and characterization of cytoskeleton-linking endoplasmic reticulum (ER) membrane protein of 63 kDa (CLIMP-63) as a novel Dicer-interacting protein. CLIMP-63 interacts with Dicer to form a high molecular weight complex, which is electrostatic in nature, is not mediated by RNA and is catalytically active in pre-microRNA processing. CLIMP-63 is required for stabilizing Dicer protein and for optimal regulation of a reporter gene coupled to the 3′ untranslated region of HMGA2 mRNA in human cells. Interacting with a portion of the luminal domain of CLIMP-63 and within minutes of its synthesis, our results suggest that Dicer transits through the ER, is glycosylated and can be secreted by cultured human cells with CLIMP-63. Our findings define CLIMP-63 as a novel protein interactor and regulator of Dicer function, involved in maintaining Dicer protein levels in human cells.
INTRODUCTION
MicroRNA-guided RNA silencing can be viewed as a succession of specialized molecular machines exerting well defined functions in the biogenesis and action of microRNAs (1). This well defined family of ∼19–24 nucleotides (nt), non-coding RNA species is known to regulate translation of specific messenger RNA (mRNA) targets (2,3). Acting through the recognition of binding sites usually located in the 3′ untranslated region (UTR) of mRNAs, microRNAs may regulate ∼60% of the genes in humans (4). Expressed from microRNA genes transcribed from the genome, microRNAs are generated upon successive processing of stem–loop structured primary microRNAs (pri-microRNAs) and microRNA precursors (pre-microRNAs) by the RNases III Drosha (5) and Dicer (6–8), respectively, as reviewed in Ouellet et al. (9).
Although the major steps and core components of the microRNA pathway have been identified and characterized, our comprehension of the molecular mechanisms underlying microRNA biogenesis and function remains imperfect, including those involving the microRNA-generating enzyme Dicer. Strategically positioned at the nucleus/cytosol interface, human Dicer is a 217-kDa multi-domain enzyme that is believed to accept its pre-microRNA substrate of nuclear origin from Exportin-5, recognize it via its central PIWI/Argonaute/Zwille (PAZ) domain and process it into microRNAs through the concerted action of its C-terminal RNase IIIa and IIIb motifs. Dicer forms a complex with transactivating response (TAR) RNA binding protein (TRBP) (10) and acts as the catalytic engine of a pre-microRNA processing complex (11,12). Dicer has also been shown to interact with the downstream microRNA effector Ago2 (13) and to transfer its microRNA products to fragile X mental retardation protein (FMRP) (14), thereby linking the biogenesis and action of microRNAs in human cells. More recently, KH-type splicing regulatory protein (KSRP) was reported to be part of a Dicer complex in HeLa cells and to interact with the terminal loop of a subset of pre-microRNA targets to promote their maturation into microRNAs (15). However, despite its central importance in microRNA biogenesis, the cellular context in which Dicer functions in RNA silencing and how Dicer itself is regulated, remains poorly defined.
In order to improve our understanding of the molecular context in which Dicer functions and how it is regulated in human cells, we sought to expand the protein interaction network of human Dicer by employing a yeast two-hybrid screening strategy. This strategy allowed the identification and characterization of cytoskeleton-linking endoplasmic reticulum (ER) membrane protein of 63 kDa (CLIMP-63) as a novel protein interactor and regulator of Dicer function, involved in maintaining Dicer protein levels and required for optimal RNA silencing in human cells.
MATERIALS AND METHODS
Cell lines, culture and transfection
The mammalian construct expressing [CLIMP-63-hemagglutinin (HA) (CLIMP-63–HA)] was prepared by PCR amplification of IMAGE clone 6084968, used as a DNA template, and cloning of the amplified fragment lacking the stop codon into the EcoRI/XhoI sites of a pcDNA3.1 vector harboring a C-terminal HA epitope. The Dicer expression vector pcDNA3.1-Flag–Dicer has been described previously (6).
HEK 293 and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine in a humidified incubator under 5% CO2 at 37°C. Transfection of the different constructs was performed by using the calcium phosphate method. Where indicated, the cells were treated with 50 μg/ml cycloheximide (Sigma) or 10 μg/ml Brefeldin A (Sigma) for 90 min.
Stable 293T-REx cell lines were generated using the pTER system (16), as described previously (17). The shRNA-mediated downregulation of Dicer and CLIMP-63 expression was induced with 2 μg/ml of doxycyclin (Dox; Sigma) for 6 days.
Protein extracts
Proteins were extracted by harvesting cells in lysis buffer (50 mM Tris–HCl, 137 mM NaCl, 1% Triton X-100, 1 mM PMSF, 1X protease inhibitor cocktail mix without EDTA, pH 8.0), as described previously (18). Lysates were incubated for 15 min on ice and centrifuged at 16 060g for 15 min at 4°C. Protein concentration was determined by the method of Bradford (19) using the Bio-Rad dye reagent, with bovine serum albumin as standard.
Immunoprecipitation
Cleared protein extracts (1 mg) derived from HEK 293 cells were incubated in the presence of anti-Dicer (5 μl) (20) or anti-CLIMP-63 (300 μl) (G1/296 Alexis Biochemicals) for 1 h at 4°C under continuous rotation. Pre-washed Protein-G agarose beads (Roche or magnetic beads from Invitrogen) were added and the incubation continued for an additional 3 h. For anti-HA immunoprecipitation, anti-HA affinity matrix (rat anti-HA 3F10 linked to agarose beads) was used. In some experiments, immunoprecipitation was performed in the presence of PhosStop inhibitors (Roche) 1×, 5 mM ethylene glycol tetraacetic acid (EGTA) or 1 mM CaCl2. The calf intestinal alkaline phosphatase (CIAP) treatment was performed by washing the beads once with lysis buffer and adding 15 U of CIAP (Roche) in CIAP buffer in a reaction volume of 100 μl for 15 min at 30°C. RNase treatments were achieved by adding 0.125 U RNase A and 5 U RNase T1 in 50 μl of RNases A/T1/V1 buffer (mirVana microRNA isolation kit, Ambion) for 15 min at 37°C. The beads were then washed four times with lysis buffer and the immune complexes were eluted by boiling in loading buffer for 5 min; the proteins were analyzed by western blot.
Mass spectrometry
The in-gel digestion and mass spectrometry (MS) analysis of selected bands stained with coomassie blue and isolated from CLIMP-63–HA immunoprecipitates were performed by the Proteomics platform of the Eastern Quebec Genomics Center, Quebec, Canada. Peptide samples were separated by online reverse-phase (RP) nanoscale capillary liquid chromatography (nanoLC) and analyzed by electrospray MS (ES MS/MS). Protein identification was confirmed if it could be established at >99% probability and contained at least two identified peptides.
Western blot
Protein extracts were analyzed by western blot using the following primary antibodies: rabbit polyclonal anti-Dicer (6,20), mouse monoclonal anti-Dicer (Abcam), mouse monoclonal anti-CLIMP-63 (Alexis Biochemicals), mouse monoclonal anti-HA (Roche), mouse monoclonal anti-actin AC-40 (Sigma), rabbit polyclonal anti-calreticulin (Affinity Bioreagents), rabbit polyclonal anti-Drosha (Upstate Cell Signalling Solutions), rabbit polyclonal anti-Coactosin-like protein (CLP) (21) and horseradish peroxidase (HRP)-conjugated Phaseolus vulgaris E (PHA-E) (USBiological). PHA-E was prepared in 3% bovine serum albumin supplemented with 1 mM MgCl2 and 1 mM CaCl2, as recommended by the manufacturer.
Gel filtration chromatography
Cleared protein extracts (100 000g for 45 min, supernatant; S100) of HEK 293 cells resuspended in lysis buffer [40 mM Tris–HCl, 137 mM NaCl, 1% (v/v) Triton X-100, 1 mM phenylmethanesulfonyl fluoride, complete protease inhibitor cocktail (Roche), pH 8.0] and filtered through a 0.22-µm filter (Millipore) were separated by gel filtration on a Superose six column (10 × 300 mm GL; GE Healthcare) using an ÄKTA FPLC system (GE Healthcare) into 400-µl fractions. The column was calibrated by using the following molecular weight size markers: Thyroglobulin (669 kDa), Ferritin (440 kDa), Aldolase (158 kDa) and Ovalbumin (43 kDa). Protein fractionation was monitored by detection at 280 nm, and the protein content of selected fractions (40 µl) was analyzed by western blot, as reported previously (22). In some experiments, equivalent amounts (360 µl) of the 400-µl fractions were subjected to anti-HA IP.
Pre-microRNA processing assay
Dicer RNase activity in selected fractions (100 µl) collected upon gel filtration chromatography was detected using randomly 32P-labeled human pre-let-7 a-3 RNA and analyzed by denaturating PAGE and autoradiography, as described elsewhere (20).
Reporter gene activity assay
Cells transfected with 0.1 μg of the psiCHECK-HMGA2 3′-UTR reporter construct, which was created by insertion of the HMGA2 3′-UTR (23) in the XhoI/NotI sites of the psiCHECK (Promega) reporter vector, or empty psiCHECK were harvested 48 h later, and Renilla luciferase (Rluc) and Firefly luciferase (Fluc) activities were measured, as described previously. Results of Rluc activity were normalized with Fluc reporter activity and expressed as percentage of the results obtained from the empty reporter construct (i.e. lacking the HMGA2 3′-UTR). The –Dox condition was set to 1 in order to exclude all other variables and allow comparison with the + Dox condition and between the different shRNA treatments.
Yeast two-hybrid experiments
The open reading frame of human Dicer (6) was inserted into pGBT9 and pACT2, and the various deletion mutants of Dicer were amplified by polymerase chain reaction (PCR) and cloned in frame into the BamHI/SalI sites of pGBT9 or EcoRI/XhoI sites of pACT2. The pGBT9-CLIMP-63 192-258 and pACT2-CLIMP-63 192-258 vectors were generated by amplification of this fragment, using IMAGE clone 6084968 as a template, and cloning into the BamHI/SalI sites of pGBT9 and the EcoRI/XhoI sites of pACT2, respectively. The integrity of the constructs was verified by DNA sequencing.
Yeast strain PJ69–4A was used for the two-hybrid cDNA library screening and assays, as described in Provost et al. (24). PJ69–4A harboring the pGBT9–Dicer vector was transformed with a human lung cDNA library, and growers on synthetic dropout (SD)/-Leu/-Trp/-Ade plates were tested for activation of their adenine, histidine and lacZ reporter genes. The interacting pACT2 plasmid was rescued from positive clones and retested to confirm the interaction prior to sequencing of the interacting cDNA inserts. For yeast two-hybrid assays, PJ69–4A cells were co-transformed with various pGBT9 and pACT2 constructs and tested for reporter gene activation 6 days later.
Confocal immunofluorescence microscopy
Double immunofluorescence staining was performed on HeLa cells grown on sterile glass coverslips and transfected with the vectors pcDNA3.1-Dicer and pcDNA3.1-CLIMP-63–HA using Lipofectamine 2000 (Invitrogen) for 18 h, essentially as described previously (21). Cells were washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 20 min and washed again with PBS. After quenching in 50 mM NH4Cl for 15 min, the cells were permeabilized in 0.1% Triton X-100. Permeabilized cells were then incubated in blocking buffer (10% FBS in PBS) for 30 min. Dicer proteins were fluorescently labeled with a mouse monoclonal anti-Dicer antibody (Abcam; dilution 1/1000) and an Alexa Fluor 488-conjugated goat anti-mouse (Molecular Probes, dilution 1/500) secondary antibody. CLIMP-63–HA protein was revealed upon staining with rabbit polyclonal anti-HA antibody (Clone Y-11, Santa Cruz Biotechnology) and an Alexa Fluor 546-conjugated goat anti-rabbit (Molecular Probes, dilution 1/500) secondary antibody. Nuclei were stained with the cell permeable far-red fluorescent DNA dye 1, 5-bis{[2-(di-methylamino)ethyl]amino}-4, 8-dihydroxyanthracene-9, 10-dione (DRAQ5). After extensive washing in PBS, the coverslips were mounted on slides with the mounting solution Prolong Gold antifade reagent (Molecular Probes). Images were acquired by a Quorum Spinning Disc Wave Fx (Quorum Technologies) microscope equipped with a Hamamatsu camera and analyzed by the Velocity 4 software.
Protease protection assays
Protease protection assays were performed based on a method described by Lorenz et al. (25). The protein content of the various fractions was analyzed by western blot.
Metabolic labeling of cultured cells
Transfected HEK 293 cells were starved in DMEM without methionine and cysteine for 30 min, pulsed with 100 μCi/ml of 35S-labeled methionine and cysteine mix (Redivue PRO-MIX L-[35S] in vitro Cell Labeling Mix, GE Healthcare) for 15 min and chased in DMEM containing 10% FBS and supplemented with 3 mM L-methionine and 1 mM L-cystenine for the indicated periods of time (26). Immunoprecipitated 35S-labeled proteins were separated by SDS–PAGE, visualized by autoradiography and quantitated by PhosphorImager acquisition and densitometry.
Extracellular Dicer analysis
293 T-REx cells were transiently transfected with pcDNA-Dicer and/or pcDNA-CLIMP-63–HA vectors for 18 h before washing the cells with PBS and changing the media for DMEM only. Mock and Dicer transfected cells were treated or not with doxycyclin (Dox) to induce short hairpin RNA (shRNA)-mediated downregulation of CLIMP-63 expression. Cells were incubated for 6 h and the cell culture medium was collected, filtered (1 μm filter) and centrifuged for 1 h at 100 000g. The pellet containing extracellular vesicles was washed with PBS and centrifuged for 1 h at 100 000g; protein extracts were prepared and analyzed by immunoblotting. The supernatant fraction (30%) was subjected to Dicer immunoprecipitation using anti-Dicer antibody (200 ng) (Abcam) and analyzed by immunoblotting, as described above.
RESULTS
Human Dicer interacts with the ER membrane protein CLIMP-63
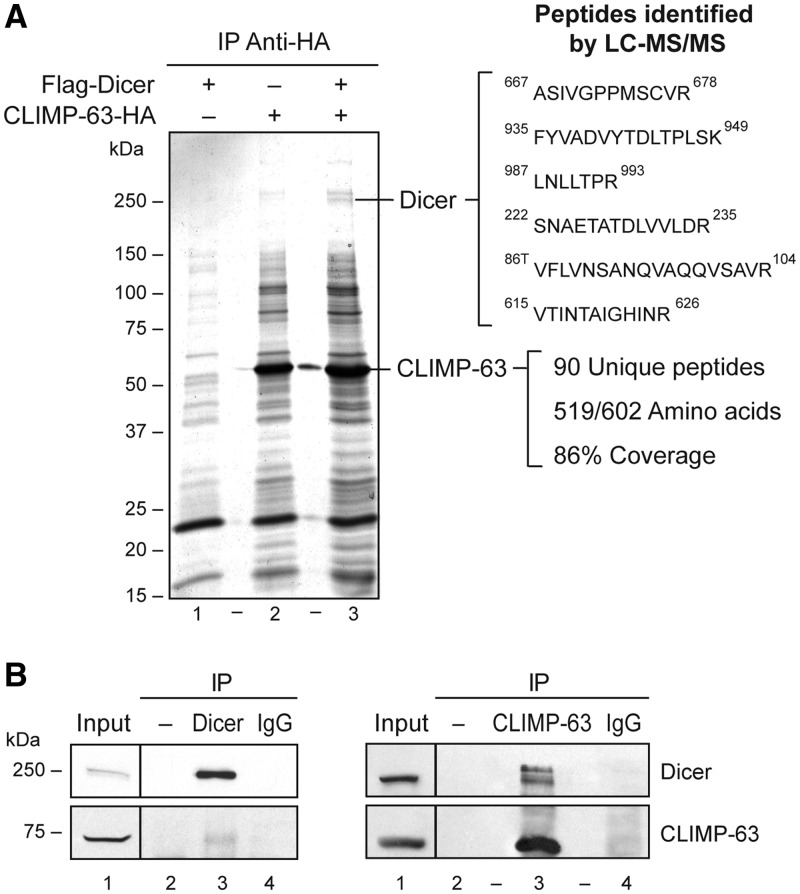
With the aim of identifying new protein interactors and modulators of Dicer, we performed a yeast two-hybrid screening of a human lung cDNA library using human Dicer as bait. Among the potential candidates, we identified several clones that encoded the CLIMP-63 protein (27). In order to confirm the interaction between Dicer and CLIMP-63 in human cells, we performed co-immunoprecipitation experiments using HEK 293 cells transiently expressing the epitope-tagged CLIMP-63–HA and/or Flag–Dicer protein. Analysis of the immunoprecipitates by electrospray mass spectrometry (ES MS/MS) confirmed the identification of CLIMP-63 as the ∼63-kDa protein species enriched in CLIMP-63–HA immunoprecipitates; more than 90 peptides, covering 86% of the amino acid sequence, were obtained (Figure 1A). Two bands migrating in the 250-kDa range, which is expected for Dicer, were also observed. Proteomic analysis of these bands confirmed the presence of Dicer in the fastest migrating band, with six unique peptides covering ∼4% of the protein (Figure 1A). The in vivo Dicer-CLIMP-63 interaction was confirmed by our ability to co-immunoprecipitate the endogenous Dicer and CLIMP-63 proteins from HEK 293 cells, reciprocally (Figure 1B).
Figure 1.
Identification of CLIMP-63 as a novel Dicer-interacting protein. (A) The indicated bands, stained with coomassie blue and isolated from CLIMP-63–HA immunoprecipitates (IP) prepared from HEK 293 cells transiently expressing Flag–Dicer and CLIMP-63–HA proteins, were analyzed by reverse-phase liquid chromatography–tandem mass spectrometry (LC–MS/MS). The identified Dicer peptides are shown. (B) Endogenous Dicer or CLIMP-63 proteins were immunoprecipitated from HEK 293 cell extracts using anti-Dicer (upper panel) or anti-CLIMP-63 (lower panel) antibody, or normal IgG (−) and the presence of Dicer and CLIMP-63 in the immune complexes was monitored by western blot.
Characterization of Dicer•CLIMP-63 complex formation
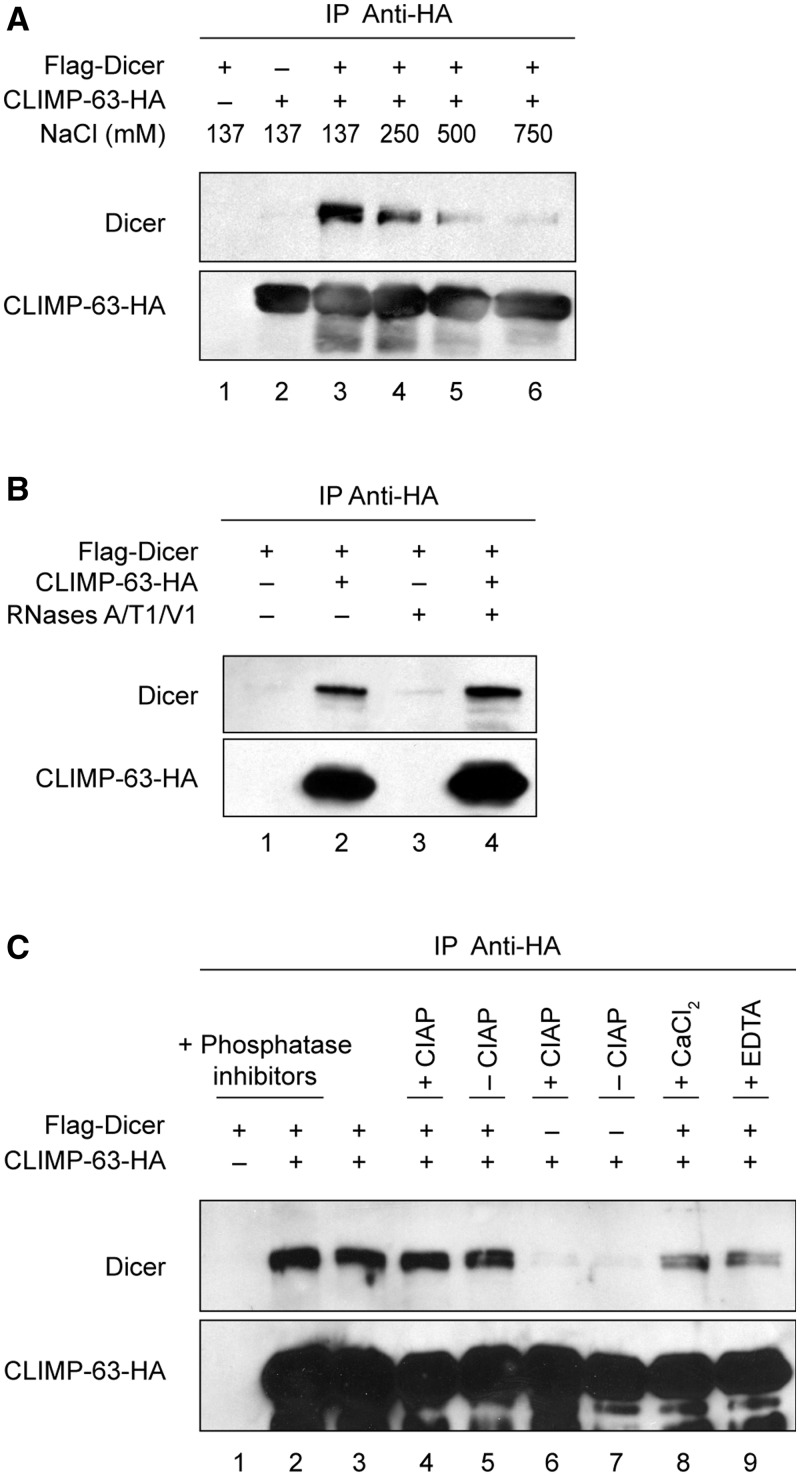
To characterize further this interaction, we exposed Flag–Dicer•CLIMP-63–HA protein complexes, derived from transiently transfected HEK 293 cells, to various conditions. As shown in Figure 2A, the association between Dicer and CLIMP-63 was gradually weakened by increasing the concentration of salt (NaCl), suggesting that the formation of the Dicer•CLIMP-63 protein complex is electrostatic, rather than hydrophobic, in nature. Because Dicer can bind to RNA and that RNA can assemble proteins that do not interact with each other, we exposed CLIMP-63–HA immune complexes to RNases A/T1/V1. This treatment had no effect, indicating that the Dicer–CLIMP-63 protein interaction is unlikely to be mediated by RNA (Figure 2B). The phosphorylation status of both Dicer (our unpublished data) and CLIMP-63 (28) proteins did not seem to influence their ability to interact together, as the presence of phosphatase inhibitors, which preserved protein phosphorylation, or calf intestinal alkaline phosphatase (CIAP), which induced protein dephosphorylation, did not modify the interaction (Figure 2C, lanes 1–7). Dicer•CLIMP-63 complex formation was also not influenced by the presence of CaCl2 or EGTA, which is a chelator of divalent cations (Figure 2C, lanes 8 and 9). These latter findings are particularly relevant, because the ER is an organelle that stocks high concentrations of calcium (29). Taken together, our results suggest that the Dicer•CLIMP-63 complex is electrostatic in nature, is independent of the phosphorylation status of the proteins and of Ca2+ and is not mediated by RNA.
Figure 2.
The Dicer•CLIMP-63 complex is electrostatic in nature and is not mediated by RNA. (A–C) CLIMP-63–HA IP, prepared from HEK 293 cells transiently expressing Flag–Dicer and/or CLIMP-63–HA proteins, were incubated in the presence of increasing concentrations of NaCl (A), in the absence or presence of RNases A/T1/V1 (B), or without or with phosphatase inhibitors, CIAP, CaCl2 or EGTA (C). The presence of Dicer and CLIMP-63 in the immune complexes was monitored by western blot.
CLIMP-63 stabilizes Dicer protein in cultured human cells
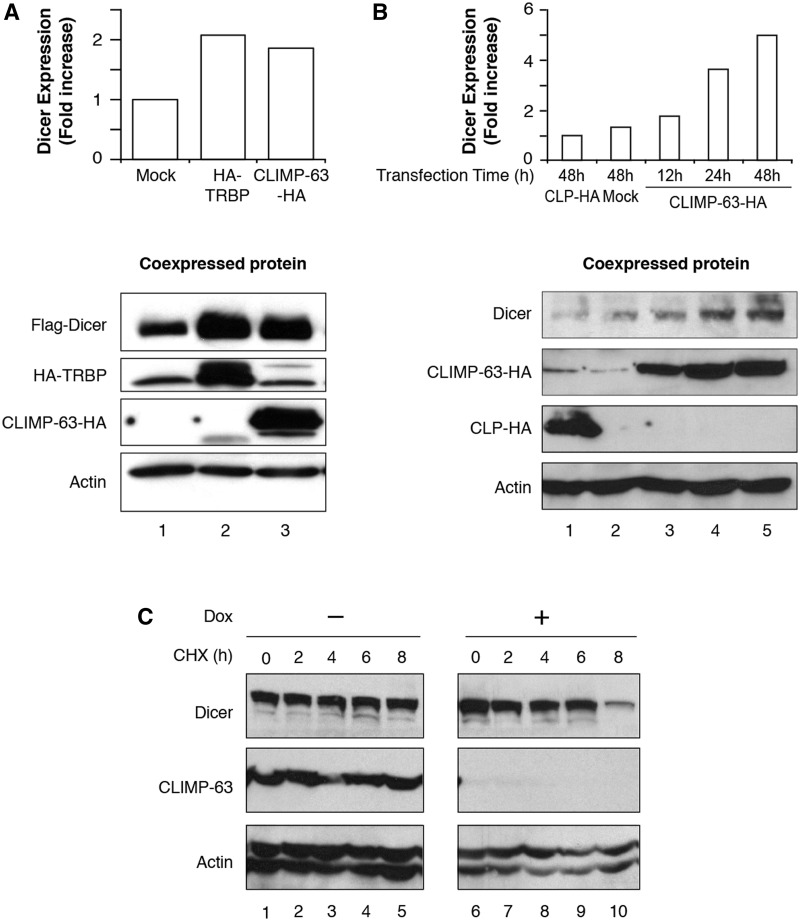
During the course of our experiments, we observed that endogenous and epitope-tagged Dicer proteins were expressed at higher levels upon CLIMP-63–HA expression. Indeed, expression of Flag–Dicer was enhanced by 2-fold when co-expressed with CLIMP-63–HA (Figure 3A, lane 3), to an extent similar to that observed upon co-expression with HA–TRBP2 (Figure 3A, lane 2). The endogenous Dicer protein also seemed to be stabilized upon CLIMP-63–HA overexpression, as a ∼5-fold accumulation of endogenous Dicer was observed 48 h after transfection, whereas no stabilizing effects were conferred upon expression of CLP–HA, used as a non-interacting protein control (Figure 3B).
Figure 3.
CLIMP-63 stabilizes Dicer protein in cultured human cells. (A and B) Protein extracts from HEK 293 cells transiently expressing HA–TRBP2, CLIMP-63–HA or mock-transfected (A), or expressing CLIMP-63 for 12, 24 or 48 h, CLP–HA or mock-transfected for 48 h (B) were analyzed by western blot (lower panels), followed by densitometric quantitation of the immunoreactive bands (upper panels). (C and D) Protein extracts derived from stable 293T-REx cells, conditionally expressing a short hairpin RNA (shRNA) directed against CLIMP-63 mRNA (shCLIMP-63-1) upon doxycyclin (Dox) induction for 6 days (C, D), and treated or not with cycloheximide (CHX) for the indicated period of time (C), or overexpressing Dicer (D), were analyzed by western blot.
In order to examine further the relative stability of both Dicer and CLIMP-63 endogenous proteins, we made use of cycloheximide. Treatment of HEK 293 cells with this known inhibitor of cellular mRNA translation had no significant effect on endogenous Dicer (Figure 3C, upper panel, lanes 1–5) and CLIMP-63 (Figure 3C, center panel, lanes 1–5) protein levels over an 8-h period, confirming that Dicer and CLIMP-63 are relatively stable in cultured human cells.
In view of a possible stabilizing effect of CLIMP-63 on Dicer protein expression, we reasoned that the depletion of CLIMP-63 would lead to a concomitant reduction in Dicer protein levels. We thus created a stable cell line expressing a shRNA directed against CLIMP-63 (shCLIMP-63-1) mRNA upon induction with doxycyclin. As shown in Figure 3C (center panel), this shRNA-based approach efficiently inhibited CLIMP-63 protein expression in HEK 293 cells. The level of Dicer proteins detected in the cells, depleted or not in CLIMP-63, were similar in the absence of cycloheximide treatment (Figure 3C, upper panel, lane 6 versus lane 1). However, treatment of CLIMP-63-depleted cells with cycloheximide induced a marked and gradual decrease in Dicer protein levels over an 8-h period (Figure 3C, upper panel, lanes 6–10). Illustrating the importance of de novo protein synthesis for maintaining Dicer protein levels in cells, these data suggest that normal Dicer protein levels may be continuously replenished by de novo protein synthesis in cells depleted of a Dicer stabilizing protein such as CLIMP-63.
Dicer and CLIMP-63 form a high molecular weight complex catalytically active in pre-microRNA processing
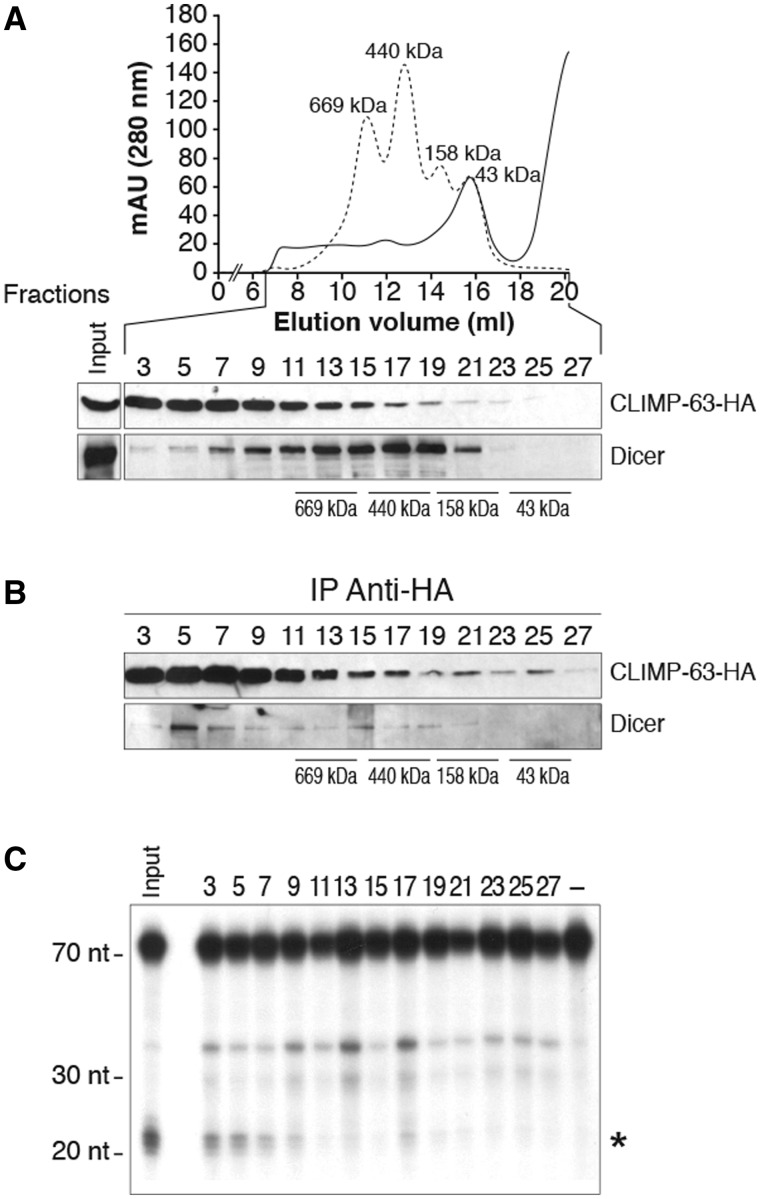
Dicer•CLIMP-63 complexes, derived from HEK 293 cells transiently expressing Flag–Dicer and CLIMP-63–HA proteins, were characterized further by gel filtration chromatography. Western blot analysis of the collected fractions revealed that Dicer and CLIMP-63 showed distinct elution profiles (Figure 4A). The 63-kDa CLIMP-63 protein was particularly enriched in fractions 3–11, corresponding to protein complexes with a molecular weight higher than ∼669 kDa (Figure 4A, upper panel), which may be explained by its ability to form oligomers (27,30). Both epitope-tagged Flag–Dicer and CLIMP-63–HA proteins exhibited a fractionation profile similar to that of the endogenous proteins (our unpublished data).
Figure 4.
Dicer and CLIMP-63 form a high molecular weight complex catalytically active in pre-microRNA processing. (A–C) Protein extracts from HEK 293 cells transiently expressing Flag–Dicer and CLIMP-63–HA proteins were separated by gel filtration chromatography and the fractions collected (40 µl) were analyzed by western blot (A). Equivalent amounts of the selected (odd) fractions (360 µl) were subjected to anti-HA IP, followed by western blot analysis of Dicer and CLIMP-63–HA proteins (B) and Dicer RNase activity assays using 32P-labeled hsa-pre-let-7a-3 RNA as substrate (C). Asterisk symbol indicates the expected let-7a-3 mature microRNA product. RNA size markers are indicated on the left. Input corresponds to an unfractionated protein sample.
The overlapping elution profiles of Dicer and CLIMP-63 prompted us to monitor the presence of Dicer•CLIMP-63 complexes in the various fractions by CLIMP-63–HA immunoprecipitation. Analysis of the anti-HA immune complexes for their protein content and Dicer activity level indicated that most of the Dicer•CLIMP-63 complexes were of relatively high molecular weight (Figure 4B, fraction 5) and catalytically active in processing a 32P-labeled pre-let-7a-3 RNA substrate (Figure 4C). These data suggest that CLIMP-63 complexes conceal a catalytically active form of Dicer in human cells.
Monitored in parallel, the Dicer-interacting TRBP2 protein could not be detected in CLIMP-63 immune complexes (Supplementary Figure S1), an observation that militates against a Dicer complex containing both CLIMP-63 and TRBP2.
Relative importance of CLIMP-63 in the microRNA-guided RNA silencing pathway
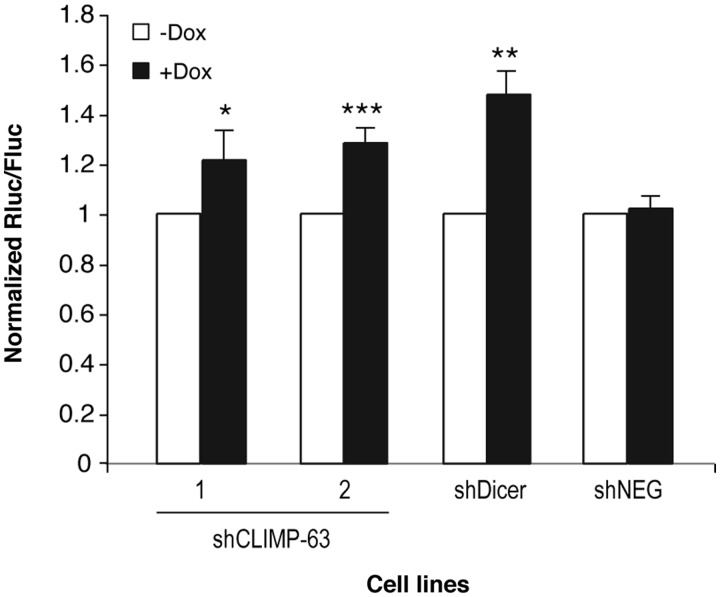
The ability of CLIMP-63 to form a complex with Dicer that is catalytically active in microRNA biogenesis prompted us to examine its relative importance in microRNA-guided RNA silencing. To address that issue, we assessed the efficiency of RNA silencing in two HEK 293 cell lines expressing different shCLIMP-63 sequences (shCLIMP-63-1 and shCLIMP-63-2), as compared with Dicer-depleted cells (shDicer). These cells were transfected with a reporter construct in which the 3′-UTR of high-mobility group A2 (HMGA2) mRNA, which is known to be regulated by let-7 microRNA recognition of up to seven let-7 binding sites (31,32), was placed downstream of the Renilla luciferase (Rluc) open reading frame in psiCHECK. As shown in Figure 5, downregulation of Dicer expression significantly altered Rluc silencing induced by the HMGA2 3′-UTR element, as compared with untreated cells or shNEG-expressing cells, thereby confirming that this regulatory element is under microRNA control in cultured human cells, as reported previously (31,32). Depletion of CLIMP-63 protein levels, using two different shRNA sequences, attenuated the level of regulation of the HMGA2 3′-UTR element, albeit to a smaller extent compared with Dicer depletion. These findings suggest that CLIMP-63 is required for optimal efficiency of microRNA-guided RNA silencing in human cells.
Figure 5.
CLIMP-63 is required for optimal gene regulation mediated by the HMGA2 3′-UTR element. (A and B) Stable 293T-REx cell lines conditionally expressing shRNAs directed against either CLIMP-63 mRNA (shCLIMP-63-1 and shCLIMP-63-2 cell lines) or Dicer mRNA (shDicer) were depleted in CLIMP-63 or Dicer protein levels upon doxycyclin (Dox) induction during 6 days (+Dox) prior to transfection with a Rluc reporter gene coupled or not with the 3′-UTR of HMGA2 mRNA. A Dox-inducible shRNA directed against a deleted region in Rluc mRNA (shNEG) was used as a negative control. Cells were harvested 48 h later for the successive measurements of Rluc and Fluc activities (n = 3 experiments, in duplicate). *P < 0.05; **P < 0.01; ***P < 0.005 versus prior Dox induction (−Dox) (Student’s t-test). Here, it is important to emphasize that the statistical analysis of our experimental data is limited and that statistical inferences only apply to the population of cells from which the data were obtained.
Depletion of CLIMP-63 protein levels in 293T-REx cells was not associated with a concomitant decrease in the level of let-7, miR-17 and miR-21 microRNAs (our unpublished data), suggesting that the effects on RNA silencing may be related to changes in the function, rather than the level, of endogenous microRNAs. However, the caveats have to be taken into account that: (i) the cells that were the most depleted in CLIMP-63, and thus most likely to exhibit a phenotype, may not have survived and been included in our analyses; (ii) changes in the level, activity or function of protein and RNA components of the microRNA pathway other than Exportin-5, which is considered as the rate-limiting step in microRNA biogenesis in human cells (33), need to fall below that threshold to be detected, a situation that may not be achieved upon incomplete CLIMP-63 depletion and (iii) the pool of Dicer proteins affected upon knockdown of CLIMP-63 may not be involved in microRNA biogenesis and may function instead as part of the microRNA effector complex, independently of microRNA levels.
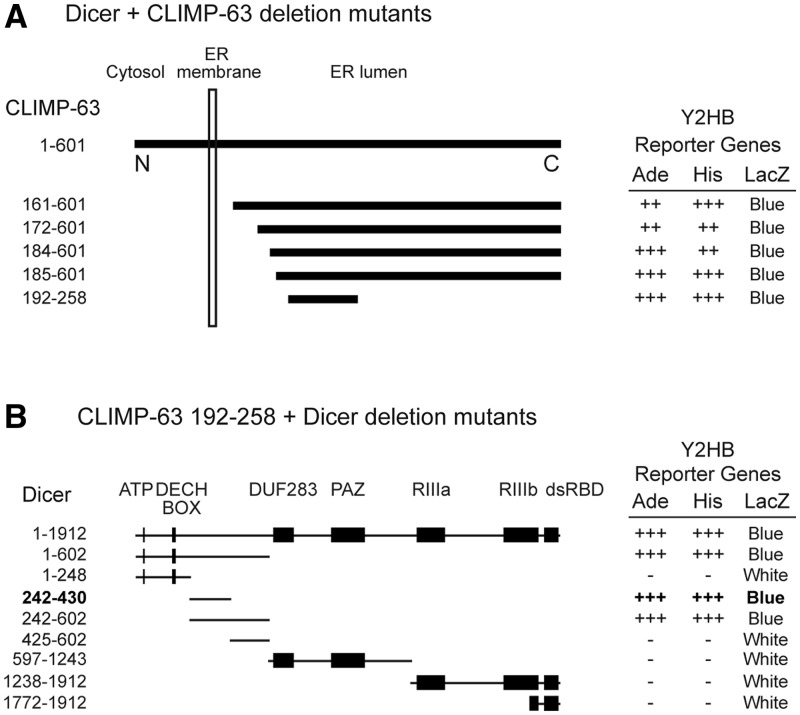
Dicer interacts with a luminal portion of CLIMP-63 via its N-terminal protein interaction domain
The yeast two-hybrid CLIMP-63 cDNA clones were found to interact with Dicer-encoded luminal fragments of the protein (Figure 6A). One of the yeast two-hybrid CLIMP-63 cDNA clones was of particular interest, as it contained an in-frame premature stop codon (TAG) in its coding sequence, resulting in a truncated form of the CLIMP-63 protein comprising amino acids 192–258 (Figure 6A). Identified serendipitously, this deletion mutant is the shortest form of CLIMP-63 capable of interacting with Dicer.
Figure 6.
Dicer interacts with a luminal portion of CLIMP-63 via its N-terminal protein interaction domain. (A) Identification of various Dicer-interacting clones encoding for CLIMP-63 in a yeast two-hybrid screen of a human lung cDNA library using human Dicer as bait. (B) Delineation of the domain of Dicer involved in mediating the Dicer–CLIMP-63 interaction. Yeast transformants were tested for activation of the adenine (Ade), histidine (His) and LacZ reporter genes.
We subsequently used this CLIMP-63 fragment to assess its ability to interact with various deletion mutants of Dicer in the yeast two-hybrid system. We were able to map the CLIMP-63 interacting domain of Dicer to an N-terminal domain comprised of the amino acids 242–430 and defined as the protein interaction domain of human Dicer (Figure 6B).
A small pool of Dicer proteins is located inside the ER lumen
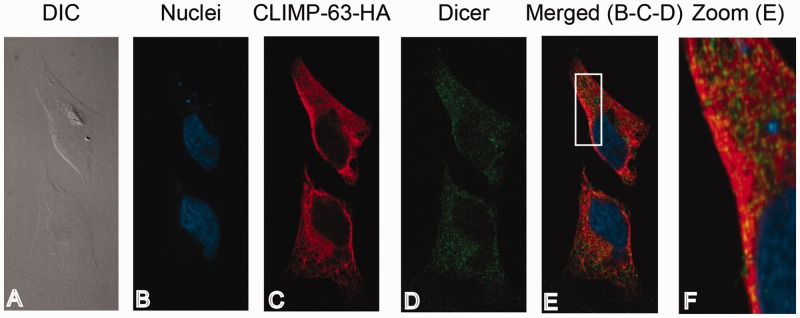
Interacting with the resident ER protein CLIMP-63, we confirmed the ER co-localization of Dicer and CLIMP-63–HA proteins in HeLa cells by confocal immunofluorescence microscopy (Figure 7), suggesting that a pool of Dicer proteins is located in the same subcellular compartment than CLIMP-63.
Figure 7.
CLIMP-63 co-localizes with Dicer proteins in cultured human cells. HeLa cells transiently transfected with pcDNA-Dicer and pcDNA-CLIMP-63–HA vectors were fixed using formaldehyde (4%) and permeabilized using Triton X-100 (1%). (A) Cells were visualized by differential interference contrast (DIC). (B) Nuclei were stained using 1,5-bis (2-(di-methylamino)ethylamino)-4,8-dihydroxyanthracène-9,10-dione (DRAQ5) (in blue). (C, D) CLIMP-63–HA protein was labeled with a polyclonal anti-HA antibody and a secondary anti-rabbit-IgG coupled to AlexaFluor 546 fluorophore (in red) (C), whereas Dicer protein was labeled using monoclonal anti-Dicer antibody and a secondary murine anti-IgG coupled to AlexaFluor 488 fluorophore (in green) (D). (E) The merged image of panels B, C and D. Panel F represents an enlarged area of the white frame shown in (E). Proteins were visualized using a confocal microscope (Quorum spinning Disc Wave Fx, Quorum Technologies) and a 63X lens.
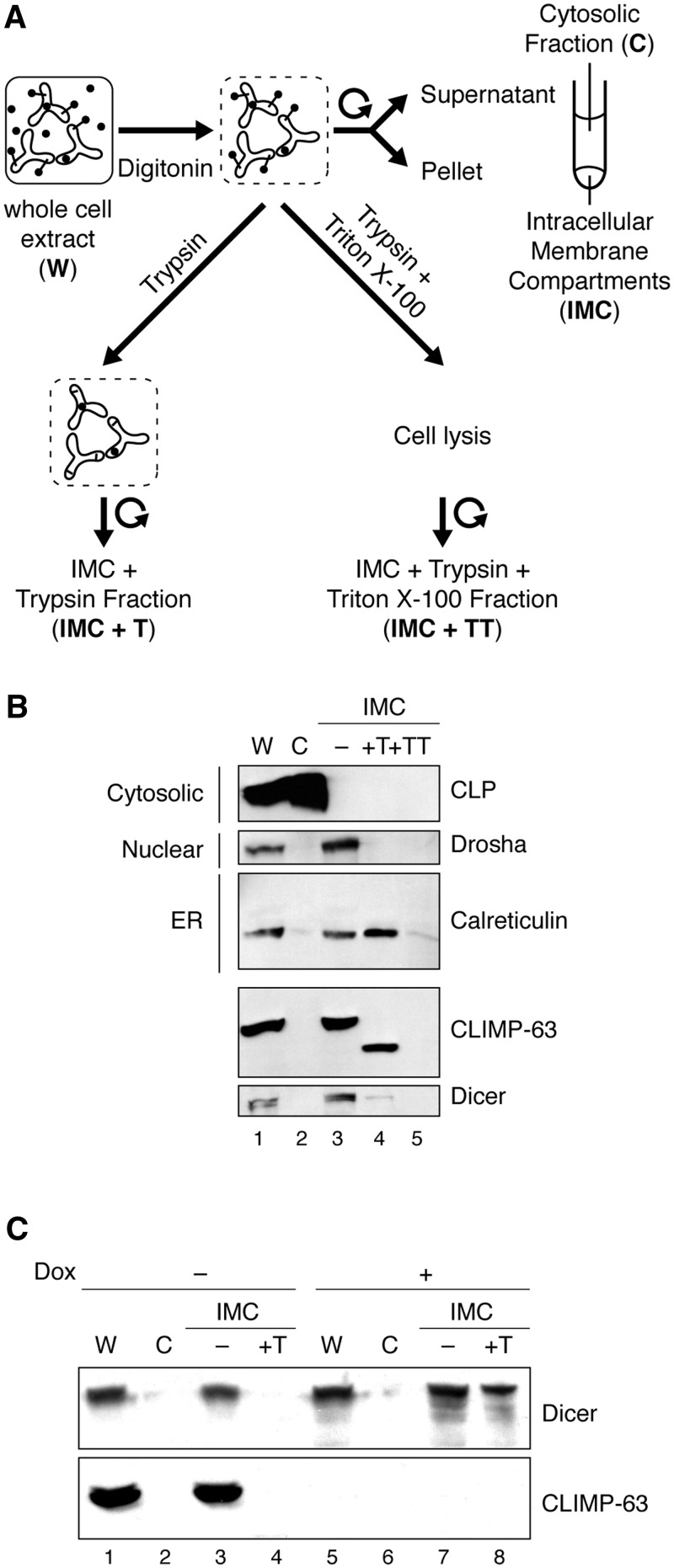
The involvement of the luminal domain of CLIMP-63 in mediating its interaction with Dicer prompted us to verify if Dicer could be found in the ER lumen. To address this question, we adapted and performed a cell-based protease protection assay (25), following the experimental scheme depicted in Figure 8A. Key to this protocol is the use of digitonin, a small toxin derived from the plant Digitalis purpurea that, when added to cells, intercalates into cholesterol-rich membranes by forming a complex with hydroxysterols, causing the plasma membrane to become permeable (34). The cytosolic content then diffuses out of the cell and small exogenous molecules, like trypsin, can diffuse into the cell and into the nucleus and degrade proteins. Notably, the permeabilizing effect of digitonin is limited to the cholesterol-rich plasma membrane. Membranes of intracellular organelles have much lower concentrations of cholesterol and are unaffected when appropriate digitonin concentrations are used (35). The intracellular membrane compartments (IMC) fraction may thus contain, in addition to the ER, intracellular membranes from mitochondria, peroxisomes, autophagosomes and the Golgi apparatus, which represents a limitation of this approach.
Figure 8.
A small pool of Dicer protein is located inside the ER lumen. (A) Schematic illustration of the protease protection assay. (B) HeLa cells were subjected to protease protection assay, and the various fractions collected were analyzed for their protein content by western blot. (C) 293T-REx cells were treated (+), or not (−), with doxycyclin (Dox) to induce shRNA-mediated downregulation of CLIMP-63 expression. The cells were harvested 6 days later, subjected to protease protection assay and analyzed as described in (B). A non-specific shRNA (shNeg) was used as a negative control.
Using the minimum concentration of digitonin necessary to permeabilize the plasma membrane sufficiently for trypsin to enter, we initially monitored the presence of cytosolic, nuclear and ER protein markers by western blot analyses of (i) the whole cell extract (W), (ii) the cytosolic (C) fraction and (iii) the IMC fraction in order to validate our experimental approach (Figure 8B).
In this bioassay, the cytosolic, filamentous actin-binding coactosin-like protein (CLP) (24) was degraded upon trypsin treatment of digitonin-permeabilized cell (Figure 8B, first panel), as expected. Present in the IMC fraction, the nuclear protein Drosha was efficiently digested by trypsin (Figure 8B, second panel, lane 4 versus lane 3), indicating permeabilization of the nuclear membrane by digitonin. Similar results were obtained when monitoring the nuclear protein PARP-1 (our unpublished data). As for the ER protein marker calreticulin, which was detected in the IMC fraction, it resisted trypsin-mediated digestion (Figure 8B, third panel), indicating that this luminal protein is protected by the compartment formed of ER membranes, and that the digitonin concentration used in our experiments was adequate.
Western blot monitoring of the ER membrane protein CLIMP-63 unveiled an increased SDS–PAGE mobility of the protein upon trypsin digestion of digitonin-permeabilized cells (Figure 8B, fourth panel, lane 4 versus lane 3). The estimated molecular weight of the remaining, trypsin-resistant CLIMP-63 peptide is consistent with the digestion of its exposed cytosolic tail of 106 amino acids. As for Dicer, most of the protein is found in the IMC fraction, and may thus be associated with IMC (Figure 8B, fifth panel, lane 3). A relatively small fraction of the protein appears to be protected from trypsin digestion by intracellular membranes (Figure 8B, fifth panel, lane 4), which is compatible with the existence of a small pool of Dicer residing inside the ER lumen. A larger proportion of trypsin-resistant Dicer protein is detected upon Dox-induced, shRNA-mediated depletion of CLIMP-63 levels (Figure 8C, upper panel, lane 8 versus lane 4), which is consistent with an accumulation of Dicer protein inside the ER lumen in the absence of CLIMP-63.
CLIMP-63 interacts with newly synthesized forms of Dicer
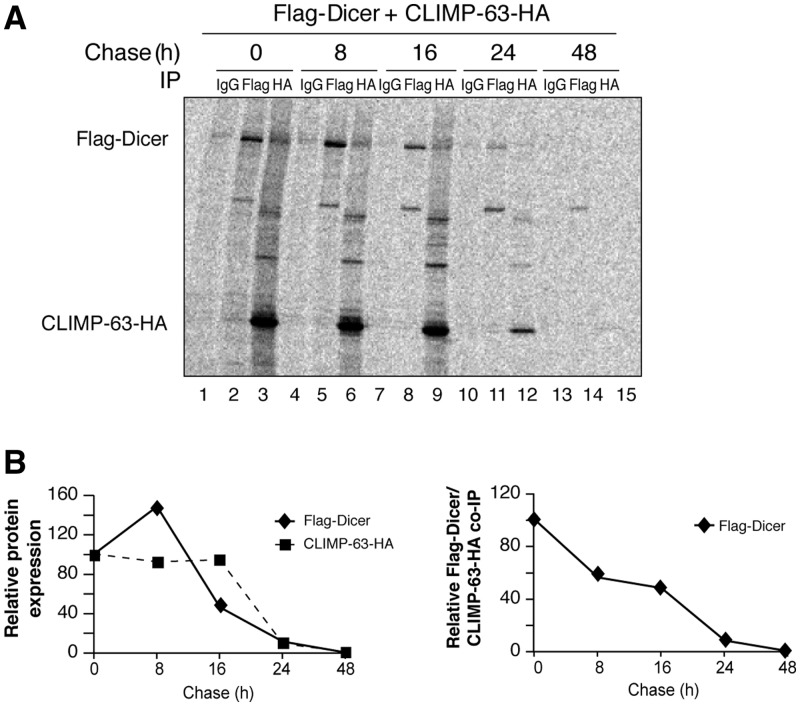
Considering (i) the ability of Dicer to interact with the luminal portion of an ER protein and (ii) the fact that several cellular proteins enter the ER to be modified co-/post-translationally, we next asked whether the small pool of Dicer present in the ER lumen represents newly synthesized forms of the protein that interact with CLIMP-63. To verify this hypothesis, we performed a pulse-chase analysis of cells metabolically labeled with 35S-methionine/35S-cysteine. We observed an efficient incorporation of 35S-Met/35S-Cys into Flag–Dicer and CLIMP-63–HA proteins following a brief 15-min pulse (at time 0 of the chase), as both proteins could be detected by autoradiographic analysis of their respective immunoprecipitates (Figure 9A, lanes 2 and 3, respectively). Chasing of the cells for 8, 16, 24 and 48 h indicated half-lifes of ∼16 h for the Flag–Dicer protein and of ∼20 h for CLIMP-63–HA (Figure 9B, left panel).
Figure 9.
CLIMP-63 interacts with newly synthesized forms of Dicer. (A, B) HEK 293 cells transiently expressing Flag–Dicer and CLIMP-63–HA were metabolically labeled with 35S-Met/35S-Cys for 15 min and chased for the indicated periods of time prior to IP. The immune complexes were analyzed by SDS–PAGE and autoradiography (A). The bands corresponding to the 35S-labeled Flag–Dicer (lanes 2, 5, 8, 11 and 14) and CLIMP-63–HA (lanes 3, 6, 9, 12 and 15) proteins were quantitated by PhosphorImager acquisition and densitometry and used to generate the left panel of Figure 5B (B). Similarly, the bands corresponding to Flag–Dicer and CLIMP-63–HA in lanes 3, 6, 9, 12 and 15 were quantified and used to generate the right panel of Figure 5B.
More importantly, newly synthesized forms of Flag–Dicer protein, labeled during the brief 15-min pulse (at time 0 of the chase), could be detected in the CLIMP–HA immunoprecipitates (Figure 9A, lane 3). Notably, a significant proportion of Flag–Dicer proteins remained associated with CLIMP-63–HA after 16 h (Figure 9A, lane 9 and Figure 9B, right panel). These results indicate that Dicer•CLIMP-63 complexes involve newly synthesized forms of Dicer and are relatively stable, suggesting that their association may modulate their stability.
Dicer transits through the ER of cultured human cells and is glycosylated
The localization of a small pool of Dicer proteins inside the ER lumen prompted us to consider the possibility and test whether Dicer can transit through the ER. For that purpose, we treated HEK 293 cells with Brefeldin A, an agent known to inhibit the ER–Golgi transport of proteins (36) and to activate the unfolded protein response (UPR), which is coordinated with the ER-associated degradation (ERAD) process to regulate the protein load at the ER (37). We observed that Brefeldin A treatment reduced Dicer protein levels, as compared to mock-treated cells (Figure 10A, upper panel), which is consistent with a scenario in which Dicer would transit through the ER and be entrapped into the ER and degraded upon Brefeldin A treatment. These findings are compatible with a previous study showing the preferential distribution of Dicer in the Golgi-reticulum area of post-mitotic neuronal cells and its altered distribution upon Brefeldin A treatment (38).
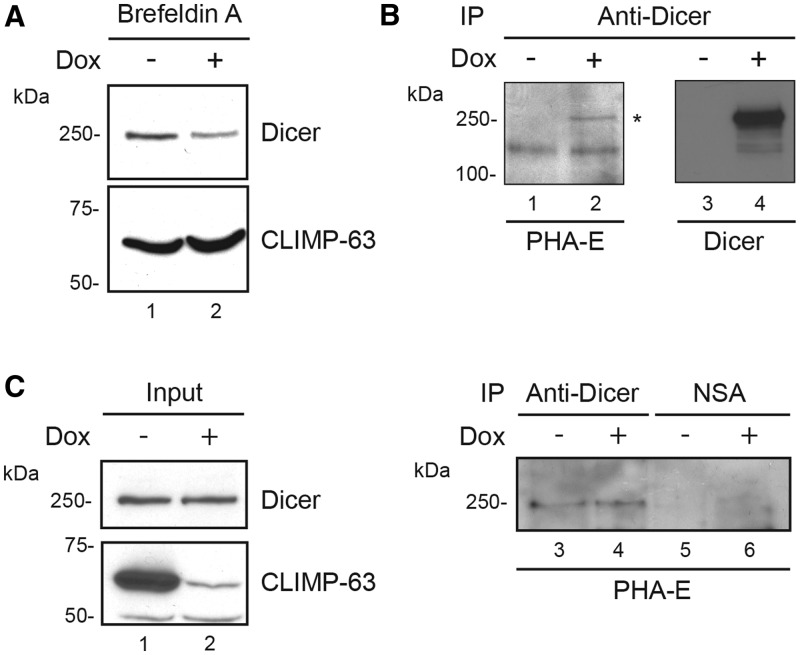
Figure 10.
Dicer transits through the ER of cultured human cells and is glycosylated. (A) 293T-REx cells, treated (+) or not (−) with doxycyclin (Dox) to induce shRNA-mediated downregulation of CLIMP-63 expression, were exposed to Brefeldin A for 18 h. Cells were harvested and protein extracts were analyzed by immunoblotting using anti-Dicer (upper panel) and anti-CLIMP-63 (lower panel) antibodies. (B) Dicer immunoprecipitates prepared from 293T-REx cells were analyzed by immunoblotting using HRP-conjugated PHA-E (left panel), a lectin that can recognize complex oligosaccharide structures or anti-Dicer antibody (right panel). Asterisk symbol indicates the expected glycosylated Dicer protein band. (C) Immune complexes from 293T-REx cells conditionnally expressing short hairpin RNA directed against CLIMP-63 mRNA (shCLIMP-63-1) following doxycyclin treatment (Dox) for 6 days were analyzed as in (B). NSA, non-specific antibody.
We obtained results similar to Brefeldin A treatment when incubating cells in the presence of tunicamycin (our unpublished data), an inhibitor of N-acetylglucosamine transferase that prevents glycosylation of newly synthesized glycoproteins (39,40). Considering that the majority of the proteins present in the ER lumen undergo glycosylation, we suspected that Dicer could be glycosylated. Bioinformatic predictions, based on the NetNGlyc 1.0 and NetOGlyc 3.1 server interfaces (http://www.cbs.dtu.dk/services/NetOGlyc/), unveiled, among others, the presence of a total of 12 putative glycosylation sites, i.e. 9 asparagines and 3 tryptophans (Supplementary Figure S2), respectively. To verify that possibility, we analyzed Dicer immunoprecipitates prepared from HEK 293 cells by immunoblotting using HRP-conjugated PHA-E, a lectin that recognizes complex oligosaccharide structures (41). We were able to detect a band corresponding to a glycosylated species of Dicer protein (Figure 10B, lane 4), suggesting that Dicer is a glycoprotein. Analysis of protein extracts derived from CLIMP-63 depleted 293T-REx cells by western blot and PHA-E staining failed to provide conclusive evidences for a role of CLIMP-63 in Dicer glycosylation (Figure 10C), although the previously reported collapse of the ER in CLIMP-63 depleted cells (42) may limit the interpretation of these data. Together, these results support the idea that Dicer is glycosylated and transits through the ER along a process important for maintaining Dicer protein levels in cultured human cells.
Dicer can be secreted by cultured human cells
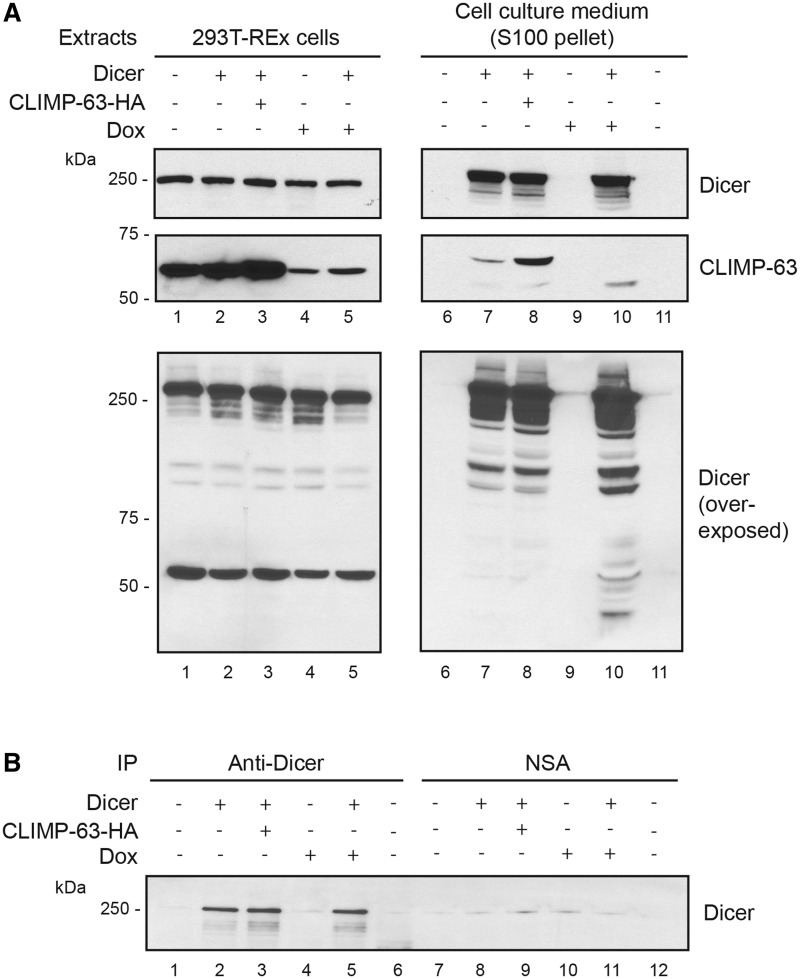
Proteins transiting through the ER usually follow the secretion pathway and are directed to the plasma membrane to be secreted. To examine whether Dicer is secreted, we analyzed the 100 000g pellet of culture medium samples collected from cells overexpressing Dicer and/or CLIMP-63, and depleted or not in CLIMP-63. We detected the presence of Dicer in the culture medium upon overexpression in 293T-REx cells (Figure 11A, upper panel, lane 7). CLIMP-63 also appeared to be secreted, but only upon Dicer overexpression (Figure 11A, center panel, lanes 7 and 8). Depletion of CLIMP-63 did not alter the level of vesicular Dicer (Figure 11A, upper panel, lane 10 versus lane 7). However, it was associated with an increase in Dicer immunoreactive bands of lower molecular weight (Figure 11A, lower panel, lane 10 versus lane 7) that may correspond to Dicer degradation products, similar to those observed in protein extracts of cells depleted in CLIMP-63 (our unpublished data), which supports further a role for CLIMP-63 in stabilizing Dicer. Considering that CLIMP-63, a resident protein of the ER, is exported from cultured human cells with Dicer, we can speculate that the Dicer-containing vesicles may originate from the secretion pathway.
Figure 11.
Dicer is secreted in the extracellular environment of cultured 293T-REx cells with CLIMP-63. 293T-REx cells were transiently transfected with pcDNA-Dicer and/or pcDNA-CLIMP-63–HA vectors (lanes 1–5) and treated (+), or not (−), with doxycyclin (Dox) to induce shRNA-mediated downregulation of CLIMP-63 expression. (A) Protein extracts prepared from the cells (left panel) and from the 100 000g pellet of the cell culture medium (right panel) were analyzed by immunoblotting using anti-Dicer (upper panel) or anti-CLIMP-63 (center panel) antibody. Overexposure of the anti-Dicer blot is shown in the lower panel. DMEM alone was used as a negative control (lanes 6 and 11). (B) Protein extracts prepared from the 100 000g supernatant of the cell culture medium were subjected to immunoprecipitation (IP) using anti-Dicer (lanes 1–6) or anti-Flag (used as a negative isotypic antibody control; lanes 7–12) antibody, and analyzed by immunoblotting using anti-Dicer antibody. Lanes 6 and 12 correspond to DMEM alone, used as a negative control.
Finally, we were able to immunoprecipitate Dicer from the 100 000g supernatant of the cell culture medium samples upon Dicer overexpression (Figure 11B), suggesting that Dicer may also be released from cultured cells either free or in complex with (an)other protein(s).
DISCUSSION
The use of a yeast two-hybrid screening approach, as a means to expand the known protein interaction network of Dicer, has the advantage: (i) to allow the detection of protein interactions that are relatively weak and/or transient in nature, which may be of biological significance and interest, (ii) to identify Dicer protein complexes of relatively low abundance and/or involving only a small pool of Dicer and (iii) to complement and extend the repertoire of Dicer-interacting proteins beyond those previously identified by the use of relatively more stringent immunoprecipitation-based strategies, such as Ago2 (11,13), TRBP (11,12) and KSRP (15).
Among the human Dicer-interacting candidates identified in our yeast two-hybrid approach were the known Dicer co-factor TRBP (11,12) (our unpublished data) and the non-glycosylated type II ER membrane protein CLIMP-63. Previously known as p63 and ubiquitously expressed in higher eukaryotes (27,43,44), CLIMP-63 is rather unique among ER proteins, in that it is excluded from the outer nuclear membrane by forming large immobile oligomers in the reticular ER (27,30). CLIMP-63 is partitioned across the ER membrane as follows: a central transmembrane domain composed of 21 amino acid residues, mainly hydrophobic, links an N-terminal cytosolic domain of 106 amino acids, which directly binds to microtubules (45) and a C-terminal luminal domain of 474 amino acids, containing more than 165 charged residues (27,43,44,46). The luminal segment of CLIMP-63 has been reported to form coiled-coil structures that can assemble into higher order oligomers, as part of a process that immobilizes the protein in the lipid bilayer of the ER (30). An integral component of the ER, CLIMP-63 can thus be viewed as an interacting protein that may anchor Dicer to this subcellular compartment, which would explain, at least in part, the perinuclear localization of Dicer previously reported in cultured human cells (6). It is worth noting that a similar enrichment of Dicer at the nuclear periphery has been observed recently in fission yeast (47).
The minimal sequence of human Dicer capable of interacting with CLIMP-63 domain is formed by amino acids 242–430. This N-terminal domain of human Dicer was also found to mediate binding to TRBP2 (our unpublished data) and thus appears to function as a protein interaction module. As suggested by our yeast two-hybrid data indicating that CLIMP-63 and TRBP2 do not interact with each other (our unpublished data), we were unable to document the presence of TRBP2 in CLIMP-63 immune complexes. Considering that CLIMP-63 and TRBP2 may share the same binding region in Dicer, these results suggest that the interactive relationship between Dicer and CLIMP-63 or TRBP2 is likely to be mutually exclusive. CLIMP-63 and TRBP2 may thus form distinct Dicer complexes that may play a role in storage and/or contribute to regulate the stability of the RNase III Dicer. This latter role would be analogous to that reported for DGCR8 in upregulating expression of the RNase III Drosha through protein stabilization (48,49).
Together with the relatively weak Dicer-CLIMP-63 co-immunoprecipitation signal, our data tend to support the relatively low abundance of cellular Dicer•CLIMP-63 protein complexes, which may have passed unnoticed and have escaped detection in studies based initially on co-immunoprecipitation approaches. As for the relatively high molecular weight of Dicer•CLIMP-63 complexes, it is compatible with the propensity of CLIMP-63 to form oligomers (27,30) and the ability of Dicer to interact with these oligomers. The fact that CLIMP-63 complexes conceal a catalytically active form of Dicer is also consistent with a previous study reporting that the CLIMP-63-interacting, N-terminal portion of Dicer is dispensible for its pre-microRNA processing activity (50).
Interestingly, Shibata et al. (51) have defined CLIMP-63 as a sheet-inducing protein that is enriched in rough ER through its association with translating membrane-bound ribosomes. Corroborating these findings, proteomic analyses of CLIMP-63 immunoprecipitates identified several proteins involved in cellular mRNA translation, including ribosomal and poly(A) binding proteins (our unpublished data). We also detected other protein components of the microRNA pathway, such as Fragile X mental retardation syndrome-related proteins 1 and 2 (our unpublished data). These Fragile X proteins form messenger ribonucleoprotein (mRNP) complexes that associate with polyribosomes and can act as a microRNA acceptor protein for Dicer and facilitate the assembly of microRNAs on specific target RNA sequences (14). Strategically anchored in the ER and linked functionally to the translational machinery (51), CLIMP-63 may thus play a role in RNA silencing by retaining a small pool of catalytically active Dicer enzymes in this specific cellular location, in close proximity to its downstream effector proteins and mRNAs to be translated under the control of Dicer-derived microRNAs.
In support to this assertion, we serendipitously mapped the Dicer-interacting region of CLIMP-63 to a 67-amino acid stretch located in the luminal domain of the protein. The salt-sensitivity of the Dicer-CLIMP-63 interaction suggests that this small stretch is exposed on the surface of CLIMP-63 oligomers and is available to mediate electrostatic interactions with Dicer. The transit of Dicer through the ER lumen may be initiated by, and imply the presence of, an ER localization signal in its amino acid sequence. These signal peptides are stretches of 7–25 hydrophobic and charged amino acids generally located at the N-terminal extremity of proteins and capable of mediating their translocation across the ER membrane (52). Inspection of the amino acids forming the N-terminal domain of Dicer revealed the presence of a stretch that shows some similarities with the signal peptide of some eukaryotic proteins (Supplementary Figure S3). Its functionality, however, remains to be experimentally validated. A recent study by Pyhtila et al. (53) reported a signal sequence-independent localization of a subpopulation of cytosolic protein-encoding mRNAs to the ER of human cells, suggesting that the presence of a signal peptide is not obligatory for the ER localization of human proteins. Collectively, our data are consistent with a model whereby the Dicer protein would enter into the ER lumen independently of, or via, a functional ER signal peptide, and be brought into close proximity of the resident ER membrane protein CLIMP-63, which would then interact with and stabilize Dicer.
An alternative explanation would have the endogenous Dicer protein interacting transiently with CLIMP-63 as it transits through the ER. This scenario would reconcile the observed decrease in Dicer protein levels in CLIMP-63-depleted, CHX-treated cells by a proportion that apparently exceeds its degree of association with CLIMP-63 or localization to the ER with the possibility that the stabilizing effects of CLIMP-63 may extend beyond protein complex formation with Dicer. However, the caveats have to be taken into account that the incomplete knockdown in CLIMP-63 protein levels and, more importantly, the previously reported collapsing of the ER compartment in CLIMP-63 knockdown cells (51), which may entrap glycosylated Dicer protein and alter its intracellular distribution and metabolism, may limit and hamper the interpretation of our data.
Together with the translocon complex, CLIMP-63 is one of the two most abundant protein constituents in ER sheets of mammalian cells (51). Depletion of CLIMP-63 was associated with a marked reduction in the distance between cisternal sheets, thereby supporting a role for CLIMP-63 as a spacer between the sheets in the ER lumen (51). Given its propensity to oligomerize, CLIMP-63 could thus assemble into parallel coiled-coil arrangements to flatten membranes and to serve as luminal ER spacers that keep individual sheets a specific distance apart (42). Therefore, it is tempting to establish a relationship between this function of CLIMP-63 and the observed accumulation of Dicer proteins in IMC fraction of human cells upon depletion of CLIMP-63, whereby Dicer may be entrapped into the collapsing ER.
When interpreting our protease protection results, the caveat has to be taken into account that we cannot (i) exclude the possible contribution of Dicer-interacting protein(s) in protecting Dicer from trypsin digestion, as described for Gerp95p (54) and (ii) rule out the possible presence of Dicer in an IMC other than the ER (e.g. mitochondria, peroxisomes, autophagosomes and the Golgi apparatus). However, it is in combination with (i) the co-immunoprecipitation of Dicer with the ER membrane protein CLIMP-63, (ii) the yeast two-hybrid data showing the interaction between Dicer and the luminal domain of CLIMP-63 and (iii) the co-localization of Dicer and CLIMP-63 in cultured human HeLa cells demonstrated by confocal immunofluorescence microscopy, that our cell-based protease protection data support a role for CLIMP-63 in the ER localization and/or transit of Dicer of human cells.
Apart from the Dicer stabilizing effects of TRBP2 (55) and [protein activator of protein kinase R (PKR) (PACT)] (50), the cellular turnover of Dicer proteins, i.e. how Dicer is synthesized, stabilized and eventually degraded, remains poorly defined. Interacting within minutes of their synthesis, Dicer protein levels are maintained by relatively stable CLIMP-63 protein complexes, raising the issue as to what happens to Dicer thereafter.
Our data obtained by the use of a lectin (PHA-E) that recognizes complex oligosaccharide structures (41) support the glycosylated nature of Dicer. Involving the enzymatic, site-specific conjugation of carbohydrate chains of various sizes and complexities, glycosylation confers important functionalities to the target proteins. For instance, some proteins do not fold correctly unless they are glycosylated first. Also, N-linked polysaccharides may confer stability to some glycoproteins, in the absence of which, the unglycosylated protein is rapidly degraded (56). Whereas the existence of a small pool of Dicer proteins inside the ER lumen and reduction in Dicer protein levels upon treatment of HEK 293 cells with Brefeldin A suggest that Dicer transits through the ER, the effects induced by tunicamycin, an inhibitor of N-acetylglucosamine transferases, support further the possibility that carbohydrate conjugation to a protein as large and complex as Dicer may contribute to its proper folding and stability in human cells. It is tempting to speculate that inhibition of glycosylation may promote misfolding of Dicer and pre-dispose it to removal by the cell, as suggested for N-glycosylation of the glycoprotein apoB-100 (57). The nature and functional implications of the carbohydrate conjugation to the Dicer protein warrants further and more detailed investigations.
We obtained data indicating that Dicer and CLIMP-63 may be released from cultured human cells, either free or in 100 000g pelletable vesicles. Although, (i) the exact nature and composition of Dicer-containing vesicles remains to be determined, the detection of Dicer in the 100 000g pellet of cell culture medium suggests a link between Dicer and the endocytic pathway. Similar findings have been reported for GW182, suggesting that the microRNA machinery is associated with multi-vesicular bodies. Indeed, GW182 was found to be exported out of the cell through the endocytic pathway along a process important for the reloading of the RISC complex with microRNAs (58,59). Overexpression of Dicer in 293T-REx cells was associated with an increase in Dicer protein levels in the culture medium, with no changes in the intracellular content of the protein, suggesting that the export of Dicer may contribute to maintain its cellular level within a physiological range. Whether CLIMP-63 plays a role in the ER-extracellular environment trafficking, in addition to its role in maintaining the ER structure (51), remains to be investigated.
Expanding the known repertoire of Dicer-interacting proteins, our study contributes to improve our understanding of the molecular mechanisms underlying the microRNA pathway by defining CLIMP-63 as a novel protein interactor and regulator of Dicer function, which (i) is involved in maintaining Dicer protein levels, (ii) is required for optimal RNA silencing in human cells, (iii) may explain the perinuclear localization of Dicer and (iii) may serve as a protein cargo for the export of Dicer from human cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3 and Supplementary Reference [60].
FUNDING
Discovery grant from Natural Sciences and Engineering Research Council of Canada (NSERC) [262938-03/08/2012 to P.P.]; doctoral studentships from Fonds de Recherche du Québec - Santé (to G.P.) and from NSERC (to M.P.P.). Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Isabelle Plante for technical assistance. The pTER vector was kindly provided by Dr Hans Clevers. P.P. is a Senior Scholar from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front. Biosci. 2008;13:2537–2547. doi: 10.2741/2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 9.Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J. Biomed. Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 11.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 14.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J. Biomed. Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and MicroRNA-328 regulate expression of mouse {beta}-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Perron MP, Landry P, Plante I, Provost P. Detection of human Dicer and Argonaute 2 catalytic activity. Methods Mol. Biol. 2011;725:121–141. doi: 10.1007/978-1-61779-046-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provost P, Doucet J, Stock A, Gerisch G, Samuelsson B, Radmark O. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem J. 2001;359:255–263. doi: 10.1042/0264-6021:3590255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat. Struct. Mol. Biol. 2009;16:961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provost P, Doucet J, Hammarberg T, Gerisch G, Samuelsson B, Radmark O. 5-Lipoxygenase interacts with coactosin-like protein. J. Biol. Chem. 2001;276:16520–16527. doi: 10.1074/jbc.M011205200. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz H, Hailey DW, Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods. 2006;3:205–210. doi: 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- 26.Zhou P. Determining protein half-lives. Methods Mol. Biol. 2004;284:67–77. doi: 10.1385/1-59259-816-1:067. [DOI] [PubMed] [Google Scholar]
- 27.Schweizer A, Ericsson M, Bachi T, Griffiths G, Hauri HP. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J. Cell Sci. 1993;104(Pt 3):671–683. doi: 10.1242/jcs.104.3.671. [DOI] [PubMed] [Google Scholar]
- 28.Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol. Biol. Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 30.Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri HP. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J. Cell Biol. 2001;153:1287–1300. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plutner H, Davidson HW, Saraste J, Balch WE. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 1992;119:1097–1116. doi: 10.1083/jcb.119.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liscum L, Munn NJ. Intracellular cholesterol transport. Biochim. Biophys. Acta. 1999;1438:19–37. doi: 10.1016/s1388-1981(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 36.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 38.Barbato C, Ciotti MT, Serafino A, Calissano P, Cogoni C. Dicer expression and localization in post-mitotic neurons. Brain Res. 2007;1175:17–27. doi: 10.1016/j.brainres.2007.07.088. [DOI] [PubMed] [Google Scholar]
- 39.Duksin D, Bornstein P. Impaired conversion of procollagen to collagen by fibroblasts and bone treated with tunicamycin, an inhibitor of protein glycosylation. J. Biol. Chem. 1977;252:955–962. [PubMed] [Google Scholar]
- 40.Takatsuki A, Tamura G. Tunicamycin, a new antibiotic. II. Some biological properties of the antiviral activity of tunicamycin. J. Antibiot. 1971;24:224–231. [PubMed] [Google Scholar]
- 41.Kaneda Y, Whittier RF, Yamanaka H, Carredano E, Gotoh M, Sota H, Hasegawa Y, Shinohara Y. The high specificities of Phaseolus vulgaris erythro- and leukoagglutinating lectins for bisecting GlcNAc or beta 1-6-linked branch structures, respectively, are attributable to loop B. J. Biol. Chem. 2002;277:16928–16935. doi: 10.1074/jbc.M112382200. [DOI] [PubMed] [Google Scholar]
- 42.Barlowe C. ER sheets get roughed up. Cell. 2010;143:665–666. doi: 10.1016/j.cell.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Schweizer A, Rohrer J, Hauri HP, Kornfeld S. Retention of p63 in an ER-Golgi intermediate compartment depends on the presence of all three of its domains and on its ability to form oligomers. J. Cell Biol. 1994;126:25–39. doi: 10.1083/jcb.126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer A, Rohrer J, Slot JW, Geuze HJ, Kornfeld S. Reassessment of the subcellular localization of p63. J. Cell Sci. 1995;108(Pt 6):2477–2485. doi: 10.1242/jcs.108.6.2477. [DOI] [PubMed] [Google Scholar]
- 45.Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweizer A, Rohrer J, Jeno P, DeMaio A, Buchman TG, Hauri HP. A reversibly palmitoylated resident protein (p63) of an ER-Golgi intermediate compartment is related to a circulatory shock resuscitation protein. J. Cell Sci. 1993;104(Pt 3):685–694. doi: 10.1242/jcs.104.3.685. [DOI] [PubMed] [Google Scholar]
- 47.Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev. Cell. 2010;18:102–113. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Heijne G. The signal peptide. J. Membr. Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 53.Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varki ACR, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories Press; 2009. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 57.Liao W, Chan L. Tunicamycin induces ubiquitination and degradation of apolipoprotein B in HepG2 cells. Biochem. J. 2001;353:493–501. doi: 10.1042/0264-6021:3530493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 59.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stryer L. Biochemistry. New York: W. H. Freeman and Company; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.