Abstract
The Mre11 complex (Mre11-Rad50-Nbs1 or MRN) binds double-strand breaks where it interacts with CtIP/Ctp1/Sae2 and ATM/Tel1 to preserve genome stability through its functions in homology-directed repair, checkpoint signaling and telomere maintenance. Here, we combine biochemical, structural and in vivo functional studies to uncover key properties of Mre11-W243R, a mutation identified in two pediatric cancer patients with enhanced ataxia telangiectasia-like disorder. Purified human Mre11-W243R retains nuclease and DNA binding activities in vitro. X-ray crystallography of Pyrococcus furiosus Mre11 indicates that an analogous mutation leaves the overall Mre11 three-dimensional structure and nuclease sites intact but disorders surface loops expected to regulate DNA and Rad50 interactions. The equivalent W248R allele in fission yeast allows Mre11 to form an MRN complex that efficiently binds double-strand breaks, activates Tel1/ATM and maintains telomeres; yet, it causes hypersensitivity to ionizing radiation and collapsed replication forks, increased Rad52 foci, defective Chk1 signaling and meiotic failure. W248R differs from other ataxia telangiectasia-like disorder analog alleles by the reduced stability of its interaction with Rad50 in cell lysates. Collective results suggest a separation-of-function mutation that disturbs interactions amongst the MRN subunits and Ctp1 required for DNA end processing in vivo but maintains interactions sufficient for Tel1/ATM checkpoint and telomere maintenance functions.
INTRODUCTION
DNA double-strand breaks (DSBs) are a driving force in genome instability, as they are a primary source of chromosome breaks that can lead to chromosome translocations or loss of chromosome fragments (1). The MRN complex, composed of Mre11, Rad50 and Nbs1 (Xrs2 in Saccharomyces cerevisiae) subunits, has crucial roles in genomic stability through its functions in DNA repair, checkpoint signaling and telomere maintenance (2). As a primary responder to DNA DSBs, the MRN complex rapidly localizes to damage sites and recruits the ATMTel1 (Ataxia telangiectasia mutated) kinase via Nbs1. ATM stimulates repair factors through phosphorylation and initiates the checkpoint response that stalls cell cycle progression during DNA repair, or activates senescence or apoptosis pathways (3). Nbs1 also recruits the DNA end-processing factor CtIPCtp1/Sae2, which along with the nuclease activity of Mre11, initiates the 5′ to 3′ resection of DNA ends that is essential for homologous recombination (HR) repair of DSBs (4–8). Extended DNA end resection is catalyzed by Exo1 exonuclease, or an activity dependent on a RecQ-family DNA helicase, Dna2 nuclease and Rmi1-Top3 topoisomerase, to generate 3′ single-stranded DNA (ssDNA) overhangs (9–13), which perpetuate the checkpoint response via the ATRRad3-ATRIPRad26 complex binding to RPA-coated ssDNA, and facilitate homology-directed repair mediated by Rad51 (14).
The loss of Mre11 or mutations that ablate its endonuclease activity cause early embryonic lethality in mice and acute clastogen sensitivity in Schizosaccharomyces pombe (8,15). Mre11 is also crucial in S. cerevisiae, although the effects of ablating its nuclease activities are weaker (16,17). Inherited hypomorphic mutations in Mre11 are associated with ataxia telangiectasia-like disorder (ATLD), a rare disease characterized by progressive cerebellar ataxia, an increase in chromosomal aberrations and sensitivity to ionizing radiation (IR) (18–20). ATLD was identified as a milder variant of a condition called ataxia telangiectasia (A-T), which results from mutations in the ATM kinase. A-T patients present with the same clinical features of ATLD, but are also afflicted with telangiectasias, immunodeficiency and a predisposition to the development of cancer. Mutations in the Nbs1 subunit cause Nijmegen breakage syndrome, which results in a characteristic microcephaly and, except for cerebellar degeneration, share the same clinical features of A-T (21,22).
Given the intimate relationship between the MRN complex and ATM in DNA repair and checkpoint signaling, it is unsurprising that mutations that impair their functions cause diseases with shared features. Mutations in MRE11 and RAD50 were recently identified in patients with Nijmegen breakage syndrome-like microcephaly (23,24), further blurring the lines that differentiate these conditions. Moreover, two brothers in Japan were recently found to have symptoms characteristic of ATLD, but additionally had mental retardation, physical malformations and subsequently were diagnosed at ages 15 and 9 years as having lung adenocarcinomas with multiple bone metastases (25). Cells from these patients had no deficiency in ATM abundance and harbored no mutations in NBS1. However, novel mutations were identified in the MRE11 genes, one causing a 27 amino acid deletion in the N-terminus of the protein, and the other a W243R missense mutation. Each allele was inherited from unaffected parents that were heterozygous for a wild-type copy of MRE11. Thus, these patients represent the 17th and 18th patients diagnosed with ATLD and the first to develop cancer.
To gain insights into the molecular effects of the W243R mutation and its implications for Mre11 functions, we have biochemically, structurally and functionally characterized corresponding mutations in human Mre11 and its orthologs. These studies indicate that the W243R mutation does not impact the intrinsic nuclease activity of Mre11 or ablate its ability to form a complex with Rad50 and Nbs1 that recruits Tel1 to DSBs, but it disturbs interactions amongst the MRN subunits resulting in defects in DNA end processing.
MATERIALS AND METHODS
Protein expression and purification
Expression and purification of P. furiosus Mre11 (residues 1-342) followed published procedures (26). The L204R mutation was introduced by Quickchange Site Directed Mutagenesis Kit (Agilent).
C-terminal tagged 6-histidine human Mre11 preparations were produced by infection of Sf9 cells in suspension culture with baculoviruses and collected after 72 h. Proteins were purified using Ni2+-NTA agarose beads (Qiagen), Superdex 200 size-exclusion column (GE Healthcare) and Hitrap Q column (GE Healthcare) done sequentially. Purified Mre11 was pooled and concentrated on a Heparin column (GE Healthcare). Immunoblotting of Mre11 was performed on nitrocellulose membranes (Whatman) using standard immunoblotting techniques. Protein concentrations were determined by comparison with a bovine serum albumin standard on a 10% polyacrylamide gel after staining with Coomassie blue. Detailed procedure is available in Supplementary Data.
Crystallization, X-ray diffraction data collection, structure determination and refinement
Mre11 crystals were grown by mixing 1 µl protein solution (20 mg/ml in 200 mM NaCl, 20 mM Tris–HCl pH 7.5, 5% glycerol) and 1 µl crystallization solution (100 mM Bicine, 10% (w/v) PEG6000, 1 M lithium chloride) (ref. cond. 7 JCSG core II suite QIAGEN). Crystals were grown at 15°C by hanging-drop vapor diffusion for 7 days. Crystals were transferred to cryoprotectant (crystallization solution plus 25% PEG200) and flash cooled in liquid nitrogen. Diffraction data were collected and refined to 2.3 Å resolution (Supplementary Table S1). Coordinates for P. furiosus Mre11-L204R crystal structure are available in the Protein Data Bank (PDB: 4HD0).
Electrophoretic mobility shift assays
Aliquots of ssDNA90, labeled with Alexa Fluor 488 at the 5′ end, and dsDNA90, labeled with Alexa Fluor 488 at the 5′ end of the upper strand were incubated with the indicated amounts of protein dimer. Substrate sequences and binding buffer composition are available in Supplementary Data. The reaction products were separated on 5% native polyacrylamide gels run in 0.5X TBE at 4°C. Labeled DNA was visualized by direct scanning of the gel with a 488-nm laser using a Typhoon 9200 (GE Healthcare). The emission signal was sorted with a 520BP40-nm filter. Quantification was performed with ImageQuant 5.2 software with error bars representing standard error from four independent experiments.
Nuclease assays
Endo- and exonuclease assays were performed with ϕX174 circular ssDNA virion DNA (New England Biolabs) or linear blunt-ended pBluescript substrates incubated with purified PfMre11 at 50°C or HsMre11 at 37°C for the indicated time points (see Supplementary Data for details). DNA was resolved on tris-acetate-EDTA agarose gels, stained with ethidium bromide and visualized by scanning of gel with a Typhoon 9200.
General S. pombe methods
Strains were generated and manipulated using standard techniques (27) and are listed in Supplementary Table S2. For DNA damage sensitivity assays, 5-fold serial dilutions of exponentially growing cells were spotted on agar plates and treated with the indicated dose of DNA damaging agents. Cultures for Rad52-red fluorescent protein (RFP) foci quantification were grown in minimal media at 25°C with data representative of the mean from quantification of three sets of ∼400 cells observed by live-cell fluorescence microscopy (error bars indicate standard deviation from the mean). Spore viability and chromatin immunoprecipitation (ChIP) experiments were performed as described previously (28) with HO endonuclease expression driven by thiamine repressible promoter, nmt41. Samples were collected at the indicated time points after removal of thiamine. Microscopic analysis confirmed cell elongation in ∼80% of cells, indicating efficient cutting of the HO site causing a cell cycle arrest before sample collection. Data shown are representative of three independent experiments.
Western blots
Whole-cell extracts were prepared from exponentially growing cells in standard NP-40-based lysis buffer. For MRN blots, proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) using 3–8% or 7% tris-acetate gels per manufacturer instructions (Life Technologies). Membranes were blotted with PAP (Sigma P1291), MYC (Convance MMS-150P), FLAG (Sigma F3165) and tubulin (Sigma T5168) antibodies. IPs were performed using Protein G or Tosylactivated Dynabeads (Life Technologies) conjugated to the indicated antibody or rabbit IgG (Sigma I5006) with whole-cell extracts representing ∼1% of the input. For H2A, cells were harvested 30 min with or without 90 Gy of IR treatment. Total protein was resolved by SDS-PAGE on 4–20% tris-glycine gels (Life Technologies). Blocking and blotting were performed with Odyssey Blocking Buffer (Li-Cor) per manufacturer instructions and incubated with a rabbit polyclonal phospho-specific histone H2A antibody (29) or total H2A (Active Motif 39235). Blots were incubated with goat anti-rabbit antibody conjugated to an infrared dye (Li-Cor 827-11081) and scanned and quantified with Odyssey Infrared Imaging System (Li-Cor) with an intensity of 4.5, subtracting median (top/bottom) background. Chk1 blots were performed as previously described (30) using 90 Gy of IR and harvested 30 minutes post treatment and were quantified using ImageJ software (http://rsb.info.nih.gov/ij/). Error bars represent standard deviation from the mean of at least three independent experiments.
Telomere Southern blot
Genomic DNA was isolated and resolved as described previously (28). Telomere probe was generated by PCR amplification of the telomere-associated sequences (TAS1) from plasmid pNSU70 (31) incorporating 85% biotinylated dCTP (TriLink N-5002) and 15% normal dCTP (see Supplementary Data for details). Membrane was blocked with Odyssey blocking solution incubated with IRdye 680 conjugated strepavidin (Li-Cor 926-68031) and scanned with Li-Cor Odyssey Infrared Imaging System at intensity 7. Image shown is representative of three independent experiments.
RESULTS
Mre11-W243R retains in vitro DNA binding and nuclease activities
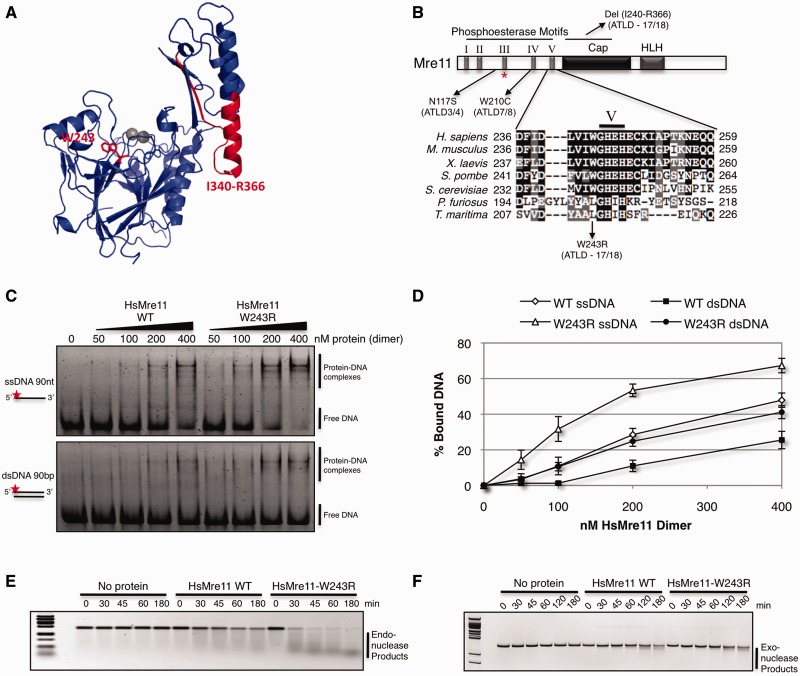
The two novel mutations in ATLD17/18 patients occur in the Mre11 core, which consists of nuclease and cap domains (25). The g.2499 G>A mutation generates an alternative mRNA splice resulting in the loss of 27 amino acids (Ile340-Arg366). This severe deletion, which removes a large part of the helix αF in the cap domain, is predicted to destabilize the protein (32). The c.727 T>C mutation replaces tryptophan-243 with arginine. This substitution maps just upstream of the well-conserved nuclease motif V in the N-terminus of Mre11 (Figure 1A and B).
Figure 1.
Human Mre11-W243R is proficient in nuclease activities. (A) Structure of human Mre11 monomer (PDB: 3T1I) with residues deleted or mutated in ATLD17/18 highlighted in red. (B) Mre11 schematic domain architecture and sequence alignment showing location of ATLD mutations and local alignment at conserved hydrophobic site W243. Mre11-W243 is adjacent to conserved nuclease motif V. HLH motif binds base of Rad50 coiled coil, forming the main interaction interface. Red asterisk (*) denotes HsMre11 histidine 129 (SpMre11-H134, PfMre11-H85) mutated in nuclease dead alleles. (C) EMSA of increasing amounts of purified HsMre11 incubated with ssDNA or dsDNA substrate labeled with Alexa Fluor 488 separated by native PAGE. (D) Quantification of free DNA from representative gel in panel D expressed as percentage of bound DNA. Error bars represent standard error of the mean of four independent experiments. (E) Endonuclease activity of HsMre11 on circular ssDNA plasmid substrate, ϕX174. Reactions were carried out at 37°C for the indicated time points. (F) Exonuclease activity of HsMre11 on linearized double-stranded plasmid DNA substrate pBluescript. Reactions were carried out at 37°C for the indicated time points.
To initiate investigations of the Mre11-W243R mutation, we used purified human Mre11 for electrophoretic mobility shift assays (EMSAs) with single-stranded and double-stranded 90-mer DNA substrates (Figure 1C). Mre11-W243R efficiently bound both substrates. In fact, it bound both DNA substrates better than wild-type Mre11 (Figure 1D). To determine whether the mutation affected ssDNA endonuclease activity, Mre11-W243R was incubated with a circular ssDNA plasmid substrate. Mre11-W243R endonuclease activity was unimpaired and appeared hyper-efficient (Figure 1E), which could possibly reflect its higher affinity for ssDNA. In the case of exonuclease assays performed with a linearized dsDNA, wild-type and mutant Mre11 had similar activity levels (Figure 1F). These data indicate that W243R may alter how Mre11 interacts with DNA, but it does not diminish its intrinsic nuclease activities.
Intact nuclease site in X-ray crystal structure of PfMre11-L204R
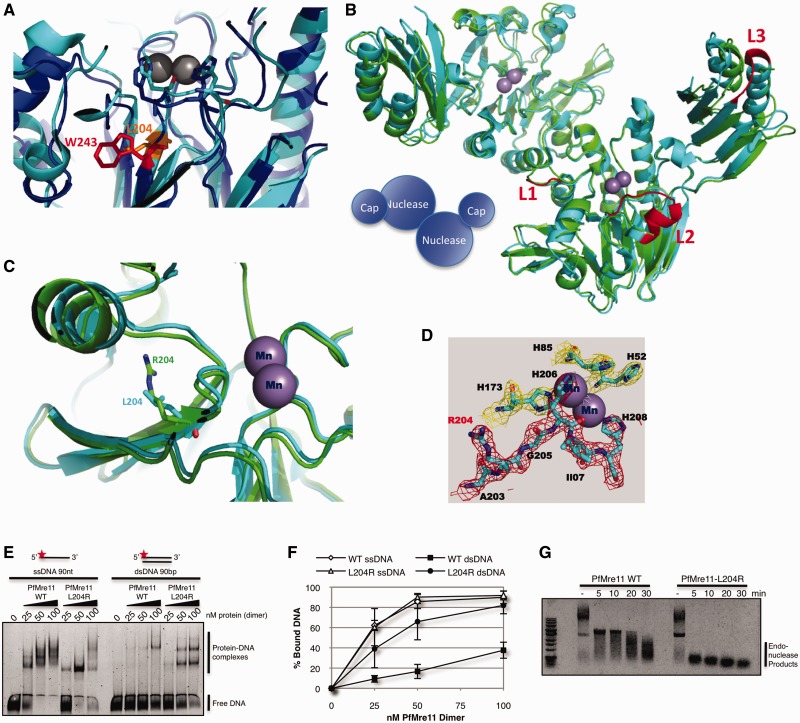
The W243R mutation does not ablate the in vitro DNA binding or nuclease activities of Mre11, even though the substitution occurs just upstream of the well-conserved nuclease motif V in the N-terminus of Mre11. To provide structural insights into the mutation, we turned to X-ray crystallography of P. furiosus Mre11. We reasoned that although there are differences in the structures of human and P. furiosus Mre11, the assembly of the nuclease site is conserved (26,32,33). Overlays of Mre11 nuclease domains sites confirmed alignments indicating that Trp243 in human Mre11 (blue) and Leu204 in PfMre11 (cyan) occupy analogous positions within the hydrophobic core of the active site (Figure 2A). We determined the X-ray crystal structure of PfMre11-L204R (1-342), comprising the nuclease and cap domains, at 2.3 Å resolution (Supplementary Table S1). Superimposition of PfMre11-L204R dimer (green) with wild-type PfMre11 dimer (cyan) reveals that the mutant maintains its overall structure and its ability to dimerize (Figure 2B). The active site core containing the L204R mutant backbone is assembled similarly to wild-type (Figure 2C), and 2Fo–Fc difference electron density map reveals that nuclease-critical residues, such as His52 and His85 required for exonuclease activity and stabilization of the transition state (for both exo- and endonuclease activities), respectively (8), are unaffected by L204R (Figure 2D). Indeed, consistent with our HsMre11-W243R DNA binding and nuclease assays (Figure 1), purified PfMre11-L204R maintained in vitro DNA binding and endonuclease activities (Figure 2E–G).
Figure 2.
X-ray crystal structure of P. furiosus Mre11-L204R. (A) Detailed view of nuclease core of HsMre11 (PDB: 3T1I) (blue) superimposed with PfMre11 (PDB: 1II7) (cyan) with Trp243 (human, red) and Leu204 (Pyrococcus, orange) highlighted. (B) Structure of PfMre11-L204R dimer (PDB: 4HD0) (green) superimposed on wild-type PfMre11 dimer (PDB: 1II7) (cyan). Mn+2 ions in active site are shown as purple spheres. Unfolded (disordered) loops in PfMre11-L204R are highlighted in red on the wild-type PfMre11 structure (C) Details of active site of wild-type PfMre11 and PfMre11-L204R. (D) 2Fo-Fc electron density map showing the PfMre11-L204R active site. (E) EMSA of increasing amounts purified PfMre11 incubated with single- or double- stranded DNA labeled with Alexa Fluor 488 (red star) separated by native PAGE. (F) Quantification of free DNA from representative gel in panel A expressed as percentage of bound DNA. Error bars represent standard error of the mean of four independent experiments. (G) Endonuclease activity of PfMre11 on circular ssDNA plasmid substrate, ϕX174. Reactions were carried out at 50°C for the indicated time points. Lanes marked—are DNA alone.
Interestingly, we observed that L204R causes the misfolding of three loops (red) in one subunit of PfMre11 (Figure 2B and Supplementary Figure S1), likely due to the torsional stress induced by the charged substitution. These loops are implicated in DNA recognition and binding by in silico DNA docking studies (Supplementary Figure S2) and by the different migration of DNA–protein complexes in EMSAs compared with wild-type Mre11 (Figure 2E). Interestingly, loop L3 maps to the same helix αF of the cap domain that is truncated in the del(I340-R366) allele of ATLD17/18 (Supplementary Figure S2) that also forms an interface with Rad50 (34,35). Collectively, these data suggest that neither HsMre11-W243R nor PfMre11-L204R disrupt the architecture of the Mre11 nuclease site or its intrinsic nuclease activities, but are more likely to alter how the MRN complex binds DNA and interacts with Rad50 and possibly Nbs1.
S. pombe mre11-W248R mutant is hypersensitive to DNA damage
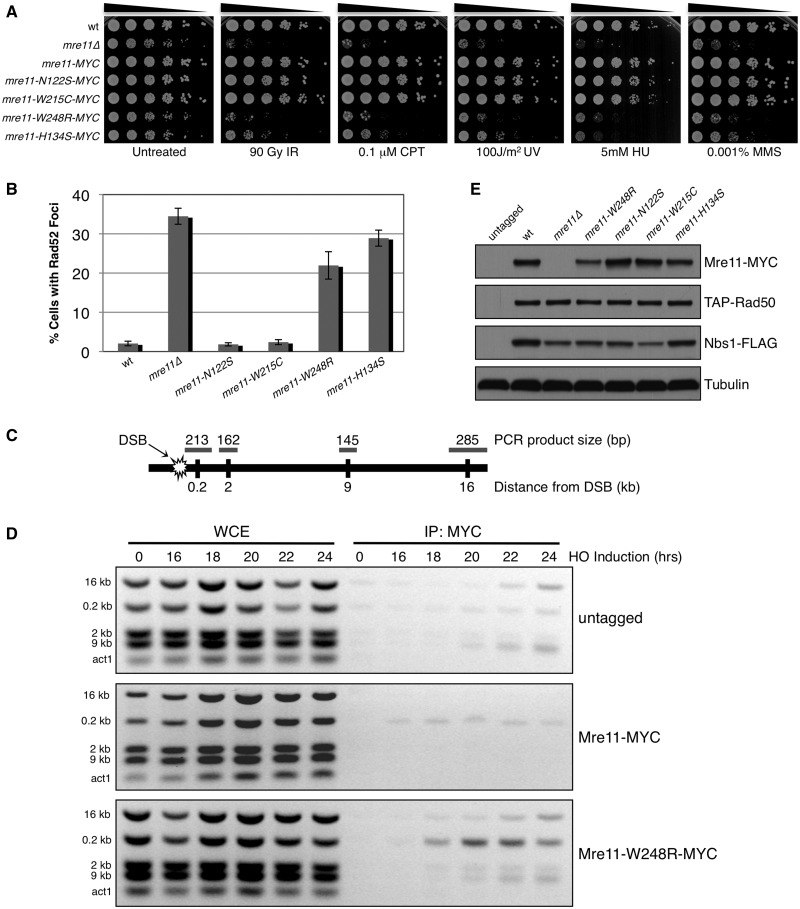
To characterize Mre11-W243R in vivo, we introduced the analogous mutation into a 13MYC-tagged genomic copy of the fission yeast S. pombe Mre11 (Rad32). The un-mutated mre11+-13MYC strain was as resistant to DNA damage as wild-type, suggesting that the MYC tag did not alter Mre11 function (Figure 3A). The mre11-N122S and -W215C alleles, corresponding to human N117S and W210C from ATLD3/4 and ATLD7/8 respectively, were largely insensitive to DNA damaging agents. These results confirm previous studies (36). In contrast, mre11-W248R cells displayed acute clastogen sensitivity, approaching the level of mre11Δ (Figure 3A). As seen with mre11Δ, the W248R mutant formed smaller colonies that consisted of many elongated cells (Supplementary Figure S3), which indicates a defect in repairing spontaneous DNA damage, leading to activation of the checkpoint that delays the onset of mitosis. Interestingly, the mre11-W248R mutant was even more sensitive to DNA damage than the mre11-H134S nuclease-dead mutant (8).
Figure 3.
Analysis of Mre11 ATLD site mutant in S. pombe. (A) 5-fold serial dilutions of Mre11 ATLD and nuclease dead (H134S) mutants were spotted and treated with indicated dose of genotoxic agents. The MYC-tagged Mre11 behaved similarly to the wild-type strain, indicating that the fusion does not alter protein function. IR = ionizing radiation, CPT = camptothecin, UV = ultraviolet radiation, HU = hydroxyurea, MMS = methyl methanesulfonate. (B) Percentage of cells with one or more spontaneously occurring Rad52-RFP focus. At least 1200 cells were analysed for each strain with error bars representing standard deviation from the mean of three experiments. (C) Schematic of chromosome I containing HO break site used in ChIP experiments indicating relative distances and expected sizes assayed by multiplex PCR. (D) ChIP of untagged, Mre11-MYC and Mre11-W248R-MYC samples collected at indicated time points after removal of thiamine from the media, leading to HO endonuclease induction. Small increases in ChIP signals are present around the HO site in the untagged controls (E) Expression levels of endogenous MRN subunits in Mre11 mutant backgrounds. Tubulin is a loading control.
Increased Rad52 foci in mre11-W248R mutant
To confirm that mre11-W248R cells were elongated owing to the presence of DNA damage, we analysed the formation of Rad52 (Rad22) foci in exponentially growing cells. Rad52 forms bright foci while mediating assembly of Rad51 (Rhp51) onto ssDNA during HR repair (37,38). Approximately 35% of mre11Δ cells had spontaneous Rad52-RFP foci, compared with only 2% in wild-type (Figure 3B). The mre11-W248R and mre11-H134S cells also had elevated levels of Rad52 foci at 22% and 29%, respectively. These results indicate increased genome instability in mre11-W248R and mre11-H134S cells from elevated levels of spontaneously occurring DNA damage and/or defects in its repair. These effects contrast with mre11-N122S and mre11-W215C, which did not differ from wild-type.
Increased retention of Mre11-W248R at a DSB
To address whether Mre11-W248R binds DSBs in vivo, we performed ChIP at a site-specific DSB generated by HO endonuclease. Multiplex PCR detected a strong Mre11-W248R signal directly adjacent (0.2 kb) to the break site, in comparison with the modest enrichment of wild-type Mre11 (Figure 3C and D). Inactivation of Mre11 nuclease activities or elimination of Ctp1 increases the ChIP signal of MRN subunits at the HO DSB, suggesting that DNA end processing by the MRN-Ctp1 complex is required for its departure from the DSB (5,30). Thus, the increased enrichment of Mre11-W248R at HO DSB indicates a defect in DNA end processing by MRN complex in vivo.
Immunoblotting indicated that Mre11-W248R abundance was reduced ∼50% relative to wild-type or other ATLD alleles (Figure 3E). This reduction may enhance the mre11-W248R phenotype; however, it is unlikely to be its primary cause. All MRN subunits are haplosufficient for meiosis, which places a heavy burden on the DSB repair machinery. Also, all Mre11-ATLD alleles reduce the abundance of Nbs1, including those that cause little or no phenotype (Figure 3A and E). Indeed, the hyper-enrichment of Mre11-W248R at the HO DSB suggests that its abundance is not limiting, at least for low levels of DNA damage (Figure 3D). The increased HO DSB ChIP signal of Mre11-W248R implies that it forms a complex with Rad50 that properly localizes in the nucleus.
Resection-dependent checkpoint signaling defect in mre11-W248R mutant
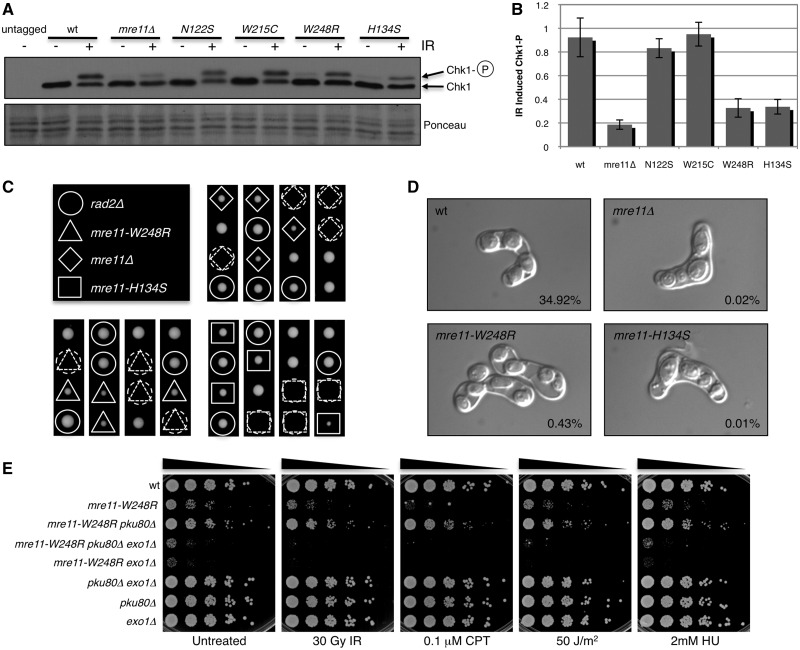
DSB resection leads to formation of ssDNA coated with RPA, which recruits Rad3ATR-Rad26ATRIP complex, which then phosphorylates Chk1 kinase (39,40). Accordingly, mutations that impair DNA end processing also reduce Chk1 phosphorylation in response to DNA damage (10,15,30). As our data indicated a DSB-processing defect in mre11-W248R cells, we investigated Chk1 phosphorylation. Immunoblotting revealed a substantial defect in IR-induced formation of phospho-Chk1 in mre11-W248R cells (Figure 4A and B). This defect was similar to that in mre11-H134S nuclease dead cells. IR-induced Chk1 phosphorylation was not significantly decreased in N122S and W215C mutants, which is consistent with their resistance to DNA damage (Figure 3A).
Figure 4.
Defective DNA processing by Mre11-W248R in vivo. (A) Chk1 phosphorylation induced by 90 Gy of IR in Mre11 mutants. Ponceau straining shows loading. (B) Quantitation of ratio of phosphorylated Chk1 over unphophorylated form from representative blot in panel A with basal (-IR) phosphorylation subtracted to specifically reflect IR induced Chk1 activation. Error bars indicate standard deviation from the mean of four independent experiments. (C) Tetrad dissection of spores from cross of rad2Δ (human FEN1 flap endonuclease) to mre11Δ (top right), mre11-W248R (bottom left) and mre11-H134S (bottom right) mutants. Dashed lines indicate predicted genotype. (D) Representative asci from a cross of opposite mating types of indicated mutants. Resulting spore viability is indicated on the bottom right. (E) Mre11-W248R is rescued by deletion of Pku80 in an Exo1 dependent manner. 5-fold serial dilutions of indicated strains were plated and treated with indicated genotoxic agents.
Notably, there was a small increase of Chk1 phosphorylation in mre11Δ, mre11-W248R and mre11-H134S cells detected before IR treatment (Figure 4A), which is consistent with the increase in spontaneous Rad52 foci and elongated cells (Figure 3B and Supplementary Figure S3) in these mutants. It is likely that defects in repair of spontaneous DNA damage results in sustained checkpoint activation in a subset of cells.
mre11-W248R mutant fails to restore collapsed replication forks
HR proteins, including the MRN complex and Ctp1, are essential to restore collapsed replication forks, which are a primary endogenous source of DSBs (28,41). To address whether mre11-W248R cells can repair collapsed replication forks, we tested for genetic interactions with rad2Δ, which eliminates the FEN1/Rad27 flap endonuclease ortholog that is required for Okazaki fragment processing (42). Unprocessed Okazaki fragments collapse replication forks, creating a critical requirement for HR repair. Tetrad dissections revealed a synthetic lethal interaction between rad2Δ and mre11-W248R, as was also observed with mre11Δ and mre11-H134S (Figure 4C). These data indicate that DNA end-processing activities of Mre11 are essential to restore collapsed replication fork by HR repair.
Meiotic defect of mre11-W248R mutant
Mre11 is essential for repair of DNA ends that have covalently attached proteins, such as when Rec12 (Spo11) generates programmed DSBs in meiosis. Rec12 removal from the 5′ DNA end requires Mre11 endonuclease activity and Ctp1 (43–47). To assess the effect of Mre11-W248R in meiosis, we crossed opposite mating types and analysed asci morphology and spore viability. Asci from a wild-type mating contained four evenly sized spores (Figure 4D). In contrast, matings with mre11-W248R, mre11Δ and mre11-H134S backgrounds produced asci with aberrant spore morphology, varying in shape and size, which is indicative of unequal chromosome segregation. Accordingly, spore viability in the mutants was drastically reduced (Figure 4D), indicating that these mutations prevent proper meiosis.
Eliminating Ku suppresses Mre11-W248R
Ku complex, consisting of the Ku70/Ku80 heterodimer, binds DSBs to promote non-homologous end joining repair. Eliminating Ku can partially rescue MRN and Ctp1 mutants (8,10,28,48–50). This rescue depends on Exo1 exonuclease, which can replace the MRN-Ctp1 resection activity but relies on it to displace Ku from DNA ends. We found that the slow growth and DNA damage sensitivity of the mre11-W248R mutant is also partially rescued by eliminating Ku (Figure 4E), indicating that Mre11-W248R is unable to efficiently initiate DNA end processing in vivo. This rescue requires Exo1, which also plays an important role in the extended resection of DNA ends required for homologous recombination (10). Accordingly, mre11-W248R mutants in combination with exo1Δ result in a substantial decrease in cell viability both prior to and after treatment with DNA damaging agents. This is consistent with our previous observation with mre11Δ and mre11-H134S cells (8), and reflects the requirement of the nuclease activity of the MRN complex in concert with Exo1 in the early steps of HR repair of DNA damage.
mre11-W248R mutation alters interaction with Rad50
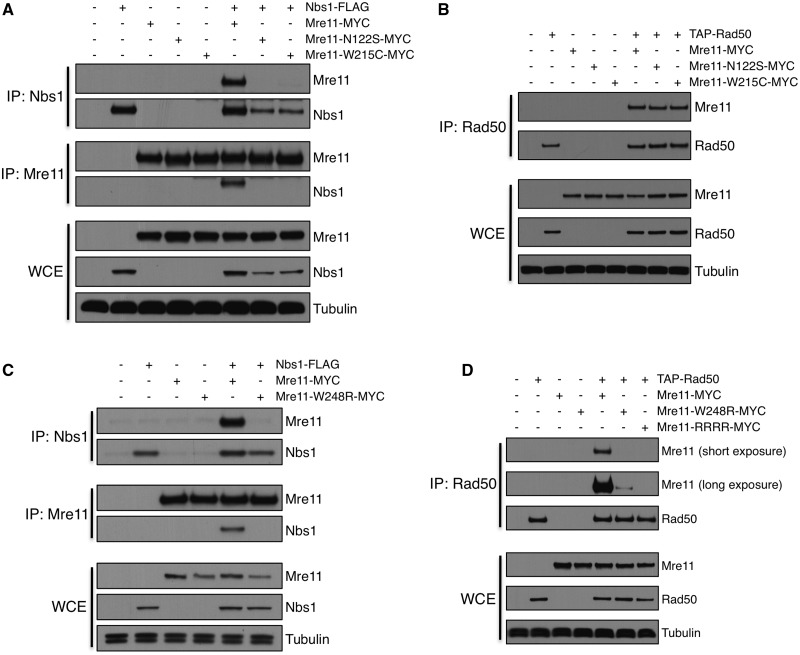
Mre11-N117S and W210C mutations impair co-immunoprecipitation (co-IP) of Mre11 with Nbs1 in mammalian cells (19,20,51). To investigate the effects of SpMre11-W248R, we performed reciprocal co-IP experiments between MRN subunits. As expected, wild-type Mre11 and Nbs1 efficiently and reciprocally co-IP (Figure 5A). In contrast, reciprocal Mre11-Nbs1 co-IPs were strongly diminished in Mre11-N122S and -W215C backgrounds, even though these mutations did not cause sensitivity to DNA damage (Figure 3A). In the Discussion section, we address how mutations can impair co-IPs without causing DNA repair defects. The reduced interactions of Mre11-N122S and W215C with Nbs1 are consistent previous data (36) and with structural evidence indicating that these residues may impact Nbs1 interaction regions (32). These mutations did not affect co-IPs with Rad50 (Figure 5B). Co-IPs also showed that Mre11-W248R similarly reduced interactions with Nbs1 (Figure 5C). However, in contrast to Mre11-N122S and W215C, Mre11-W248R also reduced co-IP efficiency with Rad50 (Figure 5D). We also examined Mre11-CL454RR CV479RR (Mre11-RRRR), which mutates the Helix-Loop-Helix (HLH) motif near the C-terminus of Mre11 (Figure 1B) that forms the dominant Mre11-Rad50 interaction interface (34,35,52,53). As predicted by the Mre11-Rad50 structures, we were unable to co-IP Mre11-RRRR with Rad50 (Figure 5D), which correlates with the acute sensitivity of mre11-RRRR to DNA damaging agents (52). Our results indicate that mutations that model ATLD alleles decrease stability of the Mre11-Nbs1 interactions as assayed by co-IPs, but that SpMre11-W248R is unique in that it also impairs interactions with Rad50.
Figure 5.
Mre11 ATLD mutations partly destabilize MRN interactions in S. pombe. (A) Reciprocal co-IP of Nbs1-FLAG, Mre11-N122S-MYC and Mre11-W215C-MYC. Proteins were precipitated and detected with FLAG, MYC or tubulin antibodies. (B) Co-IP of TAP-Rad50 with Mre11-N122S and Mre11-W215C mutants. Proteins were pulled down with IgG and detected with PAP, MYC or tubulin antibodies. (C) Reciprocal co-IP of Nbs1-FLAG and Mre11-W248R-MYC. Proteins were precipitated and detected using FLAG, MYC or tubulin antibodies. (D) Co-IP of TAP-Rad50 and Mre11-W248R-MYC. Mre11-RRRR (CL454RR CV479RR) mutates Rad50 binding domain at C-terminus of Mre11 (HLH). Proteins were pulled down with IgG and detected with PAP, MYC or tubulin antibodies.
Tel1 (ATM) checkpoint signaling is unaffected by Mre11-W248R
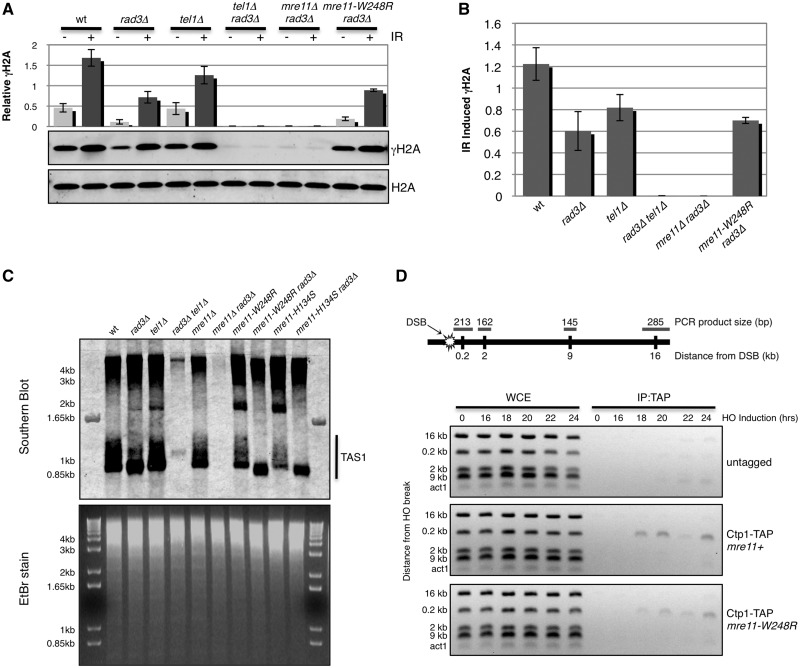
Along with its functions in DNA repair, the MRN complex also recruits the checkpoint kinase ATMTel1 to DNA lesions through interactions with the C-terminus of Nbs1 (3,54,55). ATMTel1 activity therefore depends on the assembly of MRN complex and its capacity to bind DSBs. Thus, phosphorylation of ATMTel1 substrates provides an in vivo readout for these events. The best characterized substrate of Tel1 in fission yeast is histone H2A (analogous to H2AX in mammals), which is also phosphorylated by Rad3ATR (56,57). Therefore, Tel1 activity can be specifically assayed by performing experiments in a rad3Δ background. As expected, IR-induced formation of phospho-H2A (γH2A) was partially maintained in rad3Δ and tel1Δ single mutants but was abolished in the rad3Δ tel1Δ double mutant (Figure 6A and B). γH2A was absent in mre11Δ rad3Δ cells, showing that Mre11 is required for Tel1 activity at DSBs (3). Importantly, γH2A was formed in mre11-W248R rad3Δ cells (Figure 6A and B). Thus, despite the co-IP data indicating destabilized interactions amongst the MRN subunits in lysates from mre11-W248R cells, our in vivo functional assays indicate that the MRN complex is sufficiently stable to recruit Tel1 to DSBs.
Figure 6.
Mre11-W248R is proficient in Tel1 activation but partially defective for Ctp1 recruitment. (A) Immunoblot of phosphorylated histone H2A (γH2A) and total H2A with or without treatment with 90 Gy of IR. Graph represents γH2A expression relative to total H2A with error bars representing standard deviation from the mean of three independent experiments. (B) γH2A levels induced specifically by IR treatment determined by subtracting basal levels of γH2A from IR-treated shown in Panel A. (C) Southern blot probing for telomere-associated sequences (TAS1) from genomic DNA isolated from indicated genetic backgrounds and digested with EcoRI (top panel). Ethidium bromide (EtBr)-stained gel shows DNA loading (bottom panel). (D) Chromatin immunoprecipitation of Ctp1-TAP to an HO-induced DNA double-strand break in mre11+ and mre11-W248R backgrounds. Schematic of chromosome I containing HO break site and the relative distances and expected DNA sizes assayed by multiplex PCR is shown on top.
Mre11-W248R retains telomere maintenance functions
The MRN complex also recruits Tel1 to chromosome ends, where it shares an overlapping activity with Rad3 in maintaining telomeres (2,58–61). Therefore, to independently assess whether the mre11-W248R mutation affects the ability of the MRN complex to bind DNA ends and recruit Tel1, we performed Southern blot analysis of genomic DNA isolated from S. pombe and probed for telomere associated sequences (TAS1) (31). Our results demonstrate that unlike the tel1Δ rad3Δ or mre11Δ rad3Δ strains, which showed no signal, the mre11-W248R rad3Δ had a TAS1 signal, indicating the presence of intact telomeres (Figure 6C). The same result was observed for mre11-H134S rad3Δ cells. From these results, we conclude that Tel1 can localize to telomeres and is active in the mre11-W248R and mre11-H134S backgrounds.
Reduced Ctp1 localization at a DSB in mre11-W248R cells
Collectively, these data suggest that the mre11-W248R mutation disturbs interactions amongst the MRN subunits that are required for efficient DNA end processing, but it retains interactions that are sufficient for recruiting Tel1 to perform checkpoint signaling and telomere maintenance functions. DNA end processing by MRN complex requires Ctp1, whereas Tel1 activity is independent of Ctp1. Indeed, Tel1 checkpoint signaling is increased in ctp1Δ cells because of increased retention of MRN and Tel1 at DSBs (30). To address whether interactions with Ctp1 are disturbed by the mre11-W248R mutation, we performed ChIP analysis at the HO DSB site. This analysis revealed that the Ctp1 signal at HO DSB in mre11-W248R cells was reduced compared with wild-type (Figure 6D). This occurred even though Mre11 had the opposite pattern—its signal at the DSB was increased in mre11-W248R cells (Figure 3D). These data indicate that an inability to efficiently recruit Ctp1 contributes to the DNA end-processing defect in mre11-W248R cells.
DISCUSSION
In this study, we have investigated the properties of Mre11-W243R, a mutation found in two brothers with a particularly severe form of ATLD. Unlike prior cases of ATLD, these ATLD17/18 patients also developed cancer. The question arises as to how the biochemical consequences of Mre11-W243R differ from other ATLD alleles.
X-ray crystallography studies of SpMre11 bound to a portion of Nbs1 predicts that the equivalent residue Trp248 may affect both nuclease activity and Nbs1 binding, as it forms the hydrophobic core of the structural region, linking the nuclease active site to an Nbs1 contact site (32). Our data establish that HsMre11-W243R retains full in vitro DNA nuclease activities when assayed without the other MRN subunits (Figure 1). In this regard, W243R differs from mutations that directly alter critical residues in the nuclease active site, such as mutations corresponding to HsMre11-H129N or SpMre11-H134S, which have severe effects in mice and fission yeast (8,15). Our new X-ray crystal structure and biochemical assays show that the mutation in PfMre11 analogous to W243R does not alter the architecture of the highly conserved active site nor impair nuclease activity (Figure 2), providing insight as to why HsMre11-W243R also retains nuclease activity. However, our in vivo studies with SpMre11-W248R indicate defects in DNA end processing, which are likely explained by an inability to form a fully functional complex with Rad50, Nbs1 and Ctp1, which is required for DNA end processing in vivo.
The structure of SpMre11-Nbs1 has also shed light on the effect of ATLD mutations on Mre11 binding to Nbs1. Mre11-N122S derived from ATLD3/4 maps to eukaryotic specific loops bridging the Mre11 dimers that make direct contact with the Nbs1-NFKxFxK motif (32). Mre11-W215C (ATLD7/8) and Mre11-W248R (ATLD 17/18) are predicted to alter structural elements that affect the other Nbs1 interaction region along the side of the Mre11 nuclease core. These mutations all destabilized Mre11 binding to Nbs1 when assessed using purified proteins in gel filtration assays in the absence of Mn2+ (32).
Accordingly, we found that SpMre11-N122S, -W215C and -W248R, all compromised Mre11-Nbs1 interactions in co-IP assays from cell lysates (Figure 5). This effect was also observed for a mutation in the Nbs1-NFKxFxK motif, Nbs1-K522A, which mutates the same interaction interface as Mre11-N122S (Supplementary Figure S4). Yet, only Mre11-W248R has a strong phenotype in vivo. A similar effect was observed in budding yeast, where ScMre11-R76A, -R76K, -D109G and -N113S, all compromised Mre11-Xrs2/Nbs1 interactions as assessed by co-IP; yet, only ScMre11-N113S cells were sensitive to DNA damaging agents (32). Thus, an impaired Mre11-Nbs1 co-IP does not necessarily correlate with a DNA repair defect in vivo. It appears that co-IP can be a particularly stringent measurement of protein interactions that does not necessarily reflect whether subunits of the MRN complex are able to form a functional complex in vivo.
Interestingly, the repair defects of budding yeast mre11-N113S cells were attributed to defects in nuclear localization that could be rescued by the fusion of a nuclear localization signal (32). This is unlikely to be the case for the analogous allele in S. pombe, as mre11-N122S cells are largely insensitive to DNA damaging agents (Figure 3A) (36). A nuclear localization defect is also unlikely to explain mre11-W248R phenotypes, as our ChIP data showed efficient binding to a DSB (Figure 3D). Indeed, the persistent binding of SpMre11-W248R at a DSB is especially informative, as it is strikingly similar to mutations that impair the DNA end-processing activity of MRN complex without destroying the overall integrity of the complex. Notably, we have observed that the Mre11-H134S nuclease-dead allele has these properties (5). Moreover, as observed with Mre11-W248R, we found that the H134S mutation causes strong sensitivity to DNA damaging agents, an acute meiotic defect and synthetic lethality with a mutation that eliminates Rad2 (FEN1), without blocking the ability of the MRN complex to recruit active Tel1ATM to DSBs, or to maintain telomeres. This same constellation of effects is seen with elimination of Ctp1, which is essential for the DNA end-processing activity of the MRN complex in vivo (10,28,30).
Our histone H2A phosphorylation and telomere maintenance assays established that Tel1 ATM activity is maintained in mre11-W248R cells (Figure 6). We performed these assays in cells lacking Rad3ATR, which allowed us to specifically assess the activity of Tel1. From these results, we conclude that components of the MRN complex interact sufficiently well in mre11-W248R cells to form a stable complex that can bind DSBs and recruit Tel1ATM. The same results were obtained with the nuclease-dead mre11-H134S allele in the rad3Δ background (Figure 6C) (30), which is consistent with studies in mammalian cells indicating that Mre11 nuclease activity is dispensable for ATM activation in response to DNA damage or telomere deprotection (15).
The synthetic lethality of Mre11-W248R with a mutation that eliminates Rad2 (FEN1) supports their complementary roles in protecting or processing forks and is consistent with recent results showing Mre11 can protect or degrade stalled forks in human cells depending on BRCA1/2 partner status (41,62,63). Both Mre11 and FEN1-DNA complex structures indicate that DNA is unpaired for endonuclease cleavage, and FEN1 structures imply that correct positioning of DNA duplex is critical not only for specific DNA opening and nuclease activity, but also for productive interaction with protein partners (64).
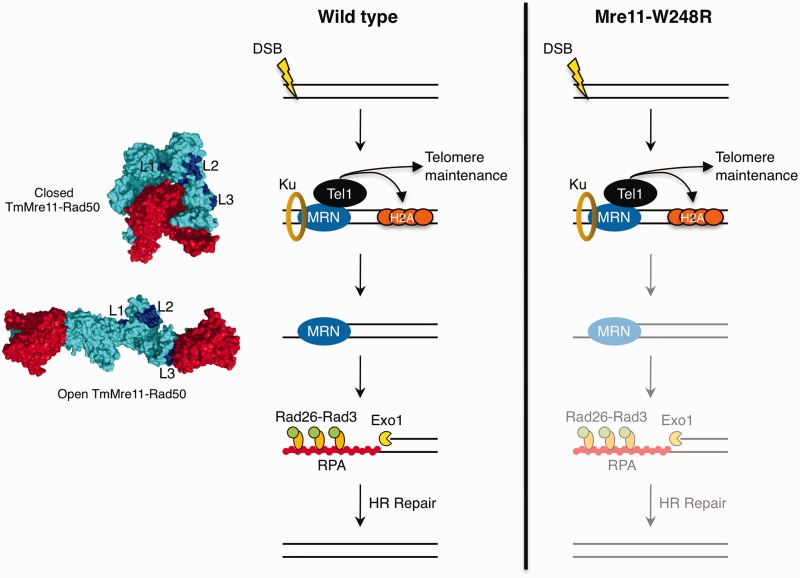
Unlike other alleles that model ATLD mutations, the mre11-W248R allele causes acute sensitivity to DNA damage, increased spontaneous Rad52 foci, synthetic lethality with loss of Rad2/FEN-1, a strong defect in Chk1 activation in response to IR and an inability to complete meiosis (Figures 3 and 4). Yet, Mre11-W248R associates with DSBs in vivo and forms an MRN complex that recruits active Tel1ATM (Figures 3 and 6). In these respects, Mre11-W248R is very similar to the H134S nuclease dead allele; yet, Mre11-W248R clearly retains its intrinsic nuclease activities. What then accounts for the mre11-W248R phenotypes? One clear distinction between W248R and the other ATLD alleles (i.e. N122S and W215C) is the defective co-IP with Rad50 (Figure 5). Recent structures of Mre11-Rad50 in bacterium and archaea have shown that ATP binding and hydrolysis induces large conformational changes that have implications on the nuclease activity of Mre11 (34,35,53). ATP hydrolysis leads to the ‘opening’ of the MR complex, exposing the nuclease sites of Mre11 to DNA through conformational changes that cause an interaction interface to form between the helix αF in the cap domain of Mre11 and the lobe of Rad50 (34,35). Interestingly, our structure of PfMre11-L204R causes the misfolding of αF (loop L3) required for this interface (Figure 7). Although it remains to be shown whether the Mre11-Rad50 conformational changes or the misfolding of loop L3 are conserved in the eukaryotic MRN complex, this would explain the compromised interaction we observed between Mre11-W248R and Rad50 by co-IP. An inability to properly interface with Rad50, or to correctly position DNA, could explain why Mre11-W248R mutants are proficient for binding DNA damage sites and enabling Tel1 signaling, yet shows defects in DNA end processing (Figure 7). Fittingly, mutations that perturbed the interaction between Methanococcus jannaschii Mre11 cap domain and Rad50 lobe result in decreased endonuclease activity (53).
Figure 7.
Model for impact of Mre11-W248R on early steps of DSB repair. Surface structures of Thermotoga maritima Mre11 (cyan)-Rad50 (red) in closed (left, top) (PDB: 3THO) and open (left, bottom) (PDB: 3QG5) conformations with equivalent L1, L2 and L3 loops highlighted (dark blue). MRN recruitment to DSB ends facilitates Tel1/ATM signaling to histone H2A and telomeres. Mre11 nuclease activity (with Ctp1) initiates DNA end processing, releasing Ku from DNA ends and generating a resected DNA end that is no longer a preferred substrate for Ku binding. Extended resection is facilitated by Exo1 and RecQ helicases, exposing long stretches of ssDNA that is coated by RPA. Rad26-Rad3 binds RPA to facilitate Chk1-dependent checkpoint signaling. In mre11-W248R mutants, MRN is able to detect and engage DSB sites and facilitate Tel1/ATM-dependent signaling, but is defective in DSB end processing.
Our data also suggest that defective interactions of the MRN complex with Ctp1 may contribute to the mre11-W248R phenotypes. Ctp1 localization at HO DSB is decreased in these cells, even though Mre11 retention is increased (Figures 3D and 6D). Furthermore, the Ctp1 signal at HO DSB is actually increased in nuclease-defective mre11-H134S cells, which otherwise have a spectrum of phenotypes that are similar to mre11-W248R cells (5). Defective interactions between Mre11 and Nbs1 could explain defects in Ctp1 localization at a DSB in mre11-W248R cells, as Ctp1 recruitment requires an interaction with Nbs1 (4,5), but Tel1 activity also requires an interaction with Nbs1 (54,55), and our data show that Tel1 signaling is intact. It is possible that efficient recruitment of Ctp1 to DSBs requires interactions with both Mre11 and Nbs1.
The acute DNA damage-sensitive phenotypes caused by the mre11-W248R mutation in fission yeast raises the question of whether the equivalent W243R substitution has comparable effects in humans. The structural and mechanistic effects are likely similar, although the W243R mutation may have weaker consequences in the context of the human MRN complex. It is also formally possible that the g.2499 G>A allele, which eliminates 27 residues in the cap domain, is a milder hypomorphic allele, although structural considerations predict the opposite.
The increased foci of Rad52 (which has parallel functions to human BRCA2) and inability to repair collapsed replication forks in mre11-W248R cells are strongly indicative of genome instability in fission yeast. Although it remains to be established whether the exceedingly rare pediatric cancers in ATLD17/18 patients were directly connected to the Mre11 deficiencies, the correlations are nonetheless striking and support a role for Mre11 in preventing cancer-causing genome instability, as was first demonstrated for ATM and Nbs1.
ACCESSION NUMBERS
PDB: 4HD0.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online: Supplementary Tables 1–2, Supplementary Figures 1–4, Supplementary Methods and Supplementary References [65–68].
FUNDING
National Institutes of Health [CA77325 and GM59447 to P.R., CA117638 to J.T. and P.R.]; Skaggs Institute for Chemical Biology (to D.M.); National Cancer Institute [P01 CA092584 to C.W. and J.A.T.]; Netherlands Organization for Scientific Research [VICI 700.56.441 to C.W.]; X-ray diffraction technologies at the SIBYLS beamline 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory, are supported in part by the U.S. Department of Energy program Integrated Diffraction Analysis Technologies (IDAT). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sari van Rossum-Fikkert for help with production and purification of human protein and Taema Bajo and Devon Nieto for technical support. We acknowledge Yoshiki Yamada, Santiago Cavero, Nicholas Rhind and Antony M. Carr for providing S. pombe strains used in this study, Christophe Redon for the generous gift of γH2A antibody, and Daun Clizbe (Li-Cor) for advice and reagents used with the Southern blot.
REFERENCES
- 1.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 2.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell. Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair. 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das D, Moiani D, Axelrod HL, Miller MD, McMullan D, Jin KK, Abdubek P, Astakhova T, Burra P, Carlton D, et al. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J. Mol. Biol. 2010;397:647–663. doi: 10.1016/j.jmb.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimitou EP, Symington LS. DNA end resection–unraveling the tail. DNA Repair (Amst.) 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst.) 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogh BO, Llorente B, Lam A, Symington LS. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics. 2005;171:1561–1570. doi: 10.1534/genetics.105.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delia D, Piane M, Buscemi G, Savio C, Palmeri S, Lulli P, Carlessi L, Fontanella E, Chessa L. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum. Mol. Genet. 2004;13:2155–2163. doi: 10.1093/hmg/ddh221. [DOI] [PubMed] [Google Scholar]
- 19.Fernet M, Gribaa M, Salih MA, Seidahmed MZ, Hall J, Koenig M. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Hum. Mol. Genet. 2005;14:307–318. doi: 10.1093/hmg/ddi027. [DOI] [PubMed] [Google Scholar]
- 20.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 21.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 22.Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto Y, Miyamoto T, Sakamoto H, Izumi H, Nakazawa Y, Ogi T, Tahara H, Oku S, Hiramoto A, Shiiki T, et al. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair. 2011;10:314–321. doi: 10.1016/j.dnarep.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am. J. Hum. Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchisaka N, Takahashi N, Sato M, Kikuchi A, Mochizuki S, Imai K, Nonoyama S, Ohara O, Watanabe F, Mizutani S, et al. Two brothers with ataxia-telangiectasia-like disorder with lung adenocarcinoma. J. Pediatr. 2009;155:435–438. doi: 10.1016/j.jpeds.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 26.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 27.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell. Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limbo O, Porter-Goff ME, Rhind N, Russell P. Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks. Mol. Cell. Biol. 2011;31:573–583. doi: 10.1128/MCB.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 32.Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mockel C, Lammens K, Schele A, Hopfner KP. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter-Goff ME, Rhind N. The role of MRN in the S-phase DNA damage checkpoint is independent of its Ctp1-dependent roles in double-strand break repair and checkpoint signaling. Mol. Biol. Cell. 2009;20:2096–2107. doi: 10.1091/mbc.E08-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 38.Du LL, Nakamura TM, Moser BA, Russell P. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell Biol. 2003;23:6150–6158. doi: 10.1128/MCB.23.17.6150-6158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 41.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA. Unpairing and gating: sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem. Sci. 2012;37:74–84. doi: 10.1016/j.tibs.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 44.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 45.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ataxia-telangiectasia-like disorder mutant complexes. J. Biol. Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- 52.Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 55.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura TM, Moser BA, Russell P. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161:1437–1452. doi: 10.1093/genetics/161.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 60.Moser BA, Chang YT, Kosti J, Nakamura TM. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 2011;18:1408–1413. doi: 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamazaki H, Tarumoto Y, Ishikawa F. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012;26:241–246. doi: 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du LL, Moser BA, Russell P. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J. Biol. Chem. 2004;279:38409–38414. doi: 10.1074/jbc.M403326200. [DOI] [PubMed] [Google Scholar]
- 66.Mandell JG, Roberts VA, Pique ME, Kotlovyi V, Mitchell JC, Nelson E, Tsigelny I, Ten Eyck LF. Protein docking using continuum electrostatics and geometric fit. Protein Eng. 2001;14:105–113. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- 67.Roberts VA, Pique ME. Definition of the interaction domain for cytochrome c on cytochrome c oxidase. III. Prediction of the docked complex by a complete, systematic search. J. Biol. Chem. 1999;274:38051–38060. doi: 10.1074/jbc.274.53.38051. [DOI] [PubMed] [Google Scholar]
- 68.Roberts VA, Pique ME, Hsu S, Li S, Slupphaug G, Rambo RP, Jamison JW, Liu T, Lee JH, Tainer JA, et al. Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase. Nucleic Acids Res. 2012;40:6070–6081. doi: 10.1093/nar/gks291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.