Abstract
Heterochromatic regions represent a significant portion of the mammalian genome and have been implied in several important cellular processes, including cell division and genomic stability. However, its composition and dynamics remain largely unknown. To better understand how heterochromatin functions and how it is organized within the context of the cell nucleus, we have developed molecular tools allowing the targeting of virtually any nuclear factor specifically to heterochromatic regions and, thereby, the manipulation, also in a temporally controlled manner, of its composition. To validate our approach, we have ectopically targeted MeCP2 chromatin binding deficient Rett mutants to constitutive heterochromatic regions and analyze its functional consequences. We could show that, once bound to their endogenous target regions, their ability to re-organize higher order chromatin structure is restored. Furthermore, a temporally controlled targeting strategy allowed us to monitor MeCP2-mediated chromatin rearrangements in vivo and to visualize large-scale chromatin movements over several micrometers, as well as heterochromatic foci fusion events. This novel strategy enables specific tethering of any protein to heterochromatin and lays the ground for controlled manipulation of its composition and organization.
INTRODUCTION
Heterochromatin represents a significant but mostly unexplored portion of the mammalian genome. Although its composition and architecture are largely unknown it has been implied in several fundamental cellular processes, including cell division, genomic stability and expression.
One of the most conserved hallmarks of heterochromatic regions is a high density of cytosine methylation. Methylation of cytosine residues at CpG dinucleotides is recognized and read in mammals by the methyl CpG binding domain (MBD) protein family. This protein family comprises five members MBD1-4 and MeCP2. All members (except for MBD3) have a conserved MBD domain that targets them to methylated DNA (1) and consequently accumulate at constitutive heterochromatin in vivo (2). We have previously shown that MeCP2, the founding member of the MBD protein family, induces aggregation of pericentric heterochromatin in a dose dependent manner in vivo with the MBD domain being necessary and sufficient for this function (3). MeCP2 has also been shown to cluster polynucleosomes in vitro (4–6). Mutations within the MECP2 gene have been linked to the neurological disease Rett syndrome (RTT), a post-natal disorder with an incidence of 1/10 000 female birth (7,8). Whereas missense mutations are mostly clustered within the MBD domain, nonsense mutations occur frequently after the MBD domain. We have recently analyzed the effect of 22 MBD missense mutations on MeCP2 ability to bind and cluster heterochromatin and could show that half of the mutants were significantly affected in their heterochromatin binding and two-third of all mutants exhibited significantly decreased clustering ability of pericentric heterochromatin (9). The majority of mutants tested were affected in both, their binding to and clustering of heterochromatin (9). As most drastic examples, two mutants (MeCP2 R111G and F155S) showed the lowest binding to heterochromatin resulting in their mislocalization to nucleoli and concomitantly lacked any capability to aggregate pericentric heterochromatin (9). These findings raised the question whether in such Rett mutations heterochromatin clustering is impaired as a consequence of the inability to bind methylated DNA or whether these mutants are independently affected in heterochromatin clustering.
In this study, we have developed and validated molecular tools to target proteins to heterochromatin regions and follow their impact on heterochromatin composition, architecture and dynamics in living cells. This approach allowed us to discriminate whether Rett mutations affecting chromatin binding consequently decrease the ability of the MeCP2 mutant to aggregate heterochromatin, or alternatively whether forcing Rett mutants to bind to chromocenters can rescue their clustering ability. The addition of an estrogen receptor domain to our targeting tool further enabled us to temporally control the intra-nuclear re-localization of a MeCP2 Rett mutant. Thus, we were able to monitor in living cells the dynamics of large-scale chromatin reorganization. The targeting of Rett mutants to pericentric heterochromatin constitutes a very clear and feasible in vivo assay to reveal the effect (or phenotype) of various Rett mutations on MeCP2 clustering properties independently of chromatin binding. Furthermore, these novel molecular tools are readily applicable to other chromatin regulators to dissect their functional effects on heterochromatin dynamics and organization.
MATERIALS AND METHODS
Expression plasmids
Expression vectors encoding GFP-tagged fusions of human wild-type or mutant MeCP2 cDNA cloned into the pEGFP-C1 vector are described in (10). pGBP-MaSat was cloned by introducing the PCR-amplified sequence of the polydactyl zinc finger major satellite binder (11) into a plasmid encoding the GFP-binder nanobody (12,13) using XbaI restriction sites. To create the estrogen-controlled nuclear switch, we introduced a PCR-amplified estrogen receptor (ER) fragment (14) using an EcoRI restriction site between GFP-binding protein (GBP) and MaSat, resulting in pGBP-ER-NLS-MaSat (GERM). To make the nuclear switch detectable by fluorescence microscopy, we used the Clontech vector pEGFP.N1, replaced the eGFP by mCherry and then introduced a fragment coding for the GBP-ER-MaSat, resulting in pGBP-ER-NLS-MaSat-mCherry (GERM-cherry). GERM-cherry was made cytoplasmic by deleting the N-terminus of MaSat, which contains a nuclear localization signal (NLS), by restriction with PvuI and NheI, followed by treatment with Klenow polymerase large fragment, to create blunt ends, and re-ligation.
Cell culture, transfection and staining
C2C12 mouse myoblasts were cultured as described (15). For fixed cell work cells grown on glass coverslips in six well dishes were transfected with PEI (poly-ethyleneimine; 1 mg/ml in ddH2O, neutralized with HCl). For that, 200 µl serum-free Dulbecco’s modified Eagle’s medium (DMEM) with 4 µg DNA were added to 200 µl serum-free DMEM with 12 µl of PEI. After mixing and incubation for 15 min at room temperature, the solution was added to the cells dropwise and the culture incubated at 37°C over night. Transfected cells were fixed with a formaldehyde (FA) gradient: cells were rinsed in 0.1% FA, incubated 2 min in 1% FA, followed by 20 min incubation in 4% FA. Nuclear DNA was counterstained for 10 min in 1 µg/ml DAPI (4′-6′-diamidino-2-phenylindol) and samples were mounted in Vectashield antifade medium (Vector Laboratories, Burlingame, CA). For live cell imaging and fixed time-lapse analysis, cells were transfected using Amaxa nucleofection as previously described (16).
Microscopy and image analysis
For chromocenter counting, cells were fixed and fluorescent images were collected using either an UltraVIEW VoX spinning disc confocal system mounted on a Nikon Ti microscope equipped with an oil immersion Plan-Apochromat 60x/1.45 NA objective lens (pixel size in XY = 111 nm, Z-step = 0.3 µm) or a Zeiss Axioplan2 wide-field epifluorescence microscope. Image stacks from the Zeiss Axioplan2 microscope (pixel size in XY = 104 nm, Z-step = 0.5 µm) were acquired with a 63x Plan-Apochromatic NA 1.4 oil immersion phase contrast objective lens and a PCO Sensicam QE cooled CCD camera.
The image stacks were analyzed using a semi-automated approach as described in (9). Chromocenters were counted in DAPI to avoid bias due to heterochromatin targeting efficiency. The distributions of chromocenter numbers were tested for statistical significance by the two-tailed t-test.
Live cell microscopy
Transfected cells were plated on a glass bottom p35 dish and grown in estrogen-free medium under otherwise standard conditions. Cells were selected and imaged in estrogen-free medium. Immediately after addition of 1 µM tamoxifen, 3D stacks were acquired at 20-min intervals for up to 12 h. 1 µM tamoxifen was added to the medium every 4 h. Time lapse imaging was carried out on a UltraVIEW VoX spinning disc confocal system (PerkinElmer, UK) in a closed live cell microscopy chamber (ACU control, Olympus, Japan) heated to 37°C, with 5% CO2 and 60% air humidity control, mounted on a Nikon Ti microscope (Nikon, Japan). Image acquisition was performed using a 60x/1.45 NA Planapochromat oil immersion objective lens. Images were obtained with a cooled 14-bit EMCCD camera (C9100-50, CamLink). Maximum intensity projections or single Z-slices were assembled into galleries and videos and annotated using ImageJ (http://rsb.info.nih.gov/ij/ 6 August 2012, date last accessed).
RESULTS AND DISCUSSION
Based on our recent findings that many Rett missense mutations within the MBD domain of MeCP2 affect both MeCP2 heterochromatin binding and chromatin clustering (9), we aimed to discriminate whether a Rett MBD mutation severely affecting MeCP2 chromatin binding independently and additionally impairs MeCP2 heterochromatin aggregation or whether the decreased chromatin aggregation property of certain Rett mutants is simply a consequence of their impaired binding to heterochromatin.
To artificially recruit the mutant proteins to pericentric regions we made use of the previously described polydactyl zinc finger protein MaSat. This protein specifically binds to major satellite DNA repeats (11), which are highly enriched in pericentric heterochromatin. Like MeCP2, the artificially synthesized MaSat protein is mostly accumulated at mouse pericentric regions (3,9,11,16), also known as chromocenters.
We generated an expression construct encoding the major satellite binding protein MaSat fused to the GBP, an antibody fragment with high affinity to GFP in vivo and in vitro (12,13).
The fusion protein GBP-MaSat is able to recruit a methyl cytosine binding deficient GFP-labeled MeCP2 Rett mutant to chromocenters
To test whether the GBP-MaSat protein is capable of targeting to chromocenters GFP-tagged proteins that do not show any binding to pericentric heterochromatin on their own, we co-transfected C2C12 mouse myoblasts with expression constructs encoding GBP-MaSat and the GFP-labeled MeCP2 Rett mutant R111G. We fixed the cells after 24 h and performed staining with the A/T selective dye DAPI to highlight pericentric regions. As previously shown, MeCP2 R111G is severely affected in its chromocenter binding and aggregation ability in vivo leading to its nuclear distribution with additional miss-localization to the nucleoli (9). In contrast to co-expression of GBP with MeCP2 R111G, co-expression of GBP-MaSat and GFP-MeCP2 R111G resulted in strong accumulation of the MeCP2 Rett mutant R111G at the chromocenters. This observation was further confirmed by the colocalization of the GFP and DAPI signals (Figure 1A and B).
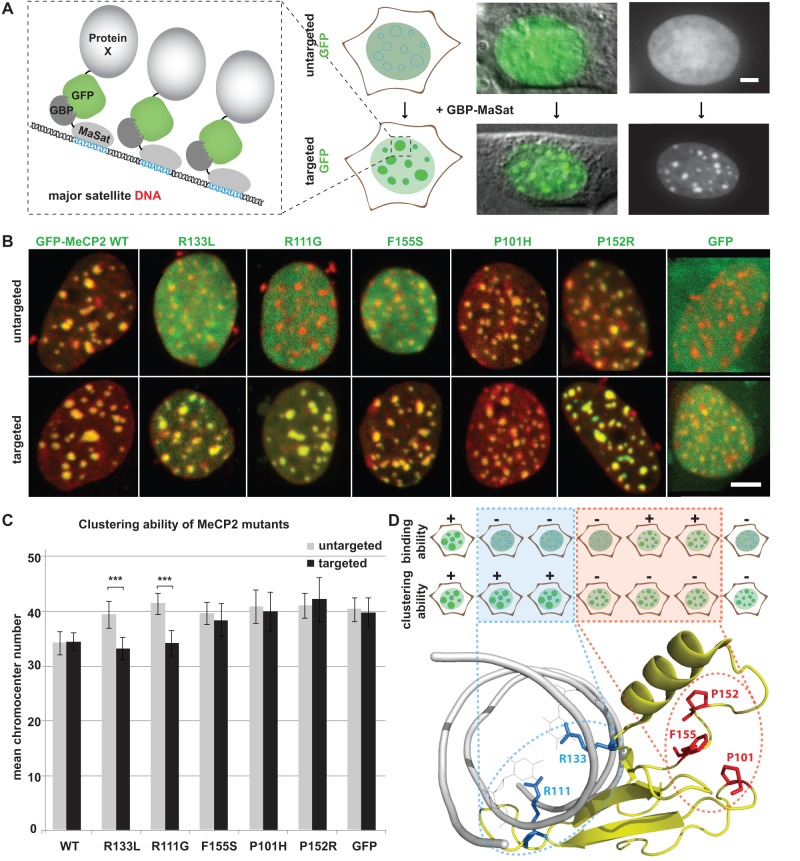
Figure 1.
Specific Rett mutations affect the chromatin binding ability of MeCP2. (A) Left: schematic representation of the targeting assay. The fusion protein composed of the major satellite DNA binding polydactyl zinc-finger protein (MaSat) linked to the GBP recruits nuclear GFP-labeled proteins to major satellite DNA repeats within pericentric heterochromatic regions. Targeted GFP-tagged proteins are visible as multiple green fluorescent spots throughout the nucleus, corresponding to chromocenters. In absence of the fusion protein GBP-MaSat, the GFP signal is distributed throughout the nucleus showing no accumulation at chromocenters. Upon co-expression of both, GFP and the fusion protein GBP-MaSat, the GFP protein is targeted to chromocenters through the interaction between its GFP-tag and the GBP linked to MaSat. Right: exemplary cells expressing GFP-tagged protein showing the clear re-distribution of the GFP signal when GBP-MaSat is co-expressed. (B) C2C12 mouse myoblasts were seeded on coverslips and co-transfected with expression vectors coding for GBP-MaSat and GFP or GFP-tagged MeCP2 wt or one of the MeCP2 Rett mutants bearing missense mutation as indicated. Z-stacks of fixed nuclei with similar expression levels of the GFP-tagged protein were imaged. Maximum intensity projections of representative cells co-expressing wt MeCP2, different MeCP2 Rett mutants or GFP and either the GBP alone (untargeted) or GBP-MaSat (targeted). GFP fusion proteins are shown in green, DAPI stained DNA in red. Scale bars: 5 µm. (C) The bar graph shows mean numbers of chromocenters of cells expressing the indicated proteins. Error bars represent 95% confidence interval (CI). Experiments were repeated two times with at least 30 cells per construct each time. Asterisks represent statistically significant difference P < 0.001. (D) Top: summarizing scheme of mouse cells expressing GFP-labeled wt MeCP2 or a Rett mutant bearing the missense mutation as indicated. Pericentric heterochromatin is illustrated as round spots distributed throughout the nucleus. The upper row illustrates the untargeted state and therefore the intrinsic chromatin binding ability of GFP-tagged wt or mutant MeCP2. The lower row stands for the targeted state and concomitantly for the cluster ability of the different GFP-labeled MeCP2 constructs obtained by co-expression with the fusion protein GBP-MaSat. The chromocenter binding and clustering potentials of the corresponding GFP-labeled MeCP2 constructs are indicated using + or − . Bottom: X-ray structure of MeCP2 MBD domain (displayed in yellow) interacting with methylated DNA (shown in gray) (PDB accession code 3C2I) (17). Structural data were displayed and annotated using PyMOL software (http://pymol.sourceforge.net/ 6 August 2012, date last accessed). Amino acids crucial for MeCP2 clustering of pericentric heterochromatin are highlighted in red, residues directly interacting with methylated DNA are shown in blue.
Some Rett mutations exclusively affect the chromatin binding ability but do not influence the chromocenter aggregation property of MeCP2
Having recently shown that many Rett mutants with missense mutations within their MBD domain are affected in both, their ability to bind and to aggregate heterochromatin (9), we now wanted to elucidate whether the impairment in the clustering ability is a direct consequence of such mutations or rather an indirect effect of the severe binding deficiency.
For this reason, we chose the MeCP2 R133L mutant, representing the group of Rett mutations that strongly impair the binding function and additionally decrease to some extent the chromocenter clustering ability of the protein. We further selected the MeCP2 R111G and MeCP2 F155S mutants that showed the most drastic phenotypes with almost completely impaired chromatin binding and clustering, resulting in an overall nuclear, as well as nucleolar, localization. In addition, we took the MeCP2 P101H and P152R mutants as examples for mutations predominantly affecting heterochromatin aggregation (9). We co-transfected C2C12 cells with constructs coding for the GBP-MaSat protein and either GFP-labeled wild-type MeCP2, one of the representative mutants or GFP alone. We used confocal fluorescence microscopy to determine the localization of the different proteins and their chromocenter clustering ability, as well as to visualize the pericentric regions after staining with the A/T selective DNA dye DAPI (Figure 1B and C). In order to address the degree of chromocenter clustering, we scored the number of chromocenters in cells expressing one of the GFP-labeled fusion proteins or GFP alone.
All GFP-tagged proteins co-expressed with the GBP-MaSat protein efficiently accumulated at chromocenters and colocalized with the heterochromatic regions highlighted by DAPI staining. In contrast, the Rett mutants MeCP2 R111G, F155S and R133L, as well as GFP, co-expressed with the GBP protein alone showed little to no chromocenter accumulation and significantly decreased chromocenter clustering ability in comparison to wild-type MeCP2 (Figure 1B and C), in agreement with previous studies (9).
In the case of the MeCP2 R133L mutant, showing strongly impaired chromatin binding, as well as reduced chromocenter aggregation in the absence of the GBP-MaSat protein, the improved binding to heterochromatin via GBP-MaSat rescued its chromocenter clustering ability to a level comparable to that of wild-type MeCP2 (Figure 1B and C). These results indicate that the R133L mutation solely affects MeCP2 chromatin binding and has no influence on the heterochromatin clustering function of the protein. That the amino acid R133 directly interacts with the 5-methyl cytosine of the DNA, as evident from the crystal structure of MeCP2 MBD domain [Figure 1D; (9,17)], strongly supports our findings and underlines the importance of the R133 amino acid for 5-methyl cytosine binding and consequently MeCP2 accumulation to heterochromatin. Importantly, our results for the first time show that the amino acid R133 is not involved in heterochromatin aggregation.
Similar to MeCP2 R133L, the degree of chromocenter clustering of MeCP2 R111G co-expressed with the GBP-MaSat protein was significantly increased and comparable to the chromocenter aggregation function of wild-type MeCP2 (Figure 1B and C). Analogous to the amino acid R133, also the amino acid R111 directly forms an interaction with the 5-methyl cytosine of DNA, as seen on the 3D structure of the MBD domain of MeCP2 (9,17). It is, therefore, not surprising that both amino acids, R133 as well as R111, seem to be exclusively responsible for MeCP2 5-methyl cytosine binding and do not seem to exert any additional effect on MeCP2 heterochromatin aggregation function.
In the case of the Rett mutants MeCP2 P101H and P152R, the ability to bind chromatin was independent of whether GBP-MaSat was co-expressed or not. Even in the absence of the GBP-MaSat, the MeCP2 P101H and P152R mutants show comparable chromatin binding to wild-type MeCP2 but exhibit a severely impaired chromocenter aggregation function in comparison to wild-type MeCP2. This clustering deficiency was not altered by co-expression of GBP-MaSat (Figure 1B and C). These observations reinforce the fact that these mutations exclusively affect the clustering ability of pericentric heterochromatin. With the help of the 3D-structure of the MBD domain (Figure 1D; (17)), we already described that the amino acids P101 and P152 are located far away from the 5-methyl cytosine interacting pocket and proposed that they might be involved in the formation of interactions with chromatin proteins thus connecting heterochromatin fibers (9).
In contrast to the Rett mutants MeCP2 R133L and R111G, but comparable to the mutants MeCP2 P101H and MeCP2 P152R, the aggregation function of pericentric heterochromatin of the MeCP2 F155S mutant was not rescued upon co-expression of the GBP-MaSat protein and consequent targeting of the MeCP2 F155S mutant to chromatin (Figure 1B and C). This fact clearly indicates that the F155S mutation simultaneously and independently strongly alters both properties of MeCP2, chromatin binding and clustering. Importantly, the amino acid F155 is located within the loop containing the amino acid P152, near the loop including amino acid P101 and forms a likely interaction pocket with these residues. It is possible that the amino acid F155 might be, besides P101 and P152, also involved in interactions to chromatin proteins responsible for aggregation of chromocenters. The additional defect of the MeCP2 F155S mutant to bind to chromatin might be explained by the possibility that the amino acid F155 also plays a crucial role in the positioning of the nearby alpha helix that is located in close proximity to amino acid R133, exerting direct contact to the 5-methyl cytosine of the DNA. The F155S mutation might, therefore, severely alter the orientation of amino acids important in chromatin binding to a degree of misfolding impairing the 5-methyl cytosine binding ability of the protein, as described previously (18,19).
Temporal and spatial monitoring of large-scale chromatin reorganization induced by MeCP2 R111G re-localization
As described in Figure 1, the artificial targeting of MeCP2 R111G to chromocenters revealed a chromocenter clustering potential of the MeCP2 R111G mutant similar to that of wild-type MeCP2. The fact that this MeCP2 mutant is not constitutively bound to heterochromatin, but on the contrary can only cluster chromocenters when targeted via the GBP-MaSat fusion protein, provides an excellent opportunity to control and visualize the dynamics of chromatin reorganization in vivo. To this end, we designed a further development of the GBP-MaSat fusion protein that enabled us to temporally control targeting of GFP-tagged proteins to heterochromatin. To achieve this goal, we introduced a domain of the ER into our targeting tool, thereby creating a GBP-ER-NLS-MaSat (GERM) fusion protein. The ER domain contains a NLS that is exposed only upon hormone binding (14,20), which causes a dramatic conformational change and normally the re-localization of the ER from the cytoplasm into the nucleus. Our construct, however, contains a second, constitutively active NLS at the N-terminus of the MaSat, and is, therefore, imported into the nucleus even in the absence of estrogen. Interestingly, in the absence of estrogen, the conformation of the fusion protein GBP-ER-NLS-MaSat is such that the major satellite binding site of MaSat is blocked. On the other hand, estrogen binding results in a conformational change so that MaSat’s binding site becomes exposed and the GBP-ER-NLS-MaSat fusion protein quickly relocalizes within the nucleus to the chromocenters (Figure 2A). To be able to visualize its localization in vivo, we additionally fused the GBP-ER-MaSat construct to a C-terminal mCherry fluorescent protein.
Figure 2.
Spatio-temporal monitoring of the targeted nuclear redistribution of MeCP2 R111G mutant and subsequent large-scale chromatin rearrangements. (A) Schematic outline of the chromocenter targeting assay for temporal controlled recruitment of GFP-labeled proteins to pericentric heterochromatin. Left side: co-expression of the fusion protein GBP-ER-NLS-MaSat-mCherry (GERM) and GFP-labeled nuclear protein (exemplified by the Rett mutant MeCP2 R111G) in the absence of tamoxifen results in homogeneous nuclear distribution of both GERM and the GFP-labeled MeCP2 R111G mutant. Right side: upon hormone binding, the GERM re-locates to pericentric heterochromatin, enriched in major satellite DNA repeats, and consequently recruits the GFP-tagged Rett mutant to chromocenters. Visualization of the nucleus over time gives information regarding the chromocenter aggregation potential and dynamics of the mutant targeted to chromocenters. (B) Selected time points of living cells expressing GERM (red) and GFP-MeCP2 R111G (green). Cells were imaged over time prior to and following tamoxifen addition. 3D confocal images were obtained every 20 min for over 10 h. Scale bars: 2 µm (top) and 5 µm (bottom). (C) C2C12 mouse myoblasts were seeded on coverslips and double transfected with vectors coding for GERM and the GFP-tagged MeCP2 R111G mutant or GFP alone. Cells were fixed and DAPI stained before (t = 0), as well as 2 h (t = 2 h), 8 h (t = 8 h) and 16 h (t = 16 h) after tamoxifen addition. To address the chromocenter clustering ability of MeCP2 R111G and GFP upon time, 3D-stacks of fluorescent images of transfected were recorded in the DAPI channel. Graphs show mean numbers of chromocenters of cells expressing the proteins as indicated. Error bars represent 95% CI. Experiments were repeated two times resulting in at least 47 examined cells per construct. Asterisks represent statistically significant difference: * for P < 0.05, ** for P < 0.01 and *** for P < 0.001.
To create a fusion protein that would shuttle between the nucleus and the cytoplasm, we further removed the NLS signal located in the N-terminus of MaSat resulting in a construct predominantly located in the cytoplasm in the absence of tamoxifen. However, a small fraction of GBP-ER-MaSat-mCherry leaked into the nucleus even in the absence of estrogen. Strikingly, already these minimal amounts of GBP-ER-MaSat were sufficient to efficiently target the GFP-tagged MeCP2 R111G to chromocenters even in the absence of tamoxifen, preventing the application of this construct for controlled induction of chromatin targeting of proteins (data not shown). We, therefore, chose the nuclear switch GBP-ER-NLS-MaSat as an optimal tool to manipulate and monitor heterochromatin in a temporally controlled manner.
Co-expression of GBP-ER-NLS-MaSat and the GFP-tagged MeCP2 R111G Rett mutant resulted in a homogeneous nuclear distribution of both fusion proteins in most of the cells. Upon addition of the estrogen analogue tamoxifen, the GBP-ER-NLS-MaSat quickly started to accumulate at pericentric heterochromatin, reaching a maximum accumulation within 1–2 h. Importantly, using live cell microscopy we observed that, closely following the re-localization of GBP-ER-NLS-MaSat to chromocenters, a clear accumulation of the MeCP2 R111G mutant to pericentric regions took place (Figure 2A and B). Besides binding of the GBP-ER-NLS-MaSat fusion protein to the chromocenters and subsequent targeting of MeCP2 R111G to the latter, we were able to monitor fusion of adjacent heterochromatic regions over time (Figure 2B and Supplementary Movie S1). Strikingly, such fusion events were preceded by the movement of the large heterochromatic structures over several micrometers until they eventually encountered and finally fused into one chromocenter (Supplementary Movie S1). These data represent the direct visualization of the MeCP2-mediated large-scale chromatin rearrangement in vivo. Moreover, our temporally controlled intra-nuclear targeting tool allows the study of chromatin reorganization dynamics in vivo upon tethering of any chromatin modifier to these heterochromatic regions.
To examine whether the observed chromocenter fusion events might reflect the rescued chromatin clustering capability of MeCP2 R111G, we again transfected C2C12 cells with plasmids coding for GBP-ER-NLS-MaSat-mCherry and MeCP2 R111G or GFP alone. Before addition of tamoxifen and upon 2, 8 and 16 h of tamoxifen treatment, cells were fixed and stained with DAPI to highlight heterochromatic regions. To assess the degree of the chromocenter clustering ability of the MeCP2 R111G mutant over time, cells were imaged using an epifluorescent microscope in the GFP, RFP and DAPI channel and the number of chromocenters in cells overexpressing GBP-ER-NLS-MaSat-cherry and the MeCP2 R111G Rett mutant or GFP alone was quantified (using the DAPI channel). Before tamoxifen treatment, the MeCP2 R111G mutant showed weak to no chromocenter aggregation ability resulting in its severe miss-localization to the nucleoli in many of the cells. Strikingly, after 2 as well as after 8 h of tamoxifen addition and consequent targeting of MeCP2 R111G to heterochromatin, the chromocenter aggregation ability of this mutant was already prominent in comparison to the tamoxifen untreated state. The degree of rescue of the heterochromatin clustering potency of MeCP2 R111G reached its maximum after 16 h of tamoxifen treatment (Figure 2C). In contrast to cells overexpressing the GBP-ER-NLS-MaSat-mCherry fusion protein and the MeCP2 R111G Rett mutant, cells overexpressing GBP-ER-NLS-MaSat-mCherry with GFP alone and treating such cells with tamoxifen did not exhibit any increase in chromocenter aggregation upon 16 h of tamoxifen treatment, ruling out the possibility that the major satellite binding fusion protein and/or GFP and/or tamoxifen treatment induce clustering of pericentric heterochromatin independently from MeCP2 (Figure 2C).
Besides tethering GFP-labeled MeCP2 and MeCP2 Rett mutants to heterochromatin, we further want to highlight the flexibility provided by our targeting assay to artificially recruit any GFP-labeled nuclear protein to pericentric heterochromatin. For instance, GFP-labeled enzymatic chromatin modifiers such as histone acetylases, histone deacetylases and poly(ADP-ribose)polymerases can easily be recruited to heterochromatin using our assay to alter the post-translational state of pericentric heterochromatin and study the effects on any chromatin-related process. Especially, the co-expression of our nuclear switch GBP-ER-NLS-MaSat-mCherry with a GFP-tagged chromatin modifier allows the spatial and temporal monitoring of the effect of the latter on heterochromatin and is a powerful tool particularly suitable for live cell microscopy.
Using the synthetic polydactyl zinc-finger protein MaSat to tether nuclear proteins to chromatin ensures that the targeting tool per se does not exert any effect on the state of the chromocenters and the cell in general. Therefore, the observed effect mediated through targeting of the GFP-labeled nuclear protein is exclusively due to the nuclear protein of interest, such as, for example, the MeCP2 mutants, as we further validated by the targeting of GFP alone.
Our assay, therefore, provides the novel opportunity of targeting nuclear proteins to chromocenters with the help of the major satellite binding proteins GBP-MaSat and GBP-ER-NLS-MaSat. By that, this targeting method lays the ground for controlled manipulation of chromatin organization and provides an important tool to obtain a deep insight into the function of any protein and its effect on heterochromatin and thus contributes to our general understanding of chromatin architecture and its regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Movie S1.
FUNDING
Deutsche Forschungsgemeinschaft [DFG Ca198/7 to M.C.C. (in part)]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank H. Leonhardt and Chromotek for providing the GBP expression constructs, P. Weber and A. Lehmkuhl for experimental help, B. Bertulat and K. L. Jost for plasmids, as well as A. Rapp for discussions and advice.
REFERENCES
- 1.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 3.Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 5.Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 6.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 8.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal N, Becker A, Jost KL, Haase S, Thakur BK, Brero A, Hardt T, Kudo S, Leonhardt H, Cardoso MC. MeCP2 Rett mutations affect large scale chromatin organization. Hum. Mol. Genet. 2011;20:4187–4195. doi: 10.1093/hmg/ddr346. [DOI] [PubMed] [Google Scholar]
- 10.Kudo S, Nomura Y, Segawa M, Fujita N, Nakao M, Schanen C, Tamura M. Heterogeneity in residual function of MeCP2 carrying missense mutations in the methyl CpG binding domain. J. Med. Genet. 2003;40:487–493. doi: 10.1136/jmg.40.7.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindhout BI, Fransz P, Tessadori F, Meckel T, Hooykaas PJ, van der Zaal BJ. Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res. 2007;35:e107. doi: 10.1093/nar/gkm618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 14.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 15.Casas-Delucchi CS, Brero A, Rahn HP, Solovei I, Wutz A, Cremer T, Leonhardt H, Cardoso MC. Histone acetylation controls the inactive X chromosome replication dynamics. Nat. Commun. 2011;2:222. doi: 10.1038/ncomms1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casas-Delucchi CS, van Bemmel JG, Haase S, Herce HD, Nowak D, Meilinger D, Stear JH, Leonhardt H, Cardoso MC. Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res. 2012;40:159–169. doi: 10.1093/nar/gkr723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Gierasch LM, Woodcock CL. Rett syndrome-causing mutations in human MeCP2 result in diverse structural changes that impact folding and DNA interactions. J. Biol. Chem. 2008;283:20523–20534. doi: 10.1074/jbc.M803021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh RP, Nikitina T, Horowitz-Scherer RA, Gierasch LM, Uversky VN, Hite K, Hansen JC, Woodcock CL. Unique physical properties and interactions of the domains of methylated DNA binding protein 2. Biochemistry. 2010;49:4395–4410. doi: 10.1021/bi9019753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.