Abstract
Disrupting the interaction between primase and helicase in Escherichia coli increases Okazaki fragment (OF) length due to less frequent primer synthesis. We exploited this feature to increase the amount of ssDNA at the lagging strand of the replication fork that is available for λ Red-mediated Multiplex Automatable Genome Engineering (MAGE). Supporting this concept, we demonstrate that MAGE enhancements correlate with OF length. Compared with a standard recombineering strain (EcNR2), the strain with the longest OFs displays on average 62% more alleles converted per clone, 239% more clones with 5 or more allele conversions and 38% fewer clones with 0 allele conversions in 1 cycle of co-selection MAGE (CoS-MAGE) with 10 synthetic oligonucleotides. Additionally, we demonstrate that both synthetic oligonucleotides and accessible ssDNA targets on the lagging strand of the replication fork are limiting factors for MAGE. Given this new insight, we generated a strain with reduced oligonucleotide degradation and increased genomic ssDNA availability, which displayed 111% more alleles converted per clone, 527% more clones with 5 or more allele conversions and 71% fewer clones with 0 allele conversions in 1 cycle of 10-plex CoS-MAGE. These improvements will facilitate ambitious genome engineering projects by minimizing dependence on time-consuming clonal isolation and screening.
INTRODUCTION
High-throughput genome engineering requires the ability to cheaply and efficiently generate exact genomic DNA sequences. In this way, de novo genome synthesis (1,2) is an attractive approach for generating designer organisms. However, the incomplete understanding of genome structure and function poses a significant risk of designing non-viable genomes. Therefore, it is essential to test many designs. For example, a single-nucleotide DNA synthesis error in the completely de novo synthesized Mycoplasma mycoides chromosome caused a frameshift in dnaA that prevented the transplanted genome from surviving (2). As de novo synthesis becomes commonly used for creating genomes with novel or altered functionalities, the risk of generating non-viable genomes will increase. Multiplex Automatable Genome Engineering (MAGE) is a powerful alternative strategy for engineering genomes in vivo. MAGE simultaneously introduces several synthesized DNA oligonucleotides (oligos), resulting in the efficient modification of the Escherichia coli chromosome. This technique relies on phage λ Redβ recombinase, which binds to ssDNA oligos, protecting them from ssDNA exonucleases and facilitating their annealing to the lagging strand of the replication fork (3). This highly efficient process generates a diverse heterogenic population, which converges toward a fully modified isogenic population after many cycles of recombination with non-degenerate oligo pools. Generating a heterogenic population has been harnessed for directed evolution of biosynthetic pathways (4) and extensive cycling toward isogenic populations has been used to remove all 314 TAG stop codons in subsets across 32 E. coli strains (5). By integrating evolution with engineering, MAGE combinatorially explores a broad pool of viable and non-viable mutations. Since MAGE edits the genome in vivo, non-viable mutations never accumulate in the population. Yet, while this attribute of in vivo genome engineering enables increasingly ambitious genome designs, the ability of MAGE to efficiently generate those designs is often a limiting factor.
Several advances have already enhanced λ Red-mediated recombination from its initial ∼0.2% singleplex allele replacement (AR) frequency up to ∼30% (4). Thus far, the predominant approach for improving Redβ-mediated AR has been to optimize oligo design. Such advances include targeting oligos to the lagging strand of the replication fork (6), evading mismatch repair using modified nucleotides (7), minimizing oligo secondary structure and optimizing homology lengths (4), blocking oligo degradation with 5′ phosphorothioate bonds (4) and avoiding sequences with high degrees of off-target homology elsewhere in the genome (5). Additionally, removing the mismatch repair protein MutS to avoid reversion of mutated alleles (8) was a key innovation, but little other strain engineering was reported until recently. Such strain engineering could significantly augment the power of MAGE. For instance, by removing the potent ExoI, ExoVII, ExoX, RecJ and Redα exonucleases, we were able to shift the AR frequency distribution toward a more highly modified population (Mosberg, J.A., Gregg, C.J., et al., in review). These results indicate that intracellular ssDNA oligo availability is a limiting factor for MAGE and that phosphorothioate protection alone cannot eliminate nuclease-mediated degradation of oligos.
Recently, a new strategy (co-selection MAGE or CoS-MAGE) has been developed to engineer highly modified cells. This strategy uses an oligo that repairs a broken selectable marker (e.g. antibiotic resistance gene) to enhance AR frequency of nearby non-selectable alleles (9,10). CoS-MAGE enhances the average multiplex AR frequency ∼4-fold by selecting for cells that take up MAGE oligos and that have a permissive replication fork in the desired region of the genome (9). Additionally, this approach selects against cells that do not take up oligos during electroporation, as it removes the population that does not revert the selectable allele.
The fact that CoS-MAGE is most effective for oligos targeted in close proximity to the selectable marker suggests that replication fork position and accessibility are limiting factors in Redβ-mediated recombination (9). Thus, we reasoned that we could improve AR frequencies by manipulating replication fork dynamics to increase the amount of ssDNA on the lagging strand of the replication fork. Since Okazaki fragment (OF) size can be modulated by the frequency of OF primer synthesis by DnaG primase (11), we hypothesized that attenuating the interaction between DnaG primase and the replisome would increase the amount of accessible ssDNA on the lagging strand of the replication fork and enhance multiplex AR frequencies (Figure 1). Tougu et al. (12) have reported E. coli primase variants with impaired helicase binding, resulting in less-frequent OF initiation, but normal replication fork rate, priming efficiency and primer utilization during in vitro replication. These variants, K580A and Q576A, resulted in in vitro OFs that were ∼1.5- and 8-fold longer, respectively, than those initiated by wild-type DnaG (13). Therefore, these variants were used to explore whether increasing accessible ssDNA on the lagging strand can improve multiplex AR frequency.
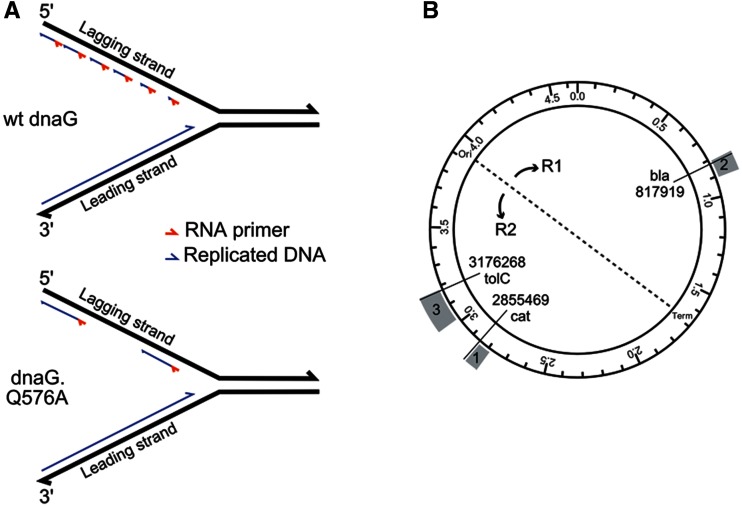
Figure 1.
Effect of dnaG attenuation on replication fork dynamics. (A) Schematic showing the replication fork in E. coli, including the leading and lagging strands undergoing DNA synthesis. DnaG synthesizes RNA primers (red) onto the lagging template strand, which in turn initiate OF synthesis (blue) by PolIII. Compared with wt DnaG primase, the variants tested in this study have lower affinities for DnaB helicase (12). Since the DnaG–DnaB interaction is necessary for primase function, primer synthesis occurs less frequently, thereby exposing larger regions of ssDNA on the lagging template strand (13). (B) A schematic representing the E. coli MG1655 genome with the origin (oriC) and terminus (T) of replication indicated, splitting the genome into Replichore 1 and Replichore 2. Each oligo set converts 10 TAG codons to TAA codons within the genomic regions indicated in gray. Co-selection marker positions are denoted by radial lines.
In this work, we demonstrate that accessible ssDNA on the lagging strand of the replication fork is a limiting factor for multiplex AR, and that disrupting the interaction between DnaG primase and DnaB helicase significantly improves multiplex AR frequencies. We further describe the creation of an optimized strain for CoS-MAGE, which combines approaches to increase intracellular oligo concentration and to expose accessible ssDNA on the lagging strand of the replication fork. This strain demonstrates greatly improved CoS-MAGE performance, and provides a foundation for genome engineering projects of a much more ambitious scope.
MATERIALS AND METHODS
Supplementary Table S1 presents a full list of DNA oligos used in this work. All oligos were ordered with standard purification and desalting from Integrated DNA Technologies. Cultures were grown in LB-Lennox media (LBL; 10 g tryptone, 5 g yeast extract, 5 g NaCl per 1 l water).
Strain creation
Oligo-mediated λ Red recombination was used to generate all mutations as described below. All of the strains used in this work were generated from EcNR2 (4) (E. coli MG1655 ΔmutS::cat Δ(ybhB-bioAB)::[λcI857 N(cro-ea59)::tetR-bla]). Strain Nuc5-.dnaG.Q576A was generated by recombining oligo dnaG_Q576A into strain Nuc5- (EcNR2 xonA-, recJ-, xseA-, exoX- and redα-; Mosberg, J.A., Gregg, C.J., et al., in review). EcNR2. DT was created by deleting the endogenous tolC gene using the tolC.90.del recombineering oligo (5). EcNR2.T.co-lacZ was created by recombining a tolC cassette (T.co-lacZ) into the genome of EcNR2.DT, upstream of the lac operon. CoS-MAGE strains were prepared by inactivating a chromosomal selectable marker (cat, tolC or bla) using a synthetic oligo. Clones with a sensitivity to the appropriate antibiotic or sodium dodecyl sulphate (SDS) (14) were identified by replica plating. The growth rate of strains EcNR2, EcNR2.dnaG.K580A and EcNR2.dnaG.Q576A are approximately equivalent, whereas Nuc5-.dnaG.Q576A has a doubling time that is only ∼7% longer than the others.
Generating dsDNA recombineering cassettes
The T.co-lacZ dsDNA recombineering cassette was generated by polymerase chain reaction (PCR) using primers 313 000.T.lacZ.coMAGE-f and 313 001. T.lacZ.coMAGE-r (Supplementary Table S1). The PCR was performed using KAPA HiFi HotStart ReadyMix, with primer concentrations of 0.5 µM and 1 µl of T.5.6 (5) used as template (a terminator was introduced downstream of the stop codon in the tolC cassette). PCRs (50 µl total) were heat activated at 95°C for 5 min, then cycled 30 times at 98°C (20 sec), 62°C (15 sec) and 72°C (45 sec). The final extension was at 72°C for 5 min. The Qiagen PCR purification kit was used to isolate the PCR products (elution in 30 µl deionized water (dH2O)). Purified PCR products were quantitated on a NanoDrop™ ND1000 spectrophotometer and analyzed on a 1% agarose gel with ethidium bromide staining to confirm that the expected band was present and pure.
Performing λ Red recombination
λ Red recombinations of ssDNA and dsDNA were performed as previously described (4,15). Briefly, 30 µl from an overnight culture was inoculated into 3 ml of LBL and grown at 30°C in a rotator drum until an OD600 of 0.4–0.6 was reached (typically 2–2.5 h). The cultures were transferred to a shaking water bath (300 rpm at 42°C) for 15 min to induce λ Red, then immediately cooled on ice for at least 3 min. For each recombination, 1 ml of culture was washed twice in ice cold dH2O. Cells were pelleted between each wash by centrifuging at 16 000 rcf for 20 sec. The cell pellet was resuspended in 50 µl of dH2O containing the DNA to be recombined. For recombination of dsDNA PCR products, 50 ng of PCR product was used. Recom-bination using dsDNA PCR products was not performed in Nuc5- strains, since λExo is necessary to process dsDNA into a recombinogenic ssDNA intermediate prior to β-mediated annealing (15,16). For experiments in which a single oligo was recombined, 1 µM of oligo was used. For experiments in which sets of 10 or 20 recombineering oligos were recombined along with a co-selection oligo, 0.5 µM of each recombineering oligo and 0.2 µM of the co-selection oligo were used (5.2 µM total for 10-plex and 10.2 µM total for 20-plex). A BioRad GenePulser™ was used for electroporation (0.1 cm cuvette, 1.78 kV, 200 Ω, 25 µF), and electroporated cells were allowed to recover in 3 ml LBL in a rotator drum at 30°C for at least 3 h before plating on selective media. For MAGE and CoS-MAGE experiments, cultures were recovered to apparent saturation (5 or more hours) to minimize polyclonal colonies (this was especially important for strains based on Nuc5-, which exhibit slow recovery after λ Red induction/electroporation). MAGE recovery cultures were diluted to ∼5000 cfu/ml, and 50 µl of this dilution was plated on non-selective LBL agar plates. To compensate for fewer recombinants surviving the co-selection, CoS-MAGE recovery cultures were diluted to ∼1E5 cfu/ml and 50 µl of this dilution was plated on appropriate selective media for the co-selected resistance marker (LBL with 50 µg/ml carbenicillin for bla, 20 µg/ml chloramphenicol for cat or 0.005% w/v SDS for tolC). Leading-targeting CoS-MAGE recovery cultures were diluted to ∼5E6 cfu/ml before plating.
Analyzing recombination
GalK activity was assayed by plating recovered recombination cultures onto MacConkey agar supplemented with 1% galactose as a carbon source. Red colonies were scored as galK+ and white colonies were galK−. LacZ activity was assayed by plating recovery cultures onto LBL agar + IPTG/X-Gal (Fisher ChromoMax IPTG/X-Gal solution). Blue colonies were scored as lacZ+ and white colonies were lacZ−.
Kapa 2G Fast ReadyMix was used in colony PCRs to screen for correct insertion of dsDNA selectable markers. PCRs had a total volume of 20 µl, with 0.5 µM of each primer. These PCRs were carried out with an initial activation step at 95°C for 2 min, then cycled 30 times at 95°C (15 sec), 56°C (15 sec), 72°C (40 sec), followed by a final extension at 72°C (90 sec).
Allele-specific colony PCR (ascPCR) was used to detect the dnaG_K580A and dnaG_Q576A mutations. Multiplex allele-specific colony PCR (mascPCR) (17) was used to detect the 1–2 bp mutations generated in the MAGE and CoS-MAGE experiments. Each allele is interrogated by two separate PCRs—one with a forward primer whose 3′ end anneals to the wild-type allele, and the other with a forward primer whose 3′ end anneals to the mutated allele (the same reverse primer is used in both reactions). Primers are designed to have a Tm ∼ 62°C, but a gradient PCR is necessary to optimize annealing temperature (typically between 63°C and 67°C) to achieve maximal specificity and sensitivity for a given set of primers. A wild-type allele is indicated by amplification only in the wt-detecting PCR, whereas a mutant allele is indicated by amplification only in the mutant-detecting PCR. For mascPCR assays, primer sets for interrogating up to 10 alleles are combined in a single reaction. Each allele has a unique amplicon size (100, 150, 200, 250, 300, 400, 500, 600, 700 and 850 bp). Template is prepared by growing monoclonal colonies to late-log phase in 150 µl LBL and diluting 2 µl of culture into 100 µl dH2O. Typical mascPCR reactions use KAPA2GFast Multiplex PCR ReadyMix and 10X Kapa dye in a total volume of 10 µl, with 0.2 µM of each primer and 2 µl of template. These PCRs were carried out with an initial activation step at 95°C (3 min), then cycled 27 times at 95°C (15 sec), 63–67°C (30 sec; annealing temperature optimized for each set of mascPCR primers) and 72°C (70 sec), followed by a final extension at 72°C (5 min). All mascPCR and ascPCR assays were analyzed on 1.5% agarose/EtBr gels (180 V, duration depends on distance between electrodes) to ensure adequate band resolution.
We performed at least two independent replicates for all strains with each oligo set in CoS-MAGE experiments. All replicates for a given strain and oligo set were combined to generate a complete data set. Polyclonal or ambiguous mascPCR results were discarded from our analysis. Mean number of alleles replaced per clone were determined by scoring each allele as 1 for converted or 0 for unmodified. Data for EcNR2 and Nuc5- are from Mosberg, J.A., Gregg, C.J., et al. (in review), as experiments for these manuscripts were planned and performed together. Given the sample sizes tested in the CoS-MAGE experiments (n > 47), we used parametric statistical analyses instead of their non-parametric equivalents, since the former are more robust with large sample sizes (18). We used a one way ANOVA to test for significant variance in CoS-MAGE performance of the strains (EcNR2, EcNR2.dnaG.K580A, EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A) for a given oligo set. Subsequently, we used a Student’s t-test to make pairwise comparisons with significance defined as P < 0.05/n, where n is the number of pairwise comparisons. Here, n = 15 as this data set was planned and collected as part of a larger set with 6 different strains although only EcNR2, EcNR2.dnaG.K580A, EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A are presented here. As such, significance was defined as P < 0.003 for the analyses presented in Figures 3 and 5. Statistical significance in Figures 3 and 5 are denoted using a star system where * denotes P < 0.003, ** denotes P < 0.001, and *** denotes P < 0.0001. In the case of the experiment comparing EcNR2 and EcNR2.dnaG.Q576A using leading targeting oligos (Supplementary Figure S1), we tested for statistical significance using a single t-test with significance defined as P < 0.05.
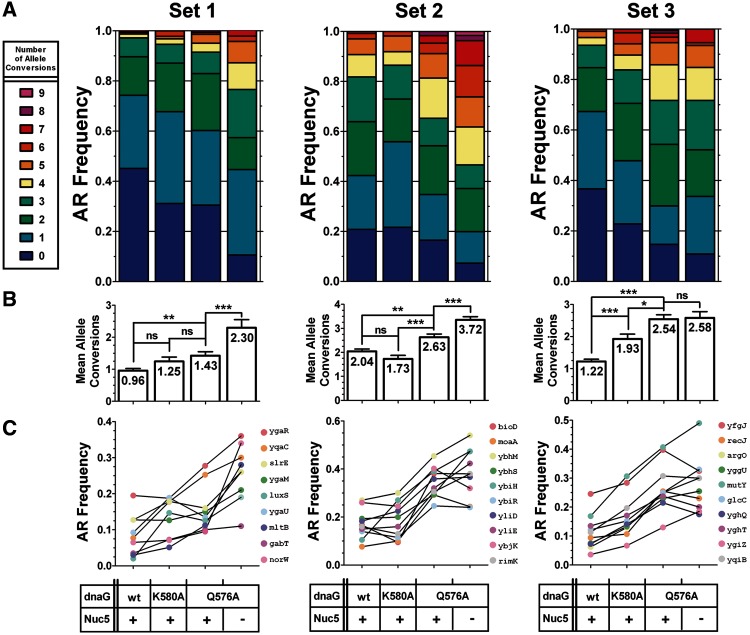
Figure 3.
DnaG variants improve CoS-MAGE Performance. EcNR2, EcNR2.dnaG.K580A, EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A were tested for their performance in CoS-MAGE using 3 sets of 10 oligos as described in Figure 1B. For each set, all 10 alleles were simultaneously assayed by mascPCR in recombinant clones after 1 cycle of CoS-MAGE. (A) The data are presented using stacked AR frequency plots, which show the distribution of clones exhibiting a given number of allele conversions. (B) Mean number of alleles converted for each strain are shown with P-values indicated above the bars. Statistical significance is denoted using a star system where * denotes P < 0.003, ** denotes P < 0.001 and *** denotes P < 0.0001. The data are presented as the mean (reported numerically inside each bar) ± SEM. (C) AR frequencies for each individual allele are shown for all tested strains. Overall, the relative performance of each strain was Nuc5-.dnaG.Q576A > EcNR2.dnaG.Q576A > EcNR2.dnaG.K580A > EcNR2. This trend reflects an improvement commensurate with the severity of primase attenuation (i.e. the Q576A variant has more severely disrupted primase and larger OFs than the K580A variant). Furthermore, Nuc5-.dnaG.Q576A combines the benefits of the DnaG Q576A variant and the benefits of the inactivation of five potent exonucleases (Mosberg, J.A., Gregg, C.J., et al., in review). For Set 1: EcNR2, n = 319; EcNR2.dnaG.K580A, n = 93; EcNR2.dnaG.Q576A, n = 141; Nuc5-.dnaG.Q576A, n = 47. For Set 2: EcNR2, n = 269; EcNR2.dnaG.K580A, n = 111; EcNR2.dnaG.Q576A, n = 236; Nuc5-.dnaG.Q576A, n = 191. For set 3: EcNR2, n = 327; EcNR2.dnaG.K580A, n = 136; EcNR2.dnaG.Q576A, n = 184; Nuc5-.dnaG.Q576A, n = 92.
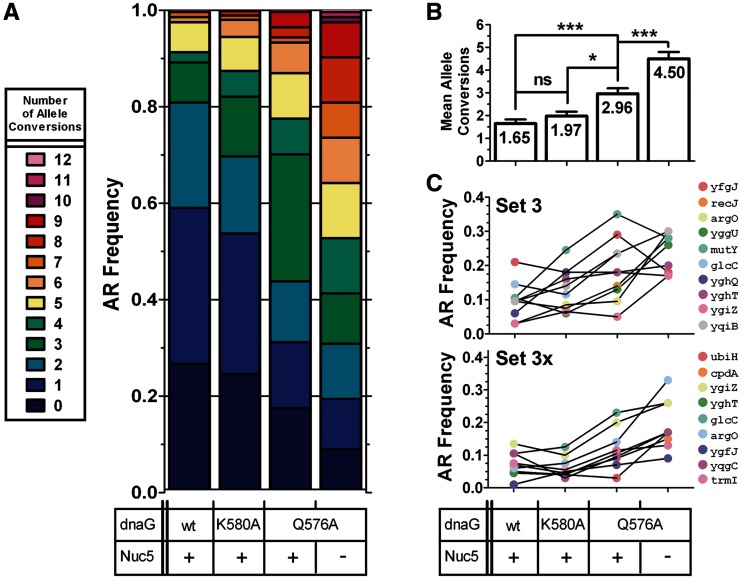
Figure 5.
Testing DnaG variants with a 20-plex CoS-MAGE oligo set. EcNR2, EcNR2.dnaG.K580A, EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A were tested for their performance in CoS-MAGE using an expanded set of 20 oligos (Sets 3+3X). Genotypes of recombinant clones were assayed by mascPCR after one cycle of CoS-MAGE (ygfT could not be assayed by mascPCR). (A) AR frequency distributions. (B) Mean number of alleles converted ± SEM, with p-values indicated above the bars. Statistical significance is denoted using a star system where * denotes P < 0.003, ** denotes P < 0.001 and *** denotes P < 0.0001. (C) Mean individual AR frequencies. As seen with the smaller oligo sets, the dnaG variants reduce the number of clones with zero conversions and increase the average number of conversions per clone. Nuc5-.dnaG.Q576A strongly outperforms all other strains, with a mean of 4.50 alleles converted and fewer than 10% of clones having zero conversions. Notably, Nuc5-.dnaG.Q576A has strongly improved performance with Sets 3 + 3X compared with Set 3, whereas EcNR2.dnaG.Q576A does not. EcNR2, n = 96; EcNR2.dnaG.K580A, n = 113; EcNR2.dnaG.Q576A, n = 95; Nuc5-.dnaG.Q576A, n = 96.
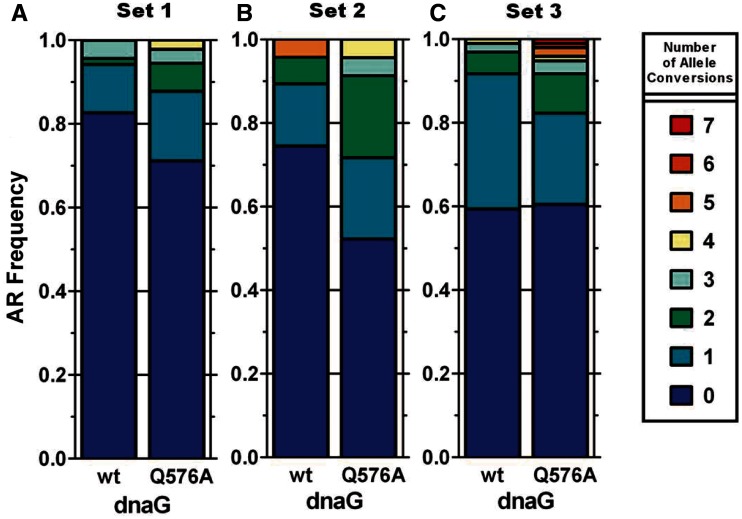
For the experiment in which 10 oligos were targeted within lacZ, recombinants were identified by blue/white screening. The frequency of clones with 1 or more alleles replaced (number of white colonies / total number of colonies) was determined for every replicate. For white colonies only, a portion of the lacZ gene was amplified with primers lacZ_jackpot_seq-f and lacZ_jackpot_seq-r (Supplementary Table S1), using KAPA HiFi HotStart ReadyMix as described above. PCR purified (Qiagen PCR purification kit) amplicons were submitted to Genewiz for Sanger sequencing in each directions using lacZ_jackpot_seq-f and lacZ_jackpot_seq-r. Combined, the two sequencing reads for each clone interrogated all 10 alleles (i.e. unmodified or mutant sequence). Three replicates of recombinations and blue/white analysis were performed to ensure consistency, but only one replicate was sequenced (n = 39 for EcNR2 and n = 55 for EcNR2.dnaG.Q576A). Mean number of alleles replaced per clone were determined as described above. We tested for statistically significant differences in mean allele conversion between the strains using a Student’s t-test with significance defined as P < 0.05. Statistical significance in Figure 4C is denoted using a star system where *** denotes P < 0.0001.
Figure 4.
Placing all targeted alleles within one OF does not cause a bimodal distribution for recombination frequency. EcNR2 and EcNR2.dnaG.Q576A were tested for their performance in CoS-MAGE using a set of 10 non-overlapping oligos that introduce 10 premature stop codons in the first 1890 bp of lacZ. The targeted region of the genome is likely to be small enough to be frequently encompassed within a single OF in EcNR2.dnaG.Q576A. After 1 cycle of CoS-MAGE, LacZ− recombinant clones were Sanger sequenced to assay all 10 alleles. Recombinations were performed in triplicate to estimate the frequency of white colonies (lacZ−), but sequencing was only performed on a single replicate. (A) EcNR2.dnaG.Q576A (n = 715, 5.33:1) exhibited a significant increase in the lacZ−:lacZ+ ratio compared with EcNR2 (n = 485, 1.46:1). (B) EcNR2.dnaG.Q576A exhibited an AR distribution similar to those observed with Sets 1–3 (which span 70, 85 and 162 kb, respectively). (C) Compared with EcNR2, EcNR2.dnaG.Q576A exhibited a higher mean number of alleles converted (unpaired t-test, ***P < 0.0001). For EcNR2, n = 39, and for EcNR2.dnaG.Q576A, n = 55. (D) Compared with EcNR2, AR frequencies increased for 9 out of 10 individual alleles in EcNR2.dnaG.Q576A. The alleles are represented by their positions in lacZ (e.g. ‘+61’ means that this oligo introduces a nonsense mutation by generating a mismatch at the 61st nucleotide of lacZ). Taken together, all of these results demonstrate improved CoS-MAGE in EcNR2.dnaG.Q576A compared with EcNR2, but no significant enhancement was obtained from targeting all oligos to a single putative OF.
RESULTS
Impaired primase activity enhances multiplex AR frequency
It is generally accepted that Redβ mediates annealing of exogenous DNA to the lagging strand of the replication fork prior to extension as a nascent OF (3,15,16,19). Therefore, we sought to increase the amount of ssDNA on the lagging strand by disrupting the ability of DnaG primase to initiate OFs. Prior work (13) has shown that DnaG K580A and Q576A mutations increase OF length in vitro by ∼1.5- and 8-fold, respectively (see Supplementary Table S2 for further explanation).
To investigate whether longer OFs could improve MAGE, we compared the performance of EcNR2, EcNR2.dnaG.K580A and EcNR2.dnaG.Q576A. Three sets of recombineering oligos (designed in (5) to convert TAG codons to TAA and renamed herein for clarity as Sets 1–3) were used in order to control for potential oligo-, allele-, region- and replichore-specific effects (5). The genomic regions targeted by these oligo sets are indicated in Figure 1B. The AR distribution improved for EcNR2.dnaG.Q576A, as reflected by the increase in mean number of alleles converted per clone per MAGE cycle (Figure 2). These results were encouraging, so we used CoS-MAGE (9) to augment the observed effects. In this experiment, each of the three oligo sets was paired with a co-selection oligo which restored the function of a nearby mutated selectable marker (cat for Set 1, bla for Set 2 and tolC for Set 3). In order to improve on the current best practices for CoS-MAGE, we also introduced the dnaG.Q576A mutation into Nuc5-, a strain previously shown to have improved CoS-MAGE properties (Mosberg, J.A., Gregg, C.J., et al., in review). EcNR2. dnaG.Q576A robustly outperformed EcNR2, yielding a significantly increased mean number of alleles converted (mean ± std. error of mean) for Set 1 (Figure 3B, left panel, 1.43 ± 0.12 versus 0.96 ± 0.07, **p = 0.0003), Set 2 (Figure 3B, middle panel, 2.63 ± 0.13 versus 2.04 ± 0.10, **p = 0.0003) and Set 3 (Figure 3B, right panel, 2.54 ± 0.14 versus 1.22 ± 0.07, ***p < 0.0001). In agreement with the previous observation for MAGE without co-selection, EcNR2.dnaG.Q576A exhibited an increased AR distribution for all three oligo sets in CoS-MAGE (Figure 3A). Furthermore, EcNR2.dnaG.K580A (intermediate-sized OFs) appears to have intermediate performance between EcNR2 (normal OFs) and EcNR2.dnaG.Q576A (longest OFs). This suggests that OF length correlates with AR frequency, and supports our hypothesis that exposing more ssDNA at the lagging strand of the replication fork enhances Redβ-mediated annealing.
Figure 2.
DnaG variants improve MAGE performance. EcNR2 (wt) and EcNR2.dnaG.Q576A (Q576A) were tested for their MAGE performance without co-selection (4) using 3 sets of 10 oligos as described in Figure 1B. For each set, all 10 alleles were simultaneously assayed by mascPCR after one cycle of MAGE. The data are presented using stacked AR frequency plots, which show the distribution of clones exhibiting a given number of allele conversions. Compared with EcNR2 (A, Set 1, n = 69; B, Set 2, n = 47; C, Set 3, n = 96), EcNR2.dnaG.Q576A exhibited fewer clones with zero conversions for Set 1 (A, n = 90) and Set 2 (B, n = 46), but not for Set 3 (C, n = 96). In all three sets, EcNR2.dnaG.Q576A displayed more clones with 2 or more allele conversions.
Visualizing AR frequency for individual alleles in all three sets (Figure 3C) reinforces the relationship between OF size and MAGE performance. Compared with EcNR2, the K580A variant trends toward a modest increase in individual AR frequency, whereas the Q576A variant starkly improves AR frequency. Finally, the Nuc5-.dnaG.Q576A strain yielded the highest observed AR frequencies for all oligo sets, suggesting a combined effect of decreasing oligo degradation through nuclease inactivation and increasing the amount of exposed target ssDNA at the lagging strand of the replication fork. Interestingly, EcNR2.dnaG.Q576A strongly outperformed Nuc5- for Set 3 (***P < 0.0001), whereas EcNR2.dnaG.Q576A performance was not significantly different from that of Nuc5- for Sets 1 (p = 0.33) and 2 (p = 0.26) (Tables 1 and 2). This suggests that the relative importance of replication fork availability and oligo protection can vary for MAGE targets throughout the genome, possibly due to oligo and/or locus-specific effects that have not yet been elucidated. Since both factors are important, combining impaired primase mutants with nuclease knockouts should reliably improve CoS-MAGE performance.
Table 1.
Summary of mean number of alleles converted per clone for each MAGE oligo set
| Set | EcNR2 | Nuc5- | EcNR2.dnaG.Q576A | Nuc5-.dnaG.Q576A |
|---|---|---|---|---|
| Mean ± SEM (n) | Mean ± SEM (n) | Mean ± SEM (n) | Mean ± SEM (n) | |
| 1 | 0.96 ± 0.07 (319) | 1.58 ± 0.10 (257) | 1.43 ± 0.12 (141) | 2.30 ± 0.25 (92) |
| 2 | 2.04 ± 0.10 (269) | 2.89 ± 0.19 (142) | 2.63 ± 0.13 (236) | 3.72 ± 0.17 (191) |
| 3 | 1.22 ± 0.07 (327) | 1.61 ± 0.12 (139) | 2.54 ± 0.14 (184) | 2.59 ± 0.19 (92) |
The mean number of alleles converted per clone, SEM and sample size (n) were compared for EcNR2, Nuc5-, EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A. Nuc5- and EcNR2.dnaG.Q576A had statistically equivalent performance for Sets 1 and 2, whereas EcNR2.dnaG.Q576A strongly outperformed Nuc5- for Set 3. Nuc5-.dnaG.Q576A consistently outperformed all other strains. Data for EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A were determined in this work. Data for EcNR2 and Nuc5- are from Mosberg, J.A., Gregg, C.J., et al. (in review).
Table 2.
CoS-MAGE AR performance of modified strains (presented as fold change from EcNR2)
| Metric | Set | Nuc5- | E2.dnaG. Q576A | Nuc5-.dnaG. Q576A |
|---|---|---|---|---|
| Average | 1 | 1.65 | 1.49 | 2.40 |
| 2 | 1.41 | 1.29 | 1.82 | |
| 3 | 1.32 | 2.08 | 2.12 | |
| Average | 1.46 | 1.62 | 2.11 | |
| 5+ Conversions | 1 | 5.28 | 3.96 | 10.18 |
| 2 | 2.65 | 2.01 | 4.11 | |
| 3 | 1.07 | 4.20 | 4.52 | |
| Average | 3.00 | 3.39 | 6.27 | |
| 0 Conversions | 1 | 0.67 | 0.68 | 0.24 |
| 2 | 0.58 | 0.79 | 0.35 | |
| 3 | 0.71 | 0.40 | 0.30 | |
| Average | 0.65 | 0.62 | 0.29 |
The fold improvement was calculated as (strain performance)/(EcNR2 performance), where performance refers to the average number of allele conversions per clone, or the fraction of clones with 5+ or 0 conversions. These metrics were the average of individual metrics for Oligo Sets 1, 2 and 3. In all three categories, Nuc5-.dnaG.Q576A exhibited an effect that was roughly an additive combination of the effects yielded in Nuc5- and EcNR2.dnaG.Q576A. Data for EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A were determined in this work. Data for EcNR2 and Nuc5- are from Mosberg, J.A., Gregg, C.J., et al. (in review).
OF location is not a major determinant of available ssDNA on the lagging strand of the replication fork
Given the significant enhancement of CoS-MAGE performance in EcNR2.dnaG.Q576A, we sought to determine whether localizing all 10 targeted alleles to a single-putative OF would result in ‘jackpot’ recombinants with all 10 alleles converted. We hypothesized that nascent OFs sometimes obstructed target alleles, leading to a non-accessible lagging strand. According to this hypothesis, successful replacement of one allele would indicate permissive OF localization, greatly increasing the chance that other alleles occurring within the same OF could be replaced. Therefore, we speculated that the larger OF size in EcNR2.dnaG.Q576A might allow many changes to occur within 1 large OF. Therefore, we designed 10 MAGE oligos that introduce inactivating nonsense mutations into a region spanning 1829 bp of lacZ. Despite their close proximity, all 10 alleles were spaced far enough apart that their corresponding MAGE oligos would not overlap. Given the difference in average OF sizes between strains, it is unlikely for all 10 alleles to be located in the same OF in EcNR2, but quite likely that all 10 alleles would be located in the same OF in EcNR2.dnaG.Q576A. A tolC cassette (T.co-lacZ) was installed ∼50 kb upstream of lacZ for efficient co-selection. Prior to use, this cassette was inactivated using the tolC-r_null_mut* oligo. Since the placement of these mutations is not compatible with mascPCR analysis, we used Sanger sequencing for analysis of white colonies. Blue colonies were scored as having zero conferred mutations. For EcNR2, 59% of the clones were white with 1.24 ± 0.23 (mean ± SEM, standard error of the mean) conversions per clone, whereas 84% of the EcNR2.dnaG.Q576A clones were white with 2.52 ± 0.25 allele conversions per clone (Figure 4A and C). Although EcNR2.dnaG.Q576A exhibits more mean allele conversions in CoS-MAGE than EcNR2 (***p < 0.0001), the magnitude of this improvement (Figure 4B) is comparable with those observed for Sets 1–3 (Figure 3) where non-selectable oligos were spread across 70, 85 and 162 kb, respectively. Moreover, ‘jackpot’ clones with 7 + converted alleles were not frequently observed for EcNR2.dnaG.Q576A using this oligo set. Thus although replication fork position is relevant, OF placement is not the predominant limiting factor for multiplex AR. Other important factors could include target site occlusion by single-stranded binding proteins or the availability of oligos, Redβ or host factors.
Improved strains have larger optimal oligo pool size for multiplex AR
A MAGE oligo pool size of ∼10 was found to be most effective in prior studies (5). However, given the enhanced Redβ-mediated recombination in our dnaG (this work) and Nuc5- (Mosberg, J.A., Gregg, C.G., et al., in review) strains, we tested whether an expanded set of oligos would lead to more alleles converted in average and top clones. Therefore, we designed 10 additional MAGE oligos (Set 3X) that swapped synonymous AGA and AGG codons in alleles within the same region targeted by the Set 3 oligos. The ygfT allele (Set 3X) was not successfully assayed by mascPCR, so a maximum of 19 ARs could be detected out of the 20 conversions attempted. One round of CoS-MAGE using the combined oligo Sets 3 and 3X with tolC as a selectable marker improved AR frequency in all strains (Figure 5A). The mean number of alleles converted (and fold increase over 10-plex means for Set 3 alone) per clone are as follows: 1.65 (1.35-fold) for EcNR2, 1.97 (1.02-fold) for EcNR2.dnaG.K580A, 2.96 (1.17-fold) for EcNR2.dnaG. Q576A and 4.50 (1.74-fold) for Nuc5-.dnaG.Q576A (Figure 5B). Notably, Nuc5-.dnaG.Q576A exhibited the greatest improvement with the expanded oligo set, suggesting that preventing oligo degradation is important when the intracellular concentration of each individual oligo is low. Longer OFs then increase the probability that scarce oligos will find their genomic target. This observation assumes that a limited number of oligos are internalized during electroporation, which is consistent with the fact that the mole fraction of an oligo in a multiplex experiment affects its relative AR frequency at saturating oligo concentrations (9). Notably, the Set 3X oligos yielded lower recombination frequencies compared with the Set 3 alleles that converted TAG to TAA codons, and Nuc5-.dnaG.Q576A strongly elevated the AR frequency of these alleles (Figure 5C). Nuc5-.dnaG.Q576A exhibited the largest number of simultaneous allele conversions reported to date in a single recombination (tolC plus 12 additional alleles converted). Although CoS-MAGE in Nuc5-.dnaG.Q576A was able to simultaneously convert an unprecedented number of alleles, the lack of clones with ARs near the maximum of 19 suggests that CoS-MAGE is approaching a practical maximum for oligo pool complexity, where further increases in oligo pool size may not substantially improve AR frequency or increase the mean number of alleles converted.
Disrupting DnaG primase activity enhances leading strand recombination
Since DnaG primase synthesizes RNA primers only at the lagging strand of the replication fork, we expected its alteration to have minimal effect on Redβ-mediated annealing to the leading strand. To examine this hypothesis, we tested oligos designed to target the Set 3 alleles on the leading strand (reverse complements of the Set 3 oligos described above). The tolC-reverting co-selection oligo was also re-designed to target the leading strand so that the correct strand would be co-selected. Although the number of tolC-reverted co-selected recombinants were few, of the tolC+ clones, EcNR2 gave 0.85 ± 0.13 allele conversions per clone (mean ± std. error of the mean, n = 88), whereas EcNR2.dnaG.Q576A gave 1.39 ± 0.18 conversions (n = 91), which was significantly different (*p = 0.018). Similar to lagging targeting Set 3, we observed a reduction in zero conversion events for EcNR2.dnaG.Q576A, as well as a broadening of the distribution of total allele conversions per clone and a greater maximum number of alleles converted (Supplementary Figure S1A). Thus, leading-targeting CoS-MAGE yields recombination frequencies nearly within two-fold of those attained with lagging-targeting CoS-MAGE (1.22 ± 0.07 versus 2.54 ± 0.14 for EcNR2 and EcNR2.dnaG.Q576A, respectively). Furthermore, contrary to our expectations, EcNR2.dnaG.Q576A exhibited significantly enhanced AR frequency over EcNR2 at 9 out of 10 alleles on the leading strand (Supplementary Figure S1C). Interestingly, using leading targeting oligos, the co-selection advantage quickly diminished with distance (Supplementary Figure S1B, top panel). In contrast, co-selection using lagging targeting oligos increases the AR frequency of other alleles spanning a large genomic distance (∼0.5 Mb; (9)), as observed for the lagging-targeting Set 3 oligos (Supplementary Figure S1B, bottom panel).
Disrupting DnaG primase activity enhances deletions but not insertions
MAGE is most effective at introducing short mismatches, insertions and deletions, as these can be efficiently generated using λ Red-mediated recombination without direct selection (4). However, large deletions and gene-sized insertions are also important classes of mutations that could increase the scope of applications for MAGE. For example, combinatorial deletions could be harnessed for minimizing genomes (20) and efficient insertions could increase the ease of building biosynthetic pathways by removing the need for linking inserted genes to selectable markers (14,21–23). Large deletions require two separate annealing events often spanning multiple OFs, but large insertions should anneal within the same OF, as the heterologous portion loops out and allows the flanking homologies to anneal to their adjacent targets (15,16). Maresca et al. (16) have demonstrated that the length of deletions have little effect on Redβ-mediated recombination, but that insertion frequency is highly dependent on insert size (presumably due to constraints on λExo-mediated degradation of the leading-targeting strand and not the lagging-targeting strand). Therefore, we investigated whether diminishing DnaG primase function would enhance deletion and/or insertion frequencies.
Based on the ssDNA intermediate model for λRed recombination (15,16), we expected enhanced deletion frequency in EcNR2.dnaG.Q576A especially for intermediate-sized deletions (500 bp – 10 kb), since less frequent priming would increase the probability of both homology regions being located in the same OF. Therefore, we designed three oligos that deleted 100, 1149 or 7895 bp of the genome, including a portion of galK. In addition to galK, oligo galK_KO1.7895 deleted several non-essential genes (galM, gpmA, aroG, ybgS, zitB, pnuC and nadA). The recombined populations were screened for the GalK- phenotype (white colonies) on MacConkey agar plates supplemented with galactose as a carbon source. EcNR2.dnaG.Q576A significantly outperformed EcNR2 for the 100 bp (*p = 0.03) and 1149 bp (*p = 0.03) deletions, but there was no difference detected between the two strains for the 7895 bp deletion (p = 0.74, Supplementary Figure S2). The lack of improvement using galK_KO1.7895 may be due to reduced target availability if the two homology sites are split across two or more OFs even in EcNR2.dnaG.Q576A.
Finally, if λExo degradation most strongly impacts λ Red-mediated insertions of large cassettes, modifying the replisome should not significantly impact their insertion frequency. Therefore, we quantified the insertion frequency of a selectable kanamycin resistance cassette (lacZ::kanR, 1.3 kb) targeted to lacZ. Insertion of lacZ::kanR (4,15) in 3 replicates yielded recombination frequencies of 1.81E-04 ± 6.24E-05 in EcNR2 versus 1.28E-04 ± 4.52E-05 in EcNR2.dnaG.Q576A (p = 0.30 by unpaired t-test). Therefore, modifying DnaG primase function does not appear to significantly affect λ Red-mediated gene insertion.
DISCUSSION
MAGE is a powerful technique that can be used to generate combinatorial sets of designed mutations in a population (4) and/or modify hundreds of alleles in a single strain (5). We have engineered optimized strains for multiplex genome engineering in an effort to streamline extensive genome editing. Previously, we showed that converting a selectable allele in the vicinity of multiple non-selectable alleles enriches the candidate pool for highly modified clones (9). Additionally, we demonstrated that exonucleases are capable of degrading single-stranded MAGE oligos even when these oligos are protected using phosphorothioate bonds (Mosberg, J.A., Gregg, C.J., et al., in review). Inactivating ExoI, ExoVII, ExoX, RecJ and λExo significantly enhanced multiplex AR frequencies (Mosberg, J.A., Gregg, C.J., et al., in review). This showed that intracellular MAGE oligos are a limiting factor in Redβ-mediated recombination. In the current work, we demonstrate that available ssDNA on the lagging strand of the replication fork is another limiting factor that can be increased by disrupting the interaction between DnaG primase and DnaB helicase on the replisome.
In order to increase ssDNA on the lagging strand of the replication fork, we introduced two known mutations in primase (DnaG)—K580A and Q576A. These mutations have been shown in vitro to increase OF size by interrupting the primase–helicase interaction on the replisome (13). Based on the measurements of Tougu et al. (13), we estimate that the K580A mutation increases OF length by ∼1.5-fold and the Q576A mutation increases OF length by ∼8-fold (Supplementary Table S2). EcNR2.dnaG.K580A and EcNR2.dnaG.Q576A exhibited significant increases in the mean number of alleles converted and decreases in the proportion of clones with zero non-selectable alleles converted. Furthermore, the strongest enhancement was observed in EcNR2. dnaG.Q576A (the variant with the longest OFs of the strains reported herein), with an intermediate enhancement observed in EcNR2.dnaG.K580A (the variant with intermediate-sized OFs). This relationship between recombination frequency and OF length further supports the model in which Redβ mediates annealing at the lagging strand of the replication fork (3,15,16,19), and our hypothesis that ssDNA on the lagging strand of the replication fork is a limiting factor during this process. With this in mind, we unsuccessfully attempted to generate a DnaG Q576A/K580A double mutant, suggesting that such an extensive manipulation of the DnaG C-terminal helicase interaction domain (24) was lethal.
Our results indicate that intracellular concentrations of MAGE oligos and the accessibility of their genomic targets are both limiting. To further increase the number of simultaneous mutations that can be generated by CoS-MAGE, it is helpful to understand whether the AR frequency is limited predominantly by the number of oligos that enter the cytoplasm, or whether kinetics are also relevant. Since a maximum of 9 ARs was observed for the 10-oligo sets compared to a maximum of just 12 ARs for the 20-oligo set, oligo uptake may be limiting. However, the fact that primase modulation—in addition to nuclease inactivation—enhances AR frequency underscores the kinetic constraints regarding Redβ-mediated annealing. Each missed opportunity to anneal (i) increases the number of wt alleles in the population due to replication and (ii) decreases the number of MAGE oligos available, via dilution (cell division) and degradation (nucleases). Increasing the concentration of each reactant (i.e. intracellular oligos and accessible genomic targets) would increase the kinetics of annealing. Therefore, the number of intracellular oligos may limit the maximum number of possible mutations, but kinetics appear to be a significant force limiting the population-wide AR frequency average.
Interestingly, the nuclease-deficient Nuc5- strain (Mosberg, J.A., Gregg, C.J., et al., in review) performed statistically similarly to the EcNR2.dnaG.Q576A strain for Sets 1 and 2, whereas EcNR2.dnaG.Q576A strongly outperformed the nuclease-deficient strain for Set 3 (Tables 1 and 2; see also Mosberg, J.A., Gregg, C.J., et al., in review). While oligo design parameters such as type of designed mutation (4), oligo length (4), oligo secondary structure (4) and off-target genomic homology (5) are major determinants of AR frequency, our results highlight the relevance of genomic context. This has previously been difficult to demonstrate, but is apparent from the discrepancy in performance of the same oligo sets tested in our Nuc5- (Mosberg, J.A., Gregg, C.J., et al., in review), EcNR2.dnaG.Q576A, and Nuc5-.dnaG.Q576A strains. For example, different regions may have different replication fork speed or priming efficiency. These factors could locally modulate OF length, thus affecting Redβ-mediated AR frequency (although replication fork speed did not appear to be a major factor in vitro (13)). Therefore, increasing the region that must be replicated by a single OF may profoundly increase AR frequency for oligos targeting such regions. Alternatively, certain oligos may be more susceptible to nuclease degradation, so removing the responsible nucleases would disproportionately improve AR frequency for such oligos. With this in mind, we tested whether combining primase modification and nuclease removal would enhance MAGE performance more than either strategy used individually. Indeed, Nuc5-.dnaG.Q576A consistently performed the best (Figures 3 and 5) of all tested strains. Therefore, the two disparate strategies can be combined for a larger and more robust MAGE enhancement.
To explore the extent to which OF localization impacts CoS-MAGE performance, we tested whether placing 10 oligos within a single putative OF would yield subpopulations of unmodified (few alleles converted) and ‘jackpot’ (most alleles converted) recombinants. However, CoS-MAGE using the densely clustered lacZ oligos (Figure 4) produced a similar AR distribution to the ones observed for Sets 1–3 (Figure 3), which target regions of the genome spanning several putative OFs. Since mutations within a single putative OF behaved similarly to mutations spread across many OFs, nascent OF placement does not appear to be a critical determinant of multiplex AR frequency. A number of hypotheses could explain why the expected ‘jackpots’ are not observed. Most likely, MAGE oligos are limiting due to degradation and/or lack of uptake. Thus, it is possible that most cells lack some of the oligos necessary for generating a majority of the desired mutations. Additionally, OF extension may occur too fast for all of the MAGE oligos to anneal before the OF occludes their targets. Still another explanation could be that ssDNA binding proteins occlude ssDNA on portions of the lagging strand, rendering these regions non-accessible for Redβ-mediated annealing. Finally, it is also possible that several MAGE oligos annealed within a single OF could destabilize lagging strand synthesis, leading to selection against highly modified ‘jackpot’ clones. Indeed, Corn and Berger (25) hypothesize that DnaG primase has evolved to only initiate synthesis when multiple DnaG units are bound to DnaB Helicase, as OF synthesis away from the replisome could be detrimental. Since polIIIlag dissociates from the replisome after completing an OF (26), the rapid and repeated dissociation of polIIIlag caused by multiple nearby MAGE oligos could inhibit lagging strand synthesis as the replisome proceeds beyond the target region. In the absence of the rest of the replisome, a cytosolic PolIII holoenzyme alone can synthesize 1.4 kb on a ssDNA template primed by 30 nt DNA oligos (27), but this activity is considerably diminished compared to that of an intact replisome. Therefore, if OFs are not completed when the replisome is in close proximity, this could result in persisting ssDNA that could destabilize the chromosome and/or cause lesions when the next replication fork passes through.
We also investigated whether targeting a greater number of alleles would increase the resulting number of conversions in our enhanced strains (Figure 5). Although the mean number of alleles converted (mean ± std. error of the mean) increased from 2.59 ± 0.19 with 10-oligo Set 3 to 4.50 ± 0.30 (1.74-fold) with 20-oligo Sets 3 + 3X for Nuc5-.dnaG.Q576A, the mean number of alleles converted for EcNR2.dnaG.Q576A only increased from 2.54 to 2.96 (1.17-fold). The superior enhancement for the nuclease-depleted Nuc5-.dnaG.Q576A strain suggests that the intracellular oligo concentration is a limiting factor for highly multiplexed MAGE (>10 alleles targeted). Therefore, enhancing DNA uptake and/or preservation may be a fruitful means of further improving MAGE. However, the greater multiplexibility of Nuc5-.dnaG.Q576A could also be due to the 10 new Set 3X oligos being more responsive to decreased exonuclease degradation than to increased lagging strand ssDNA availability. Additionally, there may be other limiting factors such as insufficient Redβ or unidentified host proteins. Although there is no known precedent for limiting amounts of λ Red proteins during recombination (28), our novel ability to attain 12 simultaneous non-selectable ARs (Figure 5A) shows that our improved strains are in uncharted territory for probing the limits of λ Red recombination.
Given that DnaG primase acts solely on the lagging strand of the replication fork, we expected that the primase modifications would only enhance lagging strand recombination. Therefore, the performance of leading-targeting CoS-MAGE in our strains was surprising, as EcNR2.dnaG.Q576A significantly outperformed EcNR2 (*p = 0.018). Furthermore, while the total number of tolC+ recombinants was far smaller (∼102-fold) for leading-targeting CoS-MAGE, the AR frequency of non-selectable alleles in these recombinants was still quite impressive, especially in extremely close proximity to the selectable allele. This suggests that one leading strand recombination event strongly correlates with multiple additional recombinations. Two possible explanations for the superior performance of EcNR2.dnaG.Q576A in leading-targeting CoS-MAGE are that (i) an impaired primase–helicase interaction increases accessible leading strand ssDNA or (ii) infrequent Redβ-mediated strand invasion initiates a new replication fork that travels in the opposite direction and swaps which strand is the lagging strand.
There is strong support for primase function affecting the dynamics of replication on both the lagging and leading strands (26,27,29). Lia et al. (26) observed phases in which OF synthesis is faster than helicase progression at the replication fork, alternating with phases in which helicase progression outstrips the rate of OF synthesis by PolIIIlag. These results demonstrate that DnaB-PolIIIlead does not progress at the same instantaneous speed as PolIIIlag (26). Furthermore, Yao et al. (29) showed that the velocity of leading-strand synthesis decreases during lagging strand synthesis, while its processivity increases. Perhaps less frequent primase–helicase binding leads to transient asynchrony of the helicase and PolIIIlead. Given that PolIII tends to release from the replication fork more readily than does DnaB helicase (29), a transiently increased fork rate and decreased PolIIIlead processivity could exacerbate such an asynchrony, creating a leading strand trombone loop similar to those observed during lagging strand synthesis. However, the effects of lagging strand synthesis on leading strand replication have been historically difficult to demonstrate in experiments beyond single-molecule studies (29). Given that instantaneous changes in replication dynamics appear to occur on timescales relevant to Redβ-bound oligo recombination, it is conceivable that snapshots of exposed ssDNA on the leading strand template could be recorded by measuring rates of leading-targeting AR. Single-molecule analysis of the Q576A variant could explore this hypothesis.
Alternatively, Redβ has been reported to facilitate strand invasion in vitro (30). If this also occurs in vivo, such strand invasion would produce a D-Loop that could act as a new origin of replication (31). Therefore, invasion of one leading–targeting MAGE oligo could initiate a replication fork traveling in the opposite direction. In the reverse orientation, the leading strand would become the lagging strand so that upstream oligos would become lagging targeting and much more likely to recombine. This could lead to the highly modified clones that we observed during leading-targeting CoS-MAGE (Supplementary Figure S1). If this is the case, the non-selectable alleles would be upstream of the tolC selectable marker. Since co-selection is most effective downstream of the selectable marker (9), this may explain why co-selection enhancements decay rapidly with distance on the leading strand.
In this manuscript, we have identified available ssDNA on the lagging strand of the replication fork as a limiting factor in multiplex genome engineering. Compared with a standard recombineering strain (EcNR2), EcNR2.dnaG.Q576A displays on average 62% more alleles converted per clone, 239% more clones with 5 or more allele conversions and 38% fewer clones with 0 allele conversions in a given round of CoS-MAGE with 10 synthetic oligos (Table 2). We used this strategy to build on our recent advances (Mosberg, J.A., Gregg, C.J., et al. in review), generating the Nuc5-.dnaG.Q576A strain, which has extended OFs and also lacks five potent exonucleases. These modifications exploited two distinct mechanisms that together increased the robustness and potency of CoS-MAGE, enabling an average of 4.50 and a maximum of 12 ARs in single cells exposed to a pool of 20 different synthetic AR oligos (Figure 5). Additionally, 48% of recombinants had 5 or more ARs and only 8% had 0 modified non-selectable alleles. Furthermore, in a given round of CoS-MAGE with 10 synthetic oligos, Nuc5-.dnaG.Q576A displays on average 111% more alleles converted per clone, 527% more clones with 5 or more allele conversions and 71% fewer clones with 0 allele conversions in comparison with EcNR2 (Table 2). This improvement in MAGE performance will be highly valuable for increasing the diversity explored during the directed evolution of biosynthetic pathways (4) and for enabling the rapid generation of desired genotypes involving tens to hundreds of ARs (5).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1 and 2 and Supplementary Reference [32].
FUNDING
Department of Energy Genomes to Life Center [DE-FG02-02ER63445]; US Department of Defense NDSEG Fellowship (to M.J.L.); National Institutes of Health [P50 HG005550 to G.C.W., in part]. Funding for open access charge: Department of Energy Genomes to Life Center [DE-FG02-02ER63445].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank John Aach, Harris Wang, Farren Isaacs and Nikolai Eroshenko for helpful discussions, and Sara Vassallo for technical assistance.
REFERENCES
- 1.Smith HO, Hutchison CA, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phi X174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 3.Ellis HM, Yu DG, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl Acad. Sci. USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li XT, Costantino N, Lu LY, Liu DP, Watt RM, Cheah KS, Court DL, Huang JD. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 2003;31:6674–6687. doi: 10.1093/nar/gkg844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HH, Xu G, Vonner AJ, Church GM. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–7347. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr PA, Wang HH, Sterling B, Isaacs FJ, Lajoie MJ, Xu G, Church GM, Jacobson JM. Enhanced multiplex genome engineering through cooperative oligonucleotide co-selection. Nucleic Acids Res. 2012:1–11. doi: 10.1093/nar/gks455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang HH, Kim H, Cong L, Jeong J, Bang D, Church GM. Genome-scale promoter engineering by coselection MAGE. Nat. Meth. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J. Biol. Chem. 1992;267:4045–4053. [PubMed] [Google Scholar]
- 12.Tougu K, Marians KJ. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J. Biol. Chem. 1996;271:21391–21397. doi: 10.1074/jbc.271.35.21391. [DOI] [PubMed] [Google Scholar]
- 13.Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 14.DeVito JA. Recombineering with tolC as a selectable/counter-selectable marker: remodeling the rRNA operons of Escherichia coli. Nucleic Acids Res. 2008;36:e4. doi: 10.1093/nar/gkm1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosberg JA, Lajoie MJ, Church GM. Lambda Red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 2010;186:791–799. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maresca M, Erler A, Fu J, Friedrich A, Zhang YM, Stewart AF. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol. Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HH, Church GM. Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods Enzymol. 2011;498:409–426. doi: 10.1016/B978-0-12-385120-8.00018-8. [DOI] [PubMed] [Google Scholar]
- 18.Jekel JF, Katz DL, Elmore JG. Epidemiology, Biostatistics, & Preventative Medicine. 2 edn. Philadelphia: W.B. Saunders; 2001. [Google Scholar]
- 19.Erler A, Wegmann S, Elie-Caille C, Bradshaw CR, Maresca M, Seidel R, Habermann B, Muller DJ, Stewart AF. Conformational adaptability of Red beta during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol. 2009;391:586–598. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 20.Posfai G, Plunkett G, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 21.Blomfield IC, Vaughn V, Rest RF, Eisenstein BI. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 22.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tashiro Y, Fukutomi H, Terakubo K, Saito K, Umeno D. A nucleoside kinase as a dual selector for genetic switches and circuits. Nucleic Acids Res. 2011;39:e12. doi: 10.1093/nar/gkq1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakley AJ, Loscha KV, Schaeffer PM, Liepinsh E, Pintacuda G, Wilce MCJ, Otting G, Dixon NE. Crystal and solution structures of the helicase-binding Domain of Escherichia coli primase. J. Biol. Chem. 2005;280:11495–11504. doi: 10.1074/jbc.M412645200. [DOI] [PubMed] [Google Scholar]
- 25.Corn JE, Berger JM. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 2006;34:4082–4088. doi: 10.1093/nar/gkl363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lia G, Michel B, Allemand J-F. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science. 2012;335:328–331. doi: 10.1126/science.1210400. [DOI] [PubMed] [Google Scholar]
- 27.Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama M, Ohara O. Improvement of recombination efficiency by mutation of Red proteins. Biotechniques. 2005;38:917–924. doi: 10.2144/05386RR02. [DOI] [PubMed] [Google Scholar]
- 29.Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc. Natl Acad. Sci. 2009;106:13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rybalchenko N, Golub EI, Bi B, Radding CM. Strand invasion promoted by recombination protein β of coliphage λ. Proc. Natl Acad. Sci. USA. 2004;101:17056–17060. doi: 10.1073/pnas.0408046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asai T, Kogoma T. D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J. Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc. Natl Acad. Sci. 1968;59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.