Abstract
MicroRNA (miRNA) biogenesis is tightly regulated by numerous proteins. Among them, Dicer is required for the processing of the precursor (pre-)miRNAs into the mature miRNA. Despite its critical function, the mechanisms that regulate Dicer expression are not well understood. Here we report that the RNA-binding protein (RBP) AUF1 (AU-binding factor 1) associates with the endogenous DICER1 mRNA and can interact with several segments of DICER1 mRNA within the coding region (CR) and the 3′-untranslated region (UTR). Through these interactions, AUF1 lowered DICER1 mRNA stability, since silencing AUF1 lengthened DICER1 mRNA half-life and increased Dicer expression, while overexpressing AUF1 lowered DICER1 mRNA and Dicer protein levels. Given that Dicer is necessary for the synthesis of mature miRNAs, the lowering of Dicer levels by AUF1 diminished the levels of miRNAs tested, but not the levels of the corresponding pre-miRNAs. In summary, AUF1 suppresses miRNA production by reducing Dicer production.
INTRODUCTION
In mammalian cells, post-transcriptional processes are regulated by two main types of factors, RNA-binding proteins (RBPs) and non-coding RNAs. RBPs govern pre-mRNA splicing as well as mRNA processing, transport, storage, stability and translation (1–3). Through their influence on protein expression patterns, RBPs regulate cellular processes including differentiation, survival, senescence, and the responses to stress and immune signals (4–8). Among the large family of RBPs, translation and turnover regulatory(TTR)-RBPs (9) associate primarily with 5′ and 3′ untranslated regions (UTRs) of target mRNAs and modulate their cytoplasmic fate. TTR-RBPs often interact with AU-rich elements (AREs), but they may also have affinity for other sequences [e.g. U-, GU-, C- or G-rich sequences (10–14)], and they mainly regulate the turnover and the translation of target mRNAs. For example, Hu/elav proteins (HuR, HuB, HuC and HuD) and nucleolin enhance mRNA stability and modulate translation, whereas T-cell-restricted intracellular antigen-1 (TIA-1) and TIA-1-related protein TIAR typically suppress translation but can also promote the stability of a subset of mRNAs (10,15–18). In contrast, the RBPs KH-homology splicing-regulatory protein (KSRP), tristetraprolin (TTP), butyrate response factor-1 (BRF1) and CUG triplet RNA-binding protein 1 (CUG-BP) generally promote target mRNA decay (14,19–21).
The TTR-RBP AU-binding factor 1 (AUF1), also known as heterogeneous nuclear ribonucleoprotein D (hnRNPD) promotes the decay of many target mRNAs, but it was also reported to enhance the stability and translation of some target transcripts (22–25). Alternative splicing of the AUF1 pre-mRNA gives rise to four isoforms (p37, p40, p42 and p45); although all of them contain two RNA-recognition motifs (RRMs), they each exhibit different affinity for target transcripts and have distinct influence on their post-transcriptional fate (26). The promotion of mRNA degradation by AUF1 was linked to the AUF1-mediated recruitment of mRNAs to the exosome and the proteasome, multiprotein complexes specialized in 3′→5′ exoribonuclease activity and proteolysis, respectively (27,28). AUF1 target mRNAs encode proteins implicated in processes such as cell-cycle progression (e.g. cyclin D1, p21, c-Myc), apoptosis (e.g. Bcl-2) and the stress response (e.g., Gadd45α, ATF3) (25,26,29). Additionally, overexpression of AUF1 triggered the development of sarcomas (30) and high AUF1 levels were detected in numerous malignancies, including cancers of the breast, skin, thyroid and liver (reviewed in (25)). Mice lacking AUF1 had an exacerbated inflammatory response, revealing a further role for AUF1 in inflammatory diseases (31).
During recent studies to identify AUF1 target mRNAs en masse (29), we discovered that AUF1 had affinity for DICER1 mRNA, the transcript that encodes the protein Dicer. A cytoplasmic RNase III-type endoribonuclease, Dicer binds short precursor (pre)-microRNAs (∼70-nt long) and assists with their processing into mature microRNAs (miRNAs, ∼22-nt in length) (32). MiRNAs constitute an important class of non-coding (nc)RNAs that regulate gene expression post-transcriptionally. They function most commonly by associating with target mRNAs with partial complementarity, causing reduced stability and/or translation of the target mRNAs. Through its influence on miRNA biosynthesis, Dicer influences cell-cycle progression, senescence, stem cell maintenance and tumorigenesis (33,34). Dicer-null mice showed lethality early in embryonic development due to the depletion of the stem cell population (35). Despite its important roles in cellular homeostasis, the mechanisms that control Dicer expression are virtually unknown. At the transcriptional level, Dicer expression is positively regulated by Tap63 in mice (36) and post-transcriptionally it is negatively regulated by let-7 and miR-103/107 (36–39). Therefore, we investigated the possible effect of AUF1 on Dicer production. After establishing that AUF1 associated with multiple segments of the DICER1 mRNA, including parts of the coding region (CR) and the 3′UTR, we discovered that AUF1 lowered DICER1 mRNA stability and confirmed this finding by studying heterologous reporters. This regulation was further reflected on the inverse correlation in AUF1 and Dicer levels in cancer and normal tissues, with cancer tissues showing relatively higher AUF1 and lower Dicer, whereas in normal tissues AUF1 levels were lower and Dicer levels higher. The AUF1-mediated reduction of Dicer led to the selective decrease in the abundance of numerous miRNAs without parallel declines in the corresponding pre-miRNAs. In summary, AUF1 lowers DICER1 mRNA stability, in turn reducing Dicer abundance and the levels of mature miRNAs.
MATERIALS AND METHODS
Cell culture, transfection, small RNAs and plasmids
HeLa cells were cultured in Dulbecco’s modified essential medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum and antibiotics. HCT116 cells were cultured in McCoy’s 5A medium (Invitrogen) supplemented with 10% fetal bovine serum and antibiotics. Control small interfering RNA (Ctrl siRNA), AUF1 siRNA and Dicer siRNA directed to DICER1 3′UTR were from Qiagen; Dicer siRNA directed to the Dicer CR was from Santa Cruz. Plasmid pEGFP expressed enhanced green fluorescent protein (EGFP); plasmid pEGFP-DICER1(3′), the DICER1 3′UTR reporter construct, was made by inserting cDNA corresponding to the DICER1 3′UTR cDNA into pEGFP-C1 (BD Bioscience); plasmid pcDNA-Dicer (pFRT/TO/FLAG/HA-DEST DICER), spanning only the DICER1 CR but not the DICER1 3′UTR, was from Addgene. All plasmids and siRNAs were transfected with Lipofectamine-RNAiMAX or Lipofectamine-2000 (Invitrogen). When comparing the expression of EGFP reporter constructs, EGFP protein signals were quantified in all lanes, and fold differences in EGFP protein levels in Ctrl siRNA relative to AUF1 siRNA were calculated for each plasmid group; fold differences were subsequently compared between plasmid transfection groups.
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer [10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS and 1 mM dithiothreitol], separated by electrophoresis in SDS-containing polyacrylamide gels, and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). Incubations with primary antibodies to detect Dicer, AUF1, EGFP (Santa Cruz Biotech) or β-actin (Abcam), were followed by incubations with the appropriate secondary antibodies conjugated with horseradish peroxidase (HRP) (GE Healthcare) and by detection using enhanced luminescence (GE Healthcare).
Immunohistochemistry
Immunohistochemistry was performed on tumor and normal tissue arrays (US Biomax, Inc., Rockville, MD). The array slides were subjected to heat-induced epitope retrieval, incubation with primary antibodies [monoclonal anti-Dicer antibody (Abcam) and polyclonal anti-AUF1 antibody (Millipore)] used at 1:2000 dilution. Signals were detected using the LSAB+ system (Dako).
RNA analysis
Total RNA was prepared from whole cells using Trizol (Invitrogen) and from RNP IP samples as explained below. After reverse transcription (RT) using random hexamers and SSII reverse transcriptase (Invitrogen) for mRNA or using QuantiMir RT Kit (System Biosciences) for precursor and mature miRNA (details below) and U6 snRNA, the abundance of transcripts was assessed by real-time, quantitative (q)PCR analysis using the SYBR green PCR master mix (Kapa Biosystems) and gene-specific primer sets (below). RT-qPCR analysis was performed on Applied Biosystems model 7300 and 7900 instruments. To measure the abundance of mature and precursor miRNAs, the QuantiMir detection kit (System Biosciences) was used. Briefly, all cellular RNA was polyadenylated using Poly(A) polymerase at 37°C for 10 min, whereupon the oligo-dT adaptor was added to the reaction and annealing was allowed to proceed for 5 min at 60°C. After RT, mature miRNAs were detected with forward primers that hybridized with the miRNAs. Pre-miRNAs were detected using forward primers that specifically hybridized with the pre-miRNA (but not the mature miRNA). In both cases, a reverse universal primer was used for qPCR amplification.
The forward and reverse primers were TGCACCACCAACTGCTTAGC and GGCATGGACTGTGGTCATGAG for GAPDH mRNA, and TTCCTCACCAATGGGTCCTTT and GCTTCAAGCAGTTCAACCTGAT for DICER1 mRNA. Specific forward primers were CTATACAATCTACTGTCTTTC for miR-let-7a, CTATACAACCTACTGCCTTCCC for miR-let-7b, TAGAGTTACACCCTGGGAGTTA for miR-let-7c, CTATACGACCTGCTGCCTTTCT for miR-let-7d, CTATACGGCCTCCTAGCTTTCC for miR-let-7e, CTATACAATCTATTGCCTTCCC for miR-let-7f, CTGTACAGGCCACTGCCTTGC for miR-let-7g, TTGCATAGTCACAAAAGTGATC for miR-153, TAAGGCACGCGGTGAATGCC for miR-124, TAAAGTGCTGACAGTGCAGAT for miR-106b, AAAAGTGCTTACAGTGCAGGTAG for miR-106a, TACAGTACTGTGATAACTGAA for miR-101, CAAAGTGCTTACAGTGCAGGTAG for miR-17-5p, TAAAGTGCTTATAGTGCAGGTAG for miR-20a, TAGCAGCACGTAAATATTGGCG for miR-16, TACCCTGTAGATCCGAATTTGTG for miR-10a, AAAGTGCATCCTTTTAGAGTGT for miR-519a, CAAAAATCTCAATTACTTTTGC for miR-548c-3p, GTCCAGTTTTCCCAGGAATCCCT for miR-145, TGAAACATACACGGGAAACCTC for miR-494, TTCCCTTTGTCATCCTATGCCT for miR-203 and CAGTGCAATGTTAAAAGGGCAT for miR-130a, CAAAAACCACAGTTTCTTTTGC for miR-548d-3p, CATCTTACCGGACAGTGCTGGA for miR-200a, for AGGCAGTGTAGTTAGCTGATTGC for miR-34c-5p, TGGCTCAGTTCAGCAGGAACAG for miR-24 and AAAGTGCTTCCTTTTAGAGGGT for miR-520c-3p. The precursors of let-7b, c and e were detected using the following primers, respectively; CTATACAACCTACTGCCTTCCCTG, CAACCTTCTAGCTTTCCTTGGAGC and TACGGCCTCCTAGCTTTCCCCAGG. Small nuclear RNA U6, primer sequence CACCACGTTTATACGCCGGTG, was used for normalization.
For mRNA stability analysis, transfected cells were treated with 2 µg/ml actinomycin D and the levels of DICER1 mRNA in each transfection group were measured by RT-qPCR from total RNA using DICER1-specific primer pairs (see above). GAPDH mRNA was measured as a control transcript encoding a housekeeping protein. The levels of DICER1 and GAPDH mRNAs were normalized to 18S rRNA levels and plotted as the percentage of mRNA remaining compared with the levels of the same mRNA at time zero.
Northern blot analysis
Northern blot analysis was performed as described previously (40,41). Briefly, total RNA was prepared by using miRNeasy (Qiagen) and separated using precast TBE-Urea gel (Invitrogen). After transfer, Nylon membranes (iBlot, Invitrogen) were UV-crosslinked and hybridized with DNA oligonucleotides complementary to miRNA or U6 that had been end-labeled with 32P-ATP. Images were acquired with a Typhoon Scanner (GE Healthcare). The probes used were AACCACACAACCTACTACCTCA to detect let-7, CTGTTCCTGCTGAACTGAGCCA to detect miR-24 and AAAATATGGAACGCTTCACGA to detect U6.
Immunoprecipitation of RNP complexes
Endogenous mRNA-protein RNPs (ribonucleoproteins) were precipitated from HeLa cytoplasmic lysates. Lysates were prepared in a buffer containing 10 mM Hepes, 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% IGEPAL, 2 mM DTT, 50 U/ml RNase out and protease inhibitors. Lysates were incubated (1 h, 4°C) with a suspension of protein-A Sepharose beads precoated with anti-AUF1or rabbit IgG. Beads were washed with NT2 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% IGEPAL) and then incubated with NT2 buffer containing RNase-free DNase I (15 min, 30°C), washed with NT2 buffer and further incubated in NT2 buffer containing SDS and Proteinase K to digest proteins bound to the beads. RNA was extracted using phenol and chloroform, precipitated in the presence of glycoblue and used for further analysis.
Biotin pulldown analysis
PCR fragments containing the T7 RNA polymerase promoter sequence were used as templates for in vitro transcription as previously described (10). Biotinylated transcripts were incubated with cytoplasmic lysates (100 μg lysate, 3 μg biotinylated RNA) for 30 min at room temperature, and complexes were isolated with streptavidin-coated magnetic Dynabeads and analysed using western blot analysis to detect AUF1.
Primers used to prepare biotinylated DICER1 mRNA transcripts (NM_177 438.2) are listed below. After purification of the template PCR products, biotinylated transcripts were synthesized using MaxiScript T7 kit (Ambion) and whole-cell lysates (200 μg per sample) were incubated with 3 μg of purified biotinylated transcripts for 30 min at room temperature, whereupon complexes were isolated with Streptavidin-coupled Dynabeads (Invitrogen). AUF1 present in the pulldown material was assessed by western blot analysis as described (10). To synthesize biotinylated transcripts, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence [(T7), CCAAGCTTCTAATACGACTCACTATAGGGAGA]. Primers used to prepare templates were as follows: (T7)CGGAGGCGCGGCGCAGGCT and TCATCCAGTGTTTCTTTCATTGC for 5′UTR, (T7)ATGAAAAGCCCTGCTTTGCA and CCACTCAACGCTTTCAAACT for CR-A, (T7)TAAACCATATGAGCGACAGC and AAGTTGAGTTCATCAGGTAAAG for CR-B, (T7)GAATGGTTTTAACTACACCTT and CTGTAAGATCTGCTGAAACT for CR-C, (T7)CCTGGTAAGCTCCACGTT and CGCTATGCTTTTGTCAGCAAT for CR-D, (T7)CTTGCACACTGAGCAGTG and TCAGCTATTGGGAACCTGA for CR-E, (T7)AACCGCTTTTTAAAATTCAAAAC and CTAAGGGTAAAGGTGCTG for 3′UTR-A, (T7)TACTTATTTAAGAAGCAAAACAC and CAGGAATCAAGAGAATCC for 3′UTR-B, (T7)CCCAGTGTTACGGGATT and ATTTTAAAAGACAATTACAGGAG for 3′UTR-C, (T7)TCTAACACTCCTGTAATTGTC and GAACAGACGATAACTTTATTGG for 3′UTR-D.
RESULTS
AUF1 interacts with Dicer mRNA
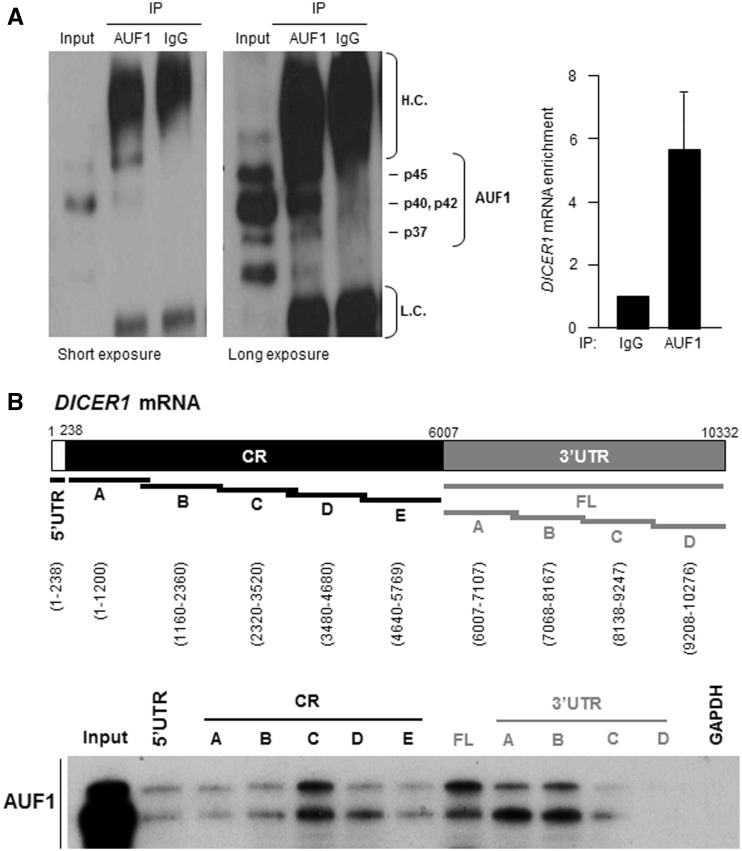
To directly test if AUF1 interacts with DICER1 mRNA, human cervical carcinoma (HeLa) cell lysates were used in ribonucleoprotein (RNP) immunoprecipitation (RIP) assays using an anti-AUF1 antibody under conditions that preserved RNP integrity. As seen in Figure 1A, p40 and p42 are typically the most abundant AUF1 isoforms and run as a single middle band; long and short exposures of the AUF1 western blot analysis are shown, in order to visualize all AUF1 isoforms present in the IP reaction. The RNA present in the IP was isolated and subjected to RT and quantitative real-time (q)PCR analysis to monitor the levels of DICER1 mRNA in the AUF1 IP relative to the control (IgG) IP. As shown in Figure 1A (graph), DICER1 mRNA was enriched more than 5-fold in AUF1 IP samples compared with IgG IP samples, indicating that DICER1 mRNA is a part of AUF1 RNP complexes.
Figure 1.
AUF1 binds DICER1 mRNA. (A) Left, AUF1 was detected by IP followed by western blot analysis. The four AUF1 isoforms are indicated; p40 and p42 generally migrate together. H.C., immunoglobulin heavy chain; L.C., immunoglobulin light chain. Long and short exposures of the western blot are provided. Right, RNP IP analysis of AUF1 interaction with DICER1 mRNA. After IP of HeLa cell lysates, DICER1 mRNA was detected by RT-qPCR analysis and its levels were compared with those present in control IgG IP. The levels of GAPDH mRNA in each IP group were used to normalize for sample input. Data (means and standard deviation, S.D.) are representative of three independent experiments. (B) Top, schematic of the DICER1 mRNA, including the 5′UTR, CR and 3′UTR; the biotinylated RNAs synthesized for use in biotin pulldown analysis are shown as black or gray lines, and the nucleotide positions amplified are indicated. Bottom, biotinylated RNA was incubated with HeLa cell lysates and the interaction of AUF1 with the biotinylated RNAs (DICER1 mRNA segments and housekeeping control GAPDH 3′UTR) was assessed by western blot analysis. Input, 5 μg of HeLa whole-cell lysate.
We also studied if endogenous AUF1 was capable of binding different recombinant fragments of DICER1 mRNA by using the biotin pulldown assay. Biotinylated RNAs spanning the DICER1 5′UTR, CR and 3′UTR were synthesized (Figure 1B, top) and their interaction with AUF1 was studied after incubating them with HeLa cytoplasmic lysates and using streptavidin-coated beads to pulldown the resulting RNP complexes. By western blot analysis to identify AUF1 present in the complex, we did not detect any association between AUF1 and a negative control (GAPDH RNA) which does not bind AUF1. AUF1 showed only modest association with biotinylated RNAs containing DICER1 5′UTR; of the five segments spanning the DICER1 CR, AUF1 showed stronger association with fragment C. The full-length 3′UTR of DICER1 mRNA (FL) was divided into four fragments; two of these (A and B) showed more interaction with AUF1 than did fragments C and D (Figure 1B, bottom). Earlier en masse identification of AUF1 targets (29) predicted the existence of four putative AUF1 sites in the 3′UTR of the DICER1 mRNA [at positions 7272-7318 (within fragment B), 8366-8398, 8563-8595 (both within fragment C), and 9780-9812 (within fragment D) in variant NM_177438]. However, binding instead mapped primarily to fragments A and B, and only modest binding to C, but there was no binding to fragment D.
These findings indicate that AUF1 associates with the endogenous DICER1 mRNA. In addition, as seen for other TTR-RBPs, which can interact with multiple segments of a target mRNA, including the CR and 3′UTR (42,43), AUF1 was capable of interacting with different parts of the DICER1 mRNA.
AUF1 regulates Dicer expression by reducing DICER1 mRNA stability
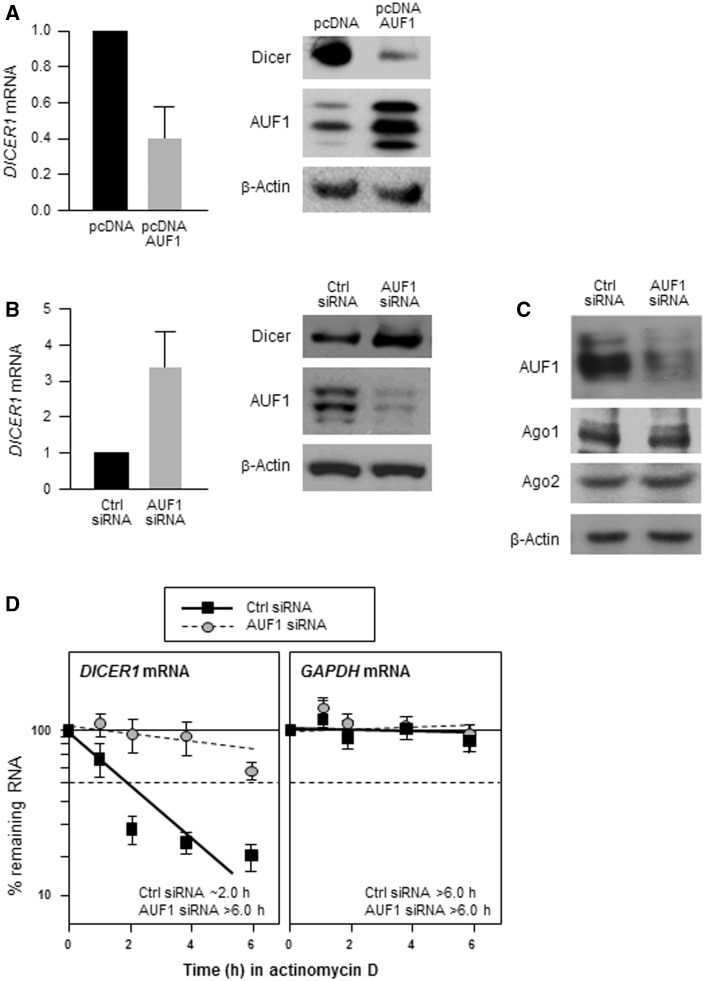
We tested the functional consequences of the association between AUF1 and DICER1 mRNA by modulating AUF1 abundance in HeLa cells and monitoring its influence on Dicer expression. Overexpression of all four AUF1 isoforms significantly decreased the levels of endogenous Dicer protein and DICER1 mRNA (Figure 2A). Conversely, silencing AUF1 by using specific AUF1-directed small interfering (si)RNA, markedly increased Dicer protein and DICER1 mRNA levels compared with what was seen in control (Ctrl siRNA) transfected cells (Figure 2B), although AUF1 silencing did not affect the levels of two other proteins implicated in miRNA function, Argonaute (Ago)1 or Ago2 (Figure 2C). These data support the notion that AUF1 suppresses Dicer expression by lowering DICER1 mRNA levels.
Figure 2.
AUF1 decreases Dicer expression. (A) Forty-eight hours after transfection with a control plasmid (pcDNA) or with plasmids expressing each of the four AUF1 isoforms, the levels of AUF1, Dicer and β-actin were assessed by western blot analysis (left) and the levels of DICER1 mRNA by RT-qPCR (right). (B) Forty-eight hours after transfecting HeLa cells with either Ctrl siRNA or AUF1-directed siRNA, lysates were prepared to assess the levels of AUF1, Dicer and loading control β-actin by western blot analysis (left) and DICER1 mRNA and normalization transcript GAPDH mRNA by RT-qPCR (right). (C) Western blot analysis of the levels of AUF1, Ago1, Ago2 and loading control β-actin in HeLa cells processed as explained in (B). (D) Forty-eight hours after transfection as described in panel (B), HeLa cells were treated with actinomycin D, and RNA was collected at the times indicated. RT-qPCR analysis of the levels of DICER1 and GAPDH mRNAs, after normalization to 18S rRNA levels, was used to determine the half-life of each mRNA, defined as the time needed to reach 50% of its original abundance at time 0 h (dashed line). Data in (A, B and D) are the means (±S.D.) of three independent experiments.
Given that modulating AUF1 changed DICER1 mRNA abundance (Figure 2A and B) and that a prominent function of AUF1 is to promote target mRNA decay, we investigated if AUF1 reduced DICER1 mRNA stability. Forty-eight hours after transfection of HeLa cells with Ctrl or AUF1-directed siRNAs, the cells were treated with actinomycin D to block de novo transcription and the half-life of DICER1 mRNA was measured by studying the rate of DICER1 mRNA clearance. As shown, DICER1 mRNA was markedly more stable in AUF1-silenced cells, with half-lives increasing from ∼2 h to >6 h; in contrast, the half-life of the housekeeping GAPDH mRNA was not influenced by AUF1 silencing (Figure 2D). In sum, AUF1 promotes the degradation of DICER1 mRNA.
The AUF1-mediated DICER1 mRNA decay elements reside within both the DICER1 CR and 3′UTR
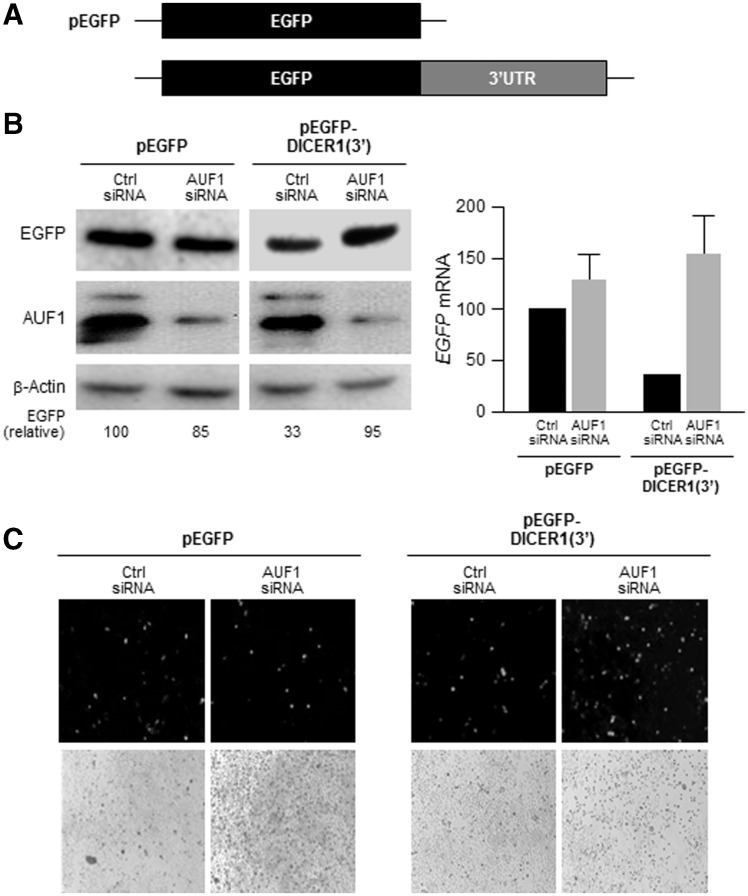
To investigate if AUF1 regulates DICER1 mRNA stability through the region where it binds more extensively (DICER1 3′UTR), we prepared the heterologous reporter construct pEGFP-DICER1(3′) (Figure 3A). Expression of the heterologous reporter protein EGFP from EGFP mRNA (encoded by the CR of the parent control vector, pEGFP) was compared to EGFP expressed from CR of the chimeric EGFP-DICER1(3′) mRNA. By western blot analysis, EGFP protein levels expressed from pEGFP were unaltered by AUF1 silencing; in contrast, the levels of EGFP protein expressed from pEGFP-DICER1(3′) were substantially higher after silencing AUF1 (Figure 3B left). These changes in EGFP protein abundance were mirrored by changes in the expression of encoding transcripts [EGFP and EGFP-DICER(3′) mRNAs, Figure 3B right] as well as by the green fluorescence of the transfected cultures (Figure 3C). These findings indicate that there are negative regulatory elements within the 3′UTR of DICER1 mRNA and that AUF1 elicits at least part of its repressive actions through this region.
Figure 3.
AUF1 regulates Dicer expression through the DICER1 3′UTR. (A) Schematic of the parent reporter plasmid pEGFP and the pEGFP-DICER1(3′) reporter plasmid bearing the DICER1 3′UTR. (B) HeLa cells were transfected with a reporter construct, together with either Ctrl siRNA or AUF1 siRNA; 48 h after transfection, the levels of EGFP, AUF1 and β-actin were assessed by western blot analysis (left) and the levels of EGFP and EGFP-DICER(3′) mRNAs (normalized to GAPDH mRNA levels and represented as % of EGFP mRNA levels in Ctrl siRNA-trasfected cells) were assessed by RT-qPCR analysis (right). (C) Fluorescence microscopy to visualize GFP in cells transfected as in (B). The data (means and ±S.D.) are representative of three independent experiments.
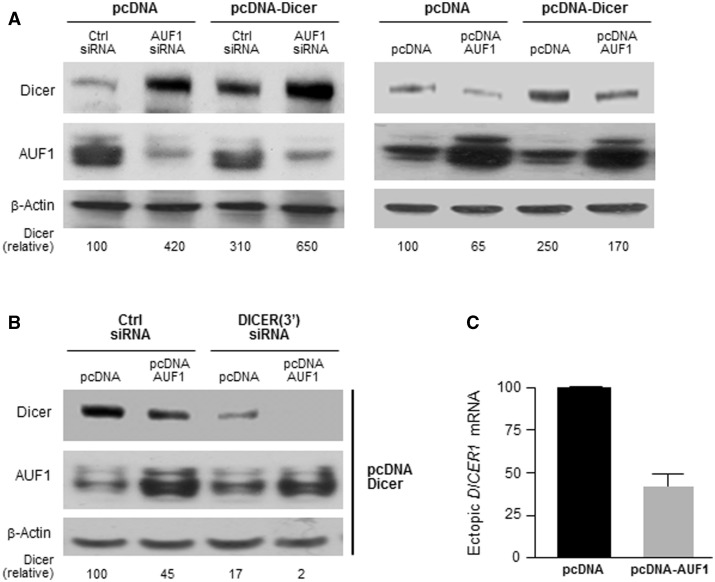
Since AUF1 was also found to interact with the DICER1 CR, we investigated whether AUF1 might elicit its effects via this region as well. In HeLa cells, expression of DICER1 CR by transfection of an expression vector lacking DICER1 3′UTR sequences (pcDNA-Dicer, Materials and Methods) was increased in AUF1-silenced cells (Figure 4A, left), while AUF1 overexpression lowered expression of the ectopic protein (Figure 4A, right). Since HeLa cells express endogenous Dicer, we confirmed the effect of AUF1 on DICER1 CR by silencing endogenous Dicer using siRNAs directed to the 3′UTR of the endogenous DICER1 mRNA [DICER(3′) siRNA]. Under these conditions, AUF1 still had a repressive influence on the expression of the Dicer CR (Figure 4B). That DICER(3′) siRNA effectively reduced endogenous Dicer levels was evidenced by the fact that Dicer signals (endogenous and ectopic) were virtually undetectable (2% of control cells; lane 4 in Figure 4B). If AUF1 had reduced Dicer expression only through the DICER 3′UTR, all of the plasmid-expressed Dicer would have remained detectable in lane 4. These results further supported the notion that AUF1 was capable of repressing Dicer production through the DICER1 CR. To study if AUF1 lowered DICER1 CR levels, we performed RT-qPCR analysis of the ectopically expressed transcript. The protein encoded by pcDNA-Dicer has a hemagglutinin (HA) tag; while we were unable to detect the HA tag by western blot analysis (not shown), the HA tag RNA sequence was present in the mRNA transcribed from the Dicer-expressing plasmid and was readily detectable by RT-qPCR. As indicated in Figure 4C, the levels of the ectopic DICER1 mRNA were lower after AUF1 overexpression, supporting the idea that the interaction of AUF1 with the DICER1 CR also contributed to the destabilization of the endogenous DICER1 mRNA.
Figure 4.
AUF1 regulates Dicer expression through the DICER1 CR. (A) HeLa cells were transfected with the plasmids and siRNAs shown; 48 h later, the levels of Dicer, AUF1 and β-Actin were assessed by western blot analysis. Dicer signals were quantified by densitometry and represented as a percentage of Dicer levels in the control group. (B and C) HeLa cells were transfected with a plasmid that expressed the Dicer CR (pcDNA-Dicer) and the endogenous Dicer was silenced using a DICER1 3′UTR-directed siRNA. The consequences of AUF1 overexpression on Dicer protein expression levels were assessed by western blot analysis (B), and the consequences on DICER1 mRNA levels were measured by RT-qPCR analysis (C). Data in panel (C) represent the means (±SD) of three independent experiments.
Inverse patterns of Dicer and AUF1 expression in a tumor survey
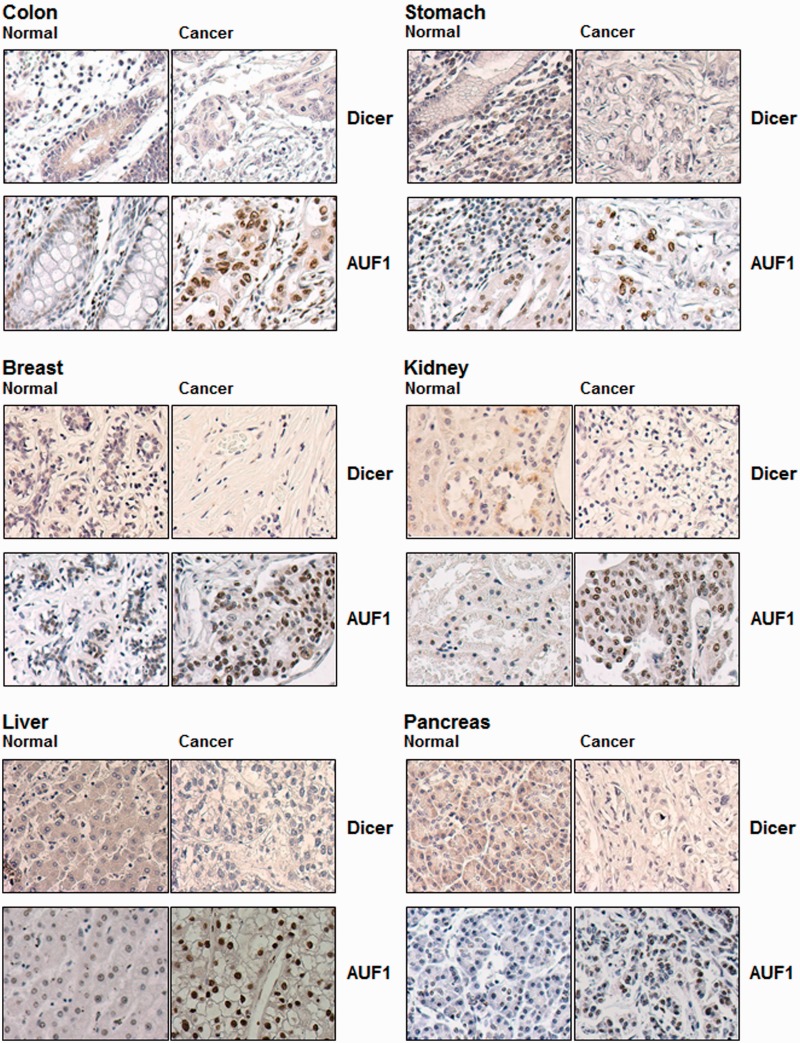
To compare systematically the expression patterns of AUF1 and Dicer, we analysed tissue arrays containing multiple pairs of samples of normal and tumor tissues. Immunohistochemical analysis of AUF1 and Dicer expression revealed a general pattern of Dicer staining that was high in normal tissues and low in cancer tissues; conversely, AUF1 levels were low in normal tissues and high in cancer tissues (Figure 5). As shown for normal and cancer tissues from colon, stomach, breast, kidney, liver and pancreas, Dicer signals were typically cytoplasmic, while AUF1 signals localized in both the nucleus and the cytoplasm.
Figure 5.
AUF1 and Dicer expression in cancer and normal tissues. The expression levels and distribution of AUF1 and Dicer in tissue arrays containing pairs of normal and cancer samples was assessed by immunohistochemistry. Representative pairs are indicated.
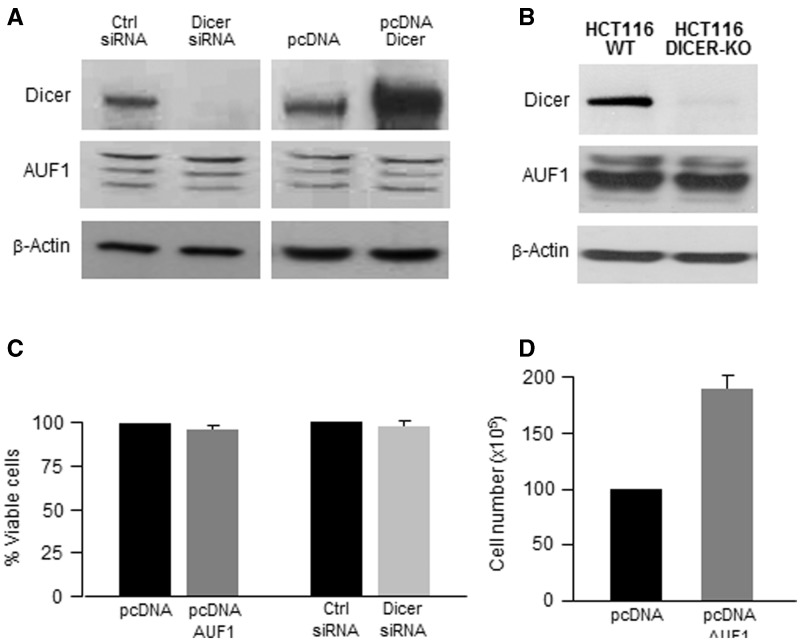
Several lines of evidence (Figures 2 and 4) suggested that upregulation of AUF1 might be partially responsible for the correlative decrease in Dicer levels in cancer tissues (Figure 5). However, the converse regulation was also possible, namely that Dicer might regulate AUF1 expression, perhaps via the numerous (>180) miRNAs predicted to bind the AUF1 3′UTR (not shown). However, as shown in Figure 6A, by 48 h after overexpressing or silencing Dicer, AUF1 abundance remained essentially unchanged. Similarly, in colon carcinoma HCT116 cells that expressed Dicer (HCT116 WT), AUF1 levels were the same as in Dicer-null HCT116 cells (HCT116 DICER-KO; Figure 6B), further supporting the view that AUF1 expression was not likely under direct control by Dicer.
Figure 6.
Effect of Dicer on AUF1 expression and cell survival, effect of AUF1 on cell survival and proliferation. (A) HeLa cells were transfected with the siRNAs and plasmids shown; 48 h later, the levels of Dicer, AUF1 and β-actin were studied by western blot analysis. (B) The levels of AUF1, Dicer and loading control β-Actin were studied in HCT116 colon cancer cells that were either wild-type (WT) or Dicer-null (DICER-KO). (C) The number of viable cells (excluding trypan blue) was calculated in cultures that were prepared as explained in panel (A). (D) The proliferation of HeLa cells was assessed by direct cell counts 48 h after transfecting the plasmids indicated. In (C and D), data represent the means ± S.D. from three independent experiments.
Given the finding that tumor tissues expressed high AUF1 levels and low Dicer levels, and given the above-mentioned links of AUF1 and Dicer to tumorigenesis, we asked whether these proteins might influence cell survival, an important trait of cancer cells. Forty-eight hours after silencing Dicer (Figure 6A) or overexpressing AUF1 (Figure 2A), neither transfection group showed appreciable toxicity, as determined by counting trypan blue-negative cells (Figure 6C). In contrast, AUF1 overexpression robustly promoted cell proliferation (Figure 6D), suggesting that AUF1 could favor the division of cancer cells. The proliferative influence of AUF1 may contribute to the tumorigenic phenotype, although future studies are needed to investigate whether AUF1 influences other cancer traits, including angiogenesis, invasion, metastasis, inhibition of senescence and evasion of immune recognition.
AUF1 influences miRNA levels
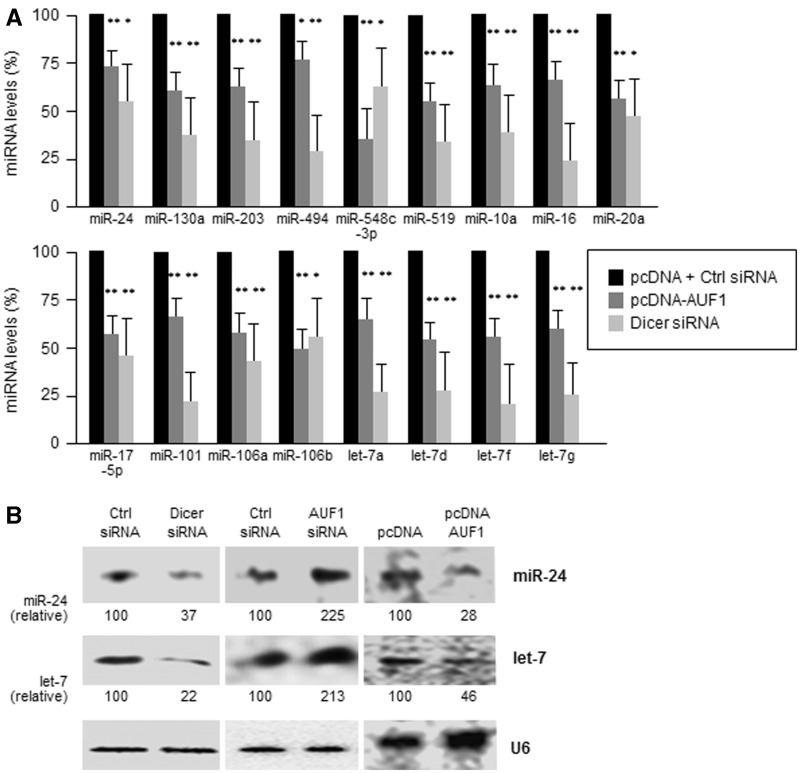
Since Dicer promotes miRNA maturation, we investigated the influence of AUF1-modulated Dicer on the abundance of miRNAs. First, under conditions of reduced Dicer levels achieved by overexpressing AUF1 (Figure 2B) or by siRNA-mediated silencing of Dicer (Figure 6A), we monitored the abundance of several miRNAs. In AUF1-overexpressing HeLa cells (which expressed lower Dicer levels) as well as in cells in which Dicer was silenced, all of the miRNAs tested showed lesser abundance compared with those measured in the control population (Figure 7A). In addition, for abundant endogenous miRNAs (miR-24 and let-7), these differences were confirmed by northern blot analysis. As shown in Figure 7B, miRNA levels were higher when AUF1 was silenced and were lower when AUF1 was overexpressed and when Dicer was silenced. We attempted to study other miRNAs, but they were below the detection level of the assay.
Figure 7.
AUF1 overexpression decreases miRNA levels. (A) Forty-eight hours after transfection with pcDNA, AUF1 overexpression plasmids (described in Figure 2A), and Ctrl or Dicer siRNAs (as in Figure 6A), the levels of several miRNAs were measured using RT-qPCR and normalized to U6. Data in (A) are the means ± S.D. from three independent experiments; *P < 0.05; **P < 0.01. (B) Forty-eight hours after transfection of the plasmids and siRNAs shown, the levels of miR-24, let-7 and loading control U6 were assessed by northern blot analysis. Data in (B) are representative of three independent experiments.
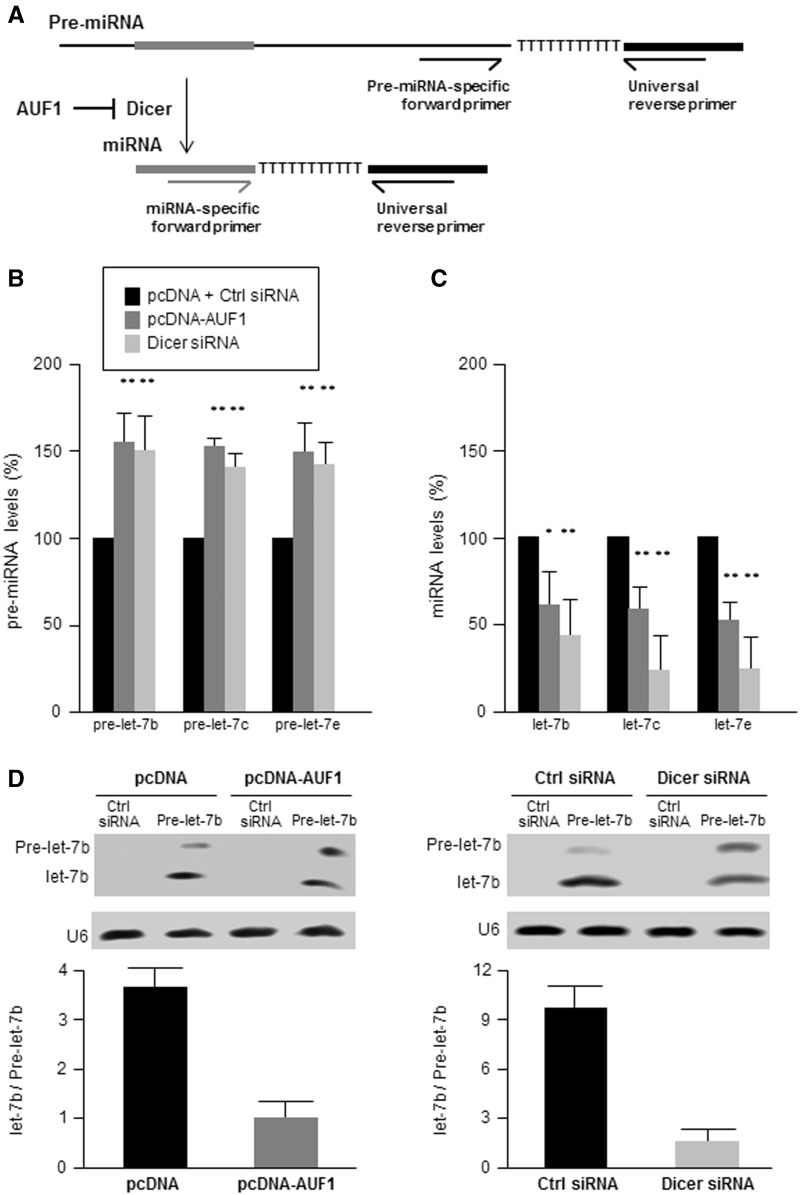
Second, we examined if this reduction reflected an impairment in the processing of pre-miRNA into mature miRNAs due to lower Dicer levels. To this end, we measured the levels of pre-miRNA and mature miRNA for three members of the let-7 family of miRNAs using the strategy outlined in Figure 8A. Briefly, both miRNAs and pre-miRNAs were quantified using the QuantiMiR RT kit (System Biosciences). All cellular RNA was first polyadenylated and an oligo-dT adaptor was added to the reaction. After RT, mature miRNAs were detected by using a forward primer which hybridized with the miRNAs, while precursor miRNAs (pre-miRNAs) were detected using forward primers specifically designed to hybridize within the pre-miRNA but not the mature miRNA. In both cases, a reverse universal primer was used for qPCR amplification (a more detailed description of the method is provided in Supplementary Figure S1). As shown in Figure 8B, the levels of pre-let-7b, pre-let-7c and pre-let-7e were slightly higher in cells with lower levels of Dicer. In contrast, in HeLa cells overexpressing AUF1 or expressing low Dicer levels, mature miRNA levels were less abundant than those measured in the control populations (Figure 8C). To visualize the influence of Dicer on miRNA processing, pre-let-7 was transfected into HeLa cells and the relative levels of pre-let-7/let-7 were studied by northern blot analysis. As shown in Figure 8D, overexpression of AUF1 (which lowered Dicer), led to decreased let-7/pre-let-7 ratios, just as did Dicer silencing. Collectively, these results indicate that AUF1 associates with the DICER1 mRNA in several regions, including its 3′UTR and CR, and lowers DICER1 mRNA stability, in turn decreasing Dicer levels and the levels of mature miRNAs.
Figure 8.
AUF1 overexpression selectively decreases miRNA processing from pre-miRNA. (A) Schematic representation of the experimental design to detect pre-miRNA and mature miRNA. (B and C) Forty-eight hours after transfection with pcDNA, AUF1 overexpression plasmids (described in Figure 2A), and Ctrl or Dicer siRNAs (as in Figure 6A), the levels of several pre-miRNAs (B) and mature miRNAs (C) were measured using RT-qPCR and normalized to U6. (D) Northern blot analysis of HeLa cells transfected with Pre-let-7b and with the plasmids and siRNAs shown. Forty-eight hours later, the levels of Pre-let-7b and let-7b were quantified and the extent of let-7 processing was represented as the ratio of let-7 to Pre-let-7 signals. Data in (B–D) are the means ± SD from three independent experiments; *P < 0.05; **P < 0.01.
DISCUSSION
We have identified the RBP AUF1 as a regulator of Dicer expression. In line with its role in promoting the degradation of many mRNAs (25,26), AUF1 associated with the DICER1 mRNA and lowered its half-life. Although the predicted AUF1 sites on the DICER1 3′UTR were in segment B (1 site), in segment C (2 sites) and in segment D (1 site), the segments that showed interaction of AUF1 with biotinylated DICER1 3′UTR transcripts were primarily A and B. This discrepancy likely reflects the fact that computational prediction of RBP targets through RIP and microarray analysis (29) does not identify the full complement of sites on interacting mRNAs, while it can identify sites that are not in vivo target sites. We have undertaken PAR-CLIP analysis of AUF1 RNPs in order to identify comprehensively all sites of AUF1 interaction with target transcripts, as well as to establish whether the different AUF1 isoforms bind DICER1 mRNA in distinct manners (unpublished). The AUF1-elicited reduction in Dicer expression in turn resulted in diminished maturation of miRNAs and lower levels of miRNAs in the cell. Thus, through its impact upon Dicer expression levels and hence on miRNA abundance, AUF1 is capable of influencing gene expression patterns far beyond the interaction of AUF1 directly with its target mRNAs.
AUF1 was found to be upregulated in numerous cancers compared with untransformed tissues (25) and was confirmed to be more highly expressed in the tumor tissues than the normal tissues studied here (Figure 5). AUF1 might elicit anti-tumorigenic roles as a destabilizing agent for the mRNAs encoding the anti-apoptotic protein Bcl-2 and the proliferative protein cyclin D1 (44,45), and can also suppress the expression of pro-inflammatory factors (e.g. IL-6, GM-CSF, iNOS, COX-2) by binding to the mRNAs that encode them, repressing their production, and thereby suppressing a pro-transformation state (46); reviewed in reference 25). However, most of AUF1 effects on gene expression, including changes in AUF1 abundance, subcellular localization and post-translational modification, appear to support a pro-tumorigenic function for AUF1. Genetic overexpression of AUF1 (p37) in mice led to the development of sarcomas in different tissues, linked to overexpression of cyclin D1 (30). Additionally, AUF1 is overexpressed in the cytoplasm of some tumors, including human hepatocellular carcinoma, where it displays elevated levels of methionine adenosyltransferase 1A (MAT1A) and methionine adenosyltransferase 2A (47), human thyroid carcinomas, and mouse lung tumors (48,49). In contrast, AUF1 is mainly localized in the nucleus of melanoma tissues, where it fails to bind the IL10 mRNA and thus permits IL10 mRNA to be stable; this effect was significant because accumulation of IL-10 was linked to the evasion of immune recognition by melanoma cells (50). In breast carcinoma cells, AUF1 shows increased binding to a number of mRNAs linked to the transformation of epithelial cells (51) and in anaplastic large cell lymphoma, hyperphosphorylation of AUF1 was associated to the stabilization of many target mRNAs, particularly several mRNAs that encode cyclins (52). However, in many of these examples, the specific influence of AUF1 on cancer-related gene expression patterns was not identified. Thus, we propose that the lowering of Dicer expression by AUF1 may be a shared feature among tumors.
Although Dicer itself is not a tumor suppressor gene, a decline in the levels of mature miRNAs is a hallmark of cancer. Indeed, the heightened levels of oncoproteins, and proteins involved in angiogenesis, invasion, metastasis and proliferation have been linked to the reduced levels of miRNAs. Some studies show higher Dicer expression levels in ovarian and prostate cancers (53,54); however, many other reports have documented declines in Dicer abundance in tumors. In non-small-cell lung carcinoma, Dicer levels were lower in areas of invasion and advanced carcinomas, and reduced DICER mRNA abundance was associated with poor patient survival (55,56). Similarly, in breast, liver, ovarian and bladder cancers, DICER1 mRNA levels were significantly lower than in non-cancer tissues (57–60); a further association was noted between Dicer protein levels and tumor stage, decreased survival, and poor prognosis (56,61).
The impact of AUF1 on the establishment and/or progression of cancer traits is poorly understood. The discovery that it represses Dicer expression and thereby contributes to lowering miRNA levels constitutes one important dimension of the influence of AUF1 in malignancy. Future work is needed to uncover the full spectrum of AUF1 regulation and function in cancer and how AUF1 modulates cancer-related protein expression patterns.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
National Institute on Aging-Intramural Research Program of the National Institutes of Health. Funding for open access charge: National Institute on Aging-Intramural Research Program, National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jennifer L. Martindale for technical assistance.
REFERENCES
- 1.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 3.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 4.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 5.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apponi LH, Corbett AH, Pavlath GK. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011;32:652–658. doi: 10.1016/j.tips.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keene JD, Lager PJ. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- 8.Masuda K, Abdelmohsen K, Gorospe M. RNA-binding proteins implicated in the hypoxic response. J. Cell. Mol. Med. 2009;13:2759–2769. doi: 10.1111/j.1582-4934.2009.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM, Selimyan R, Martindale JL, Yang X, Carrier F, et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011;39:8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J. Interferon Cytokine Res. 2005;25:1–10. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- 12.Kim HH, Gorospe M. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Mol. Cell. 2008;29:151–152. doi: 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galban S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 17.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- 19.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 20.Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 21.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 23.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zucconi BE, Wilson GM. Modulation of neoplastic gene regulatory pathways by the RNA-binding factor AUF1. Front. Biosci. 2011;17:2307–2325. doi: 10.2741/3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 28.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 29.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 31.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 33.Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srikantan S, Marasa BS, Becker KG, Gorospe M, Abdelmohsen K. Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle. 2011;10:751–759. doi: 10.4161/cc.10.5.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 36.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl Acad. Sci. USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 40.Yoon JH, Choi EJ, Parker R. Yoon Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 2010;189:813–827. doi: 10.1083/jcb.200912019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. HuR suppresses lincRNA-p21 to promote target mRNA translation. Mol. Cell. 2012;24:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee EK, Gorospe M. Coding region: the neglected post-transcriptional code. RNA Biol. 2011;8:44–48. doi: 10.4161/rna.8.1.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Niolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 is a bcl-2 A+U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J. Biol. Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. [DOI] [PubMed] [Google Scholar]
- 45.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Down-regulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marx J. Inflammation and cancer: the link grows stronger. Science. 2004;306:966–968. doi: 10.1126/science.306.5698.966. [DOI] [PubMed] [Google Scholar]
- 47.Vázquez-Chantada M, Fernández-Ramos D, Embade N, Martínez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnel RH, et al. HuR/Methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol. Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 49.Kiledjian M, DeMaria CT, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brewer G, Saccani S, Sarkar S, Lewis A, Pestka S. Increased interleukin-10 mRNA stability in melanoma cells is associated with decreased levels of A+U-rich element binding factor AUF1. J. Interferon Cytokine Res. 2003;23:553–564. doi: 10.1089/107999003322485053. [DOI] [PubMed] [Google Scholar]
- 51.Mazan-Mamczarz K, Hagner PR, Dai B, Wood WH, Zhang Y, Becker KG, Liu Z, Gartenhaus RB. Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach. Cancer Res. 2008;68:7730–7735. doi: 10.1158/0008-5472.CAN-08-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A "liaison dangereuse" between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 53.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am. J. Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flavin RJ, Smyth PC, Finn SP, Laios A, O’Toole SA, Barrett C, Ring M, Denning KM, Li J, Aherne ST, et al. Altered eIF6 and Dicer expression is associated with clinicopathological features in ovarian serous carcinoma patients. Mod. Pathol. 2008;21:676–684. doi: 10.1038/modpathol.2008.33. [DOI] [PubMed] [Google Scholar]
- 55.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 56.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, Léon-Goddard S, Rimokh R, Mikaelian I, Venoux C, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br. J. Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Down-regulation of dicer expression in ovarian cancer tissues. Clin. Biochem. 2010;43:324–327. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 59.Wu JF, Shen W, Liu NZ, Zeng GL, Yang M, Zuo GQ, Gan XN, Ren H, Tang KF. Down-regulation of Dicer in hepatocellular carcinoma. Med. Oncol. 2011;28:804–809. doi: 10.1007/s12032-010-9520-5. [DOI] [PubMed] [Google Scholar]
- 60.Wu D, Tao J, Xu B, Li P, Lu Q, Zhang W. Downregulation of Dicer, a component of the microRNA machinery, in bladder cancer. Mol. Med. Report. 2012;5:695–699. doi: 10.3892/mmr.2011.711. [DOI] [PubMed] [Google Scholar]
- 61.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.