Abstract
Alternative splicing (AS) of pre-mRNAs is an important regulatory mechanism shaping the transcriptome. In plants, only few RNA-binding proteins are known to affect AS. Here, we show that the glycine-rich RNA-binding protein AtGRP7 influences AS in Arabidopsis thaliana. Using a high-resolution RT–PCR-based AS panel, we found significant changes in the ratios of AS isoforms for 59 of 288 analyzed AS events upon ectopic AtGRP7 expression. In particular, AtGRP7 affected the choice of alternative 5′ splice sites preferentially. About half of the events are also influenced by the paralog AtGRP8, indicating that AtGRP7 and AtGRP8 share a network of downstream targets. For 10 events, the AS patterns were altered in opposite directions in plants with elevated AtGRP7 level or lacking AtGRP7. Importantly, RNA immunoprecipitation from plant extracts showed that several transcripts are bound by AtGRP7 in vivo and indeed represent direct targets. Furthermore, the effect of AtGRP7 on these AS events was abrogated by mutation of a single arginine that is required for its RNA-binding activity. This indicates that AtGRP7 impacts AS of these transcripts via direct interaction. As several of the AS events are also controlled by other splicing regulators, our data begin to provide insights into an AS network in Arabidopsis.
INTRODUCTION
Pre-mRNAs arising from the same genomic locus can be processed into multiple transcript isoforms by variable use of splice sites, combining different regions of the transcripts (1). This process, alternative splicing (AS), increases proteome complexity through the generation of protein isoforms with variable domain composition from pre-mRNAs from the same gene. AS also regulates expression at the level of transcript stability, where AS can generate transcripts with premature termination codons (PTCs) that are degraded via the nonsense-mediated decay (NMD) pathway (2).
In metazoa, cis-regulatory elements have been identified (splicing enhancers and splicing suppressors) that are located in exons or introns and influence AS through interaction with trans-acting proteins to determine splice site choice and where the spliceosome will assemble (3). The main families of splicing factors are the serine/arginine-rich (SR) proteins and heterogenous nuclear ribonucleoproteins (hnRNPs). SR proteins are characterized by one or two RNA recognition motifs (RRMs) and the SR domain that engages in protein–protein interactions (4,5). hnRNPs contain diverse types of RNA-binding domains including RRMs and KH domains (6,7). These AS regulators either activate or repress selected splice sites dependent on the position of their binding site and interactions with other factors (8). In addition to these general regulators, specific proteins affect AS of a defined subset of transcripts (9).
In the higher plant Arabidopsis thaliana, 61% of intron-containing genes undergo AS under regular growth conditions (10). AS is influenced by environmental factors including temperature stress and pathogen attack (11–13) and therefore this number is likely to be a lower limit. Intron retention is the most common AS event accounting for 40–50% of the AS events in Arabidopsis; in humans only 5% of the AS events correspond to retained introns (14–17). However, recent analysis of AS in Arabidopsis showed that on a transcript level rather than the level of individual AS events, intron retention had much less impact reflecting the likelihood that many annotated intron retention events are derived from partially spliced transcripts (10). Skipping of entire exons is relatively rare in Arabidopsis (8%), whereas it accounts for 58% of AS events in humans (18). This fundamental difference in the prevalent types of AS events is thought to reflect differences in gene structure (plant introns are generally much shorter than introns in animals) and in the way plants recognize introns (plant introns are often UA-rich). Our knowledge of sequence motifs that influence the choice of splice sites and the cognate protein factors is still limited and a comparative analysis of splicing regulators in plants is of major interest.
The best studied plant splicing factors are the SR protein and polypyrimidine tract-binding protein (PTB) families (5,19,20). Ectopic expression of some of the 18 SR proteins in Arabidopsis produces a range of morphological and physiological phenotypes and impacts AS of their own pre-RNAs and of a suite of other transcripts (21,22). To date, several hnRNP-like proteins have been identified in plants but an involvement in splice site control is not well documented (23). UBP1 (oligouridylate-binding protein 1) from Nicotiana plumbaginifolia is a nuclear RBP that binds to U-rich sequences in introns and untranslated regions (UTRs) (24). It enhances the splicing efficiency of otherwise inefficiently processed introns and increases steady-state abundance of reporter mRNAs that have either no or suboptimal introns. A related hnRNP-like protein RBP45 from N. plumbaginifolia also enhances intron recognition of a mini-exon reporter (25). The hnRNP-like PTB1 and PTB2 proteins show negative auto- and cross-regulation by AS of their own pre-mRNAs, where inclusion of a PTC-containing exon creates an NMD substrate (19). Downstream targets have not been identified as yet.
Plants also contain a family of small hnRNP-like glycine-rich RBPs. The AtGRP7 (A. thaliana glycine-rich RNA-binding protein 7) and AtGRP8 proteins have been linked to AS. Both contain a single RRM-type RNA-binding domain with the highly conserved RNP-2 and RNP-1 motifs and a glycine-rich stretch, and are under control of the circadian clock (26–28). Through reverse genetics, AtGRP7 has been shown to be part of an auto-regulatory feedback loop (29). Ectopic over-expression of AtGRP7 in transgenic plants leads to the use of an alternative 5′ splice site in the intron of the endogenous AtGRP7 pre-mRNA. The resulting AS isoform retains part of the intron with a PTC and is short-lived. Its degradation depends on UPF1 (UP FRAMESHIFT PROTEIN 1) and UPF3 that are part of the NMD pathway (30,31).
In this study, we show that AtGRP7 has a more global effect on AS. For this, we employed an RT-PCR system designed to analyze known AS events with high sensitivity and high resolution (10,32–35). We identified splicing events that are controlled by both AtGRP7 and its paralog AtGRP8. For several transcripts, the ratio of AS isoforms changes in opposite directions in plants constitutively over-expressing AtGRP7 or in plants lacking AtGRP7, respectively, suggesting that these transcripts may be direct AtGRP7 targets. Indeed, we showed by RNA immunoprecipitation (RIP) from whole cell extracts that seven of the identified transcripts are bound by AtGRP7 in vivo. Furthermore, the effect of AtGRP7 on AS of some of these targets is abrogated by mutation of a single arginine within the RRM required for RNA binding and in vivo function (36,37) indicating that AtGRP7 affects AS of these transcripts via direct interaction. Several of the AS events controlled by AtGRP7 are also controlled by SR proteins or the cap binding complex, either in the same direction or antagonistically. Thus, our data begin to provide insights into an AS network in Arabidopsis.
MATERIALS AND METHODS
Plant material and growth
Arabidopsis seeds were surface-sterilized and sown on agar-solidified half-strength MS (Murashige-Skoog) medium (Duchefa) supplemented with 0.5% sucrose and 0.5 g MES/I (38). Plants were grown in 16-h light/8-h dark cycles at 20°C in Percival incubators (CLF laboratories). All above-ground parts of the plants were harvested after 2 weeks at zt10 (Zeitgeber time 10, 10 h after lights on) when AtGRP7 level peaks. The genotypes used were Col-2 wild-type (wt), AtGRP7-ox plants constitutively over-expressing AtGRP7 under the control of the Cauliflower Mosaic Virus promoter (CaMV) in the Col-2 background (39), C24 wt, AtGRP7-ox in the C24 background (40), AtGRP8-ox constitutively over-expressing AtGRP8 (31) under the control of the CaMV promoter in the Col-2 background, and AtGRP7-RQ-ox plants constitutively over-expressing a mutated AtGRP7 protein with arginine 49 exchanged for glutamine (R49Q) under the control of the CaMV promoter both in the Col-2 and C24 background (36). A line without AtGRP7 expression and with reduced AtGRP8 levels was generated by crossing the atgrp7-1 T-DNA insertion mutant (41) to the RNA interference line AtGRP8i-l71 with an RNAi construct directed against AtGRP8 (39). Homozygous F2 plants were identified and designated atgrp7-1 8i. Levels of AtGRP7 and AtGRP8 protein in each of the analyzed lines are shown in Supplementary Figure S1.
RNA isolation and high resolution RT-PCR AS panel
Total RNA was extracted from above-ground tissue using TriReagent (GE Healthcare, Freiburg, Germany) and treated with the DNase kit (Qiagen, Hilden, Germany). RNA (5 µg) was reverse transcribed using M-MuLV RT and oligo(dT)18 primer. After inactivation of the enzyme, PCR was performed in a 96-well format (24 cycles). The forward primers were labeled with the fluorescent dye 6-carboxy fluorescein. Each RT-PCR reaction (1 µl) was diluted into 10 µl Hi-Di formamide (Applied Biosystems) and 0.05 µl GeneScan 500 LIZ internal size standard. The fragments were separated on an ABI3730 DNA Analyzer (Applied Biosystems) and analyzed using the GENEMAPPER Software (Applied Biosystems). The RT-PCR products with the sizes expected for the respective AS isoforms were identified. The percentage of each AS isoform relative to the sum of all relevant AS isoforms from each RT-PCR reaction was calculated using the fluorescent peak areas. The mean ratio of AS isoforms for each event was determined based on three biological replicates. Significant changes in the ratios of AS isoforms were identified from direct comparisons of values from wt and over-expression or mutant lines according to a t-test analysis (significance with P ≤ 0.05). We initially focused on transcripts that showed a significant increase or decrease with a minimum 5% difference between the means of wt plants and over-expression or mutant plants, respectively. However, some AS events showed significance even though the difference in ratios was 3–5%—the lower level was selected because we previously determined that different technical reps in the AS RT-PCR system showed a SEM of up to 3% (33).
The AS panel contains primers for ACTIN11 and RPL12c that serve as reference transcripts to allow determination of relative expression levels of all splice forms. To calculate the total transcript level, the fluorescent peak area of all relevant AS forms was normalized to the level of ACTIN11 and RPL12c, respectively. Supplementary Table S1 contains the complete list of genes and primer pairs used on the RT-PCR panel.
RNA immunoprecipitation
AtGRP7 was fused in frame to Green fluorescent protein (GFP) and expressed under the control of the AtGRP7 promoter (42) and authentic cis-regulatory sequences within the transcribed part of the gene, i.e. 5′-UTR, 3′-UTR and introns, and introduced into atgrp7-1. As a control, transgenic plants expressing GFP only under the control of the AtGRP7 promoter including 5′- and 3′-UTR were generated. Plants grown in long day conditions on agar plates for 2 weeks were vacuum-infiltrated with 1% formaldehyde for 15 min, followed by quenching with 125 mM glycine. A whole-cell extract was prepared in RIP-lysis buffer [50 mM Tris–HCl pH 7.5, 150 mM NaCl, 4 mM MgCl2, 0.1% Igepal, 0.1% SDS, 5 mM DTT, 10 mM vanadylribonucleosid complex, 100 U RiboLock™/ml (Fermentas), 1 mM phenylmethylsulfonylfluorid and protease inhibitor tablets (Roche)]. The extract was pre-cleared with Sepharose beads and subjected to immunoprecipitation with GFP-Trap® beads (Chromotek, Martinsried, Germany), hereafter called IP+. A mock immunoprecipitation was performed using Red fluorescent protein (RFP)-Trap® beads (Chromotek), hereafter called IP−. After extensive washing, coprecipitated RNAs were eluted with TriReagent from IP+ and IP− samples, respectively, and quantified in duplicates via qPCR essentially as described (43). In parallel, transcript levels were determined in RNA isolated from the extract (input). PCR primers are listed in Supplementary Table S1.
Immunoblots
Preparation of protein extracts and western-blot analysis with antibodies against AtGRP7 and AtGRP8 were done as described (44,45).
RESULTS
Identification of AS events influenced by constitutive over-expression of hnRNP-like AtGRP7
The hnRNP-like RBP AtGRP7 auto-regulates AS of its own pre-mRNA and of its closest paralog AtGRP8 (30,31). The impact of AtGRP7 on AS of additional transcripts was examined using the high resolution RT-PCR AS panel described previously (33). A total of 278 primer combinations were used to analyze 288 AS events in transcripts encoding stress-related proteins, transcription factors, RNA-binding proteins, flowering time regulators and other proteins selected from published work or databases (33) (Supplementary Table S1). The splicing patterns in 14-day-old transgenic plants constitutively over-expressing AtGRP7 (AtGRP7-ox) were compared to those in Col-2 wt plants. In AtGRP7-ox plants, a change in the ratio of splice variants >5% relative to wt plants was found for 59 events (21%) (P < 0.05) (Supplementary Table S2). Of these, 27 events were alternative 3′ splice sites, 24 were alternative 5′ splice sites, 7 events were exon skipping and 2 were intron retentions (Table 1). Forty-one percentage of the AS events affected by AtGRP7 over-expression were alternative 5′ splice sites including AtGRP8 AS at a cryptic 5′ splice site within its intron (30,31). Considering that only 23% of all AS events on the panel represent alternative 5′ splice sites, this indicates that AtGRP7 preferentially influences the choice of 5′ splice sites (P = 0.0004 determined by hypergeometric test). Only 3% of the events influenced by AtGRP7 represented intron retention, in contrast to 17% present in the panel (P = 0.0006 determined by hypergeometric test).
Table 1.
Types of AS events showing significant changes in AtGRP7-ox plants in the Col-2 background
| Types of AS events | 288 Events analyzed |
59 Events changed in AtGRP7-ox (Col-2) |
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Alternative 3′ splice site | 135 | 47 | 26 | 44 |
| Alternative 5′ splice site | 67 | 23 | 24 | 41 |
| Intron retention | 48 | 17 | 2 | 3 |
| Exon skipping | 34 | 12 | 7 | 12 |
| Cryptic intron | 4 | 1 | 0 | 0 |
The ratios of AS forms could change due to regulation of splice site usage by AtGRP7 or change indirectly as a consequence of altered steady-state abundance of individual AS forms. Therefore, transcript levels of all investigated splice variants were calculated relative to ACTIN11 and RPL12c. In the AtGRP7-ox plants, 12 of the 278 transcripts analyzed (4.3%) showed a difference in steady-state abundance of at least 2-fold (Supplementary Table S3). Of the 59 transcripts with a change in AS in AtGRP7-ox plants, only 4 also showed an altered steady-state abundance of at least 2-fold (6.8%). Thus, AtGRP7 over-expression does not significantly alter steady-state abundance of the affected transcripts (P = 0.1501 determined by hypergeometric test). We conclude that the observed changes in the ratio of individual splice variants are due to changes in splice site selection as a consequence of AtGRP7 over-expression.
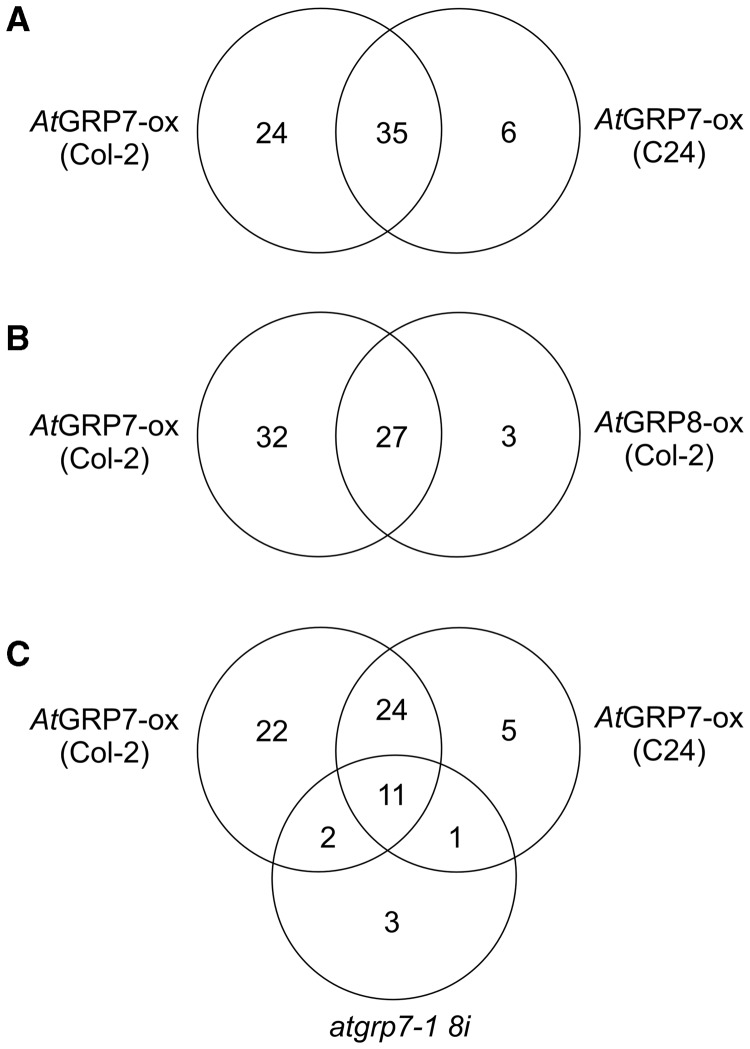
To increase confidence in the effects of AtGRP7 on AS, we tested the influence of AtGRP7 in a different genetic background. In an independent AtGRP7-ox line in the C24 background (Supplementary Figure S1), 41 of 87 events analyzed [(including the events most strongly affected in AtGRP7-ox (Col-2) plants)] were changed significantly compared to C24 wt (>5%; P < 0.05) (Figure 1A). Of these, 35 were also changed significantly in AtGRP7-ox (Col-2) (Table 2). This supports the conclusion that elevated AtGRP7 levels are responsible for the changed ratios in AS isoforms in the AtGRP7-ox lines.
Figure 1.
Changes in the ratio of AS isoforms in AtGRP7-ox, AtGRP8-ox and atgrp7-1 8i. (A) Venn diagram showing the number of splicing events with significant changes in AtGRP7-ox (Col-2) and AtGRP7-ox (C24) when compared to the Col-2 and C24 wild-types, respectively. (B) Venn diagram showing the number of splicing events with significant changes in AtGRP7-ox (Col-2) and AtGRP8-ox (Col-2). (C) Venn diagram showing the number of splicing events with significant changes in AtGRP7-ox (Col-2 and C24) and atgrp7-1 8i. The numbers represent transcripts with significant changes in the ratio of AS isoforms (>5% and P < 0.05).
Table 2.
Genes/transcripts with significant changes in the ratio of alternatively spliced isoforms in AtGRP7-ox plants
| primer pair | AGI | Description | Product sizes (bp) | Col-2 |
AtGRP7-ox (Col-2) |
P-value | C24 |
AtGRP7-ox (C24) |
P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Small prod. (%) | Large prod. (%) | Small prod. (%) | Large prod. (%) | 7-ox (Col-2) | Small prod. (%) | Large prod. (%) | Small prod. (%) | Large prod. (%) | 7-ox (C24) | ||||
| Alternative 3′SS | |||||||||||||
| 12 | At1g72320 | APUM23 (Arabidopsis pumilio 23); RNA binding | 141/150/215 | 15 | 83 | 5 | 95 | 0.0002 | 23 | 74 | 8 | 92 | 0.0192 |
| 19 | At2g32320 | tRNA His guanylyltransferase | 202/273 | 33 | 67 | 45 | 55 | 0.0047 | 31 | 69 | 39 | 61 | 0.0048 |
| 36 | At4g12790 | ATP-binding family protein | 212/338 | 45 | 55 | 25 | 75 | 0.0029 | 46 | 54 | 25 | 75 | 0.0034 |
| 49 | At5g41150 | UVH1 (ultraviolet hypersensitive 1) | 293/346 | 82 | 18 | 72 | 28 | 0.0003 | 92 | 8 | 84 | 16 | 0.0034 |
| 59 | At5g66010 | RNA binding | 105/182 | 30 | 70 | 40 | 60 | 0.0261 | 38 | 62 | 59 | 41 | 0.0016 |
| 242 | At1g60850 | ATRPAC42 (Arabidopsis thaliana RNA polymerase I subunit 42); DNA binding/DNA-directed RNA polymerase | 111/122 | 70 | 30 | 77 | 23 | 0.0311 | 73 | 27 | 79 | 21 | 0.0325 |
| 254 | At5g50240 | l-isospartyl methyltransferase | 192/201 | 63 | 37 | 73 | 27 | 0.0282 | 64 | 36 | 73 | 27 | 0.0079 |
| 273 | At3g07810 | Heterogeneous nuclear ribonucleoprotein, putative/hnRNP, putative | 126/173 | 8 | 92 | 3 | 97 | 0.0094 | 10 | 90 | 4 | 96 | 0.0372 |
| 288 | At3g12570 | FYD | 159/188 | 43 | 57 | 52 | 48 | 0.0148 | 39 | 61 | 53 | 47 | 0.0207 |
| 295 | At2g02390 | ATGSTZ1 (glutathione s-transferase 18); glutathione transferase | 181/202 | 88 | 12 | 69 | 31 | 0.0023 | 87 | 13 | 71 | 29 | 0.0016 |
| 378 | At3g62190 | DNAJ heat shock N-terminal domain-containing protein | 144/334 | 89 | 11 | 99 | 1 | 0.0075 | 85 | 15 | 98 | 2 | 0.0032 |
| Alternative 5′SS | |||||||||||||
| 72 | At2g04790 | Similar to unnamed protein product [Vitis vinifera] (GB:CAO23994.1) | 167/190 | 68 | 32 | 53 | 47 | 0.0077 | 41 | 59 | 29 | 71 | 0.0027 |
| 75 | At2g36000 | Mitochondrial transcription termination factor-related/mTERF-related | 150/254 | 31 | 69 | 14 | 86 | 0.0009 | 40 | 60 | 17 | 83 | 0.0016 |
| 87 | At4g35450 | AKR2 (ankyrin repeat-containing protein 2); protein binding | 305/350 | 78 | 22 | 59 | 41 | 0.0052 | 72 | 28 | 57 | 43 | 0.0009 |
| 90 | At4g39260 | ATGRP8/GR-RBP8 (CCR1) | 158/316 | 94 | 6 | 23 | 77 | 0.0000 | 97 | 3 | 23 | 77 | 0.0004 |
| 112 | At1g09530 | PAP3/PIF3/POC1 (phytochrome interacting factor 3); DNA binding/protein binding/transcription factor | 230/290 | 88 | 12 | 59 | 41 | 0.0000 | 90 | 10 | 66 | 34 | 0.0033 |
| 129 | At2g40830 | RHC1A (RING-H2 finger C1A); protein binding/zinc ion binding | 220/329 | 91 | 9 | 59 | 41 | 0.0012 | 91 | 9 | 63 | 37 | 0.0037 |
| 136 | At3g07740 | ADA2A (Arabidopsis adaptor 2A homolog); DNA binding/transcription factor | 139/240 | 100 | 0 | 46 | 54 | 0.0006 | 100 | 0 | 70 | 30 | 0.0063 |
| 141 | At3g51880 | HMGB1 (High mobility group B 1); transcription factor | 204/225 | 92 | 8 | 79 | 21 | 0.0471 | 92 | 8 | 87 | 13 | 0.0003 |
| 145 | At3g17609 | HYH (Hy5-homolog); DNA binding/transcription factor | 150/153/195/268 | 32 | 68 | 54 | 46 | 0.0197 | 29 | 71 | 44 | 56 | 0.0275 |
| 148 | At1g76510 | ARID/BRIGHT DNA-binding domain-containing protein | 189/212 | 28 | 72 | 16 | 84 | 0.0073 | 37 | 63 | 25 | 75 | 0.0006 |
| 149 | At2g27230 | Transcription factor-related | 208/246 | 28 | 72 | 18 | 82 | 0.0177 | 34 | 66 | 22 | 78 | 0.0060 |
| 189 | At5g43270 | SPL2 (squamosa promoter binding protein-like 2); DNA binding/transcription factor | 160/244 | 87 | 13 | 63 | 37 | 0.0021 | 87 | 13 | 69 | 31 | 0.0044 |
| 261 | At4g10100 | CNX7/SIR5; catalytic | 254/270 | 58 | 42 | 46 | 54 | 0.0007 | 58 | 42 | 45 | 55 | 0.0028 |
| 272 | At3g23900 | RNA recognition motif (RRM)-containing protein | 118/125 | 30 | 70 | 22 | 78 | 0.0008 | 30 | 70 | 23 | 77 | 0.0041 |
| 285 | At3g19840 | FF domain-containing protein/WW domain-containing protein | 171/207 | 61 | 39 | 40 | 60 | 0.0004 | 65 | 35 | 43 | 57 | 0.0084 |
| 322 | At2g33480 | ANAC041 (Arabidopsis NAC domain containing protein 41); transcription factor | 321/399 | 17 | 83 | 34 | 66 | 0.0047 | 15 | 85 | 29 | 71 | 0.0025 |
| 324 | At5g43270 | SPL2 (squamosa promoter binding protein-like 2); DNA binding/transcription factor | 186/270 | 86 | 14 | 66 | 34 | 0.0046 | 95 | 5 | 66 | 34 | 0.0158 |
| 343 | At3g29160 | AKIN11 (Arabidopsis snf1 kinase homolog 11); protein kinase | 159/307 | 30 | 70 | 3 | 97 | 0.0008 | 45 | 55 | 2 | 98 | 0.0009 |
| Exon skipping | |||||||||||||
| 181 | At5g05550 | Transcription factor | 210/308 | 64 | 36 | 74 | 26 | 0.0143 | 59 | 41 | 75 | 25 | 0.0044 |
| 196 | At3g01150 | PTB (polypyrimidine tract-binding); RNA binding | 165/268 | 91 | 9 | 97 | 3 | 0.0035 | 92 | 8 | 97 | 3 | 0.0071 |
| 226 | At4g24740 | AFC2 (Arabidopsis fus3-complementing gene 1); kinase | 143/309 | 19 | 81 | 47 | 53 | 0.0020 | 12 | 88 | 45 | 55 | 0.0004 |
| 227 | At4g24740 | AFC2 (Arabidopsis fus3-complementing gene 1); kinase | 152/343 | 30 | 70 | 73 | 27 | 0.0000 | 33 | 67 | 68 | 32 | 0.0005 |
| 380 | At5g08185 | npcRNA 78; MIR162a | 103/168 | 64 | 36 | 42 | 58 | 0.0044 | 83 | 17 | 75 | 25 | 0.0267 |
| Intron retention | |||||||||||||
| 327 | At5g59950 | RNA and export factor-binding protein, putative | 226/422 | 84 | 16 | 50 | 50 | 0.0020 | 93 | 7 | 69 | 31 | 0.0091 |
The two AS isoforms considered here are underlined.
AS pairs that only change in one of the AtGRP7-ox lines could either be false targets or undergo differential splicing in the two ecotypes themselves. When the AS ratios were directly compared between the Col-2 and C24 wt plants 61 of the 87 AS events showed no significant changes. On the other hand, 26 AS events showed significant differences (>5%; P < 0.05) between Col-2 and C24 wt (Supplementary Table S4). This differential splicing could be due to sequence variation (SNPs and indels) in splice sites, in other signals which directly affect the splicing of specific introns, and in primer binding sites or could be due to differential expression of trans-acting factors. To determine whether differences in AS in the two AtGRP7-ox lines could be explained by ecotype differences, we compared the sequences surrounding the splicing events (position −50 to +50 relative to the splice sites) between the two ecotypes (46).
In some cases, local sequence variation was detected that could potentially contribute to the different splicing patterns observed. For example, two primer pairs (#51 and #105) did not generate RT-PCR products in the C24 background and both had mismatches in one of the primers. For two primer pairs (#160 and #270), a transcript isoform observed in Col-2 was absent in C24. The sequence of the alternative 5′ splice site in #270 has a G to C mutation at position −1 in C24 that may be responsible for the difference in usage of this site between ecotypes (Supplementary Figure S2A). In contrast, for #160 there are no SNPs in the splice site sequences but there are sequence differences in the surrounding intron (Supplementary Figure S2B). Four primer pairs detected a significant difference in AS between ecotypes and in response to AtGRP7 over-expression (#72, #118, #128 and #343) (Supplementary Figure S2C–F). In all four cases, the effect of AtGRP7 over-expression was in the same direction in both backgrounds despite the different pattern in the two ecotypes. Primer pairs #171 and #179 showed differences between Col-2 and C24, and over-expression of AtGRP7 in Col-2 background shifts the pattern in the direction of the C24 wt situation. Over-expression of AtGRP7 in C24 had a smaller effect on the already elevated levels in C24 (Supplementary Figure S2G and S2H).
Overall, the majority of genes did not show significant variation in AS ratios between the two ecotypes. Where significant qualitative or quantitative variation was observed, SNPs and indels in or near splice sites may explain the differential splicing behavior.
AtGRP7 and AtGRP8 impact an overlapping set of AS events
AtGRP7 cross-regulates its paralog AtGRP8 that is 77% identical in sequence (31,40). Constitutive over-expression of AtGRP8, in turn, promotes production of the NMD-sensitive AS isoform of AtGRP7. Thus, AtGRP7 and AtGRP8 are able to bind their own and each others’ pre-mRNAs and may act similarly on downstream targets. We analyzed this by monitoring the impact of AtGRP8 over-expression on 87 events including the events most strongly affected in AtGRP7-ox (Col-2) plants. A total of 30 AS events showed a significant change in AtGRP8-ox plants including AtGRP7 AS at the cryptic 5′ splice site within its intron (Figure 1B). Of these, 27 also showed a change in AtGRP7-ox (Col-2) and over-expression of either AtGRP7 or AtGRP8 affected these AS events in the same direction (Supplementary Table S5). Thus, AtGRP7 and AtGRP8 not only cross-regulate but also share a number of downstream targets.
Inverse regulation of AS events by AtGRP7 gain-of-function and loss-of-function
The genes showing changes in AS in AtGRP7-ox plants may be direct targets of AtGRP7 or indirect targets whose changes in AS are due to the altered expression of splicing regulators. We reasoned that if a gene is a direct target the AS events should also respond to reduced levels of AtGRP7 and, moreover, the ratio of the AS forms should change in the opposite direction.
In the atgrp7-1 T-DNA insertion line that lacks AtGRP7 (41), steady-state abundance of AtGRP8 is strongly elevated (39). This is most likely due to relief from repression, as over-expression lines of AtGRP7 show little AtGRP8 expression (31,40). There is no true loss-of-function mutant in AtGRP8 that has been characterized to date. To obtain plants that lack AtGRP7 and express reduced amounts of AtGRP8, atgrp7-1 was crossed to the RNAi line AtGRP8i-l71 (39). F2 plants were identified that show an AtGRP8 protein level close to wt, indicating that the RNAi construct substantially reduced the increased AtGRP8 level in atgrp7-1 (Supplementary Figure S1). In this line, designated atgrp7-1 8i, 17 of the 87 investigated AS events (20%) changed significantly by >5% (P < 0.05) (Figure 1C). Thus, fewer AS events were affected by the reduced AtGRP7 level than when AtGRP7 was over-expressed. For 10 of the events that changed in both AtGRP7-ox (Col-2) and AtGRP7-ox (C24), as well as in the atgrp7-1 8i line, the ratio of AS isoforms in atgrp7-1 8i changed significantly in the opposite direction to AtGRP7-ox plants, suggesting these transcripts may be direct targets of AtGRP7 (Table 3). An additional five events showed a smaller, yet statistically significant change between 3% and 4.75% in the opposite direction (P < 0.05) (Table 3).
Table 3.
Genes/transcripts with changes in the AS pattern in opposite direction in AtGRP7-ox and atgrp7-1 8i plants
| Primer pair | AGI | Description | Product sizes (bp) |
grp7-1 8i |
P-value grp7-1 8i | Col-2 |
AtGRP7-ox (Col-2) |
P-value 7-ox (Col-2) |
AtGRP8-ox (Col-2) |
P-value 8-ox (Col-2) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Small product (%) | Large product (%) | Small product (%) | Large product (%) | Small product (%) | Large product (%) | Small product (%) | Large product (%) | |||||||
| 12 | At1g72320 | APUM23 (Arabidopsis pumilio 23); RNA binding | 141/150/215 | 21 | 77 | 0.0001 | 15 | 83 | 5 | 95 | 0.0002 | 7 | 93 | 0.0099 |
| 19 | At2g32320 | tRNA His guanylyltransferase | 202/273 | 27 | 73 | 0.0051 | 33 | 67 | 45 | 55 | 0.0047 | 35 | 65 | 0.2379 |
| 288 | At3g12570 | FYD | 159/188 | 33 | 67 | 0.0111 | 43 | 57 | 52 | 48 | 0.0148 | 44 | 56 | 0.5852 |
| 75 | At2g36000 | mitochondrial transcription termination factor-related/mTERF-related | 150/254 | 40 | 60 | 0.0076 | 31 | 69 | 14 | 86 | 0.0009 | 22 | 78 | 0.0098 |
| 90 | At4g39260 | ATGRP8/GR-RBP8 (CCR1) | 158/316 | 99 | 1 | 0.0060 | 94 | 6 | 23 | 77 | 0.0000 | 98 | 2 | 0.0215 |
| 343 | At3g29160 | AKIN11 (Arabidopsis snf1 kinase homolog 11); protein kinase | 159/307 | 57 | 43 | 0.0001 | 30 | 70 | 3 | 97 | 0.0008 | 11 | 89 | 0.0027 |
| 181 | At5g05550 | transcription factor | 210/308 | 50 | 50 | 0.0049 | 64 | 36 | 74 | 26 | 0.0143 | 67 | 33 | 0.2237 |
| 226 | At4g24740 | AFC2 (Arabidopsis fus3-complementing gene 1); kinase | 143/309 | 7 | 93 | 0.0004 | 19 | 81 | 47 | 53 | 0.0020 | 33 | 67 | 0.0506 |
| 227 | At4g24740 | AFC2 (Arabidopsis fus3-complementing gene 1); kinase | 152/343 | 22 | 78 | 0.0125 | 30 | 70 | 73 | 27 | 0.0000 | 53 | 47 | 0.0155 |
| 327 | At5g59950 | RNA and export factor-binding protein, putative | 226/422 | 90 | 10 | 0.0102 | 84 | 16 | 50 | 50 | 0.0020 | 68 | 32 | 0.0456 |
| 129 | At2g40830 | RHC1A (RING-H2 finger C1A); protein binding/zinc ion binding | 220/329 | 94 | 6 | 0.0441 | 91 | 9 | 59 | 41 | 0.0012 | 76 | 24 | 0.0648 |
| 171 | At5g18620 | CHR17 (chromatin remodeling factor17); DNA-dependent ATPase | 213/222 | 75 | 25 | 0.0179 | 72 | 28 | 66 | 34 | 0.0005 | 71 | 29 | 0.4350 |
| 273 | At3g07810 | heterogeneous nuclear ribonucleoprotein, putative/hnRNP, putative | 126/173 | 12 | 88 | 0.0184 | 8 | 92 | 3 | 97 | 0.0094 | 5 | 95 | 0.0108 |
| 295 | At2g02390 | ATGSTZ1 (glutathione s-transferase 18); glutathione transferase | 181/202 | 91 | 9 | 0.0165 | 88 | 12 | 69 | 31 | 0.0023 | 74 | 26 | 0.0512 |
| 322 | At2g33480 | ANAC041 (Arabidopsis NAC domain containing protein 41); transcription factor | 321/399 | 12 | 88 | 0.0226 | 17 | 83 | 34 | 66 | 0.0047 | 26 | 74 | 0.0836 |
The first 10 events show a significant change (>5%, P < 0.05). For primer pair #12, the two splice forms considered are underlined (the 141 nt form does not change). The last five events show a smaller, yet statistically significant change between 2.9% and 4.75% (P < 0.05) in atgrp7-1 8i plants in the opposite direction to AtGRP7-ox plants.
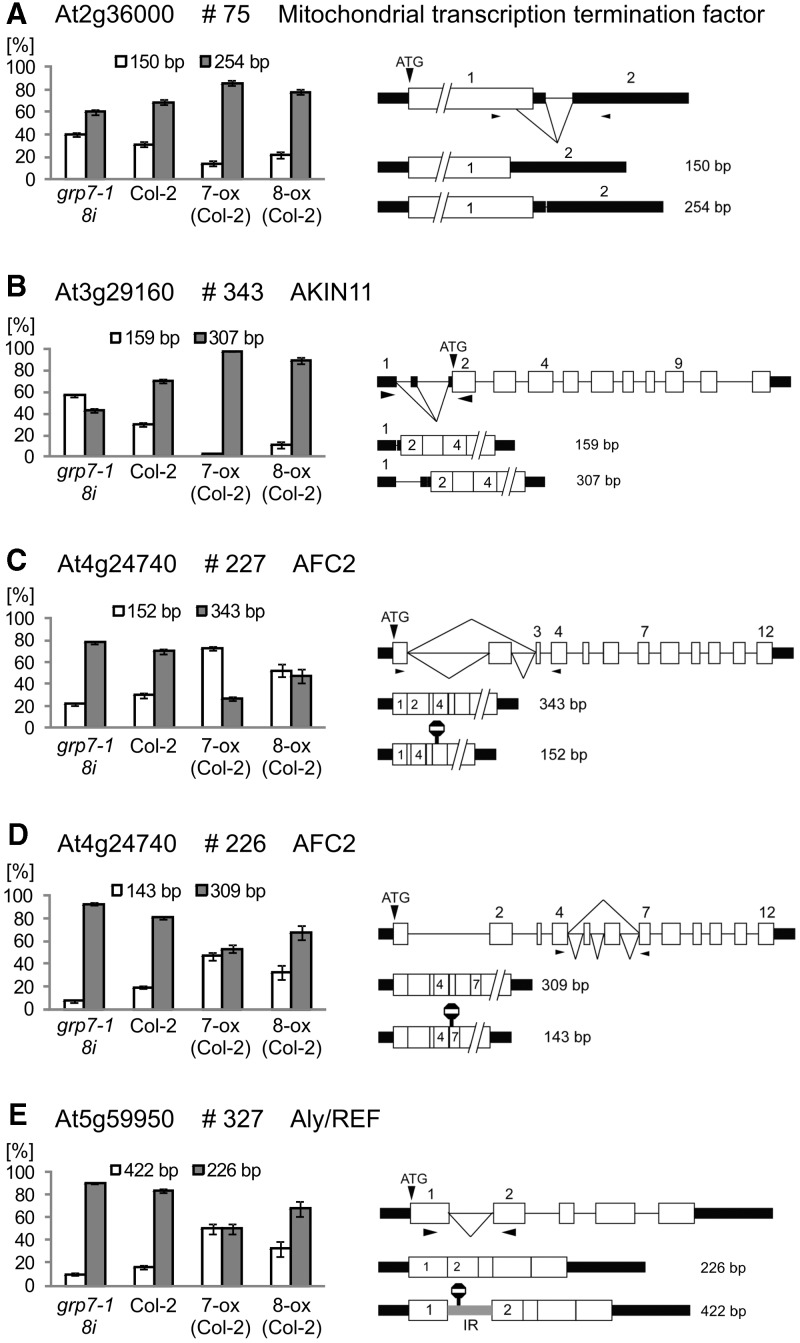
The three lines, atgrp7-1 8i, Col-2 wt and AtGRP7-ox represent a series of genotypes with increasing amounts of AtGRP7, and where the AtGRP8 level is effectively the same in wt and atgrp7-1 8i lines and AtGRP8 is virtually absent in the AtGRP7-ox line (Supplementary Figure S1). In these lines, we observed reciprocal changes in the abundance of AS isoforms (Figure 2 and Supplementary Figure S3). For example, in At2g36000, encoding a mitochondrial transcription termination factor-related protein (#75), there is AS of an intron in the 3′-UTR and lower levels of AtGRP7 give increased usage of the alternative 5′ splice site towards the end of the coding region and therefore increased levels of the shorter isoform, which could generate a C-terminally truncated protein (Figure 2A). For AKIN11 encoding a catalytic subunit of Snf1-related (SnRK1) protein kinase (#343), an increasing AtGRP7 concentration shifts the ratio in favor of a longer splice variant containing part of intron 1 due to enhanced usage of an alternative 5′ splice site in the 5′-UTR (Figure 2B). AFC2 encoding a LAMMER kinase has two different AS events: skipping of exon 2 and skipping of exons 5 and 6. Increased AtGRP7 levels cause increased skipping of exon 2 (#227), which leads to an AS isoform containing a PTC (Figure 2C). For the event detected with primer pair #226, in AtGRP7-ox plants, both isoforms are produced at similar levels, whereas in plants lacking AtGRP7 or wt plants, the variant with exons 4 through 7 predominates (Figure 2D). Thus, AtGRP7 promotes skipping of exon 2 and exons 5 and 6. For At5g59950 encoding an Aly/REF-related RNA-binding protein/export factor (#327), AtGRP7 strongly increases the proportion of the AS isoform retaining the first intron leading to introduction of a PTC (Figure 2E). For all these events, overexpression of AtGRP8 has a similar effect as overexpression of AtGRP7 (Figure 2 and Supplementary Figure S3). Taken together, we observed an increase of one AS isoform and the concomitant reduction of the other isoform dependent on the AtGRP7 dosage, and in several cases, a switch in the predominant splice form occurred between plants lacking AtGRP7 and AtGRP7-ox plants.
Figure 2.
Genes/transcripts with significant changes in the AS patterns in opposite directions in atgrp7-1 8i and AtGRP7-ox plants. (A) At2g3600 (#75) Mitochondrial transcription termination factor, (B) At3g29160 (#343) AKIN11, (C) At4g24740 (#227) AFC2, (D) At4g24740 (#226) AFC2 and (E) At5g59950 (#327) Aly/REF-like protein. On the left side of each panel, the percentage of each splice form ± SD based on three biological replicates is indicated for atgrp7-1 8i, wt, and AtGRP7-ox plants, respectively. For comparison, the data for AtGRP8-ox plants are included. On the right side of each panel, the gene and transcript structures and the AS events are shown schematically. Exons are indicated by open boxes and numbered, UTRs—black rectangles, introns—thin lines, splicing events—diagonal lines and stop signs—PTCs. The sizes of the PCR products from each splice isoform are indicated.
RNA immunoprecipitation shows direct binding of AtGRP7 to target transcripts in vivo
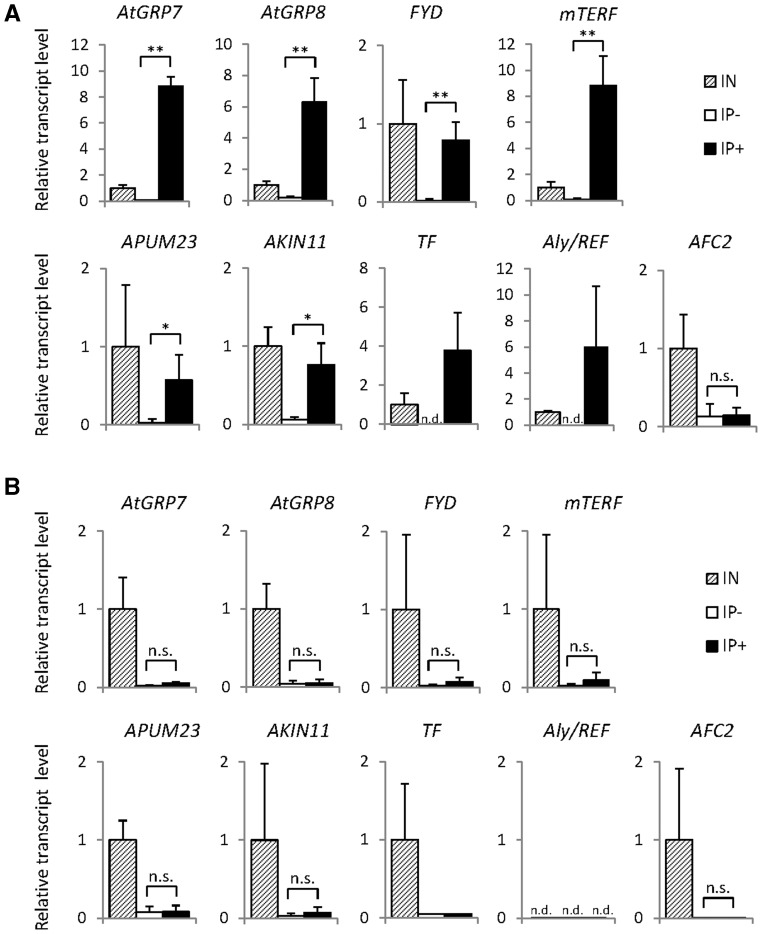
The antagonistic regulation in AtGRP7 loss-of-function and gain-of-function lines, respectively, suggests that AtGRP7 may directly interact with the affected transcripts. To study the potential in vivo association of the transcripts with AtGRP7, we established an efficient protocol to immunoprecipitate ribonucleoprotein complexes from whole cell extracts, followed by qPCR. An AtGRP7-GFP fusion protein driven by the AtGRP7 promoter and all regulatory elements in the transcribed region was expressed in the atgrp7-1 background. As proof-of-principle, we showed that the AtGRP7 transcript, a known in vitro binding substrate of AtGRP7 was efficiently precipitated with GFP-Trap® beads (IP+) but was barely detected in mock precipitates using RFP-Trap® beads (IP−) (Figure 3A). As an additional control, we performed RIP on plants expressing GFP only driven by the same regulatory elements. Only a very small amount of AtGRP7 was detected in the IP+ fraction relative to the input, and no enrichment was detected relative to IP− (Figure 3B). This demonstrates that the procedure faithfully identifies a known binding substrate.
Figure 3.
In vivo interaction of AtGRP7-GFP with candidate target transcripts. RIP was performed on plants expressing the AtGRP7-GFP fusion protein under control of the AtGRP7 promoter including 5′- and 3′-UTR and intron in atgrp7-1 (A) and transgenic plants expressing GFP under control of the AtGRP7 promoter including 5′- and 3′-UTR (B). The levels of transcripts co-precipitated in the GFP-Trap® bead precipitate (IP+), the RFP-Trap® bead mock precipitate (IP−) and in the input fractions, respectively, were determined by qRT-PCR in duplicates for At2g21660 (AtGRP7) which served as positive control, At4g39260 (AtGRP8, #90), At3g12570 (FYD, #288), At2g3600 (mTERF, #75), At1g72320 (APUM23, #12), At3g29160 (AKIN11, #343), At5g05550 [transcription factor (TF), #181], At4g24740 (AFC2, #226 and #227) and At5g59950 (Aly/REF-like RNA binding/export factor, #327). Transcript levels were normalized to PP2A and expressed relative to the input. Means ± SD are presented based on three biological and significance was tested using Student’s t-test (**P < 0.005, *P < 0.05). n.d., not detectable; n.s., not significant. In IP− from GFP plants, the transcripts #181 and #327 were not detectable and thus no statistical test was applied.
We next examined the precipitated RNA for the presence of the candidate targets (depicted in Figure 2 and Supplementary Figure S3). The AtGRP8 transcript (#90), the transcript of the mitochondrial transcription termination factor-related protein (mTERF, #75), and the FYD transcript (#288) were strongly enriched in the IP+ fraction but not in the mock precipitate (IP−) (P < 0.005) (Figure 3A). In the GFP plants, a far lower level relative to input and no difference between IP+ and IP− were detected (Figure 3B). APUM23 encoding a member of the Pumilio RBP family (#12) and AKIN11 (#343) showed a weaker yet statistically significant enrichment in IP+ versus IP− from AtGRP7-GFP plants (Figure 3A) (P < 0.05), and no enrichment in IP+ of GFP plants (Figure 3B). The transcript encoding a transcription factor (TF, #181) was enriched in IP+ but detected only once in IP−. The Aly/REF-like protein (#327) was enriched in IP+ but not detectable in IP− and the GFP plants. In several cases, levels of transcripts coprecipitated with AtGRP7-GFP appeared higher than the input level. This has been observed before and may be attributed to higher efficiency of RNA extraction and amplification from the IP compared to the total extract (47,48).
Although AFC2 (#226 and #227) was recovered from AtGRP7-GFP plants but not from GFP plants, the signals in IP+ and IP− were similar and therefore AFC either may not be a specific in vivo substrate of AtGRP7 or the interaction was too weak to be detected. The expression level of At2g32330 encoding an unknown protein (#19) was too low to allow a reliable quantification. Thus, using RIP-qPCR, we were able to confirm direct binding of AtGRP7 to seven putative target transcripts identified by their antagonistic AS behavior in the over-expression lines and the loss-of-function mutant.
Site-specific mutation of the conserved Arg49 in RNP-1 abrogates the effect of AtGRP7 on AS events
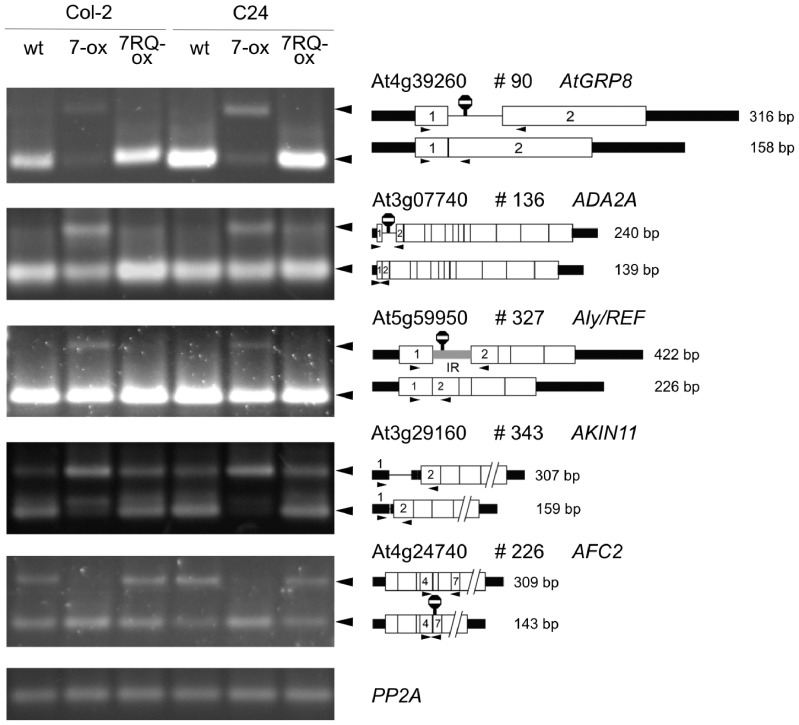
Mutation of a single arginine to glutamine within the RRM interferes with both in vitro binding of recombinant AtGRP7 to its own RNA and the impact of AtGRP7 on AtGRP7 and AtGRP8 pre-mRNA splicing in vivo (36). Therefore, we investigated the splicing pattern of selected candidate AtGRP7 targets in two independent transgenic lines constitutively over-expressing the mutant protein (AtGRP7-RQ-ox). As a control, we showed that over-expression of AtGRP7 affected AS of AtGRP8 by causing part of the intron to remain in the transcript, generating the NMD-sensitive AS isoform (Figure 4). This AtGRP7-induced change in AS of AtGRP8 was not observed in AtGRP7-RQ-ox plants (36).
Figure 4.
AS events in AtGRP7-ox lines not observed in AtGRP7-R49Q-ox plants. Left side: RT-PCR analysis of selected genes/transcripts with total RNA from AtGRP7-ox and AtGRP7-RQ-ox plants in both the Col-2 and C24 backgrounds and their respective wt plants. Arrowheads denote RT-PCR products representing AS events whose presence/absence or abundance differs between the over-expression lines with and without the R49Q mutation. Right side: gene and transcript structures and the AS events for AtGRP8 (#90), ADA2A (#136), Aly/Ref (#327), AKIN11 (#343) and AFC2 (#226) are shown schematically (see Figure 2). PP2A served as constitutive control.
For the transcriptional activator adapter ADA2A (#136), ectopic AtGRP7 expression increased the proportion of the larger splice variant compared to wt plants through increased use of an alternative 5′ splice site and inclusion of part of the intron. This effect was not seen upon ectopic expression of AtGRP7-RQ. Similarly, the larger splice form of the Aly/REF-related protein (#327) that retains the first intron appeared in AtGRP7-ox plants but not in AtGRP7-RQ-ox plants. For AKIN11 (# 343), over-expression of AtGRP7 wt protein led almost exclusively to the production of the long splice variant retaining the intron in the 5′-UTR, whereas over-expression of AtGRP7-RQ resembled wt plants. Finally, for the LAMMER kinase AFC2 (#226), the transcript where exons 5 and 6 are skipped was detected almost exclusively in AtGRP7-ox plants but not in AtGRP7-RQ-ox plants. Thus, in all of these examples, the effects observed with over-expression of authentic AtGRP7 protein are lost when the AtGRP7-RQ mutant protein is over-expressed suggesting that AS of these genes depends on the RNA-binding activity of AtGRP7.
Based on these results, we investigated further AS events that showed opposite splicing behavior in AtGRP7-ox and atgrp7 8i lines (Table 3) in the AtGRP7-RQ-ox plants using the RT-PCR panel. None of the 14 analyzed events showed a difference in AS pattern in wt and the AtGRP7-RQ-ox line in Col-2, again suggesting that these events are also dependent on the conserved arginine in the RRM (data not shown).
The influence of AtGRP7 on AS events generating NMD substrates
AS can regulate expression levels by generating transcript isoforms that are degraded by the NMD pathway. In Arabidopsis, ∼13–18% of transcripts are estimated to undergo AS-linked NMD by identifying AS forms stabilized in upf1-5 and upf3-1 mutants or upon cycloheximide (CHX) treatment of wt plants using the high resolution RT-PCR system also used here (35). Because AtGRP7 and AtGRP8 auto- and cross-regulate by binding to their pre-mRNAs to generate an NMD-sensitive AS transcript (31), we mined the data by Kalyna et al. for the behavior of the 10 pairs of splice isoforms that showed mirror-image patterns in plants with high AtGRP7 level and plants that lack AtGRP7, respectively. Seven of these events included a PTC in one AS isoform (Supplementary Table S6). In six cases, At2g32330 (unknown protein; #19), mTERF-related protein (#75), AtGRP8 (#90), AFC2 (#226 and #227) and Aly/REF-related protein (#327), elevated levels of AtGRP7 favored the production of the PTC-containing form. In four of these cases (#19, #90, #226 and #327) the PTC-containing form is stabilized in upf mutants and/or upon CHX treatment (35). For #181, the non-PTC-containing form was favored by increased levels of AtGRP7. The other three AS events with opposite AS patterns involved AS in introns in the 5′-UTR and gave rise to AS isoforms that were stabilized in upf3-1 (#12), in upf3-1 and upon CHX treatment (#343) or only upon CHX treatment (#288), respectively, although no obvious effect of an upstream open reading frame or NMD signals were found (35). Thus, AtGRP7 regulates the expression of, at least, some of its target genes by affecting NMD-sensitive AS isoforms.
DISCUSSION
Here, we identify the hnRNP-like proteins, AtGRP7 and AtGRP8, as novel splicing regulators in Arabidopsis using a high resolution RT-PCR system capable of detecting changes in AS. Constitutive over-expression of AtGRP7 caused significant changes in the ratio of AS isoforms in 21% of the 288 investigated AS events.
To study the effect on AS of AtGRP7 in detail, we used a series of lines (over-expression, wt and loss-of-function mutant) expressing AtGRP7 at different levels. Generation of the AtGRP7 knock-down was complicated by up-regulation of AtGRP8 in the atgrp7-1 mutant (39) and required it to be combined with an RNAi line reducing AtGRP8 expression. This genotype, atgrp7-1 8i, allowed us to study the effect of loss of AtGRP7 with AtGRP8 expression at a similar level to wt plants (Supplementary Figure S1). The advantage of comparing lines with different levels of AtGRP7, effectively representing a dosage series, became apparent when over-expression and loss-of-function of AtGRP7 had opposite consequences on 10 AS events, suggesting that these transcripts represent direct targets. Importantly, RIP from whole cell extracts confirmed that seven of the transcripts are indeed bound by AtGRP7 in vivo. Thus, dosage-dependent splicing behavior acts as an indicator of direct interaction of proteins affecting AS site choice. In line with this, the events with a reciprocal change in the AS ratio upon increasing or decreasing AtGRP7 levels were influenced by over-expression of the AtGRP7 wt protein but not of the mutant version where arginine of RNP-1 was exchanged for glutamine (R49Q). This mutation has been shown to impair the RNA-binding activity of recombinant AtGRP7 and the in vivo function (36,49). Thus, it is a valuable tool for future studies of the physiological consequences of AtGRP7 mis-expression on splicing of target genes identified here. Initial experiments to identify conserved sequence motifs surrounding the AS events in the direct AtGRP7 targets were not successful, presumably due to the small sample size. Moreover, current computational programs to identify conserved sequence motifs at the RNA level still have their limitations: In addition to the sequence context, structural features of the RNA are relevant (50).
In contrast to the 10 events with reciprocal changes, 3 AS events showed a significant change in the ratio of AS forms in the same direction both in atgrp7-1 8i and AtGRP7-ox plants (Supplementary Table S5). The identical effect of perturbation of AtGRP7 steady-state abundance in either direction could indicate that these AS events are controlled by a protein complex involving AtGRP7.
Loss of AtGRP7 affected fewer transcripts than elevated levels. Thus, AtGRP7 is clearly required for normal regulation of some AS events but is not limiting for others. This is likely to be due to functional redundancy with other splicing regulators such as other hnRNP proteins and, in particular, AtGRP8 that is still expressed at a low level in atgrp7-1 8i. Indeed, the interdependence of AtGRP7 and AtGRP8 makes it difficult to disentangle the effect of the paralogs on AS of downstream transcripts. Here, we show a clear overlap in AS of target transcripts which would be consistent with these proteins having similar RNA-binding properties (51,52). However, AtGRP7 also affected pre-mRNAs uniquely suggesting that its role in splice site selection is modulated by interactions with other proteins. Another feature of the interaction of AtGRP7 with pre-mRNAs is the over-representation of events involving alternative 5′ splice sites in those affected by AtGRP7. This observation is consistent with our previous results showing that elevated levels of AtGRP7 led to preferential use of an alternative 5′ splice site both within the AtGRP7 and AtGRP8 intron (30,31).
AtGRP7 and AtGRP8 levels are regulated by linked AS and NMD. For 7 of the 10 AS events with opposite splicing patterns in AtGRP7-ox and atgrp7-1 8i, a recent study has found that one of the splice isoforms is stabilized in upf mutants impaired in NMD and/or upon CHX treatment of wt plants (35). Although only a small sample of genes/transcripts, the enrichment of NMD substrates among AtGRP7 targets suggests that the impact of AtGRP7 on splicing events has functional consequences.
Finally, in corroborating putative AS targets of AtGRP7, we used independent over-expression lines in two different genetic backgrounds and found that most events were influenced by AtGRP7 over-expression in both ecotypes. Comparison of Col-2 and C24 wt plants showed that 70% of events were unaffected by the ecotype background. This demonstrates that the HR RT-PCR system can be useful in the analysis of splicing factor mutants even if these are in different ecotypes. On the contrary, almost 30% of the AS events showed significant changes (>5%, P < 0.05) between the two ecotypes. Many of these AS events had sequence variation in the form of SNPs/indels in the region of the splicing events. Thus, the panel can in turn complement transcriptome analysis of Arabidopsis accessions by high-throughput sequencing for detecting ecotype variation in AS (10,53). As the identified SNPs did not affect the splice site consensus sequences themselves, further systematic analysis of the SNPs/indels and their qualitative and quantitative effects on AS will be required to elucidate the impact of particular sequences in different sequence contexts and improve predictability of consequences of sequence variation.
Overlapping targets with other factors influencing AS
Splice site selection and assembly of the spliceosome depends on the recognition of sequences in the pre-mRNA by multiple protein factors and interactions between them. In humans, genome-wide mapping of human splicing regulatory proteins to their target RNA sequences is generating a ‘splicing code’ that will ultimately allow prediction of AS behavior of transcripts in different cells and tissues (8,18,54,55). For the most part, splicing factors will affect AS of numerous downstream target pre-mRNAs and their function will be modulated by interactions with other factors bound to the same pre-mRNA such that splicing behavior will be determined by the relative abundance and activity of different factors within a network of AS regulation (1). In plants, our knowledge of the functions of factors which affect AS is relatively limited particularly in terms of their binding sites and interactions (56–58). Information on the effects on splicing and AS of individual genes is increasing particularly for SR proteins but also for hnRNP proteins such as the PTB family, AtGRP7 and AtGRP8, and other RBPs (12,19,21,22,59–61) but virtually nothing is known about common targets of different factors.
The HR RT-PCR system has been used to examine the effects on AS of SR30, At-RS2Z33 (RSZ33) and cap-binding complex (CBC) proteins in addition to AtGRP7 and AtGRP8 (33,34). AS of RSZ33 (#21) was influenced in the same direction by over-expression of At-RS2Z33, SR30 and AtGRP7, respectively, leading to an AS form that does not produce functional protein (Supplementary Tables S2 and S5). Over-expression of SR30 altered splice site usage in At1g04400 (cryptochrome2, #2), At2g32320 (tRNA His Guanylyltransferase #19), At4g12790 (ATP-binding family protein, #36), At5g04430 (KH domain-containing protein NOVA, #42) in the same direction as over-expression of AtGRP7, and in At5g41150 (UV hypersensitive 1, #49) and At5g66010 (#59, unknown RNA-binding protein) in the opposite direction. Thus, we show here both an effect in the same direction and an antagonistic effect of SR proteins and the hnRNP-like AtGRP7, as often observed in mammals.
Mutants defective in components of the CBC also significantly affected 101 of 252 analyzed AS events (>3% change; P ≤ 0.1), implicating the CBC in the choice of AS sites (34). In the absence of complementary data on gain-of-function mutants or in vivo binding data, it is not yet clear which transcripts are direct targets. Notably, the CBC, like AtGRP7, also preferentially influences alternative 5′ splice site selection. Loss of CBP20 and/or CBP80 and the loss of AtGRP7 in atgrp7-1 8i altered splicing in the same direction in the case of At2g40830 encoding a zinc finger protein (#129) and of AFC2 (#227), suggesting that AtGRP7 and the cap binding complex have a similar effect on these AS events. In turn, for At5g18620 encoding chromatin remodeling factor 15 (#171), FYD (#288) and AKIN11 (#343) loss of CBP20 and/or CBP80 and loss of AtGRP7 shift the splicing ratio in the opposite direction, suggesting that AtGRP7 acts antagonistically to the CBC on these AS events.
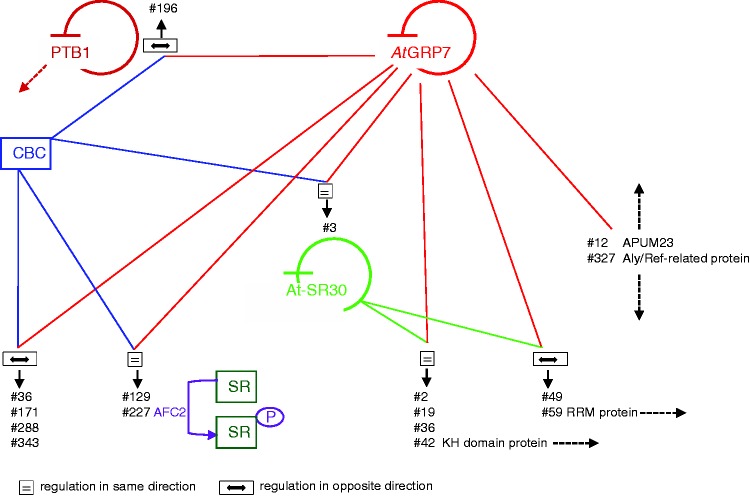
Many splicing factors in animals and plants are themselves regulated at the level of AS often involving conserved splice site sequences (62–64). AS of splicing factors that are involved in determining AS of other transcripts generates a hierarchical network of splicing regulators influencing downstream targets. For example, CBPs affect AS of PTB1, RS2Z33 and SR30 (34). Here, we show that AtGRP7 affects AS of transcripts encoding putative RBPs and predicted splicing regulators (Table 3), among those PTB1, RS2Z33 and SR30 which in turn influence splice selection of other targets. Among the targets of AtGRP7 is AFC2 encoding a LAMMER kinase. LAMMER kinases share an EHLAMMERILG motif in their catalytic subdomain X, giving rise to their name. In mammals, Drosophila and fission yeast members of the LAMMER kinase family are involved in AS through phosphorylation of SR proteins (65–67). Arabidopsis AFC and the tobacco ortholog PK12 phosphorylate SR proteins in vitro (68,69). Furthermore, heterologous expression of PK12 in transgenic Arabidopsis modulates AS of specific transcripts. For example, AtSR30 has an AS event involving alternative 3′ splice sites in intron 10 (#3) that is influenced in the same way upon expression of PK12 in transgenic Arabidopsis and over-expression of AtGRP7 or AtGRP8 (Supplementary Table S5). On the other hand, AFC2 itself is spliced into multiple AS variants, and skipping of exon 2 (#227) and of exons 5 and 6 (#226) resulting in unproductive mRNAs is promoted in AtGRP7-ox plants, pointing to a complex interaction. Skipping of exon 2 is reduced both in the cbp20 and cbp20 cbp80 (34) and atgrp7-1 8i mutants, suggesting that AtGRP7 and CBC both negatively impact exon2 inclusion.
In addition, ectopic AtGRP7 expression in Col-2 promoted skipping of exon 3 in PTB1 (#196). PTBs are hnRNP proteins that control AS of an extensive network of downstream transcripts in humans (70). In Arabidopsis, alternative inclusion of a cassette exon in PTB1 leads to a PTC and degradation via the NMD pathway (19) such that AtGRP7 favored the production of functional mRNA (Supplementary Table S2). Finally, AtGRP7 and AtGRP8 have been identified as substrates of the protein arginine methyltransferase AtPRMT5, an Arabidopsis homolog of human PRMT5 involved in methylation of histones and Sm proteins of spliceosomal small nuclear ribonucleoprotein particles (71). Mutation in AtPRMT5 leads to splicing defects in hundreds of genes, inviting the speculation that some of the PRMT5 effects may be mediated via AtGRP7 and/or AtGRP8 (32). A conceptual model of the interaction of AtGRP7 with known splicing regulators is shown in Figure 5.
Figure 5.
Conceptual model depicting common targets of known splicing regulators in Arabidopsis. The impact of AtGRP7, the cap binding complex (CBC) and At-SR30 on selected splicing events, analyzed by the RT-PCR panel, is displayed (33,34). The numbers of the primer pairs for detection of the AS events are indicated. Regulation of the ratio of splice isoforms in the same direction is indicated by ‘=’; regulation in opposite direction is indicated by a line with two arrowheads. For RNA-binding proteins that are influenced by AtGRP7, the name is indicated and dotted arrows indicate a presumed post-transcriptional regulation of yet unknown targets of these proteins. The negative autoregulation of AtGRP7, PTB1 and At-SR30 is depicted (19,31,57). ‘P’ denotes phosphorylation of SR proteins by the LAMMER kinase AFC2 (68). For clarity, the effects of AtGRP8 and At-RS2Z33 on AS events are omitted (see text for details).
In conclusion, our data indicate that the hnRNP-like proteins AtGRP7 and AtGRP8 are novel splicing regulators in Arabidopsis that affect a number of downstream targets. Both the influence of AtGRP7 on AS of other splicing regulators or RBPs and the observation that AtGRP7 shares targets with other splicing regulators like the CBC point to an extensive network of post-transcriptional regulation in Arabidopsis (18,72,73).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–3.
FUNDING
EMBO short term fellowship (to C.S.); German Research foundation [STA 653/2, SFB613 to D.S.]; Biotechnology and Biological Sciences Research Council (BBSRC) [BB/G024979/1—European Research Area network (ERA-NET) Plant Genomics (Plant Alternative Splicing and Abiotic Stress)]; EU FP6 Programme—European Alternative Splicing Network of Excellence (EURASNET) [LSHG-CT-2005-518238]; Scottish Government Rural and Environment Science and Analytical Services division (RESAS) (to J.W.S.B.). Funding for open access charge: German Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kristina Neudorf and John Fuller for expert technical assistance.
REFERENCES
- 1.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 2.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Heyd F, Lynch KW. DEGRADE, MOVE, REGROUP: signaling control of splicing proteins. Trends Biochem. Sci. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 5.Barta A, Kalyna M, Reddy AS. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell. 2010;22:2926–2929. doi: 10.1105/tpc.110.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CWJ, Valcarel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 7.Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2009;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez Y, Brown JWS, Simpson CG, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22:1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XC, Gassmann W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell. 2003;15:2333–2342. doi: 10.1105/tpc.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palusa SG, Ali GS, Reddy AS. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 13.Gulledge AA, Roberts AD, Vora H, Patel K, Loraine AE. Mining Arabidopsis thaliana RNA-seq data with Integrated Genome Browser reveals stress-induced alternative splicing of the putative splicing regulator SR45a. Am. J. Bot. 2012;99:219–231. doi: 10.3732/ajb.1100355. [DOI] [PubMed] [Google Scholar]
- 14.Wang B-B, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl Acad. Sci. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 17.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 18.Reddy ASN, Rogers MF, Richardson DN, Hamilton M, Ben-Hur A. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 2012;3:18. doi: 10.3389/fpls.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stauffer E, Westermann A, Wagner G, Wachter A. Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 2010;64:243–255. doi: 10.1111/j.1365-313X.2010.04321.x. [DOI] [PubMed] [Google Scholar]
- 20.Reddy AS, Day IS, Gohring J, Barta A. Localization and dynamics of plant splicing regulators. Plant Physiol. 2011;158:67–77. doi: 10.1104/pp.111.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalyna M, Lopato S, Barta A. Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell. 2003;14:3565–3577. doi: 10.1091/mbc.E03-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopato S, Kalyna M, Dorner S, Kobayashi R, Krainer AR, Barta A. atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999;13:987–1001. doi: 10.1101/gad.13.8.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wachter A, Rühl C, Stauffer E. The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 2012;3:81. doi: 10.3389/fpls.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambermon MH, Simpson GG, Wieczorek Kirk DA, Hemmings-Mieszczak M, Klahre U, Filipowicz W. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 2000;19:1638–1649. doi: 10.1093/emboj/19.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson CG, Jennings SN, Clark GP, Thow G, Brown JW. Dual functionality of a plant U-rich intronic sequence element. Plant J. 2004;37:82–91. doi: 10.1046/j.1365-313x.2003.01941.x. [DOI] [PubMed] [Google Scholar]
- 26.Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 1994;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staiger D. RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philos. Trans. Roy. Soc. Lond. B. Biol. Sci. 2001;356:1755–1759. doi: 10.1098/rstb.2001.0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staiger D, Heintzen C. The circadian system of Arabidopsis thaliana: forward and reverse genetic approaches. Chronobiol. Int. 1999;16:1–16. doi: 10.3109/07420529908998708. [DOI] [PubMed] [Google Scholar]
- 30.Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 31.Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 33.Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Barta A, Brown JW. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 2008;53:1035–1048. doi: 10.1111/j.1365-313X.2007.03392.x. [DOI] [PubMed] [Google Scholar]
- 34.Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, Szweykowska-Kulinska Z, Brown JW, Jarmolowski A. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010;38:265–278. doi: 10.1093/nar/gkp869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 37.Jeong H-J, Kim YJ, Kim SH, Kim Y-H, Lee I-J, Kim YK, Shin JS. Nonsense-Mediated mRNA Decay Factors, UPF1 and UPF3, Contribute to Plant Defense. Plant Cell Physiol. 2012;52:2147–2156. doi: 10.1093/pcp/pcr144. [DOI] [PubMed] [Google Scholar]
- 38.Staiger D, Apel K, Trepp G. The Atger3 promoter confers circadian clock-regulated transcription with peak expression at the beginning of the night. Plant Mol. Biol. 1999;40:873–882. doi: 10.1023/a:1006278030024. [DOI] [PubMed] [Google Scholar]
- 39.Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D. The small glycine-rich RNA-binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J. 2008;56:239–250. doi: 10.1111/j.1365-313X.2008.03591.x. [DOI] [PubMed] [Google Scholar]
- 40.Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 42.Staiger D, Apel K. Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol. Gen. Genet. 1999;261:811–819. doi: 10.1007/s004380050025. [DOI] [PubMed] [Google Scholar]
- 43.Streitner C, Hennig L, Korneli C, Staiger D. Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 2010;10:221. doi: 10.1186/1471-2229-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heintzen C, Fischer R, Melzer S, Kappeler S, Apel K, Staiger D. Circadian oscillations of a transcript encoding a germin-like protein that is associated with cell walls in young leaves of the long-day plant Sinapis alba L. Plant Physiol. 1994;106:905–915. doi: 10.1104/pp.106.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lummer M, Humpert F, Steuwe C, Schüttpelz M, Sauer M, Staiger D. Reversible photoswitchable DRONPA-s monitors nucleocytoplasmic transport of an RNA-binding protein in transgenic plants. Traffic. 2011;12:693–702. doi: 10.1111/j.1600-0854.2011.01180.x. [DOI] [PubMed] [Google Scholar]
- 46.Weigel D, Mott R. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terzi LC, Simpson GG. Arabidopsis RNA immunoprecipitation. Plant J. 2009;59:163–168. doi: 10.1111/j.1365-313X.2009.03859.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide Identification of Polycomb-Associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong B-r, Lin Y, Joe A, Guo M, Korneli C, Yang H, Wang P, Yu M, Cerny RL, Staiger D, et al. Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity. J. Biol. Chem. 2011;286:43272–43281. doi: 10.1074/jbc.M111.290122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, et al. The RNA structure alignment ontology. RNA. 2009;15:1623–1631. doi: 10.1261/rna.1601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schüttpelz M, Schöning JC, Doose S, Neuweiler H, Peters E, Staiger D, Sauer M. Changes of conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J. Am. Chem. Soc. 2008;130:9507–9513. doi: 10.1021/ja801994z. [DOI] [PubMed] [Google Scholar]
- 52.Fuhrmann A, Schoening JC, Anselmetti D, Staiger D, Ros R. Quantitative analysis of single-molecule RNA-protein interaction. Biophys J. 2009;96:5030–5039. doi: 10.1016/j.bpj.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 55.Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 56.Pertea M, Mount SM, Salzberg SL. A computational survey of candidate exonic splicing enhancer motifs in the model plant Arabidopsis thaliana. BMC Bioinformatics. 2007;8:159. doi: 10.1186/1471-2105-8-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barta A, Kalyna M, Lorkovic ZJ. Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 2008;326:83–102. doi: 10.1007/978-3-540-76776-3_5. [DOI] [PubMed] [Google Scholar]
- 58.Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 59.Zhang XN, Mount SM. Two alternatively spliced isoforms of the Arabidopsis thaliana SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009;150:1450–1458. doi: 10.1104/pp.109.138180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song H-R, Song J-D, Cho J-N, Amasino RM, Noh B, Noh Y-S. The RNA Binding Protein ELF9 Directly Reduces SUPPRESSOR OF OVEREXPRESSION OF CO1 Transcript Levels in Arabidopsis, Possibly via Nonsense-Mediated mRNA Decay. Plant Cell. 2009;21:1195–1211. doi: 10.1105/tpc.108.064774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng Y, Kato N, Wang W, Li J, Chen X. Two RNA binding proteins HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev. Cell. 2003;4:53–66. doi: 10.1016/s1534-5807(02)00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 63.Kalyna M, Lopato S, Voronin V, Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duncan PI, Stojdl DF, Marius RM, Bell JC. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol. Cell. Biol. 1997;17:5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Z, Luca M, Portillio J, Ngo B, Chang C, Wen T, Murray J, Carr A. LAMMER kinase Kic1 is involved in pre-mRNA processing. Exp. Cell Res. 2011;317:2308–2320. doi: 10.1016/j.yexcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 67.Du C, McGuffin ME, Dauwalder B, Rabinow L, Mattox W. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol. Cell. 1998;2:741–750. doi: 10.1016/s1097-2765(00)80289-0. [DOI] [PubMed] [Google Scholar]
- 68.Savaldi-Goldstein S, Sessa G, Fluhr R. The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J. 2000;21:91–96. doi: 10.1046/j.1365-313x.2000.00657.x. [DOI] [PubMed] [Google Scholar]
- 69.Savaldi-Goldstein S, Aviv D, Davydov O, Fluhr R. Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell. 2003;15:926–938. doi: 10.1105/tpc.011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B, Hu S, et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl Acad. Sci. USA. 2010;107:19114–19119. doi: 10.1073/pnas.1009669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Syed NH, Kalyna M, Marquez Y, Barta A, Brown JWS. Alternative splicing in plants - coming of age. Trends Plant Sci. 2012;17:616–623. doi: 10.1016/j.tplants.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staiger D, Green R. RNA-based regulation in the plant circadian clock. Trends Plant Sci. 2011;16:517–523. doi: 10.1016/j.tplants.2011.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.