Misleading morphological observations assigned Alexandrium catenella as local dinoflagellate responsible for HABs in Southern Chilean coasts. Our work based on molecular methods found that local Alexandrium belongs to group I of the tamarensis complex composed mainly of A. tamarense.
Abstract
Background and aims
On the basis of morphological evidence, the species involved in South American Pacific coast harmful algal blooms (HABs) has been traditionally recognized as Alexandrium catenella (Dinophyceae). However, these observations have not been confirmed using evidence based on genomic sequence variability. Our principal objective was to accurately determine the species of Alexandrium involved in local HABs in order to implement a real-time polymerase chain reaction (PCR) assay for its rapid and easy detection on filter-feeding shellfish, such as mussels.
Methodology
For species-specific determination, the intergenic spacer 1 (ITS1), 5.8S subunit, ITS2 and the hypervariable genomic regions D1–D5 of the large ribosomal subunit of local strains were sequenced and compared with two data sets of other Alexandrium sequences. Species-specific primers were used to amplify signature sequences within the genomic DNA of the studied species by conventional and real-time PCR.
Principal results
Phylogenetic analysis determined that the Chilean strain falls into Group I of the tamarensis complex. Our results support the allocation of the Chilean Alexandrium species as a toxic Alexandrium tamarense rather than A. catenella, as currently defined. Once local species were determined to belong to Group I of the tamarensis complex, a highly sensitive and accurate real-time PCR procedure was developed to detect dinoflagellate presence in Mytilus spp. (Bivalvia) samples after being fed (challenged) in vitro with the Chilean Alexandrium strain. The results show that real-time PCR is useful to detect Alexandrium intake in filter-feeding molluscs.
Conclusions
It has been shown that the classification of local Alexandrium using morphological evidence is not very accurate. Molecular methods enabled the HAB dinoflagellate species of the Chilean coast to be assigned as A. tamarense rather than A. catenella. Real-time PCR analysis based on A. tamarense primers allowed the detection of dinoflagellate DNA in Mytilus spp. samples exposed to this alga. Through the specific assignment of dinoflagellate species involved in HABs, more reliable preventive policies can be implemented.
Introduction
Harmful algal blooms (HABs) occur throughout the world and are known for their negative economic and sanitary impacts (Anderson 2009). Of particular concern are the paralytic shellfish toxins produced mainly by bloom-forming dinoflagellates in the genus Alexandrium. Over the past few decades, Alexandrium blooms have extended, covering new territories. In this sense, expansion of dinoflagellate species could be explained by ocean currents, human-induced mechanisms such as water from ballasts and global warming, climate adaptation and colonization of newly generated niches (Anderson 1989). On the other hand, it is also possible that the increasing number of blooms reported today is the result of a worldwide effort to implement new techniques to detect and prevent their negative effects (Anderson 2007). Blooms of different Alexandrium species have been reported from Japan (Kodama 2010), northwestern Mediterranean Sea (Vila et al. 2001), Australia (Hallegraeff 1998), Caribbean Sea, off the Venezuelan coast (Halstead and Schantz 1984), Brazil (Persich et al. 2006), along the American Pacific coasts, from Alaska to the Strait of Magellan (Suárez et al. 2002; Glibert et al. 2005; Hernández et al. 2005), and from north Atlantic coasts from the Gulf of St Lawrence to North Carolina (Anderson et al. 1994). Compared with the numerous studies describing Australian, North American and Japanese Alexandrium ribotypes and morphotypes, relatively few works describe their South American counterparts from the South Pacific (Lilly 2003).
Historically, Alexandrium species were described based on microscopic observations of morphological features including plate patterns, cell size and shape, and secondary characteristics such as chain formation. Unfortunately, these morphological traits have often proven insufficient for identifying species, leading to confusion concerning the distribution, ecology and toxicity within this genus (Lilly et al. 2005). When morphological features are questionable for taxonomic identification, they must be combined with molecular data for accurate species definition (Hansen et al. 2003).
Sequence variation analyses have been accepted to be a valid methodology for an accurate species description. This is even clearer in the Alexandrium genus, which has been partially reclassified on the basis of molecular genetic data, as the taxonomic value of only morphological characters proved to be insufficient for the tamarensis complex (Leaw et al. 2005). A good example is the taxonomic trait ‘presence or absence of the ventral pore’, used to discriminate between A. affine and Alexandrium tamarense. This trait would not be deemed useful for species identification considering that it is homoplastic (Leaw et al. 2005).
Using the classical species definition for lineage formation (Mayr 1982), Lilly et al. (2007) recognize Alexandrium tamarense (Lebour) Balech, Alexandrium catenella (Whedon and Kofoi) Balech and Alexandrium fundyense Balech as different species. On the other hand, phylogenies of Alexandrium species have been established based on genomic sequences of the large and small subunits of ribosomal DNA (LSU and SSU rDNA, respectively) (Guillou et al. 2002; Usup et al. 2002; John et al. 2003, 2005; Murray et al. 2005; Rogers et al. 2006). Of these sequences, the D1/D2 region of the LSU rDNA has been proved to be the most suited for discrimination of closely related Alexandrium species (Ki and Han 2007). Thus, this hypervariable region has been proposed as a suitable candidate to discriminate between species with similar fidelity as Cytochrome Oxidase I gene (Sonnenberg et al. 2007). Scholin and Anderson (1994, 1996) and Scholin et al. (1994, 1995), based on DNA sequencing of the divergent D1/D2 LSU rDNA region and restriction fragment length polymorphism (RFLP) analysis of the small subunit rDNA genes, consider that they could be strains of the same species, naming them the tamarensis complex. Using these results as a starting point, Lilly et al. (2007) further established the tamarensis complex as a valid cluster, derived from a phylogenetic analysis comparing more than 126 different Alexandrium D1/D2 region sequences. Their detailed examination revealed the presence of five clades, defined as: Group I (North American), Group II (Mediterranean), Group III (Western European), Group IV (Temperate Asian) and Group V (Tasmanian).
In this study, we sequenced local Alexandrium intergenic spacer 1 (ITS1), 5.8S rDNA, ITS2 and D1–D5 hypervariable LSU rDNA regions in order to incorporate molecular data that could help define more clearly the Alexandrium species responsible for HABs in Chilean coasts. This is considering that the local South American Pacific Alexandrium species were classified as A. catenella (Muñoz 1985), mainly based on morphological traits, but without a further assessment of sequence variation. In this sense, species-specific assignment allows the implementation of a polymerase chain reaction (PCR) assay for accurate monitoring along Chilean coasts in order to prevent public health hazards and economic losses.
Methods
Cell cultures
Three different clonal Alexandrium cell cultures (ACC01, ACC02, ACC07) were kindly provided by Professor Benjamín Suárez from the Laboratorio de Toxinas Marinas, Universidad de Chile. These were collected from Canal Costa in Aysén region, Chile (45°37′60 S, 73°32′60 W), between April 1994 and March 1995, and maintained in f/2 medium (Guillard 1975) at 12 °C under a 16 : 8 h light:dark cycle and 60 μmol m−2 s−1 photon flux density. These three strains belong to the Chilean algae repository collection used in various national and international studies (Córdoba and Müller 2002; Amaro et al. 2005; Lilly et al. 2007; Montoya et al. 2010).
DNA purification from cell cultures
Cells for analysis (100 mL) were collected from each clonal culture at mid-logarithmic phase and centrifuged at 3000 g for 5 min at 4 °C. The supernatant was removed, the pellet resuspended in 500 μL of Milli-Q water, and transferred to a 1.5-mL microfuge tube. Microfuge tubes were placed in liquid nitrogen for 30 s and the cells subsequently disrupted for 1 min using an Axygen polypropylene pestle (PES-15-B-SI, Union City, CA, USA). Genomic DNA was then extracted from the pellet using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Determination of Alexandrium DNA concentration and quality
DNA concentration was measured using a fluorometer (Qubit, Sunnyvale, CA, USA) together with the Qubit dsDNA BR Assay Kit (Invitrogen, Eugene, OR, USA). The quality was evaluated based on its integrity by comparison with a 23-kb band of λ-HindIII ladder (Invitrogen, USA) in 1 % agarose (Ultrapure, Invitrogen, Barcelona, Spain) gel electrophoresis stained with ethidium bromide.
PCR amplification and sequence analysis
In order to determine the species of the three isolates, the ITS1-D5 rDNA was amplified, sequenced and then aligned with sequences from GenBank (Ki and Han 2007). All amplifications were carried out in duplicate with 1× PCR buffer, 20–50 ng of genomic DNA template, 3 mM MgCl2, 100 µM each dNTP, 0.1 µM each primer and 0.4 U of recombinant Taq DNA polymerase (MBI Fermentas, Vilnius, Lithuania) in a 10-µL reaction volume. Polymerase chain reaction primer sequences for LSU rDNA and optimized annealing temperatures are specified in Table 1. Polymerase chain reaction parameters were: 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 30 s, annealing for 30 s, extension at 72 °C for variable time spans which depended upon the size of the amplifying fragment (1000 bases min−1); and a final extension at 72 °C for 5 min. Reactions were run on a MaxyGene Gradient thermocycler (Axygen). Five microlitres of PCR products were analysed by 2 % agarose (Invitrogen, USA) gel electrophoresis according to standard methods. rDNA PCR amplification products from clones ACC01, ACC02 and ACC07 were purified from gels using the MinElute Gel Extraction Kit (Qiagen) following the manufacturer's instructions and directly sequenced in an ABI PRISM 3100. An electropherogram base quality assignment algorithm, phred (Ewing and Green 1998; Ewing et al. 1998), was used to re-analyse the sequenced fragment of all strains in order to determine intragenomic polymorphic sites.
Table 1.
Primer sequences used in amplifying the ITS1-D5 region in Alexandrium species.
| Primer name | Nucleotide sequence 5′ to 3′ | Annealing temperature (°C) | Reference |
|---|---|---|---|
| catF | cctcagtgagattgtagtgc | Between 45 and 65 | Hosoi-Tanabe and Sako (2005) |
| catR | gtgcaaaggtaatcaaatgtcc | ||
| tamF | tgcttggtgggagtgttgca | 66 | Hosoi-Tanabe and Sako (2005) |
| tamR | taagtccaaggaaggaagcatc | ||
| tamF-1 | tgagggaaatatgaaaaggac | TD 58–48 (−0.5 ) | This study |
| tamR-1 | attcggcaggtgagttgtta | ||
| tamF-2 | gaaggagaagtcgtaacaagg | TD 58–48 (−0.5 ) | This study |
| tamR-2 | caatgccaaggagtgtgac |
The annealing temperature for each primer and the study from which the sequences were obtained are listed.
Additional confirmation of species identification was achieved by amplifying the extracted DNA using four microsatellite primers specific to A. catenella (Nagai et al. 2005; Table 2) or A. tamarense (Alpermann et al. 2006; Table 2). The amplification conditions were the same as those provided in the original publications describing the assays.
Table 2.
Primer sequences used in amplifying species-specific microsatellite genomic regions in A. catenella and A. tamarense.
| Primer | Nucleotide sequence 5′ to 3′ | Annealing temperature (°C) | Species | Reference |
|---|---|---|---|---|
| Acat02-F | caagtgaactaaatccgct | 60 | A. catenella | Nagai et al. (2005) |
| Acat02-R | aaaacggaatgtttatgtgc | |||
| Acat16-F | tgtctttcttcctgcctgcctt | 60 | A. catenella | Nagai et al. (2005) |
| Acat16-R | ttcaccccagcgaagccattatg | |||
| Acat20-F | aggagaaaagtgatgcatctcagcaa | 60 | A. catenella | Nagai et al. (2005) |
| Acat20-R | aatcctgtggatgatggaaggtactg | |||
| Acat44-F | tgccccataagggttcttccaga | 60 | A. catenella | Nagai et al. (2005) |
| Acat44-R | gacagtggtattgcaaacccaacggat | |||
| ATB1-F | cgcctgctcgagaaaaga | 53 | A. tamarense | Alpermann et al. (2006) |
| ATB1-R | ttgggggacagttgagtttc | |||
| ATB8-F | cagggtagccgatcaaacac | TD 61–54 (−0.3) | A. tamarense | Alpermann et al. (2006) |
| ATB8-R | cttccatcgccttgcatact | |||
| ATD8-F | caacactggaagcgtgctaa | TD 61–54 (−0.3) | A. tamarense | Alpermann et al. (2006) |
| ATD8-R | cccatgcgctacctcttaca | |||
| ATF11-F | agcagcgcggcgggagatt | TD 68.5–61 (−0.3) | A. tamarense | Alpermann et al. (2006) |
| ATF11-R | acctgcggctgcgacacgact |
The annealing temperature for each primer, the species and the study from which the sequences were obtained are listed.
TD, touchdown.
Collection, analysis and association of sequences
All selected Alexandrium sequences were obtained using the keywords ‘Alexandrium LSU rDNA’ or ‘Alexandrium 28S in the GenBank database (http://www.ncbi.nlm.nih.gov) [see ADDITIONAL INFORMATION]. For a detailed species-specific analysis, two data sets were generated. The first was composed of 81 unique sequences at least 641 bp long, covering the D1/D2 region of the LSU rDNA. This group incorporated 79 Alexandrium genus species (including the local strain), and two Prorocentrum micans strains that were used as outgroups. The second set had 18 sequences at least 1776 bp long, of which 16 corresponded to tamarensis complex (including the local strain), one to A. minutum and one to A. affine; the last two were used as outgroups. For both data sets, alignments were carried out in the ClustalX2 V2.0 (Larkin et al. 2007) graphical platform.
Substitution model and associated parameter estimation
jModeltest (Posada 2008) was used to find the best substitution model and associated parameters for phylogenetic analysis in both data sets using the Akaike (Hirotugu 1974) and Bayesian (Schwarz 1978) information criteria.
Phylogenetic analysis
Bayesian analysis was implemented with MrBayes V3.2 (Ronquist et al. 2012) for the first and second data sets, and was carried out with 1 500 000 runs, with five separate initial trees, with the Markov chain Monte Carlo (MCMC) process set to four chains and 25 % of initial trees discarded as ‘burn-in’. Within each chain, samples were obtained every 100 iterations, and the values of the average deviation of split frequencies (AVSF) and potential scale reduction factor (PSRF) were obtained. These values were used to evaluate convergence of the generated trees. Additionally, maximum likelihood analysis was carried out in PhyML V3.0 (Guindon and Gascuel 2003) in order to further support taxon assignment. Analysis was started with a random tree sample (Subtree pruning and regrafting method) and 1000 bootstrap replicate runs.
Figtree V1.3.1 (Andrew Rambaut. FigTree v1.3.1 2006–2009. http://tree.bio.ed.ac.uk/software/figtree) was used for a graphical visualization and representation of PhyML and MrBayes output trees.
In order to estimate the nucleotide differences within the tamarensis complex in each data set, the values for the average number of differences per pair of sequences aligned within each group and the number of parsimonious informative sites were calculated in the MEGA 5.05 program (Tamura et al. 2011).
Alexandrium DNA detection in challenged Mytilus samples by real-time PCR
An in situ experimental protocol for dinoflagellate challenge was developed in order to implement it subsequently for Alexandrium detection in filter-feeding shellfish and water columns. The experiments were performed in quadruplicate using four experimental aquaria of 15 L, containing five mussels each (one mussel from each aquarium for each of the 5 days was used for DNA extraction).
Individuals of the edible mussel Mytilus were transported to the laboratory, where they were acclimatized for 1 week at 14 °C and seawater salinity of 30 practical salinity units. During this period, mussels were continuously fed with the microalga Isochrysis galbana at 1.5 mg L−1 using a peristaltic pump and providing constant aeration. The seawater was changed every 48 h. Following acclimation, the mussels were exposed to a contaminated diet (1.7–2.0 mg L−1; dry weight) containing 50 % toxic dinoflagellate Alexandrium strain ACC02 and 50 % I. galbana (by weight) for a period of 12 days, followed by a detoxification period of 15 days where they were fed with I. galbana. Every day the aquaria received an amount of food representing 2 % of the dry body weight of the experimental mussels (Shafee 1976), delivered continuously using a Masterflex 7519-05 peristaltic pump at the temperature and salinity cited above. One mussel from each replicate aquarium was taken on Days 2, 3, 4 and 5 of the toxic feeding cycle and on Day 15 of the detoxification cycle. Animals were immediately processed for purification of gill DNA.
To determine the possibility of detecting Alexandrium DNA in challenged mussel tissue, we used real-time PCR. Experiments were run in triplicate, for each sample, on an Eco real-time PCR System (Illumina, San Diego, CA, USA) using Quantace SensiMix HRMtm kit (Bioline, London, UK). Reaction conditions were: 1 × SensiMix HRM buffer, 0.6 μL of EvaGreen dye, 0.5 μM primers tamF and tamR (Hosoi-Tanabe and Sako 2005), and 100 ng of challenged Mytilus spp. gill DNA in a 10-μL final reaction volume. The PCR protocol cycling was: a 10-min initial activation step at 94 °C, followed by 40 cycles of 94 °C for 30 s, 55.3 °C for 30 s and 72 °C for 30 s. A PCR amplification product of 235 bp (Fig. 1) obtained with tamF and tamR was purified and sequenced in order to corroborate specificity.
Fig. 1.
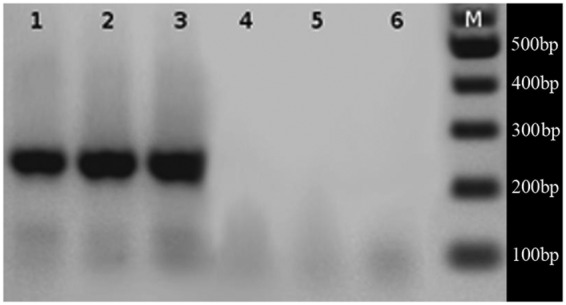
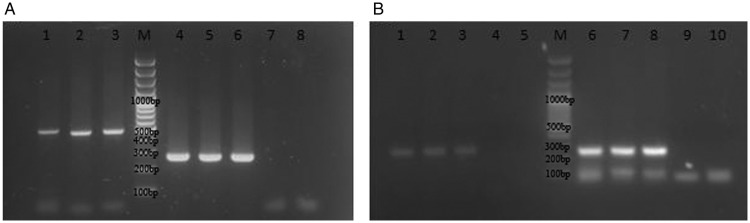
Amplification of local Alexandrium strains ACC01, ACC02 and ACC07 using species-specific primers; A. tamarense (lanes 1, 2 and 3) and A. catenella (lanes 4, 5 and 6). M = 100-bp DNA size marker. Species-specific amplification in the rDNA region using A. catenella and A. tamarense primers were carried out in a MaxiGene Gradiente thermocycler (Axygen) in 1× PCR buffer, 20–50 ng of genomic DNA template, 3 mM MgCl2, 100 µM each dNTP, 0.1 µM each primer and 0.4 U of TopTaq DNA polymerase (Fermentas) in a 10-µL reaction volume. Five microlitres of each PCR product were analysed in a 2 % agarose gel. A Fermentas GeneRuller™ 100-bp DNA ladder was used for size estimation of amplified fragments.
DNA purification from challenged Mytilus gill tissue
Mussels were randomly taken from each of the four aquaria (replicates) on each day of sampling. Animals were dissected alive and 30 g of drained gill tissue were used as the starting material for DNA purification with DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's protocol. Each sample of purified DNA was stored at −20 °C. As explained below, the gill was considered as a useful source of Alexandrium DNA as particulate materials, such as unicellular organisms, tend to accumulate in this organ (Jørgensen 1996; Riisgard et al. 1996) and other tissues such as the hepatopancreas do not provide DNA with the integrity needed for this type of study.
Results
PCR amplification of microsatellite genomic regions
The results of the species-specific PCR of eight microsatellite genomic regions were consistent in all three isolates, amplifying only those directed towards A. tamarense and not to A. catenella (Fig. 2). No changes could be observed with the A. catenella set of primers despite the numerous protocol modifications of PCR conditions.
Fig. 2.
Alexandrium tamarense microsatellite amplifications of the three local strains with (A) specific primers ATB8 (lanes 1, 2 and 3) and ATD8 (lanes 4, 5 and 6). Lanes 7 and 8 correspond to controls with primers ATB8 without DNA. (B) Specific amplification with primers ATB1 (lanes 1, 2 and 3) and ATF11 (lanes 6, 7 and 8). Lanes 4–5 and 9–10 are controls without DNA for primer sets ATB1 and ATF11, respectively. M = 100-bp DNA size marker. Species-specific microsatellite amplifications using A. catenella and A. tamarense primers were carried out in a MaxiGene Gradiente thermocycler (Axygen) in 1× PCR buffer, 20–50 ng of genomic DNA template, 3 mM MgCl2, 100 µM each dNTP, 0.1 µM each primer and 0.4 U of TopTaq DNA polymerase (Fermentas) in a 10-µL reaction volume. Five microlitres of each PCR product were analysed in a 2 % agarose gel. A Fermentas GeneRullerTM 100-bp DNA ladder was used for size estimation of amplified fragments.
Analysis based on ITS1-D5 LSU rDNA sequences
Sequence analysis and evaluation
Electropherogram profiles from the ITS1-D5 region of the rDNA of strains ACC01, ACC02 and ACC03 were analysed with the phred algorithm in order to discard the presence of pseudogenes or intragenomic rDNA polymorphisms (IRP). Only bases with scores over 30 (sequencing error probability 1/1000) were considered for further analysis. As no nucleotide differences were obtained for this region between the three local Alexandrium strains, only one sequence, Ach01, was used as a representative of them (NCBI accession no. JN657223).
Substitution model and associated parameter evaluation
Analysis by jModeltest estimated that the GTR + Γ was the best substitution model for later Bayesian and maximum likelihood analysis. If a given model was not an option in MrBayes, the following least restrictive model was used (e.g. GTR).
Phylogenetic analysis
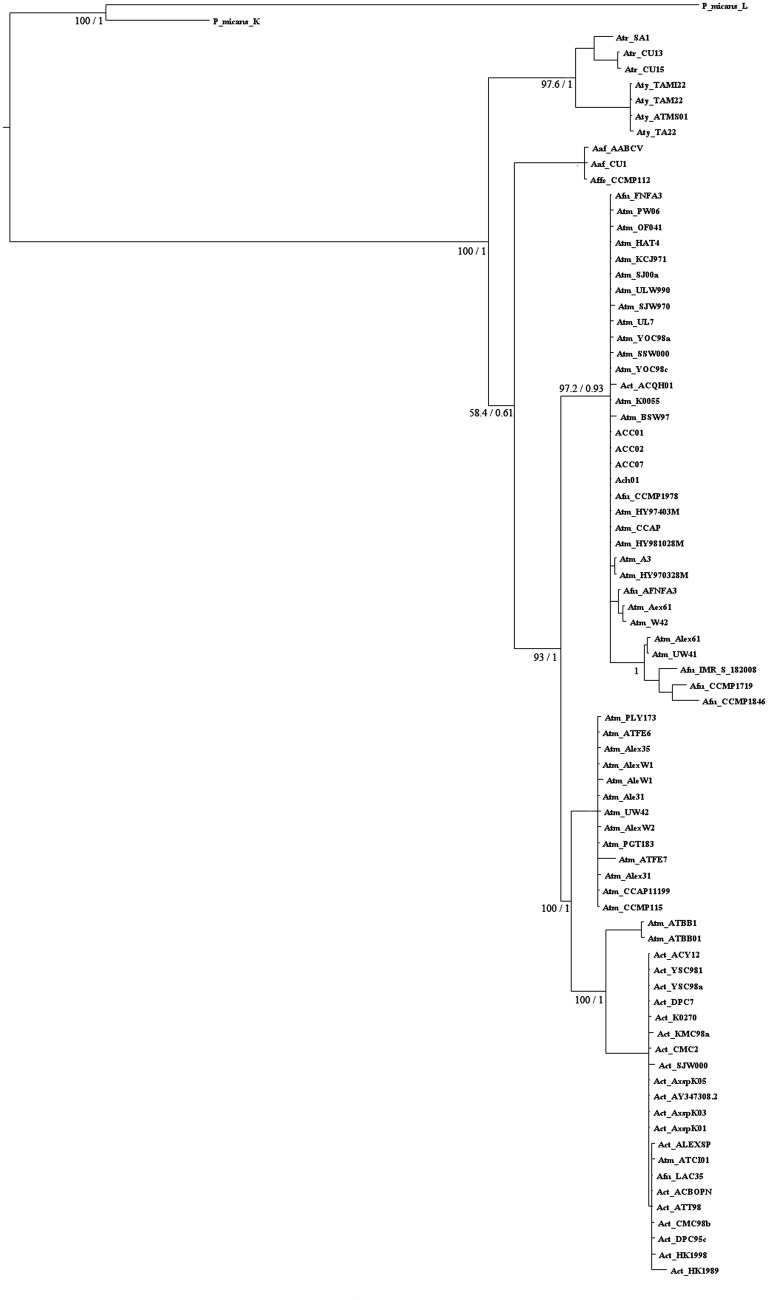
Using 81 sequences of length 641 bp, all aligning in the same D1/D2 LSU rDNA region, from different species of the genus Alexandrium and two strains of P. micans, the phylogenetic distribution of local Alexandrium strains could be estimated (Fig. 3). Convergence of Bayesian trees was evaluated through AVSF and PSRF. Values were less than 0.01 and ∼1 ±0.005, respectively, suggesting that the distribution reached a stationary phase in both data sets. Additionally, bootstrap values and the logarithm of the likelihood score of the optimal tree (−4229.43342) extracted by maximum likelihood analysis were indicative of a precise tree. The results indicate that the local Alexandrium strain is in Group I, dominated by A. tamarense, and is grouped with other previously sequenced Alexandrium strains from Chilean waters (ACC01, ACC02 and ACC07; Fig. 3).
Fig. 3.
Phylogenetic tree of 80 Alexandrium species and strains (Atm, A. tamarense; Act, A. catenella; Afu, A. fundyense; Amn, A. minutum; Aaf, A. affine; Atr, A. tropicale; Aty, A. tamiyanavichii; P. micans, Prorocentrum micans) and local Alexandrium species Ach01. Sequences were obtained from the GenBank database using the keywords ‘Alexandrium LSU rDNA’ or ‘Alexandrium 28S’. Phylogenetic trees were generated for an alignment of 81 sequences of 641 bp in the D1/D2 region through Bayesian inference in MrBayes V3.2 and maximum likelihood (ML) in PhyML. Bayesian analysis was carried out with 1 500 000 runs, with five separate initial trees. Convergence was checked through PSRF and AVSF. FigTree V1.3.1 was used as a visual representation of output trees. For ML analysis these were carried out with 1000 bootstrap replicates.
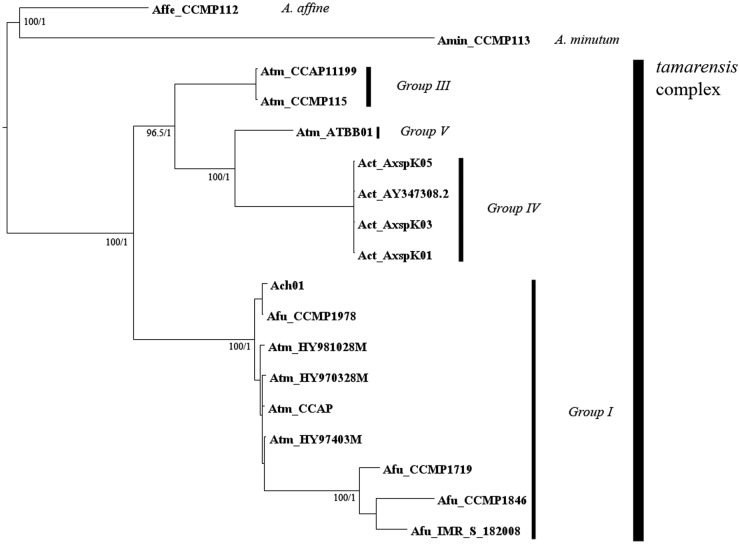
In order to achieve further resolution of the tamarensis complex, 18 sequences longer than 1776 bp, from different species and strains of the tamarensis complex, located in the rDNA region were analysed (Fig. 4). The total alignment involved ITS1, 5.8S rDNA, ITS2 and 1185-bp of the D1–D5 regions of the 28S rDNA (Ki and Han 2007). Sequences of A. minutum and A. affine were selected as outgroup species for the tamarensis complex.
Fig. 4.
Phylogenetic tree of 17 Alexandrium species and strains (Atm, A. tamarense; Act, A. catenella; Afu, A. fundyense; Amn, A. minutum; Aaf, A. affine) and local Alexandrium species Ach01. Sequences were obtained from the GenBank database using the keywords ‘Alexandrium LSU rDNA’ or ‘Alexandrium 28S’. Phylogenetic trees were generated for an alignment of 18 sequences of 1776 bp in the ITS1-D5 region through Bayesian inference in MrBayes V3.2 and maximum likelihood (ML) in PhyML. Bayesian analysis was carried out with 1 500 000 runs, with five separate initial trees. Convergence was checked through PSRF and AVSF. FigTree V1.3.1 was used as a visual representation of output trees. For ML analysis these were carried out with 1000 bootstrap replicates.
For maximum likelihood analysis, the logarithm of the likelihood score of the optimal tree was −6591.25984. As expected, the results were again consistent and the local Alexandrium strain was allocated to Group I of the tamarensis complex.
Topologies for trees generated through maximum likelihood and Bayesian inference were the same, and had high bootstrap and posterior probability values (Figs 3 and 4). Group generation was carried out analysing clades formation and the previous literature. Comparing the structure of both trees, using 18 or 81 sequence alignments, the same clades were formed.
In order to obtain information on the variability of the amplified region for both data sets, the average number of differences per pair of sequences aligned and the number of parsimonious sites were measured. For the first data set, values were 8.37 bp and 130, respectively. On the other hand, for the second data set, the average number of differences per pair of sequences aligned was 52.03 bp and the number of parsimonious sites was 298. Considering the 1185-bp segment covering only the LSU rDNA region, the amount of parsimonious informative sites and the average number of differences per pair of sequences aligned decreases from 298 to 174 and from 52.02 to 35 bp, representing a fall of 42 and 32 %, respectively, with respect to the whole amplified region.
Alexandrium DNA detection in challenged Mytilus samples by real-time PCR
From each aquarium, DNA from the gill tissues of five different randomly picked Mytilus challenged in vivo with Alexandrium strain ACC02 were used to detect the presence of A. tamarense through a real-time PCR assay with species-specific primers. We obtained similar and positive results for the samples, which were extracted between Days 2 and 5 of the toxicfeeding phase, with Ct values ranging from 21 to 22. On the other hand, all samples extracted on Day 15 of the detoxification phase gave no specific amplification, indicating that there was no detectable Alexadrium DNA in the Mytilus samples (Table 3). Replicates of Mytilus samples challenged in parallel in four independent aquaria showed the same pattern, confirming the detection of Alexandrium in Mytilus gills during the toxic phase. Through amplicon melt analysis and direct sequencing of the 235-bp PCR-amplified fragment, the region corresponded to the expected specific sequence within the D1/D2 LSU rDNA domain, according to Hosoi-Tanabe and Sako (2005).
Table 3.
Detection of Alexandrium DNA in gill tissue samples from challenged Mytilus through days 2 to 27 by q-PCR.
| Sample | Challenge periods (days) | Ct |
|---|---|---|
| 1 | 2a | 21.0 |
| 2 | 3a | 21.2 |
| 3 | 4a | 21.1 |
| 4 | 5a | 22.0 |
| 5 | 15b | NA |
Mytilus were exposed to a contaminated diet (1.7–2.0 mg L−1; dry weight) containing 50 % toxic dinoflagellate Alexandrium strain ACC02 and 50 % I. galbana (by weight) for a period of 12 days, followed by a detoxification period of 15 days, where they were fed with I. galbana. Animals were dissected alive on Days 2, 3, 4 and 5 of the toxic phase and Day 15 of the detoxification phase, and 30 g of drained gill tissue were used as the starting material for DNA purification with the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's protocol. Real-time PCR assays using species-specific A. tamarense primers were carried out in extracted DNA from Mytilus in order to determine the possibility of detecting Alexandrium DNA in challenged mussel tissue. NA, no amplification.
aDays in toxic phase.
bDays in detoxification phase.
Discussion
The study of HABs has become increasingly important given the recent rise in the number and frequency of toxic events, and associated adverse impacts on public health, fisheries and ecosystem services (Anderson 1989; Cassis et al. 2002). This threat has led to extensive investigation on how to develop international standards for the detection of toxins in seafood and on the implementation of expanded monitoring programmes for toxic algae. One of the most frequently used methods for the evaluation of toxins in HAB episodes worldwide has been the mouse bioassay. Unfortunately, this technique does not always provide accurate estimates of toxicity (Fernández et al. 2002) and requires considerable resources and time. Moreover, this technique has been questioned with regard to animal welfare and is prohibited in some countries. In this context, increasing efforts have been focused on monitoring the toxic algae directly to predict their occurrence and better allocate sampling effort, particularly with regard to toxin analysis.
In this paper, we present phylogenetic analyses using ITS1-D5 rDNA sequence data which demonstrate that the Chilean strains analysed belong to Group I in the tamarensis complex, rather than to the A. catenella grouping (Scholin et al. 1994; Medlin et al. 1998; Higman et al. 2001; Lilly et al. 2007). The first studies concerning Chilean HABs carried out 40 years ago, based on morphological observations, identified A. catenella as the dominant Alexandrium species (Muñoz 1985). This identification has never been questioned or assessed by more accurate methods such as genomic sequencing. The phylogenetic analysis carried out in this study clearly indicates that the local Alexandrium species belongs to the Group I ribotype. This group is mainly composed of A. tamarense, in contrast to Group IV in which the predominant species is A. catenella. Similarly, phylogenetic trees based on Alexandrium toxin variability showed that strains from Argentina, Brazil, Chile and Uruguay belonged to the same clade, paralleling the Group I results (Montoya et al. 2010). As red tide blooms have been present since the 19th century in Chile and Brazilian, Uruguayan and Argentinian events are more recent, it has been proposed that toxic episodes in Eastern South America could be due to the expansion of Chilean Alexandrium species (Lilly et al. 2007). Even more, Uruguayan and Brazilian strains have been classified as A. tamarense in Group I, consistent with our findings in relation to the fact that Chilean species, supporting the hypothesis that the local strain was missclassified as A. catenella.
Very recently, Miranda et al. (2012) discussed the validity of using direct amplification sequences of the rDNA subunits as a suitable method for strain differentiation in Alexandrium species, due to the existence of paralogue genes. In this respect, base quality discrimination did not give evidence of paralogue sequences. On the other hand, Ki and Han (2007), eliminating paralogue sequences for their analysis, found 39 parsimony informative sites within the D1–D5 LSU rDNA region in five different Alexandrium species. Our study found a much higher value of parsimony variable sites (174) when our local sequence was aligned with published sequences, which probably contained paralogue DNA regions. It is unlikely that this considerable difference could be explained by increased mutation rates in the sequenced regions. Thus, this result agrees with Miranda et al. (2012), who suggest that the great diversity in Group I of the tamarensis complex could be due to the lack of an accurate discrimination of paralogue sequences.
The sequence analysis facilitated the use of a specific and highly sensitive real-time PCR assay to detect local Alexandrium. Owing to the ability of filter-feeding molluscs to capture and concentrate phytoplankton, by pumping water through their gill filaments, we tested the possibility of detecting dinoflagelate DNA in this organ of challenged mussels. Preliminary experimental results showed, for the first time, the implementation of a practical test to detect these algae in gill tissue extracted from mussels challenged under laboratory conditions. Currently, we are working on the implementation of this test in field samples in order to detect traces of Alexandrium in seawater. It would be very useful to count with methods to detect traces of dinoflagellates, in order to predict massive algal blooming through constant monitoring of red tide episodes, thus preventing human consumption of toxic filter-feeding shellfish.
Conclusions and forward look
Traditionally, South American Pacific HABs have been assigned to A. catenella, based only on morphological evidence that has proven to be an unreliable indicator of species identification within the A. tamarense complex (Lilly et al. 2007).
This study was focused on the molecular identification of the Alexandrium species that causes paralytic shellfish poisoning in Chilean coasts (Hernández et al. 2005). Phylogenetic analyses based on sequence data and species-specific PCR assays targeting LSU rDNA and microsatellite regions, all demonstrate that cultures isolated from Chilean coasts belong to the tamarensis complex Group I and are not A. catenella (Figs 1 and 2, Tables 1 and 2, Hosoi-Tanabe and Sako 2005).
As not all A. tamarense are toxic, we are currently developing a real-time PCR assay based on primer pairs that target signature nucleic acid sequences of genes involved in toxin production. Our goal is to set up a new technique for early, sensitive and accessible HAB detection in order to avoid the important financial damage and public health issues.
Additional information
The following additional information is available in the online version of this article –
Sequences used for phylogenetic analyses.
File 1: DNA sequences used in this study. Strain assignment, morphospecies, origin and accession number in GenBank are given when available.
Accession numbers
The Ach01 (NCBI accession no. JN657223) sequence was uploaded to GenBank.
Sources of funding
This work was funded by the Corporación del fomento de la producción (Corfo) of Chile through the INNOVA Chile Project #07CN13PPD-240.
Contributions by the authors
A.J. and G.F. did the sequence analysis, phylogenetic analysis, PCR experiments and writing of the manuscript. M.A., P.O., J.E.T. and J.M.N. contributed with the challenge of the Mytilus spp. samples with local Alexandrium species. V.M. developed the idea, provided further inputs during the project and contributed to the financial means for carrying out the project.
Acknowledgements
We thank M. Córdova and B. Suárez-Isla for providing Alexandrium cultures.
Conflict of interest statement
None declared.
Literature cited
- Alpermann TJ, John UE, Medlin LK, Edwards KJ, Hayes PK, Evans KM. Six new microsatellite markers for the toxic marine dinoflagellate Alexandrium tamarense. Molecular Ecology Notes. 2006;6:1057–1059. [Google Scholar]
- Amaro AM, Fuentes MS, Ogalde SR, Venegas JA, Suárez-Isla BA. Identification and characterization of potentially alagal-lytic marine bacteria strongly associated with the toxic dinoflagellate Alexandrium catenella. Journal of Eukaryotic Microbiology. 2005;52:191–200. doi: 10.1111/j.1550-7408.2005.00031.x. [DOI] [PubMed] [Google Scholar]
- Anderson DM. Toxic algal blooms and red tides: a global perspective. In: Okaichi T, Anderson DM, Nemoto T, editors. Red tides: biology, environmental science, and toxicology. New York: Elsevier; 1989. pp. 11–16. [Google Scholar]
- Anderson DM. 2007. The ecology and oceanography of harmful algal blooms: multidisciplinary approaches to research and management. UNESCO IOC Technical Series 74. [Google Scholar]
- Anderson DM, Kullis DM, Doucette GJ, Gallagher JC, Balech E. Biogeography of toxic dinoflagellates in the genus Alexandrium from the northeastern United States and Canada. Marine Biology. 1994;120:467–478. [Google Scholar]
- Anderson DM. Approaches to monitoring, control and management of harmful algal blooms (HABs) Ocean and Coastal Management. 2009;52:342–347. doi: 10.1016/j.ocecoaman.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassis D, Muñoz PS, Avaria S. Variación temporal del fitoplancton entre 1993 y 1998 en una estación fija del seno Aysén, Chile (45°26′S 73°00′W) Revista de Biología Marina y Oceanografía. 2002;37:43–65. [Google Scholar]
- Córdoba JS, Müller I. Use of PCR and partial sequencing of the large-subunit rRNA gene to identify Alexandrium catenella (Dinophyceae) from the south of Chile. Harmful Algae. 2002;1:343–350. [Google Scholar]
- Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fernández ML, Míguez A, Cacho E, Martínez A, Diogéne J, Yasumoto T. Bioensayos con mamíferos y ensayos bioquímicos y celulares para la detección de ficotoxinas. In: Sar EA, Ferrario ME, Reguera B, editors. Floraciones algales nocivas en el Cono Sur Americano. Madrid: Instituto Español de Oceanografía; 2002. pp. 77120–120. [Google Scholar]
- Glibert PA, Anderson DM, Gentien PG, Granéli E, Selner KG. The global complex phenomena of harmful algal blooms. Oceanography. 2005;18:136–147. [Google Scholar]
- Guillard R. In: Culture of phytoplankton for feeding marine invertebrates. Smith WL, Chanley MH, editors. New York: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- Guillou L, Nézan E, Cueff V, Erard-Le Denn E, Cambon-Bonavita MA, Gentien P, Barbier G. Genetic diversity and molecular detection of three toxic dinoflagellate genera (Alexandrium, Dinophysis and Karenia) from French coasts. Protist. 2002;153:223–238. doi: 10.1078/1434-4610-00100. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. PhyML: a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hallegraeff GM. Transport of toxic dinoflagellates via ships’ ballast water: bioeconomic risk assessment and efficacy of possible ballast water management strategies. Marine Ecology Progress Series. 1998;168:297–309. [Google Scholar]
- Halstead BW, Schantz EJ. Paralytic shellfish poisoning. Vol. 79. Geneva: World Health Organization; 1984. pp. 1–59. [PubMed] [Google Scholar]
- Hansen G, Daugbjerg N, Franco JM. Morphology, toxin composition and LSU rDNA phylogeny of Alexandrium minutum (Dinophyceae) from Denmark, with some morphological observations on other European strains. Harmful Algae. 2003;2:317–335. [Google Scholar]
- Hernández C, Ulloa J, Vergara JA, Espejo R, Cabello F. Vibrio parahaemolyticus infections and algal intoxications as emergent public health problems in Chile. Revista Médica de Chile. 2005;133:1081–1088. doi: 10.4067/s0034-98872005000900013. [DOI] [PubMed] [Google Scholar]
- Higman WA, Stone DM, Lewin JM. Sequence comparison of toxic and non-toxic Alexandrium tamarense (Dinophyceae) isolates from UK waters. Journal of Phycology. 2001;40:256–262. [Google Scholar]
- Hirotugu A. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- Hosoi-Tanabe S, Sako Y. Species-specific detection and quantification of toxic marine dinoflagellates Alexandrium tamarense and A. catenella by real-time PCR assay. Marine Biotechnology. 2005;7:506–514. doi: 10.1007/s10126-004-4128-4. [DOI] [PubMed] [Google Scholar]
- John U, Fensome RA, Medlin LK. The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense ‘species complex’ (Dinophyceae) Molecular Biology and Evolution. 2003;20:1015–1027. doi: 10.1093/molbev/msg105. [DOI] [PubMed] [Google Scholar]
- John U, Medlin LK, Groben R. Development of specific rRNA probes to distinguish between geographic clades of the Alexandrium tamarense species complex. Journal of Plankton Research. 2005;27:199–204. [Google Scholar]
- Jørgensen CB. Bivalve filter feeding revisited. Marine Ecology Progress Series. 1996;142:287–302. [Google Scholar]
- Ki JS, Han MS. Informative characteristics of 12 divergent domains in complete large subunit rDNA sequences from the harmful dinoflagellate genus, Alexandrium (Dinophyceae) Journal of Eukaryotic Microbiology. 2007;54:210–219. doi: 10.1111/j.1550-7408.2007.00251.x. [DOI] [PubMed] [Google Scholar]
- Kodama M. Paralytic shellfish poisoning toxins: biochemistry and origin. Aqua-BioScience Monographs. 2010;3:1–38. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leaw CP, Lim PT, Ng BK, Cheah MY, Ahmad A, Usup G. Phylogenetic analysis of Alexandrium species and Pyrodinium bahamense (Dinophyceae) based on theca morphology and nuclear ribosomal gene sequence. Phycologia. 2005;44:550–565. [Google Scholar]
- Lilly EL. Massachusetts, USA: 2003. Phylogeny and biogeography of the toxic dinoflagellate Alexandrium. PhD Thesis. [Google Scholar]
- Lilly EL, Halanych KM, Anderson DM. Phylogeny and biogeography of the toxic dinoflagellate Alexandrium. Harmful Algae. 2005;4:1004–1020. [Google Scholar]
- Lilly EL, Halanych KM, Anderson DM. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae) Journal of Phycology. 2007;43:1329–1338. [Google Scholar]
- Mayr E. The growth of biological thought: diversity, evolution and inheritance. Cambridge, MA: Harvard University Press; 1982. [Google Scholar]
- Medlin LK, Lange M, Wellbrock U, Donner G, Elbrachter M, Hummert C, Luckas B. Sequence comparisons link toxic European isolates of Alexandrium tamarense from the Orkney Islands to toxic North American stocks. European Journal of Protistology. 1998;34:329–335. [Google Scholar]
- Miranda LN, Zhuang Y, Zhang H, Lin S. Phylogenetic analysis guided by intragenomic SSU rDNA polymorphism refines classification of ‘Alexandrium tamarense’ species complex. Harmful Algae. 2012;16:35–48. [Google Scholar]
- Montoya N, Fulco VK, Carignan MO, Carreto JL. Toxin variability in cultured and natural populations of Alexandrium tamarense from southern South America—evidence of diversity and environmental regulation. Toxicon. 2010;56:1408–1418. doi: 10.1016/j.toxicon.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Muñoz P. Revisión taxonómica de los dinoflagelados de Chile. Revista de Biología Marina y Oceanografía. 1985;21:31–60. [Google Scholar]
- Murray S, Jurgensen MF, Ho SYW, Patterson DJ, Jermiin LS. Improving the analysis of dinoflagellate phylogeny based on rDNA. Protist. 2005;156:269–286. doi: 10.1016/j.protis.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Nagai S, Sekino M, Matsuyama Y, Itakura S. Development of microsatelitte markers in the toxic dinoflagellate Alexandrium catenella (Dinophyceae) Molecular Ecology Notes. 2005;6:120–122. [Google Scholar]
- Persich GR, Kulis DM, Lilly EL, Anderson DM, Garcia VMT. Probable origin and toxin profile of Alexandrium tamarense (Lebour) Balech from southern Brazil. Harmful Algae. 2006;5:36–44. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Riisgard HU, Larsen PS, Nielsen NF. Particle capture in the mussel Mytilus edulis: the role of latero-frontal cirri. Marine Biology. 1996;127:259–266. [Google Scholar]
- Rogers JE, Leblond JD, Moncreiff CA. Phylogenetic relationship of Alexandrium monilatum (Dinophyceae) to other Alexandrium species based on 18S ribosomal RNA gene sequences. Harmful Algae. 2006;5:275–280. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholin CA, Anderson DM. Identification of group and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). I. RFLP analysis of SSU rRNA genes. Journal of Phycology. 1994;30:744–754. [Google Scholar]
- Scholin CA, Anderson DM. LSU rDNA-based RFLP assays for discriminating species and strains of Alexandrium (Dinophyceae) Journal of Phycology. 1996;32:1022–1035. [Google Scholar]
- Scholin CA, Herzog M, Sogin M, Anderson DM. Identification of group and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae) II. Sequence analysis of a fragment of the LSU rRNA gene. Journal of Phycology. 1994;30:999–1011. [Google Scholar]
- Scholin CA, Hallegraeff GM, Anderson DM. Molecular evolution of the Alexandrium tamarense ‘species complex’ (Dinophyceae): dispersal in the North American and West Pacific regions. Phycologia. 1995;34:72–85. [Google Scholar]
- Schwarz GE. Estimating the dimension of a model. Annals of Statistics. 1978;6:461–464. [Google Scholar]
- Shafee MS. Studies on the various allometric relationships in the intertidal green mussel, Perna viridis Linnaeus of Ennore estuary, Madras. Indian Journal of Fisheries. 1976;23:1–9. [Google Scholar]
- Sonnenberg R, Nolte AW, Tautz D. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Frontiers in Zoology. 2007;4:6–18. doi: 10.1186/1742-9994-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez BA, López A, Hernández C, Clement A, Guzmán L. Impacto económico de las floraciones de microalgas nocivas en chile y datos recientes sobre la ocurrencia de veneno amnésico de los mariscos. In: Sar EA, Ferrairo ME, Reguera B, editors. Floraciones Algales Nocivas en el Cono Sur Americano. Pontevedra, Spain: Instituto Español de Oceanografía, Mos; 2002. pp. 259–268. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usup G, Pin LC, Ahmad A, Teen LP. Phylogenetic relationship of Alexandrium tamiyavanichii (Dinophyceae) to other Alexandrium species based on ribosomal RNA gene sequences. Harmful Algae. 2002;1:59–68. [Google Scholar]
- Vila M, Camp J, Garcés E, Masó M, Delgado M. High resolution spatio-temporal detection of potentially harmful dinoflagellates in confined waters in the NW Mediterranean. Journal of Plankton Research. 2001;23:497–514. [Google Scholar]