The transcription factor T-bet drives the differentiation of NKp46-expressing IL-22–producing innate lymphoid cells
Abstract
Interleukin (IL)-22–producing innate lymphoid cells (ILCs; ILC22) comprise a heterogeneous population of cells that are dependent on the transcription factor retinoid-related orphan γt (RORγt) and are critical for barrier function of the intestinal mucosa. A distinct ILC22 subset expresses the natural cytotoxicity receptor NKp46 (NKp46+ ILC22); however, the factors that contribute to the generation of this population versus other subsets are largely unknown. Herein, we show that T-bet (encoded by Tbx21) was highly expressed in NKp46+ ILC22, a feature shared by all NKp46+ cells present in the intestine but not by other IL-22–producing populations. Accordingly, the absence of T-bet resulted in loss of NKp46+ ILC22 in the intestinal lamina propria. The residual NKp46+ ILC22 present in Tbx21−/− mice showed a marked reduction of Rorγt expression and impairment in IL-22 production. Generation and functions of gut NK1.1+ cells were also altered. Bone marrow chimera experiments revealed a cell-intrinsic requirement for T-bet in these subsets and competitive reconstitution experiments revealed roles for T-bet in multiple ILC subsets. Thus, T-bet has a general importance for ILC in the gut and plays a selective and critical role in the generation of NKp46+ ILC22.
Gut homeostasis is maintained by a delicate equilibrium between commensal flora and immune cells (Hooper and Macpherson, 2010; Molloy et al., 2012). In this environment, specialized adaptive and innate immune cells with distinct profiles of cytokine production are critical for host defense and immune regulation. Paralleling the complexity of the different CD4+ Th subsets with respect to signature cytokines, a growing family of innate lymphoid cells (ILC) has been identified (Spits and Di Santo, 2011). Besides NK cells, the prototypical type 1 ILC (ILC1) which selectively produces IFN-γ, several ILC subsets have been described. These include cells that predominantly produce IL-13 (ILC2 or nuocyte), IL-17 (ILC17), and/or IL-22 (ILC22).
IL-22 plays a key role in the early protection of the gut from pathogens, such as Citrobacter rodentium, by preserving the integrity of the epithelial layer and promoting inflammation (Sonnenberg et al., 2011). Although initially identified as a Th17 cytokine, IL-22 is produced by several innate cell populations. In both human and mice, innate IL-22 producers comprise cells with features of lymphoid tissue inducer (LTi) cells. These cells, which arise during fetal development, also produce IL-17 (Cupedo et al., 2009; Takatori et al., 2009; Sawa et al., 2010). Another relevant ILC22 subset appears in the small intestine lamina propria (siLP) shortly after birth. These cells express the NK cell marker NKp46 (referred herein as NKp46+ ILC22) and CD127 (the α chain of IL-7 receptor) but express little or no NK1.1 (killer cell lectin-like receptor subfamily B member 1C; Satoh-Takayama et al., 2008; Cella et al., 2009; Luci et al., 2009; Sanos et al., 2009). The spectrum of cytokines produced by NKp46+ ILC22 differs from LTi cells in that they do not concomitantly produce IL-17. Unlike NK cells, they also do not produce IFN-γ (Satoh-Takayama et al., 2009).
Although NKp46+ ILC22 share several traits with LTi cells and, to a lesser extent, canonical NK cells (NKp46+NK1.1+), the factors required for differentiation of NKp46+ ILC22 are incompletely understood. NKp46+ ILC22, like LTi cells, are dependent on IL-7 for development (Luther et al., 2003; Satoh-Takayama et al., 2010), but in contrast to canonical NK cells, their differentiation is IL-15 independent (Kennedy et al., 2000; Satoh-Takayama et al., 2008).
The transcription factors that promote differentiation of the different IL-22–producing populations have also yet to be fully elucidated. All ILC22 populations and NK cells are thought to derive from a common precursor that expresses the inhibitor of DNA binding 2 (Id2; Yokota et al., 1999; Satoh-Takayama et al., 2010). However, the ILC22 developmental program diverges from that of canonical NK cells in that retinoid-related orphan γt (RORγt) and the AHR (aryl hydrocarbon receptor) are required for ILC22 cells but not canonical NK cells (Sun et al., 2000; Eberl and Littman, 2003; Lee et al., 2012; Qiu et al., 2012).
The transcription factor T-bet (T-box expressed in T cells, encoded by Tbx21 gene) is important in the regulation of NK cell development and effector functions, affecting cell turnover, IFN-γ production, and trafficking (Townsend et al., 2004; Jenne et al., 2009). Recently, it has been shown that the development of an NK cell subset expressing the TNF-related apoptosis inducing ligand (Trail) is dependent on T-bet (Gordon et al., 2012).
Although the role of T-bet has been investigated in T and NK cells, its role on various ILC populations present in the gut has not been determined. In this study, we set out to determine if T-bet could also control gut NKp46+ cell development and function. Herein, we showed that T-bet was differentially expressed among the distinct ILC22 populations. In particular, NKp46+ ILC22 expressed high levels of T-bet, comparable to the other NKp46+ subsets (NK1.1+) present in the gut. Using Tbx21−/− mice, we found that T-bet played a pivotal role in the generation of NKp46+ ILC22 and CD127+ NK cells in a cell-intrinsic manner.
RESULTS AND DISCUSSION
T-bet is highly expressed by NKp46+ ILC
ILC patrolling the intestine comprise distinct populations that secrete diverse arrays of cytokines. Although the role of T-bet has been investigated in T and NK cells, its impact on various ILC populations present in the gut has not been determined. To address this question, we first assessed T-bet expression in total CD3ε− siLP lymphocytes (siLPLs) by intracellular multicolor flow cytometry analyses.
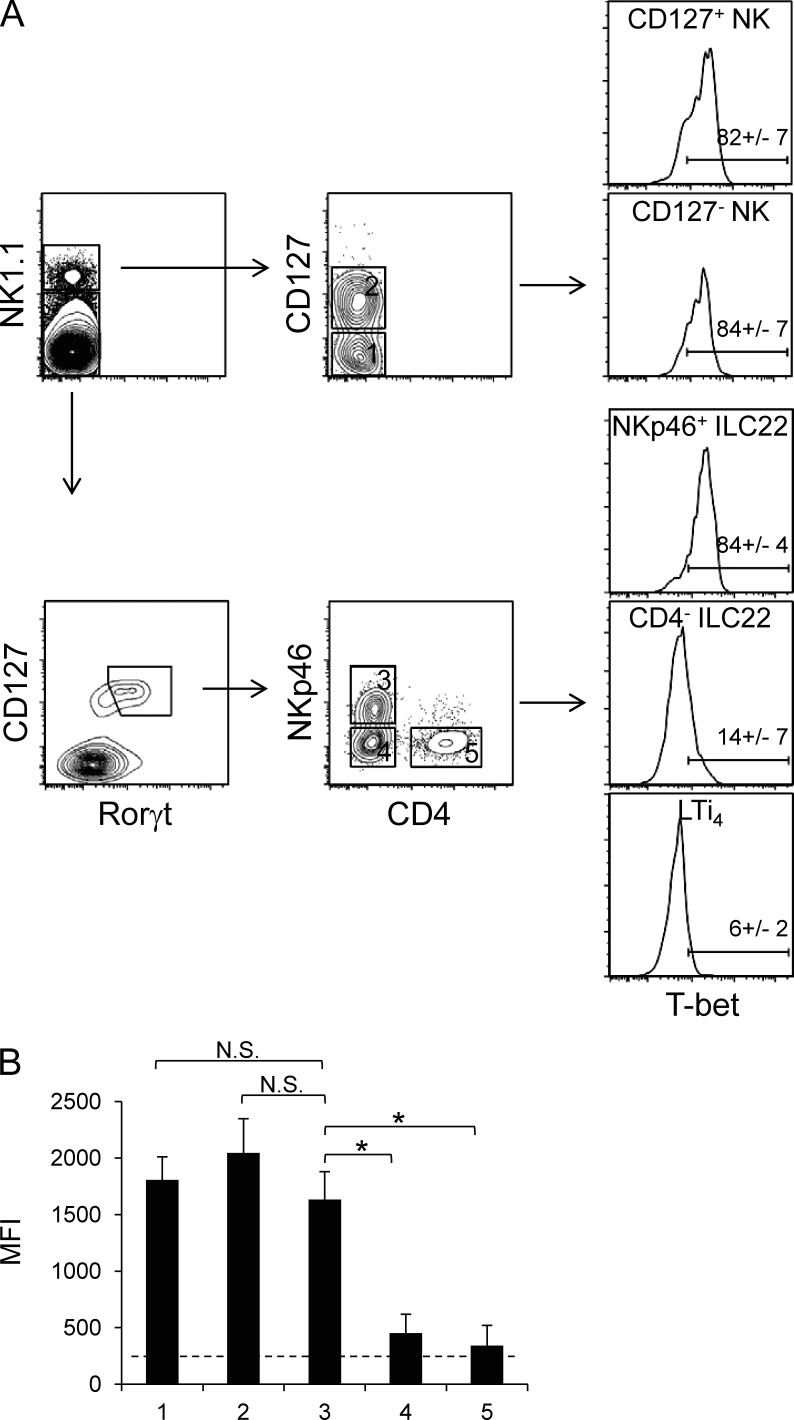
We initially focused on the CD3ε−NK1.1high cell populations residing in the siLP because these cells resemble T-bet–expressing conventional NK cells present in other organs (spleen and liver). Based on CD127 expression, it is possible to define two different NK cell subsets in the lamina propria, herein denoted CD127+ and CD127− NK cells. Notably, T-bet was highly and homogeneously expressed in both populations (Fig. 1 A, top).
Figure 1.
T-bet is differentially expressed in populations of gut innate lymphocytes. (A) T-bet expression was assessed in cell populations discriminated based on expression of NK1.1 and CD127 (IL-7 receptor α), Rorγt, NKp46, and CD4. These included populations of NK1.1+ NK cells (CD127− and CD127+), NK1.1−Rorγt+CD127+NKp46+CD4− ILC22 cells, and Rorγt+CD127+NKp46− cells (CD4+ and CD4−). A representative experiment of four performed is shown. Numbers indicate mean percentage ± SD of T-bet+ cells. (B) Bar graphs depict T-bet expression as measured by MFI (mean ± SD, n = 4; four independent experiments were performed) in the different ILC subsets analyzed. The dashed line represents background levels as determined by T-bet staining using cells from Tbx21−/− mice. T-bet levels of NKp46+ ILC22 were compared with the other subsets. Asterisks denote P < 0.05.
Aside from NK1.1, conventional NK cells can also be identified by NKp46 expression in several lymphoid (bone marrow, spleen, lymph node, and thymus) and nonlymphoid (liver and lung) organs. In addition, in the intestine, a specific NKp46+ cell subset that expresses low levels of NK1.1 is known to be an important producer of IL-22 (NKp46+ ILC22). We therefore analyzed T-bet expression in these subsets. Within CD3−NK1.1low/−Rorγt+CD127+ cells, three populations can be discerned based on CD4 and NKp46 expression: NKp46+ ILC22 (CD4−NKp46+), CD4− ILC22 (CD4−NKp46−, a heterogeneous population comprising ILC22 and LTi cells), and LTi4 (CD4+NKp46−). As shown in Fig. 1 A (bottom), although T-bet was barely detectable in CD4− ILC22 and LTi4, it was highly expressed in NKp46+ ILC22. The mean fluorescence intensity (MFI) of T-bet staining is depicted in Fig. 1 B. The data indicate that NKp46+ ILC22 expressed T-bet at comparable levels of CD127+ and CD127− NK cells. These results raise the possibility that although NKp46+ ILC22 have similar requirements for development with other ILC22 populations, T-bet could play a selective role in this subset.
Profound reduction of NKp46+ ILC22 and CD127+ NK cells in the absence of T-bet
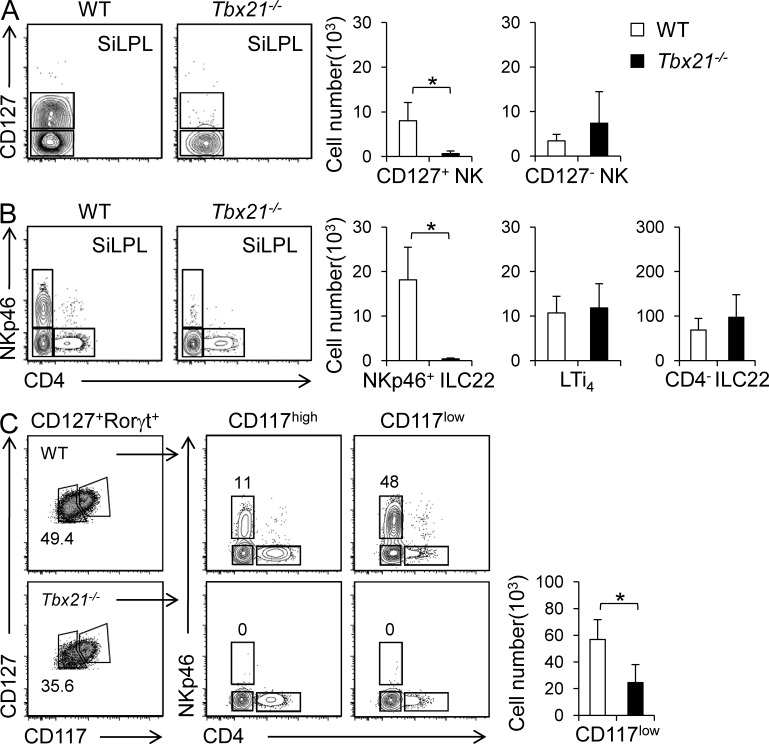
To elucidate the potential impact of T-bet in these ILC subsets, we assessed the various populations of gut innate lymphocytes in Tbx21−/− mice by applying the same gating strategy used in Fig. 1. Despite the homogeneous expression of T-bet among all CD3ε−NK1.1+ cells (CD127− and CD127+), Tbx21−/− mice showed a dramatic and selective reduction of CD127+ NK cells. In contrast, there were no significant differences in CD127− NK cells, compared with WT mice (Fig. 2 A). When we analyzed CD127+Rorγt+ ILC populations, we found no difference in the number of CD4− ILC22 or LTi4 cells. However, in agreement with the pattern of T-bet expression, we did note a striking and selective decrease in the number of NKp46+ ILC22 cells in T-bet–deficient mice (Fig. 2 B). It was possible though that T-bet was simply regulating NKp46 expression on this subset. We attacked this problem in two ways. First, we found that NKp46 expression on conventional NK cells was not affected by the absence of T-bet (unpublished data). Second, we enumerated ILC22 cells using markers independent of NKp46+. ILC22 have been shown to be enriched in the Rorγt+CD127lowCD117low population of lamina propria cells (Sawa et al., 2010 and Fig. 2 C). We observed a significant reduced proportion of this population of cells in Tbx21−/− mice, independent of reliance on NKp46 expression (Fig. 2 C). In addition, Tbx21−/− mice had normal numbers of CD45+Thy1.2+lin− cells, which contain ILC2 and lacked T-bet expression (unpublished data). Collectively, these data argue that indeed, NKp46+ ILC22 cells are preferentially affected by the loss of T-bet.
Figure 2.
NKp46+ ILC22 and CD127+ NK cells are dramatically reduced in Tbx21−/− mice. Lymphocytes were isolated from siLP of WT and Tbx21−/− mice and analyzed by flow cytometry. (A) The left panel shows representative dot plots and the right panels show absolute number of CD127+ and CD127− CD3ε−NK1.1+ populations (mean ± SD). (B) Representative dot plots and absolute number of the different ILC22 populations. Values represent mean ± SD of each indicated population (WT, five mice; Tbx21−/−, six mice). Four independent experiments were performed. Asterisks denote P < 0.05. (C) Representative dot plots of Rorγt+ ILC populations (according CD127, CD117, CD4, and NKp46 expression) and absolute number (mean ± SD) of Rorγt+CD127lowCD117low ILC in WT and Tbx21−/− mice (WT, three mice; Tbx21−/−, three mice). Two independent experiments were performed. Asterisk denotes P < 0.05.
T-bet regulates differentiation and functions of NKp46+ ILC22 and gut NK cells
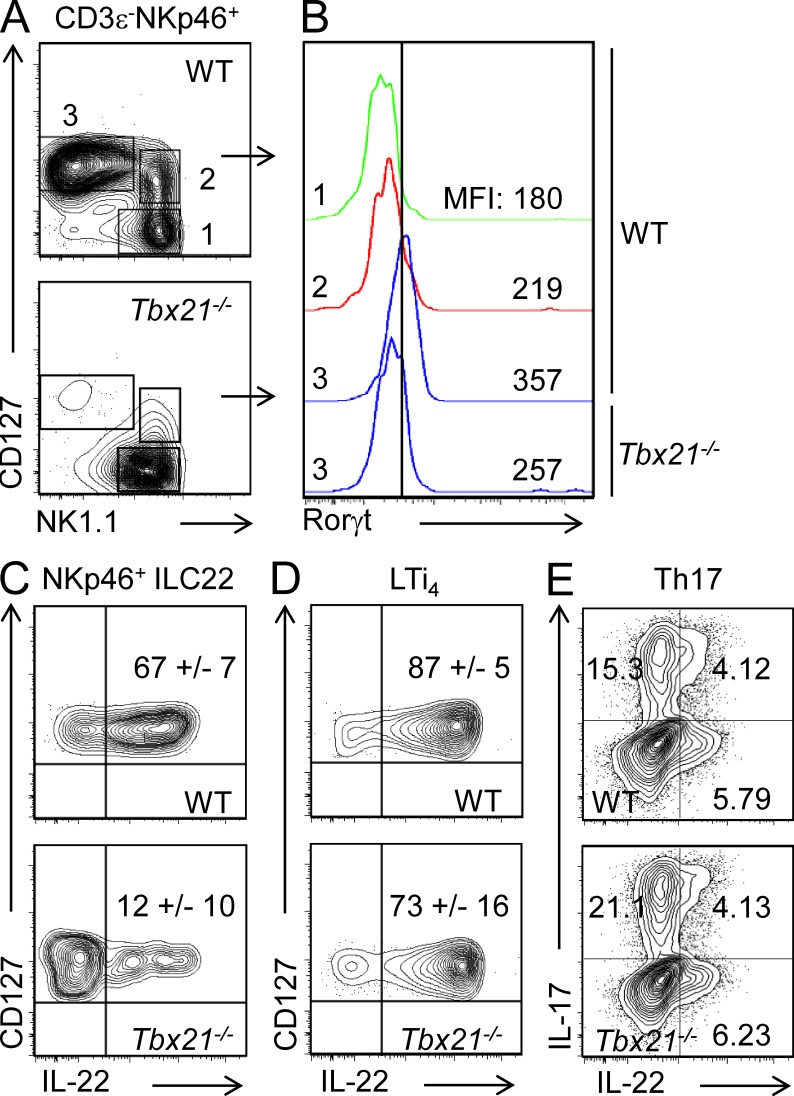
Consistent with their high levels of T-bet expression, NKp46+ ILC22 were dramatically reduced in Tbx21−/− mice. However, these cells were not completely absent. This afforded us the opportunity to determine the functional impact of T-bet in these cells. As noted above, terminal differentiation of NKp46+ ILC22 is characterized by acquisition of Rorγt expression. NKp46+ ILC22 were identified by focusing on CD3ε−NKp46+ and assessing CD127 and NK1.1 expression (Fig. 3 A). Rorγt expression was measured by intracellular staining of NKp46+ ILC22 (gated as CD127+NK1.1low/−) and, for comparison, CD127− and CD127+ NK cells, which do not express Rorγt. The absence of T-bet was associated with concomitant reduction of Rorγt in the residual NKp46+ ILC22 (Fig. 3 B).
Figure 3.
Absence of T-bet impairs Rorγt expression and IL-22 production in NKp46+ ILC22. (A) Lymphocytes were isolated from siLP of WT and Tbx21−/− mice and the different CD3ε−NKp46+ populations (according CD127 and NK1.1 expression) were evaluated in WT and Tbx21−/− mice. (B) RORγt expression levels were evaluated by flow cytometry in NKp46+ ILC22 (gated as CD3ε−NKp46+CD127+NK1.1−) from siLP of WT and Tbx21−/− mice. As negative control, RORγt expression levels in CD127− and CD127+ NK cells are also shown. The MFI of the different populations is indicated. One representative experiment out of three is shown. Three mice per group were used. (C and D) siLPL were isolated from WT and Tbx21−/− mice and stimulated for 4 h with 50 ng/ml IL-23. IL-22 production in NKp46+ ILC22 (C) and LTi4 (D) cells was analyzed by flow cytometry. (E) Naive CD4+ T cells from WT and Tbx21−/− mice were differentiated with IL-6, IL-1β, and IL-23 to generate Th17 cells as described in Materials and methods. IL-22 production was measured by intracellular staining. The numbers in the top right quadrants indicate mean ± SD of IL-22+ cells (C and D) and IL-22+ and/or IL-17+ cells (E). In C–E, at least three mice were analyzed for each condition in three different experiments.
We next investigated whether IL-22 production was also affected in the residual NKp46+ ILC22 present in T-bet–deficient mice. As previously shown, in response to IL-23, NKp46+ ILC22 from WT mice produce high levels of IL-22 (Satoh-Takayama et al., 2008; Fig. 3 C). However, the residual population of NKp46+ ILC22 present in Tbx21−/− mice showed a severe impairment in IL-22 production (Fig. 3 C). In contrast, we found that IL-23 efficiently induced IL-22 production in LTi4 regardless of whether the cells were from Tbx21−/− or WT mice (Fig. 3 D). Because IL-22 can also be a product of Th17 cells, we also asked if T-bet expression was important for the production of this cytokine in CD4+ T cells. We found that naive CD4+ T cells cultured in presence of IL-6, IL-1β, and IL-23 produced IL-17 and IL-22 (Ghoreschi et al., 2010; Fig. 3 E). However, the absence of T-bet did not influence the proportion of CD4+ T cells that produced IL-22 under these conditions. Collectively, we interpret these data to indicate that the impaired production of IL-22 in NKp46+ ILC22 expression likely reflects a maturational block, although the possibility remains that remnant cells present in Tbx21−/− mice might represent a distinct subset. Conversely, T-bet is not involved in the regulation of IL-22, in LTi4, and Th17 cells.
Cell-autonomous role of T-bet in the regulation of NKp46+ ILC differentiation
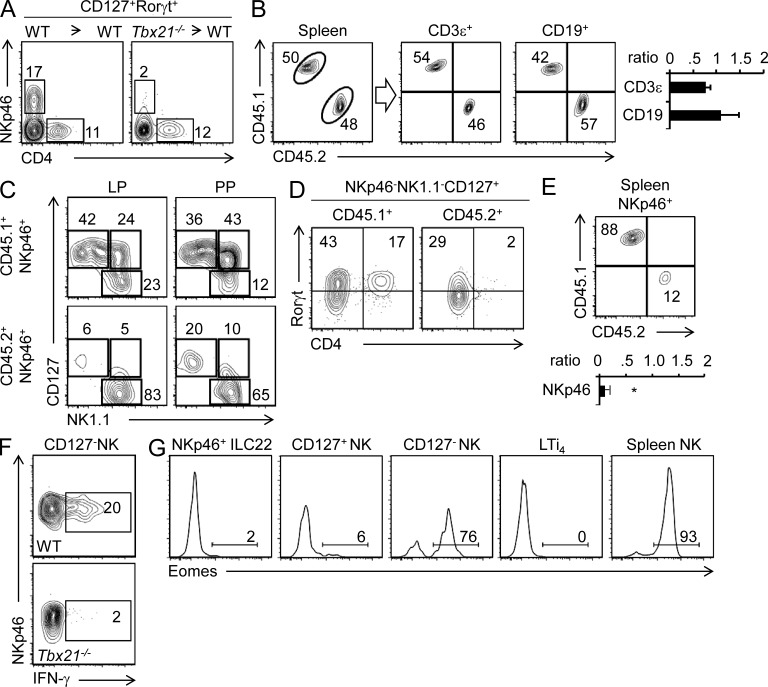
To formally establish that the phenotype we observed in Tbx21−/− mice was the result of a direct cell-intrinsic role of T-bet in NKp46+ ILC22 development, we transplanted lethally irradiated recipient mice (CD45.1) with bone marrow from WT or Tbx21−/− mice (CD45.2). After 8 wk, we found restoration of the various ILC22 subsets present in the lamina propria in mice transplanted with WT bone marrow (Fig. 4 A). In mice transplanted with T-bet–deficient bone marrow, we noted reconstitution of LTi4 cells; however, we observed that NKp46+ ILC22 cells were not present (Fig. 4 A).
Figure 4.
Intrinsic role of T-bet in the regulation of ILC22 and NK cells. (A) Lethally irradiated WT CD45.1+ mice were transplanted with bone marrow cells from WT (CD45.2+) or Tbx21−/− (CD45.2+) mice. After 8 wk, siLPLs were isolated and innate lymphoid populations were analyzed by flow cytometry. Numbers in the dot plots represent the percentage of NKp46+ ILC22 and LTi4 cells (CD45.2+). (B–D) Lethally irradiated WT CD45.1+ mice were transplanted with a 1:1 ratio of WT (CD45.1+) and Tbx21−/− (CD45.2+) bone marrow cells. In B, splenocytes were isolated and stained with the indicated antibodies. For gating on CD3ε+ and CD19+ cells, the dot plots depict percentages of CD45.2+ and CD45.1+ cell in a representative experiment. The bar graphs represent the mean ± SD (n = 5, two independent experiments) of CD45.2+/CD45.1+ ratios. In C, CD3ε−NKp46+ cells derived from WT (CD45.1+) or Tbx21−/− (CD45.2+) were analyzed based on CD127 and NK1.1 expression. Numbers in the dot plots represent the percentage of each gated subset. In D, LTi4 cells derived from WT (CD45.1+) or Tbx21−/− (CD45.2+) were analyzed (gated as CD3−/CD19−/NKp46−/NK1.1−/CD127+ cells). Numbers in the quadrants represent the percentage of each individual subset. (E) The dot plot shows percentages of CD45.2+ and CD45.1+ cells in a representative experiment and the bar graph depicts the mean ± SD (n = 5) of CD45.2+/CD45.1+ ratio, gated on CD3ε−NKp46+ cells in the spleen. *, P < 0.05. (F) LP CD127− NK cells from WT or Tbx21−/− mice were stimulated with IL-2 and IL-12 for 6 h and IFN-γ production was evaluated by flow cytometry. Numbers in the box represent the percentage of IFN-γ+ cells (WT, three mice; Tbx21−/−, three mice; three independent experiments were performed). (G) Eomes expression levels were evaluated by flow cytometry and intracellular staining in NKp46+ ILC22, CD127+ and CD127− NK cells, LTi4 cells, and splenic NK cells (CD3ε−NKp46+). Numbers represent the percentage of Eomes-positive cells. A representative experiment of three is shown. Three mice were used.
To examine the effect of T-bet deficiency in a competitive environment, we next generated mixed bone marrow chimeras, transferring bone marrow cells from WT (CD45.1) and Tbx21−/− (CD45.2) mice into CD45.1 lethally irradiated C57BL/6 host mice in a one to one ratio. We analyzed reconstitution in the spleen and found equivalent proportions of CD45.1+ and CD45.2+ lymphocytes, indicating that equal proportions of bone marrow precursors were transferred (Fig. 4 B). This ratio is maintained for total B (CD19+) and T (CD3ε+) cells. Consistent with our previous results, we found impaired generation of NKp46+ ILC22 and CD127+ NK cells in siLP and Peyer’s patches (PPs) in the absence of T-bet (CD45.2+ cells; Fig. 4 C). Moreover, the residual NKp46+ ILC22 derived from Tbx21−/− bone marrow showed lower Rorγt expression and impaired ability to produce IL-22 (unpublished data). However, in this competitive setting we also noted reductions in LTi4 cells and even conventional NK cells present in the spleen (Fig. 4, D and E). Although lamina propria CD127− NK cells were not reduced (Fig. 4 C), T-bet deficiency was associated with defective IFN-γ production in these cells (Fig. 4 F). Thus, generation and/or function of diverse NKp46+ cells is influenced by T-bet.
Eomes is another T-box transcription factor, which can drive canonical NK cell generation in absence of T-bet (Gordon et al., 2012). Consistent with the preservation of lamina propria CD127− NK cells in mixed chimeras, these cells selectively express high levels of Eomes. Other ILC populations express little or no Eomes, providing an explanation for their dependency upon T-bet (Fig. 4 G).
ILC are developmentally related immune cells that play an important role in the regulation of gut homeostasis and protection from pathogens (Pearson et al., 2012). The increasing complexity of ILC subsets to selectively produce cytokines beg the question as to what transcription factors are responsible for the generation of this diversity and how these subsets may or may not be related.
The present findings indicate that the establishment of NKp46+ ILC22 in the small intestine is tightly regulated by T-bet. The requirement of this transcription factor discriminates development of NKp46+ ILC22 from other ILC22 populations. In the residual NKp46+ ILC22 present in Tbx21−/− mice, both Rorγt expression and IL-22 production are decreased. These findings clearly warrant further exploration into the relationship of T-bet and IL-22 production. Based on the current findings, we favor the view that T-bet’s role in the regulation of IL-22 expression is likely related to impairment in differentiation. Although an alternative possibility is that it directly regulates IL-22 production, the production of this cytokine is not affected in other cells in the absence of T-bet. Ideally, one would want to tackle the problem of identifying T-bet targets in this cells using Chip-seq technology; however, the small numbers of these different cells preclude this approach at the present time.
Despite the dramatic effect of T-bet deficiency on NKp46+, its function is not restricted to this subset. Even though LTi4, like other Rorγt+ cells, is found in Tbx21−/− mice, in a competitive environment the importance of T-bet becomes apparent. This observation indicates a wider role of T-bet in the generation and survival of various ILC subsets. In support of our present observation, Tbx21−/−Rag2−/− mice develop colitis consistent with important actions on cells critical for gut homeostasis (Garrett et al., 2007). Differentiated LTi cells express low levels of T-bet, so it is tempting to speculate that T-bet might regulate the turnover of a common ILC22 precursor. It would be of interest in the future to assess this possibility using a fate-mapping approach.
T-bet has previously been reported to control NK cell turnover and the trafficking of terminally differentiated NK cells (Townsend et al., 2004; Jenne et al., 2009). In addition, Trail+ NK cells present in the liver are maintained in a T-bet–dependent manner (Gordon et al., 2012). Because the absence of T-bet also impairs generation of CD127+ NK cells, there may be a common progenitor for liver NK cells, gut CD127+ NK cells, and NKp46+ ILC22.
Like T-bet, Eomes is also a T-box family transcription factor. To some extent, its actions complement those of T-bet, driving NK cell generation when T-bet is not present (Gordon et al., 2012). In intestinal ILC populations, we found high Eomes expression only in CD127− NK cells, arguing for similarities with splenic NK cells. The high levels of Eomes expression may explain why these cells are resistant to loss of T-bet. Less obvious is why the absence of T-bet was associated with reduction of conventional splenic NK cells in a competitive bone marrow transplant and sparing of gut CD127− NK cells. It is possible that this is the consequence of effects on trafficking versus survival. Regardless, the data argue for critical roles of T-box family transcription factors in the development and function of the ILCs.
Finally, our findings reveal added complexity in the developmental pathways leading to the generation of the various ILC populations. Because NKp46+ ILC22 depend on IL-7, AHR, and Rorγt and produce IL-22, it might be argued that their development is dependent on the same factors as LTi cells. In contrast, cells that are dependent on IL-15 include conventional NK cells (CD127−) in the bone marrow and spleen, and nonconventional NK cells, found in the liver (Trail+), thymus (CD127+), and gut (CD127+/−). However, this simple dichotomous model is probably not accurate. Rather, the data presented herein along with recently published results point to a more intricate and complex developmental model. For these cells, it appears that a network of transcription factors can shape their differentiation, such that the combination of T-bet, Eomes, and Rorγt expression could determine the distinct ILC subsets. The understanding of the extrinsic and intrinsic signals responsible for inducing the differential expression of these transcription factors remains an important but unresolved issue. Elucidating how these different transcription factors can influence the specification of different ILC will be an exciting future challenge.
MATERIALS AND METHODS
Mice.
C57BL/6J, Tbx21−/−, and CD45.1 mice (The Jackson Laboratory) were backcrossed to C57BL/6 mice for at least eight generations. All animal studies were performed according to the National Institutes of Health guidelines for the use and care of live animals and were approved by the Institutional Animal Care and Use Committee of the National Institute of Arthritis, Musculoskeletal and Skin Diseases.
Cell isolation and stimulations.
Isolation of siLPL was as previously described (Sanos and Diefenbach, 2010). In brief, fat tissues and PPs were removed from small intestine. Intestine was open and cut in pieces 1 cm long and incubated (10 min, two cycles) in an HBSS solution containing 5 mM EDTA and 10 mM Hepes. Pieces were then further cut and incubated in an HBSS solution containing 0.5 mg/ml DNase I (Roche) and 0.25 mg/ml Liberase TL (Roche). Finally, the solution containing digested tissue was passed in a 100 µm cell strainer and LPLs were isolated by an 80/40% Percoll (GE Healthcare) gradient.
Lymphocytes from PPs were isolated as described (Takahashi et al., 2012). In brief, PPs were incubated for 15 min at 37°C in HBSS containing 10% FBS, 5 mM EDTA, 15 mM Hepes, pH 7.2, and 1 mM dithiothreitol. After washing, PPs were mechanically smashed for the generation of single-cell suspensions.
For IL-22 and IFN-γ intracellular staining of LPL, cells were stimulated for 4 h with 50 ng/ml IL-23 (R&D Systems) or 6 h with 1,000 U/ml IL-2 and 10 ng/ml IL-12 (R&D Systems) with the addition of brefeldin A (GolgiPlug; BD). For Th17 cell-polarizing condition, CD4+ T cells from spleens and lymph nodes from 6–8-wk-old mice were negatively selected by magnetic separation (Miltenyi Biotec). Naive CD4+CD62L+CD44−CD25− cells were then sorted by FACSAria II (BD). Sorted cells were activated with 10 µg/ml of plate-bound anti-CD3 (eBioscience) and 10 µg/ml of soluble anti-CD28 (eBioscience) in media for 3 d with 20 ng/ml IL-6 (R&D Systems), 20 ng/ml IL-1β (R&D Systems), 50 ng/ml IL-23 (R&D Systems), and 10 µg/ml IFN-γ neutralizing antibodies (BD). All cells were cultured in RPMI medium with 10% (vol/vol) FCS, 2 mM glutamine, 100 IU/ml penicillin, 0.1 mg/ml streptomycin, and 20 mM Hepes buffer, pH 7.2–7.5 (all from Invitrogen), and 2 mM β-mercaptoethanol (Sigma-Aldrich).
Flow cytometry.
For cell surface staining the following anti–mouse antibodies were used: anti–mouse CD16/CD32 (clone: 2.4G2), anti–mouse NKp46-APC, PE or FITC (29A1.4), anti–mouse CD4-FITC, PE, V500, or V450 (RM4-5), anti–mouse NK1.1-APCCy7 or PE (PK136), anti–mouse CD127-PECy7 (A7R34), anti–mouse CD3ε-PerCPCy5.5 or PE (145-2C11), anti–mouse CD45.1-FITC (A20), anti–mouse CD45.2-V500 or PE-cy7 (104), anti–mouse CD8α-PE (Ly2), anti–mouse CD11b-PE (M1/70), anti–mouse CD11c-PE (HL3), anti–mouse CD49b-PE or V450 (DX5), anti–mouse CD19-PerCp5.5 (1D3), anti–mouse Gr1-PE (RB6-8C5), anti–mouse Thy1.2-FITC (53–2.1), and anti–mouse CD117-FITC (ACK45).
Expression of cytokines and transcription factors was assessed by intracellular staining using Cytofix/Cytoperm fixation and permeabilization solution (BD) together with the following antibodies: anti–mouse IL-22–PE (1H8PWSR), anti–mouse IFN-γ–FITC (XMG1.2), anti–human/mouse T-bet–eFluor 660 (eBio4B10), and anti–human/mouse RORγt-PE (AFKJS-9). Foxp3/Transcription Factor Staining buffer set (eBioscience) was used in combination with anti–mouse Eomes-PE (Dan11mag). All the antibodies were purchased from eBioscience, BioLegend, or BD. Samples were acquired using the FACSVerse (BD) and analyzed with FlowJo software (Tree Star).
Bone marrow transfer.
Cells from bone marrow were isolated as previously described (Sciumè et al., 2011). In noncompetitive experiments, bone marrow cells (5 × 106) from WT (CD45.2) or Tbx21−/− (CD45.2) mice were injected in WT C57BL/6 mice conditioned with total body irradiation of 950 cGy. In mixed bone marrow chimera experiments, 5 × 106 cells from WT C57BL/6 (CD45.1) and Tbx21−/− (CD45.2) mice were transferred together in WT C57BL/6 (CD45.1) mice.
Statistics.
For statistical analysis, p-values were calculated with a two-tailed unpaired Student’s t test.
Acknowledgments
We thank J. Simone, J. Lay (Flow Cytometry Section, National Institute of Arthritis, Musculoskeletal and Skin Diseases [NIAMS]), and the NIAMS Laboratory Animal Care and Use staff for technical support.
This work was supported by the Intramural Research Programs of NIAMS, Istituto Pasteur-Fondazione Cenci-Bolognetti (G. Sciumè), the Japan Society for Promotion of Sciences Research Fellowship for Japanese Biomedical and Behavioral Researchers at National Institutes of Health (K. Hirahara and H. Takahashi), Pharmacology Research Associate Program, National Institute of General Medical Sciences, and National Institutes of Health (A.C. Poholek and K.L. Singleton).
The authors declare no competing financial interests.
G. Sciumè designed, performed, analyzed, and interpreted all the experiments and wrote the manuscript. K. Hirahara, H. Takahashi, A. Laurence, A.V. Villarino, K.L. Singleton, S.P. Spencer, A.C. Poholek, G. Vahedi, and Y. Kanno helped with performing experiments. Y. Kanno and Y. Belkaid contributed to the experimental design and data interpretation and made helpful suggestions. J.J. O’Shea contributed to experiment design, analyzed and interpreted all acquired data, and helped to write the manuscript.
Footnotes
Abbreviations used:
- ILC
- innate lymphoid cells
- LTi
- lymphoid tissue inducer
- MFI
- mean fluorescence intensity
- PP
- Peyer’s patch
- RORγt
- retinoid-related orphan γt
- siLP
- small intestine lamina propria
- siLPL
- siLP lymphocyte
- Trail
- TNF-related apoptosis inducing ligand
References
- Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K., Doherty J.M., Mills J.C., Colonna M. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T., Crellin N.K., Papazian N., Rombouts E.J., Weijer K., Grogan J.L., Fibbe W.E., Cornelissen J.J., Spits H. 2009. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat. Immunol. 10:66–74 10.1038/ni.1668 [DOI] [PubMed] [Google Scholar]
- Eberl G., Littman D.R. 2003. The role of the nuclear hormone receptor RORgammat in the development of lymph nodes and Peyer’s patches. Immunol. Rev. 195:81–90 10.1034/j.1600-065X.2003.00074.x [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S., Glickman J.N., Glimcher L.H. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 131:33–45 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., Reiner S.L. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Macpherson A.J. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10:159–169 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- Jenne C.N., Enders A., Rivera R., Watson S.R., Bankovich A.J., Pereira J.P., Xu Y., Roots C.M., Beilke J.N., Banerjee A., et al. 2009. T-bet–dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J. Exp. Med. 206:2469–2481 10.1084/jem.20090525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.K., Glaccum M., Brown S.N., Butz E.A., Viney J.L., Embers M., Matsuki N., Charrier K., Sedger L., Willis C.R., et al. 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15–deficient mice. J. Exp. Med. 191:771–780 10.1084/jem.191.5.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Cella M., McDonald K.G., Garlanda C., Kennedy G.D., Nukaya M., Mantovani A., Kopan R., Bradfield C.A., Newberry R.D., Colonna M. 2012. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13:144–151 10.1038/ni.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci C., Reynders A., Ivanov I.I., Cognet C., Chiche L., Chasson L., Hardwigsen J., Anguiano E., Banchereau J., Chaussabel D., et al. 2009. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10:75–82 10.1038/ni.1681 [DOI] [PubMed] [Google Scholar]
- Luther S.A., Ansel K.M., Cyster J.G. 2003. Overlapping roles of CXCL13, interleukin 7 receptor α, and CCR7 ligands in lymph node development. J. Exp. Med. 197:1191–1198 10.1084/jem.20021294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy M.J., Bouladoux N., Belkaid Y. 2012. Intestinal microbiota: shaping local and systemic immune responses. Semin. Immunol. 24:58–66 10.1016/j.smim.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C., Uhlig H.H., Powrie F. 2012. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 33:289–296 10.1016/j.it.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Qiu J., Heller J.J., Guo X., Chen Z.M., Fish K., Fu Y.X., Zhou L. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 36:92–104 10.1016/j.immuni.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanos S.L., Diefenbach A. 2010. Isolation of NK cells and NK-like cells from the intestinal lamina propria. Methods Mol. Biol. 612:505–517 10.1007/978-1-60761-362-6_32 [DOI] [PubMed] [Google Scholar]
- Sanos S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., Diefenbach A. 2009. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10:83–91 10.1038/ni.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N., Vosshenrich C.A., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29:958–970 10.1016/j.immuni.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N., Dumoutier L., Lesjean-Pottier S., Ribeiro V.S., Mandelboim O., Renauld J.C., Vosshenrich C.A., Di Santo J.P. 2009. The natural cytotoxicity receptor NKp46 is dispensable for IL-22-mediated innate intestinal immune defense against Citrobacter rodentium. J. Immunol. 183:6579–6587 10.4049/jimmunol.0901935 [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N., Lesjean-Pottier S., Vieira P., Sawa S., Eberl G., Vosshenrich C.A., Di Santo J.P. 2010. IL-7 and IL-15 independently program the differentiation of intestinal CD3−NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207:273–280 10.1084/jem.20092029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., Eberl G. 2010. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 330:665–669 10.1126/science.1194597 [DOI] [PubMed] [Google Scholar]
- Sciumè G., De Angelis G., Benigni G., Ponzetta A., Morrone S., Santoni A., Bernardini G. 2011. CX3CR1 expression defines 2 KLRG1+ mouse NK-cell subsets with distinct functional properties and positioning in the bone marrow. Blood. 117:4467–4475 10.1182/blood-2010-07-297101 [DOI] [PubMed] [Google Scholar]
- Sonnenberg G.F., Fouser L.A., Artis D. 2011. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12:383–390 10.1038/ni.2025 [DOI] [PubMed] [Google Scholar]
- Spits H., Di Santo J.P. 2011. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12:21–27 10.1038/ni.1962 [DOI] [PubMed] [Google Scholar]
- Sun Z., Unutmaz D., Zou Y.R., Sunshine M.J., Pierani A., Brenner-Morton S., Mebius R.E., Littman D.R. 2000. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 288:2369–2373 10.1126/science.288.5475.2369 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kanno T., Nakayamada S., Hirahara K., Sciumè G., Muljo S.A., Kuchen S., Casellas R., Wei L., Kanno Y., O’Shea J.J. 2012. TGF-β and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 13:587–595 10.1038/ni.2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori H., Kanno Y., Watford W.T., Tato C.M., Weiss G., Ivanov I.I., II, Littman D.R., O’Shea J.J. 2009. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 206:35–41 10.1084/jem.20072713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., Glimcher L.H. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 20:477–494 10.1016/S1074-7613(04)00076-7 [DOI] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 397:702–706 10.1038/17812 [DOI] [PubMed] [Google Scholar]