The transcriptional repressor BCL6 reduces miRNA levels in germinal center B cells to increase AID expression.
Abstract
The BCL6 proto-oncogene encodes a transcriptional repressor that is required for germinal center (GC) formation and whose de-regulation is involved in lymphomagenesis. Although substantial evidence indicates that BCL6 exerts its function by repressing the transcription of hundreds of protein-coding genes, its potential role in regulating gene expression via microRNAs (miRNAs) is not known. We have identified a core of 15 miRNAs that show binding of BCL6 in their genomic loci and are down-regulated in GC B cells. Among BCL6 validated targets, miR-155 and miR-361 directly modulate AID expression, indicating that via repression of these miRNAs, BCL6 up-regulates AID. Similarly, the expression of additional genes relevant for the GC phenotype, including SPI1, IRF8, and MYB, appears to be sustained via BCL6-mediated repression of miR-155. These findings identify a novel mechanism by which BCL6, in addition to repressing protein coding genes, promotes the expression of important GC functions by repressing specific miRNAs.
INTRODUCTION
The BCL6 proto-oncogene encodes a transcriptional repressor required for germinal center (GC) formation. In mature B cells, BCL6 expression is restricted to the GC stage by a tightly controlled transcriptional and posttranscriptional regulation (Basso and Dalla-Favera 2012). Deregulation of BCL6 expression has been implicated in lymphomagenesis via multiple mechanisms, including chromosomal translocations that prevent its transcriptional repression (Ye et al., 1995; Chen et al., 1998), defective protein degradation caused by inactivating mutations and deletions in the FBXO11 gene (Duan et al., 2012), and reduced acetylation-mediated inactivation caused by genetic alterations in acetyltransferase genes (CREBBP and EP300; Pasqualucci et al., 2011). The deregulated expression of BCL6 has been shown to contribute to lymphomagenesis in mice (Cattoretti et al., 2005).
The identification of the BCL6 transcriptional network is an essential step toward the understanding of the role of BCL6 in normal B cell development and in lymphomagenesis. Recent genome-wide analyses have identified a large number of BCL6 direct target genes and have shown that BCL6 represses critical pathways for GC development including the response to DNA damage, cell activation, and differentiation (Ci et al., 2009; Basso et al., 2010). Nonetheless, much less is known about the role of BCL6 in regulating microRNA (miRNA) expression, a mechanism by which BCL6 would be able to positively regulate, as opposed to only repress, gene expression in GC B cells.
In this paper, we identify a set of miRNAs whose expression is repressed by BCL6, leading to a positive regulation of their target genes. In particular, we characterize the direct repression of miR-155 and miR-361 by BCL6 and investigate the downstream consequences of these interactions. MiR-155 is produced from the nonprotein coding transcript of the MIR155HG (previously known as BIC) gene, which was originally identified as a recurrent site of integration of the avian leukosis virus in chicken lymphomas (Clurman and Hayward 1989; Tam et al., 1997; Tam, 2001). In B cells, miR-155 has been shown to modulate the expression of AICDA (also known as AID; Dorsett et al., 2008; Teng et al., 2008), as well as of other genes important for the GC reaction, including those involved in differentiation, such as SPI1 (Vigorito et al., 2007) and CEBPB (Costinean et al., 2009), in B cell migration, such as HGAL (Dagan et al., 2012), in TGFB1 and BMP signal transduction, such as SMAD5 (Rai et al., 2010), and in BCR and PI3K signaling, such as SHIP1 (Costinean et al., 2009; Pedersen et al., 2009). Accordingly, mice lacking miR-155 display a reduced number of GC B cells and compromised affinity maturation (Rodriguez et al., 2007; Thai et al., 2007), whereas mice engineered to constitutively express miR-155 in mature B cells show an increase in GC B cells and an enhanced antibody response (Thai et al., 2007). Conversely, much less is known about miR-361, which is embedded in the CHM gene. CHM encodes a subunit of a Rab geranylgeranyl transferase and is known for its genetic inactivation in choroideremia (van den Hurk et al., 1997), but neither CHM nor miR-361 have a defined function in B cells.
Here we show that, via direct repression of miR-155 and miR-361, BCL6 positively regulates the expression of their target genes, including AID and other factors involved in the maintenance of the GC phenotype. The results identify a broader role of BCL6 in GC formation and lymphomagenesis.
RESULTS
BCL6 transcriptionally modulates miRNA expression in GC B cells
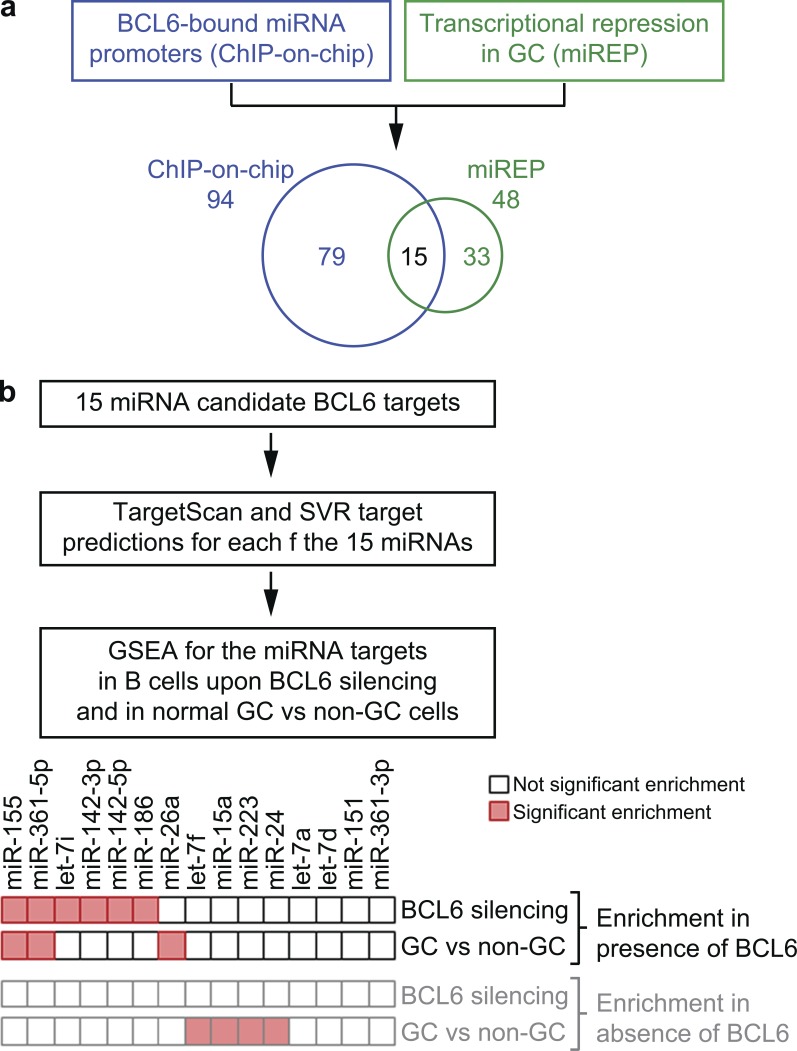
To identify BCL6 target genes in normal GC B cells, we previously used an integrated approach combining genome-wide chromatin immunoprecipitation (ChIP; ChIP-on-chip) analysis to identify promoter regions bound by BCL6, and gene expression profiling to detect protein-coding genes down-regulated in GC (Basso et al., 2010). Here, we undertook a similar approach to identify, in normal GC B cells, candidate BCL6 targets among miRNA genes. Toward this goal, ChIP-on-chip data (Basso et al., 2010) were integrated with miRNA expression profiling (Basso et al., 2009), leading to the identification of 15 miRNA down-regulated in GC B cells compared with naive and/or memory B cells and displaying evidence of BCL6 binding in their regulatory regions (Fig. 1 a).
Figure 1.
Identification of miRNAs that are candidate targets of BCL6 repression in GC B cells. (a) Identification of 15 miRNAs down-regulated in GC B cells, as detected by miRNA expression profiling (miREP), and displaying binding of BCL6 in their promoters (by genome-wide ChIP-on-chip). (b) GSEA was performed on the miRNA targets predicted by both the TargetScan and SVR algorithms. miR-155 and miR-361-5p display a significant enrichment of their targets in genes up-regulated in presence of BCL6 in a B cell line subject to BCL6 silencing and in normal GC compared with non-GC B cells.
To identify miRNAs of physiological relevance for the BCL6 program, we analyzed whether the genes computationally predicted as targets of these 15 miRNAs were dynamically connected with BCL6, i.e., up-regulated in GC B cells as a consequence of miRNA repression. Genes predicted, by both TargetScan (Lewis et al., 2005) and Miranda-mirSVR (John et al., 2004; Betel et al., 2010) algorithms, to be modulated by the 15 miRNAs were investigated by gene set enrichment analysis (GSEA; Subramanian et al., 2005) in the expression profiles of a B cell line subjected to BCL6 silencing and in the expression profiles of normal GC versus non-GC B cells (Basso et al., 2010). The results revealed that the predicted targets of 6 of the 15 miRNAs were significantly enriched among genes up-regulated concurrently with BCL6, suggesting that the negative modulation of BCL6 on the miRNAs contributes to release the expression of their targets (Fig. 1 b and Table S1). Consistent with a positive correlation between the miRNA targets and BCL6, none of the 15 miRNAs displayed enrichment of its targets among genes that are up-regulated upon BCL6 silencing (Fig. 1 b and Table S1). Among the six miRNAs highlighted by this analysis, two (miR-155 and miR-361-5p) had their targets displaying a significant enrichment among genes up-regulated in the presence of BCL6 in both the BCL6 silencing assay and in the GC versus non-GC B cell comparison, thus identifying them as the top candidates for further investigation (Fig. 1 b and Table S1). Thus, miR-155 and miR361-5p control a set of targets that are significantly enriched for genes coexpressed with BCL6, consistent with a physiological role as mediators of BCL6 activity in GC B cells.
BCL6 binds to and represses transcription of the miR-155 and miR-361 loci
To conclusively elucidate the mechanism of miR-155 and miR-361 regulation by BCL6, we first confirmed, by qChIP analysis, the binding of BCL6 detected by ChIP-on-chip at these loci in normal GC B cells. ChIP-on-chip analysis mapped BCL6 binding in intron 2 of the miR-155 host gene (MIR155HG), but its promoter region was not represented on the array used for this analysis. Investigation of the 5′ region of MIR155HG by qChIP detected an additional region of BCL6 binding within a 300 bp segment 5′ of the TSS (Fig. 2 a). Similarly, BCL6 binding was detected by ChIP-on-chip and confirmed by qChIP in the promoter of the miR-361 host gene CHM (region D; −432/+168 bp) and in its intron 9 (region E) ∼4 Kb upstream of miR-361coding region (Fig. 2 b). In conclusion, ChIP analyses showed that BCL6 is bound in normal GC B cells to the MIR155HG and CHM promoters and to intronic regions upstream of the miR-155 and miR-361 coding sequences.
Figure 2.
BCL6 binds to regulatory regions in MIR155HG and CHM loci and transcriptionally regulates miR-155 and miR-361 expression. (a) Schematic representation of the MIR155HG locus and relative fold enrichment as detected by ChIP for BCL6 followed by quantitative PCR (qChIP) analyses. The bound regions, A and B, are located in the promoter and in intron 2 of the MIR155HG locus, respectively; a region of not binding in the same locus (region C) was used as a negative control. The results are displayed as means and SD of two independent experiments, each performed in triplicates. Region A, P = 0.0006; region B, P = 0.005, Student’s t test. (b) Schematic representation of the CHM locus and relative fold enrichment as detected by qChIP analysis. The bound regions, D and E, are located in the promoter and in intron 9 of the CHM locus, respectively; a region of not binding in the same locus (region F) was used as a negative control. BCL6 binding to its own promoter (BCL6) and in the actin locus were used as positive and negative controls, respectively. The results are displayed as means and SD of two independent experiments, each performed in triplicates. Region D, P = 0.02; region B, P = 0.003, Student’s t test. (c and d) Promoter luciferase assays were performed to test the responsiveness to BCL6 repression of the bound regulatory regions in the MIR155HG (c) and in the CHM genes (d). The results are displayed as relative luciferase activity of the reporter constructs in presence of increasing amount of wild-type BCL6, or the maximum amount of its mutants (BCL6-ZF and BCL6ΔZF), compared with the reporter basal activity, normalized to renilla activity. The displayed means and SD are from three independent experiments, each performed in duplicates. Statistically significant changes were measured for the reporter constructs subjected to the highest dose of BCL6 compared with the reporter vector lacking the tested regions (empty) and to the same setting using the BCL6 mutants (region A vs. empty, P = 0.04, vs. BCL6ZF, P = 0.01, vs. BCL6ΔZF, P = 0.02; region B vs. empty, P = 0.03, vs. BCL6ZF P = 0.0004, vs. BCL6ΔZF, P = 0.0008; region D vs. empty, P = 0.5 n.s., vs. BCL6ZF, P = 0.006, vs. BCL6ΔZF, P = 0.02; region E vs. empty, P = 0.002, vs. BCL6ZF, P = 0.0004, vs. BCL6ΔZF, P = 0.001, Student’s t test). (e) miR-155 qRT-PCR performed in Raji cells engineered to inducibly (upon doxycycline treatment, Dox) express an empty vector (EV) or the BCL6ΔPEST mutant and treated as indicated. The results are displayed as miR-155 fold change relative to the αIgM/αCD40-untreated cells (mean ± SD, n = 3). The miR-155 expression changes upon stimulation are significantly different in presence of BCL6ΔPEST (P = 1.5 × 10−7, Student’s t test). (f) Detection of BCL6 (endogenous, endo; exogenous, exo) and β-actin by immunoblotting in the BCL6ΔPEST inducible cell lines displayed in e.
The BCL6-bound genomic regions included a substantial number (range 8–11) of predicted BCL6 binding sites, 92% of which appeared to be conserved in mammals (Table S2). To determine whether BCL6 binding to miR-155 and miR-361 loci could result in transcriptional repression, the relevant regions bound by BCL6 (Fig. 2, a and b) were subcloned upstream of a luciferase reporter gene, and their responsiveness to BCL6 was tested in transient transfection/reporter assays. The MIR155HG promoter region (region A) and the CHM intron 9 region (region E) displayed a significant dose-dependent BCL6 repression, whereas BCL6 mutants lacking the ability to bind DNA (BCL6ΔZF) or to recruit corepressor complexes (BCL6ZF) were impaired in their repression (Fig. 2, c and d), indicating that trans-repression was dependent on DNA binding and BCL6 corepressor complex formation. The MIR155HG intronic region (region B) and the CHM promoter (region D) were also modestly responsive to BCL6 repression activity in this reporter assay (Fig. 2, c and d). These results indicate that BCL6 can repress miR-155 and miR-361 by binding to regulatory regions in their loci.
To demonstrate that BCL6-mediated repression of its miRNA targets can occur in physiological conditions, we tested whether variations in the levels of BCL6 can affect the expression of the endogenous miRNA loci. Toward this end, we first investigated the effects of BCL6 silencing in GC-derived lymphoma cell lines characterized by high BCL6 and low miR-155 and miR-361 expression. Because no or very modest derepression of miR-155 and miR-361 was detected upon BCL6 silencing (unpublished data), we reasoned that removal of repression might be insufficient for target induction in the absence of activating stimuli. This hypothesis was supported by the observation that the expression of both miR-155 and miR-361 is increased in GC B cells located in the light zone (unpublished data), the site of extensive signaling from the GC microenvironment (Victora et al., 2010, 2012), and by previous studies on high expression of miR-155 in activated B cells but not in resting cells (van den Berg et al., 2003; Thai et al., 2007). Furthermore, previous studies have shown that miR-155 is induced by BCR signaling (van den Berg et al., 2003), whereas CD40 and BCR engagement leads to BCL6 down-regulation both at RNA and protein level (Niu et al., 1998; Basso et al., 2004). Initial experiments showed that although miR-361 was not induced by CD40 and BCR engagement in GC-derived cell lines, miR-155 expression was positively affected by these signals (unpublished data). Thus, we engineered a Burkitt lymphoma cell line (Raji) to inducibly express a BCL6 mutant (BCL6ΔPEST) insensitive to down-regulation by combined CD40/BCR signaling (Niu et al., 1998), and we tested the effect of constitutive BCL6 expression on the induction of the miRNA by the activation stimuli. CD40 and BCR engagement positively affected miR-155 expression, whereas its induction was abrogated in the presence of BCL6ΔPEST (Fig. 2, e and f), indicating that constitutive BCL6 expression negatively modulates the induction of endogenous miR-155.
Taken together, these results demonstrate that BCL6 directly trans-represses miR-155 in GC B cells. Although we cannot conclusively prove that miR-361 is a direct BCL6 transcriptional target, the binding of BCL6 in the miR-361 locus and the down-regulation observed in normal GC B cells (followed by a slight induction in the light zone GC B cells, in association with BCL6 down-regulation in the same cells) suggest that miR-361 expression is modulated by BCL6.
BCL6-mediated miRNA repression leads to positive regulation of miRNA target genes
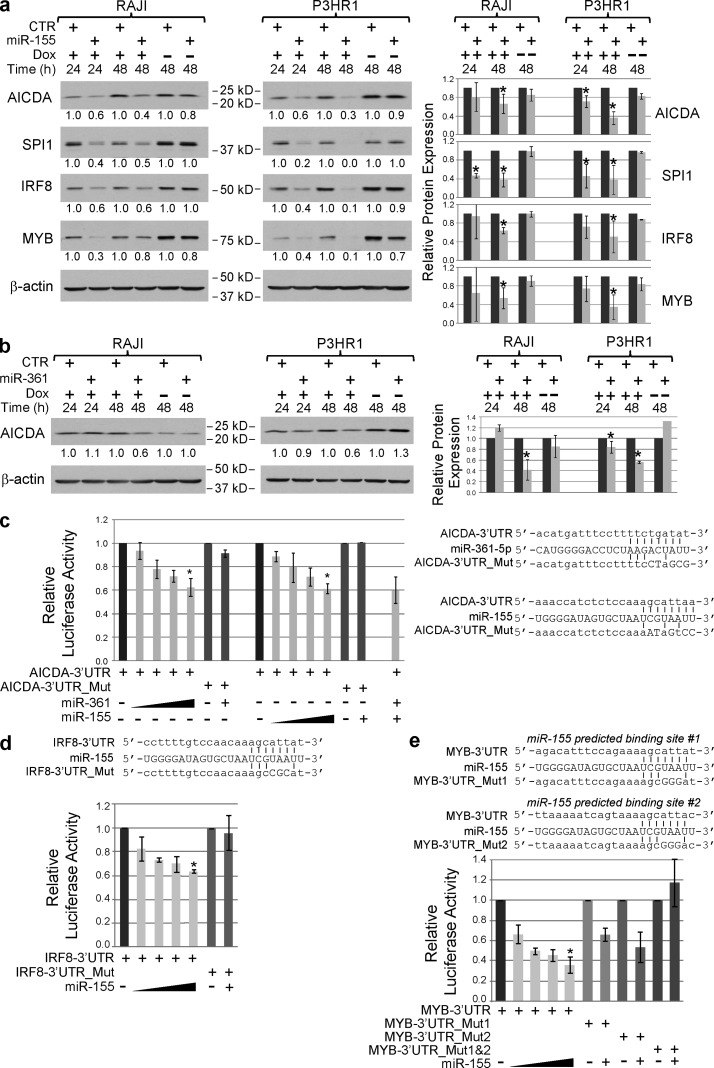
The trans-repressive activity of BCL6 on its miRNA targets becomes physiologically relevant if it is able to sustain the expression of genes that are negatively modulated by these miRNAs. To test this hypothesis, we first identified miR-155 and miR-361 candidate targets using the TargetScan (Lewis et al., 2005) and Miranda-mirSVR (John et al., 2004; Betel et al., 2010) algorithms and considered only the common predictions (Table S3). Several candidates, selected based on their relevance for GC biology, were then tested for their responsiveness to miR-155 and/or miR361 in the Burkitt lymphoma cell lines P3HR1 and Raji engineered to inducibly express miR-155 or miR-361. This approach allowed us to validate in human B cells miR-155 targets previously reported in mouse, including AID and SPI1 (Vigorito et al., 2007; Dorsett et al., 2008; Teng et al., 2008), and to identify novel GC-relevant candidates such as IRF8 and MYB (Fig. 3 a). Notably, AID was also identified as a candidate target of miR-361-5p (Fig. 3 b). As expected for miRNA targets, these candidates display a very modest down-regulation at the transcriptional level but a significant reduction in protein levels upon miRNA induction in B cells (Fig. 3, a and b). The direct inhibition of AID by both miRNAs was further demonstrated by 3′-UTR reporter assay in which the presence of the full-length AID 3′-UTR downstream of a reporter gene was associated with a dose-dependent reduction of the reporter activity in presence of miR-361 or miR-155 (Fig. 3c). Mutation of the predicted miR-155 (consistent with previous studies [Dorsett et al., 2008; Teng et al., 2008]) and/or miR361 binding sites in the AID 3′-UTR rescued the repression induced by the miRNAs (Fig. 3 c), demonstrating that both miR-155 and miR-361-5p directly target AID. The direct repression of IRF8 and MYB by miR-155 was demonstrated in an analogous assay (Fig. 4, d and e).
Figure 3.
miR-155 and miR-361 target GC-relevant genes, including AICDA. (a) The effect of induction of miR-155 by doxycycline (Dox) treatment in engineered Burkitt’s lymphoma cell lines (P3HR1 and Raji) on the expression of AICDA, SPI1, IRF8, and MYB, as detected by immunoblotting (left). The right panel represents the relative fold changes obtained in three independent experiments (mean ± SD, n = 3; *, P ≤ 0.05, Student’s t test). (b) Effects of miR-361 induction on AICDA protein levels. Normalized relative fold change in protein levels are reported below each lane. The right panel represents the relative fold changes obtained in three independent experiments (mean ± SD, n = 3; *, P ≤ 0.05, Student’s t test). (c) AICDA-3′UTR reporter assay in response to increasing amount of miR-155 and miR-361. On the right, schematic representation of the regions in the AICDA-3′UTR (AICDA-3′UTR WT) that are targeted by miR-155 and miR-361 and their alignments with the miRNA sequences. The mutations introduced in the mutant UTRs (AICDA-3′UTR Mut) are displayed below (mutated nucleotides are in capital letters). The results are displayed as relative luciferase activity of the reporter construct in presence of increasing amount of miRNA compared with its basal activity, upon normalization by renilla activity (mean ± SD, n = 3). Statistically significant changes were analyzed only for the wild type compared with the mutated UTRs in the presence of the highest dose of miRNA (miR-361, P = 0.001; miR-155, P = 1 × 10−5, Student’s t test). (d) IRF8-3′UTR reporter assay in response to increasing amount of miR-155. On top, schematic representation is shown of the region in the IRF8-3′UTR (IRF8-3′UTR WT) that is targeted by miR-155 and its alignments with the miRNA sequence. The mutations introduced in the mutant UTRs (IRF8-3′UTR Mut) are displayed below (mutated nucleotides are in capital letters). The results are displayed as relative luciferase activity of the reporter construct in presence of increasing amount of miRNA compared with its basal activity, upon normalization by renilla activity (mean ± SD, n = 3). Statistically significant changes were analyzed only for the wild type compared with the mutated UTRs in the presence of the highest dose of miRNA (*, P < 0.05, Student’s t test). (e) MYB-3′UTR reporter assay in response to increasing amount of miR-155. On top, schematic representation is shown of the regions in the MYB-3′UTR (MYB-3′UTR WT) that are targeted by miR-155 and their alignments with the miRNA sequence. The mutations introduced in the mutant UTRs (MYB-3′UTR Mut) are displayed below (mutated nucleotides are in capital letters). The results are displayed as relative luciferase activity of the reporter construct in presence of increasing amount of miRNA compared with its basal activity, upon normalization by renilla activity (mean ± SD, n = 3). Statistically significant changes were analyzed only for the wild type compared with the mutated UTRs in the presence of the highest dose of miRNA (*, P < 0.05, Student’s t test).
Figure 4.
BCL6 is coexpressed with miR-155 and miR-361 targets in normal lymphoid tissue. Immunofluorescence costaining of BCL6 and AICDA, SPI1, or IRF8 in normal lymphoid tissue. A representative GC is displayed. IRF4 is used to identify late GC cells that have down-regulated BCL6; CD20 is used as a B cell marker. Arrows point to representative cells that show coexpression (yellow arrows) or lack of expression (white arrows) for BCL6 and the miRNA targets. Scale bars: (top) 100 µm; (insets) 10 µm.
To further investigate the physiological relevance of the direct relationship between BCL6 and the targets of miR-155 and miR-361, we examined whether BCL6 and selected miRNA target genes were coexpressed in normal human lymphoid tissues by immunofluorescence analysis. Consistent with the modulation of miR-155 and miR-361 by BCL6, the miRNA targets AID, SPI1, and IRF8 were largely coexpressed with BCL6 in GC B cells (Fig. 4; coexpression of BCL6 and MYB could not be assessed because of the unavailability of suitable antibodies). Notably, the subset of B cells located in the GC light zone that down-regulate BCL6 expression and induce the plasmablastic marker IRF4 displayed absent or reduced expression of the miRNA targets (Fig. 4). Overall, these data show a direct and specific relationship in GC B cells between BCL6 and the targets of miR-361 and/or miR-155, consistent with a BCL6-mediated contribution to their expression via miRNA inhibition.
BCL6 deregulation affects the expression of miR-155 and its targets in vivo
To further investigate the relationship between BCL6 and its miRNA targets in vivo, we analyzed the expression levels of miR-155 and miR-361 in transgenic mice constitutively expressing the BCL6 oncogene in GC B cells (Iμ-HA-BCL6 mice; Cattoretti et al., 2005). In these mice, the constitutive expression of the BCL6 gene, placed under the control of the immunoglobulin Iμ promoter to mimic a translocation found in human diffuse large B cell lymphoma (DLBCL), leads to increased GC formation, perturbed post-GC differentiation, and, ultimately to DLBCL development (Cattoretti et al., 2005). Analysis of purified GC B cells from these mice at a young age (3 mo old, ∼1 yr before tumor development) revealed a significant reduction of miR-155 expression consistent with the slight increase in BCL6 levels (Fig. 5, a and b). MiR-361 expression was undetectable in GC B cells from littermate wild-type mice and, therefore, de-regulated BCL6 expression provided no further repressive effect. These results showed that in vivo de-regulation of BCL6 is associated with reduced miR-155 expression in GC B cells.
Figure 5.
BCL6 de-regulation affects the expression of miR-155 and its targets in vivo. (a) miR-155 expression in GC B cells purified from Iμ-HA-BCL6 transgenic mice compared with wild-type (WT) littermate mice as shown by qRT-PCR (P = 0.04, Student’s t test). Means and SD are calculated from five pools of wild-type and seven pools of Iμ-HA-BCL6 mice (each pool includes GC B cells isolated from two to three mice). (b) Immunoblotting for miR-155 targets in purified GC B cells isolated from three independent pools of wild-type (WT) and Iμ-HA-BCL6 littermate mice (each pool includes three mice). Relative protein fold changes, normalized by actin expression, are reported below each lane. (c) Protein (top) and mRNA (bottom) fold change relative to the wild-type mice are displayed upon normalization by actin expression (mean ± SD, n = 3). mRNA levels were detected by quantitative RT-PCR on the same samples displayed in b. (*, P < 0.05, Student’s t test).
To test whether the reduced miR-155 levels in Iμ-HA-BCL6 mice would be associated with changes in the expression of miR-155 targets, GC B cells were isolated from pools of three each of wild-type and Iμ-HA-BCL6 littermate mice and subjected to protein and RNA analyses. Protein expression of AID was indeed modestly, but significantly, increased in the Iμ-HA-BCL6 mice in the absence of changes in its mRNA levels (Fig. 5, b and c). Similarly, a significant increase in protein expression was observed for MYB and IRF8, whereas SPI1 showed a trend that did not reach statistical significance (Fig. 5, b and c).
Thus, deregulated BCL6 expression leads to transcriptional repression of miRNAs, which, in turn, is associated with the up-regulation of their target genes in vivo. Although it cannot be excluded that these relationships are more indirect and depend upon other BCL6-induced changes in GC biology, these data are consistent with a miR-155–mediated effect of BCL6.
DISCUSSION
A first result of this study is the demonstration that the BCL6 transcriptional repressor can directly target and repress miRNA expression. This function is mechanistically consistent with its demonstrated activity as a transcriptional modulator of hundreds of genes in the GC (Ci et al., 2009; Basso et al., 2010) and significantly broadens the BCL6 biological program both quantitatively and qualitatively. In fact, via repression of a specific subset of miRNAs, BCL6 can influence the expression of a much larger set of genes and therefore GC cellular pathways. In addition, the transcriptional repression of miRNAs allows BCL6 to contribute to the GC phenotype, not only through repression of direct targets but also by positively influencing the expression of a wholly distinct set of genes. Given the one-step indirect mechanism linking BCL6 to its miRNA-modulated targets, BCL6 may act to sustain, rather than to induce, the expression of those genes that are negatively regulated by the miRNAs. Although the results shown here provide a paradigm for this novel BCL6 function by identifying a subset of 15 candidate miRNA targets and by focusing on the validation of two such candidates, they represent the basis for a comprehensive investigation of the miRNA-mediated gene regulation program of BCL6.
The identification of miR-155 as a direct BCL6 target has several important implications for GC biology. First, by modulating the expression of this miRNA, BCL6 contributes to a fine tuning of its level of expression, a critical function during the GC reaction given that deregulation of miR-155 in mouse GC B cells can interfere with the selection of B cells expressing high affinity antigen receptors (Rodriguez et al., 2007; Thai et al., 2007). Second, the specific modulation of AID by miR-155 and miR-361 suggests a coordinated activity of BCL6 in sustaining high levels of AID expression in GC B cells undergoing somatic hypermutation and class-switch recombination of their immunoglobulin genes (Victora and Nussenzweig 2012). Third, the presence of transcription factors important for GC development, such as IRF8 (Lee et al., 2006), SPI1 (Garrett-Sinha et al., 1999; Vigorito et al., 2007), and MYB (Thomas et al., 2005; Lefebvre et al., 2010), among the miR-155 targets suggests that in addition to its well characterized repression functions, BCL6 contributes to the establishment and/or maintenance of the GC phenotype by sustaining specific biological programs. Notably, the expression of BCL6 and miR155 appears to be modulated coordinately as part of a broader regulatory circuit in which the stimuli that down-regulate BCL6 in the light zone of the GC, namely BCR and CD40 signaling, also induce the expression of miR-155 (Niu et al., 1998; van den Berg et al., 2003; Basso et al., 2004; Saito et al., 2007). This circuit allows the coordinated switching off of GC-specific players, such as BCL6 and AID, as well as the down-regulation of transcription factors (e.g., IRF8 and SPI1) that maintain the GC phenotype while preventing plasma cells differentiation.
Finally, these results have implications for the role of both BCL6 and miR-155 in lymphomagenesis. The observations that the MIR155HG locus is a recurrent site of integration of the avian leukosis virus in chicken lymphomas (Clurman and Hayward 1989; Tam et al., 1997; Tam, 2001) and that several types of cancer, including a subset of DLBCL, have high levels of miR-155 expression (Eis et al., 2005; Volinia et al., 2006), have led to the hypothesis that miR-155 may act as a dominant oncogene. Indeed, Eμ-miR-155 transgenic mice display a block at the pre–B cell stage of differentiation and develop acute lymphoblastic leukemia (Costinean et al., 2009), demonstrating that miR-155 de-regulation in early B cell development can be oncogenic. However, the results presented here suggest that miR-155 may have oncosuppressive functions in GC B cells, at least through its negative modulation of AID. In support of this notion, mice carrying mutations in their AID 3′UTR impairing miR-155–mediated repression display an increased number of AID-induced translocations (Dorsett et al., 2008).
Although multiple genetic and epigenetic mechanisms have been identified to deregulate BCL6 expression in DLBCL and follicular lymphoma, their common functional denominator is the prevention of the physiological suppression of BCL6 expression at the end of the GC reaction (Basso and Dalla-Favera 2012). Thus, aberrant BCL6 activity contributes, via miR-155 and miR-361 repression, to sustain the expression of AID, an event which has been shown to cause the accumulation of genetic damage and promote BCL6-driven lymphomagenesis in mice (Pasqualucci et al., 2008). Furthermore, the same mechanism supports the continued expression of transcription factors that interfere with plasma cell differentiation (Carotta et al., 2010), a second process involved in malignant transformation. Based on these observations, the miRNA-mediated function of BCL6 in sustaining gene expression may be a relevant component of its action as an oncogene in lymphomagenesis.
MATERIALS AND METHODS
Primary cells, cell lines, and treatments.
Purification of GC B cells was performed as previously reported (Klein et al., 2003) using magnetic cell sorting of mononucleated cells obtained from human tonsils. GC dark zone and light zone subpopulations were isolated from tonsillar mononucleated cells as previously reported (Victora et al., 2012). Murine GC cells were isolated from splenic mononucleated cell suspension by flow cytometric cell sorting upon staining with CD45R/B220-PercP (BD), PNA-FITC (Vector Laboratories), and CD95-PE (BD) antibodies. P3HR1 and Raji (Burkitt lymphoma cell lines) and HEK-293T (human embryonic kidney cell line) were obtained from American Type Culture Collection and maintained at the recommended concentration in IMDM and DMEM medium, respectively, supplemented with 10% fetal bovine serum and antibiotics. CD40 and BCR stimulation was performed using 5 µg/ml anti-CD40 (G28.5) and 10 µg/ml IgM (SouthernBiotech) antibodies for 24 h. P3HR1 and/or Raji cell line with inducible expression of miR-155, miR-361, or BCL6-ΔPEST were established by electroporation of the corresponding pRTS1-expressing vectors. The stable inducible cell lines were obtained upon selection with puromycin. The pRTS1 vector includes a bidirectional promoter, which allows simultaneous expression of the gene of interest along with GFP. Inducibility was assessed 24 and 48 h upon doxycycline treatment by measuring GFP levels flow cytometrically and miR-155, miR-361, or BCL6 expression by qRT-PCR and immunoblotting, respectively.
Plasmids and reporter assays.
DNA fragments encoding a mutant BCL6 lacking the PEST domain (BCL6-ΔPEST; Niu et al., 1998), as well as the miR-155– or the miR-361–encoding genomic regions were inserted in the pRTS1 doxycycline-inducible vector (Bornkamm et al., 2005). For constitutive expression, the miR-155 or the miR-361 encoding genomic regions were inserted in the pcDNA3 vector (Promega). The promoter reporter constructs were obtained by subcloning the miR155HG promoter region (region A; −350/−1 bp from the TSS) or the intronic region (region B; +10,899/+11,699 bp from the TSS) into the pGL3b vector (Promega), and the CHM promoter region (region D; −432/+168 bp from the TSS) or the intronic region (region E; −4,768/−4,168 bp from the mature miR-361 coding sequence) into the pGL4.10 vector (Promega). The plasmids expressing BCL6 wild type, or its mutants lacking the DNA binding domain (BCL6ΔZF) or the BTB/POZ domains (BCL6-ZF) were previously reported (Pasqualucci et al., 2003). The AID-3′UTR reporter construct was obtained by subcloning the full-length AID-3′UTR in the pmiRGlo vector (Promega), and mutations in the miR-155 and miR-361 binding sites were introduced using the Quick Change site-directed mutagenesis kit (Stratagene) and the primers reported in Table S4. HEK-293T cells were transiently transfected by calcium-phosphate precipitation method as previously described (Chang et al., 1996) and each transfection was performed in triplicates. Luciferase activities were measured 48 h after transfection using the Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
ChIP-on-chip and qChIP assays.
ChIP-on-chip was performed using the Human Promoter ChIP-on-chip Microarray Set (2 × 244K; Agilent Technologies), which includes probes interrogating potential regulatory regions for ∼230 miRNAs of which 32% are intergenic, whereas the remaining 68% are located in genes. ChIP was performed as previously reported (Pasqualucci et al., 2003) starting from 40 × 106 purified GC B cells and using 4 µg anti-BCL6 antibody (N3; Santa Cruz Biotechnology, Inc.) or isotype-matched polyclonal IgG (Sigma-Aldrich). DNA fragments enriched by ChIP were analyzed for expression of individual targets including BCL6 (positive control) and β-actin (negative control) using primers enlisted in Table S4 or previously reported (Basso et al., 2010) and SYBR Green PCR Master Mix (Applied Biosystems) as recommended by the manufacturer. ΔCt values were calculated for anti-BCL6 and control IgG immunoprecipitated DNA fragments relative to their input DNA and then used to calculate the ΔΔCt. Fold changes (2−ΔΔCt) observed in BCL6 versus control IgG immunoprecipitated DNA were corrected using β-actin as negative control and reported as relative fold enrichment. QPCR reactions were performed in triplicates and each experiment was repeated at least twice.
qRT-PCR.
Total RNA was isolated using the TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. 1 µg of total RNA was reverse transcribed using the Superscript First Strand Synthesis System for RT-PCR (Invitrogen) and 1/100 of cDNA was used as template for PCR amplification using the primers reported in Table S4 or previously published (Lee et al., 2006) and SYBR Green PCR Master Mix (Applied Biosystems) as recommended by the manufacturer. Actin was used as control. miRNA detection was performed using the Mature TaqMan MicroRNA assays (Applied Biosystems) for human miR-155, murine miR-155, human/murine miR-361-5p and, as control, for RNU6B according to the manufacturer’s protocol. qRT-PCR reactions were performed in triplicates and each experiment was repeated at least twice.
Immunoblotting.
Whole cell lysates were prepared as previously described (Niu et al., 1998). Proteins were analyzed by SDS-PAGE and immunoblotting using the following antibodies: anti-BCL6 (Cell Signaling Technology), anti-AICDA (mAID-2; eBioscience), anti-SPI1 (sc-352; Santa Cruz Biotechnology, Inc.), anti-IRF8 (sc-6058; Santa Cruz Biotechnology, Inc.), anti-MYB (sc-517; Santa Cruz Biotechnology, Inc.), and anti–β-actin (A5441; Sigma-Aldrich). Detection was performed using the ECL system (GE Healthcare). Images were quantitated by ImageJ software (National Institutes of Health).
Tissue immunofluorescence.
Immunofluorescence was performed on human reactive lymphoid tissue using the following antibodies: anti-BCL6 (N3; Santa Cruz Biotechnology, Inc.), anti-IRF4 (M17; Santa Cruz Biotechnology, Inc.), anti-AICDA (mAID-2; eBioscience), anti-IRF8 (sc-6058; Santa Cruz Biotechnology, Inc.), anti-CD20 (Ab-1; NeoMarkers), anti-SPI1 (sc-352; Santa Cruz Biotechnology, Inc.), PE-conjugated anti-BCL6 (BD), CyTM-3–conjugated donkey anti–rabbit IgG (DARb-Cy3; Jackson ImmunoResearch Laboratories), biotin donkey anti–goat IgG (DAGt-bio; Jackson ImmunoResearch Laboratories), biotin donkey anti–rat IgG (Southern Biotech), biotin horse anti–mouse IgG (Vector Laboratories); Alexa Fluor 350–conjugated avidin (Life Technologies), fluorescein-conjugated avidin (Life Technologies), and peroxidase-labeled avidin (Avidin-HRP; Vector Laboratories). IRF8 and SPI1 were detected using the TSA Plus Fluorescein kit (Perkin Elmer) according to the manufacturer’s instructions. Paraffin-embedded formalin-fixed 3–5 µm sections were dewaxed in xylene and rehydrated in decreasing concentration of ethanol. Antigens were unmasked by heat-induced antigen retrieval in citrate buffer, pH 6.0, or EDTA buffer, pH 8.0. Stainings were performed as previously described (Cattoretti et al., 2006). Endogenous peroxidase and biotin were blocked as reported. Primary antibodies were incubated overnight at 4°C. Stained tissues were imaged using a fluorescence microscope (Eclipse E400; Nikon).
Statistical analysis.
The statistics provided in the GSEA (including p-value, normalized p-value, and false discovery rate) were calculated using the GSEA software (Subramanian et al., 2005).
Online supplemental material.
Table S1 reports the results of the GSEA in a cell line subjected to BCL6 silencing and in normal GC versus non-GC B cells for the miRNA targets commonly predicted by Miranda-SVR and TargetScan. Table S2 displays the BCL6 binding motifs identified in the BCL6 bound regions displayed in Fig. 2. Table S3 includes the miR-155 and miR-361-5p targets predicted by both Miranda-SVR and TargetScan algorithms. The primers used in this work are listed in Table S4. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121387/DC1.
Supplementary Material
Acknowledgments
We thank David Dominguez-Sola, Gabriel D. Victora, and Michel N. Nussenzweig for suggestions and for providing RNA isolated from normal dark zone and light zone GC B cells. We thank Hongyan Tang and Tongwei Mo for mouse husbandry; and Cristina Girardi and Friederike H. Schwartz for technical support. We are grateful to Roy L. Maute for discussions and critical reading of the manuscript.
This work was supported by a Leukemia and Lymphoma Society Specialized Center for Research (SCOR) grant.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DLBCL
- diffuse large B cell lymphoma
- GC
- germinal center
- GSEA
- gene set enrichment analysis
- miRNA
- microRNA
References
- Basso K., Dalla-Favera R. 2012. Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev. 247:172–183 10.1111/j.1600-065X.2012.01112.x [DOI] [PubMed] [Google Scholar]
- Basso K., Klein U., Niu H., Stolovitzky G.A., Tu Y., Califano A., Cattoretti G., Dalla-Favera R. 2004. Tracking CD40 signaling during germinal center development. Blood. 104:4088–4096 10.1182/blood-2003-12-4291 [DOI] [PubMed] [Google Scholar]
- Basso K., Sumazin P., Morozov P., Schneider C., Maute R.L., Kitagawa Y., Mandelbaum J., Haddad J., Jr, Chen C.Z., Califano A., Dalla-Favera R. 2009. Identification of the human mature B cell miRNome. Immunity. 30:744–752 10.1016/j.immuni.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K., Saito M., Sumazin P., Margolin A.A., Wang K., Lim W.K., Kitagawa Y., Schneider C., Alvarez M.J., Califano A., Dalla-Favera R. 2010. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 115:975–984 10.1182/blood-2009-06-227017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Koppal A., Agius P., Sander C., Leslie C. 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11:R90 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G.W., Berens C., Kuklik-Roos C., Bechet J.M., Laux G., Bachl J., Korndoerfer M., Schlee M., Hölzel M., Malamoussi A., et al. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33:e137 10.1093/nar/gni137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S., Wu L., Nutt S.L. 2010. Surprising new roles for PU.1 in the adaptive immune response. Immunol. Rev. 238:63–75 10.1111/j.1600-065X.2010.00955.x [DOI] [PubMed] [Google Scholar]
- Cattoretti G., Pasqualucci L., Ballon G., Tam W., Nandula S.V., Shen Q., Mo T., Murty V.V., Dalla-Favera R. 2005. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 7:445–455 10.1016/j.ccr.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Cattoretti G., Büttner M., Shaknovich R., Kremmer E., Alobeid B., Niedobitek G. 2006. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 107:3967–3975 10.1182/blood-2005-10-4170 [DOI] [PubMed] [Google Scholar]
- Chang C.C., Ye B.H., Chaganti R.S., Dalla-Favera R. 1996. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA. 93:6947–6952 10.1073/pnas.93.14.6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Iida S., Louie D.C., Dalla-Favera R., Chaganti R.S. 1998. Heterologous promoters fused to BCL6 by chromosomal translocations affecting band 3q27 cause its deregulated expression during B-cell differentiation. Blood. 91:603–607 [PubMed] [Google Scholar]
- Ci W., Polo J.M., Cerchietti L., Shaknovich R., Wang L., Yang S.N., Ye K., Farinha P., Horsman D.E., Gascoyne R.D., et al. 2009. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 113:5536–5548 10.1182/blood-2008-12-193037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clurman B.E., Hayward W.S. 1989. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 9:2657–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S., Sandhu S.K., Pedersen I.M., Tili E., Trotta R., Perrotti D., Ciarlariello D., Neviani P., Harb J., Kauffman L.R., et al. 2009. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 114:1374–1382 10.1182/blood-2009-05-220814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan L.N., Jiang X., Bhatt S., Cubedo E., Rajewsky K., Lossos I.S. 2012. miR-155 regulates HGAL expression and increases lymphoma cell motility. Blood. 119:513–520 10.1182/blood-2011-08-370536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., McBride K.M., Jankovic M., Gazumyan A., Thai T.H., Robbiani D.F., Di Virgilio M., Reina San-Martin B., Heidkamp G., Schwickert T.A., et al. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 28:630–638 10.1016/j.immuni.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S., Cermak L., Pagan J.K., Rossi M., Martinengo C., di Celle P.F., Chapuy B., Shipp M., Chiarle R., Pagano M. 2012. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 481:90–93 10.1038/nature10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 102:3627–3632 10.1073/pnas.0500613102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha L.A., Su G.H., Rao S., Kabak S., Hao Z., Clark M.R., Simon M.C. 1999. PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity. 10:399–408 10.1016/S1074-7613(00)80040-0 [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. 2004. Human MicroRNA targets. PLoS Biol. 2:e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Tu Y., Stolovitzky G.A., Keller J.L., Haddad J., Jr, Miljkovic V., Cattoretti G., Califano A., Dalla-Favera R. 2003. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA. 100:2639–2644 10.1073/pnas.0437996100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Melchers M., Wang H., Torrey T.A., Slota R., Qi C.F., Kim J.Y., Lugar P., Kong H.J., Farrington L., et al. 2006. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 203:63–72 10.1084/jem.20051450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C., Rajbhandari P., Alvarez M.J., Bandaru P., Lim W.K., Sato M., Wang K., Sumazin P., Kustagi M., Bisikirska B.C., et al. 2010. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6:377 10.1038/msb.2010.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Niu H., Ye B.H., Dalla-Favera R. 1998. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 12:1953–1961 10.1101/gad.12.13.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Migliazza A., Basso K., Houldsworth J., Chaganti R.S., Dalla-Favera R. 2003. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 101:2914–2923 10.1182/blood-2002-11-3387 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C., III, Nussenzweig M.C., Dalla-Favera R. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Dominguez-Sola D., Chiarenza A., Fabbri G., Grunn A., Trifonov V., Kasper L.H., Lerach S., Tang H., Ma J., et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 471:189–195 10.1038/nature09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen I.M., Otero D., Kao E., Miletic A.V., Hother C., Ralfkiaer E., Rickert R.C., Gronbaek K., David M. 2009. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 1:288–295 10.1002/emmm.200900028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D., Kim S.W., McKeller M.R., Dahia P.L., Aguiar R.C. 2010. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl. Acad. Sci. USA. 107:3111–3116 10.1073/pnas.0910667107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Gao J., Basso K., Kitagawa Y., Smith P.M., Bhagat G., Pernis A., Pasqualucci L., Dalla-Favera R. 2007. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 12:280–292 10.1016/j.ccr.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. 2001. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 274:157–167 10.1016/S0378-1119(01)00612-6 [DOI] [PubMed] [Google Scholar]
- Tam W., Ben-Yehuda D., Hayward W.S. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell. Biol. 17:1490–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G., Hakimpour P., Landgraf P., Rice A., Tuschl T., Casellas R., Papavasiliou F.N. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 28:621–629 10.1016/j.immuni.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. 2007. Regulation of the germinal center response by microRNA-155. Science. 316:604–608 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- Thomas M.D., Kremer C.S., Ravichandran K.S., Rajewsky K., Bender T.P. 2005. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 23:275–286 10.1016/j.immuni.2005.08.005 [DOI] [PubMed] [Google Scholar]
- van den Berg A., Kroesen B.J., Kooistra K., de Jong D., Briggs J., Blokzijl T., Jacobs S., Kluiver J., Diepstra A., Maggio E., Poppema S. 2003. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 37:20–28 10.1002/gcc.10186 [DOI] [PubMed] [Google Scholar]
- van den Hurk J.A., Schwartz M., van Bokhoven H., van de Pol T.J., Bogerd L., Pinckers A.J., Bleeker-Wagemakers E.M., Pawlowitzki I.H., Rüther K., Ropers H.H., Cremers F.P. 1997. Molecular basis of choroideremia (CHM): mutations involving the Rab escort protein-1 (REP-1) gene. Hum. Mutat. 9:110–117 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Nussenzweig M.C. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., Nussenzweig M.C. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Dominguez-Sola D., Holmes A.B., Deroubaix S., Dalla-Favera R., Nussenzweig M.C. 2012. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 120:2240–2248 10.1182/blood-2012-03-415380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A., et al. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 27:847–859 10.1016/j.immuni.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 103:2257–2261 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B.H., Chaganti S., Chang C.C., Niu H., Corradini P., Chaganti R.S., Dalla-Favera R. 1995. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 14:6209–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.