A PI3K- and Akt-independent pathway mediated by mTORC1 regulates expression of HIF1 in CD8+ T cells and is required to sustain glucose metabolism and regulate cell trafficking.
Abstract
mTORC1 (mammalian target of rapamycin complex 1) controls transcriptional programs that determine CD8+ cytolytic T cell (CTL) fate. In some cell systems, mTORC1 couples phosphatidylinositol-3 kinase (PI3K) and Akt to the control of glucose uptake and glycolysis. However, PI3K–Akt-independent mechanisms control glucose metabolism in CD8+ T cells, and the role of mTORC1 has not been explored. The present study now demonstrates that mTORC1 activity in CD8+ T cells is not dependent on PI3K or Akt but is critical to sustain glucose uptake and glycolysis in CD8+ T cells. We also show that PI3K- and Akt-independent pathways mediated by mTORC1 regulate the expression of HIF1 (hypoxia-inducible factor 1) transcription factor complex. This mTORC1–HIF1 pathway is required to sustain glucose metabolism and glycolysis in effector CTLs and strikingly functions to couple mTORC1 to a diverse transcriptional program that controls expression of glucose transporters, multiple rate-limiting glycolytic enzymes, cytolytic effector molecules, and essential chemokine and adhesion receptors that regulate T cell trafficking. These data reveal a fundamental mechanism linking nutrient and oxygen sensing to transcriptional control of CD8+ T cell differentiation.
The differentiation of effector CTLs requires that naive T cells undergo clonal expansion and reprogram their transcriptome to express the key cytolytic effector molecules that mediate the CD8+ T cell immune response. Moreover, a striking feature of CD8+ T cells is that they massively increase glucose uptake as they respond to an immune challenge and differentiate to cytolytic effectors (Fox et al., 2005; Maciver et al., 2008). They also switch from metabolizing glucose primarily through oxidative phosphorylation to using the glycolytic pathway. Glycolysis requires that T cells switch on and sustain expression of rate-limiting glycolytic enzymes such as hexokinase 2, phosphofructokinase 1, pyruvate kinases, and lactate dehydrogenase and also requires that T cells can sustain high levels of glucose uptake by maintaining expression of the glucose transporter Glut1. In this context, it has been reported that relatively high levels of exogenous glucose are required to sustain the transcriptional program of CTLs (Cham and Gajewski, 2005; Cham et al., 2008).
During CD8+ T cell differentiation, the glycolytic switch is initiated by antigen receptors and co-stimulatory molecules but is then sustained by inflammatory cytokines such as IL-2. This cytokine controls the transcriptional program that determines CD8+ T cell differentiation and promotes effector CTL differentiation at the expense of memory cell formation (Kalia et al., 2010; Pipkin et al., 2010). In many cells, growth factors and cytokines control glucose metabolism via signaling pathways controlled by phosphatidylinositol-3 kinase (PI3K) signals and the serine/threonine kinase Akt (also called protein kinase B). However, although PI3K and Akt direct the transcriptional program of CTLs, they are not required for the TCR-mediated initiation of glucose uptake nor are they required for IL-2 to sustain glucose uptake and glycolysis (Macintyre et al., 2011). Rather, this role is controlled by a PI3K-independent mechanism involving PDK1 (phosphoinositide-dependent kinase 1; Macintyre et al., 2011).
In this context, in CD4 T cells, the serine kinase mTORC1 (mammalian target of rapamycin complex 1) can control glucose metabolism via regulation of HIF1 (hypoxia-inducible factor 1) complexes (Shi et al., 2011). In CD8+ T cells, it has been recently reported that the initial glycolytic switch induced in response to antigen receptor triggering is mediated by c-myc and is independent of HIF1 (Wang et al., 2011). It thus remains to be determined whether the mTORC1–HIF1 pathway plays any role in controlling CD8+ T cell metabolism. Nevertheless, mTORC1 does play an essential role in CD8+ T cells to integrate inputs from nutrients, antigen, and cytokine receptors to control T cell differentiation (Powell and Delgoffe, 2010). For example, inhibition of mTORC1 activity in effector CD8+ T cells can divert these cells to a memory fate (Araki et al., 2009). Moreover, mTORC1 signaling controls expression of cytolytic effector molecules in CTLs (Rao et al., 2010) and dictates the tissue-homing properties of these cells by regulating the expression of chemokine and adhesion receptors (Sinclair et al., 2008). However, the molecular mechanisms used by mTORC1 to control CD8+ T cell differentiation are not fully understood; neither are the signaling processes that activate mTORC1. Here it is pertinent that mTORC1 activity in CD8+ T cells is proposed to be controlled by PI3K and Akt (Rao et al., 2010). If this model were correct, then the PI3K–Akt independence of glucose metabolism in CD8+ T cells would argue against a role for mTORC1 in CD8+ T cell metabolism. The caveat is that models proposing PI3K control of mTORC1 activity in T cells are based on experiments with the PI3K inhibitors wortmannin and LY294002, drugs which have very well-documented off-target effects; of most concern is that they can directly inhibit mTOR catalytic function (Brunn et al., 1996). The possibility thus remains that mTORC1 is a key regulator of glucose metabolism in CD8+ T cells but is activated via PI3K–Akt-independent pathways.
Accordingly, the focus of the present study is the regulation and role of mTORC1 in CD8+ T cells. The data establish that PI3K- and Akt-independent mechanisms mediated by PDK1 control mTORC1 activity in CD8+ T cells. They also expose that a PI3K- and Akt-independent pathway mediated by PDK1 and mTORC1 controls expression of HIF1 transcriptional complexes in CD8+ T cells. This mTORC1–HIF1 pathway is required to sustain glucose metabolism and glycolysis in effector CTLs and strikingly functions to couple mTORC1 to a diverse transcriptional program that controls expression of cytolytic effector molecules and essential chemokine and adhesion receptors that regulate T cell trafficking.
RESULTS
mTORC1 regulates glucose uptake and glycolysis in TCR- and IL-2–stimulated CD8+ T cells
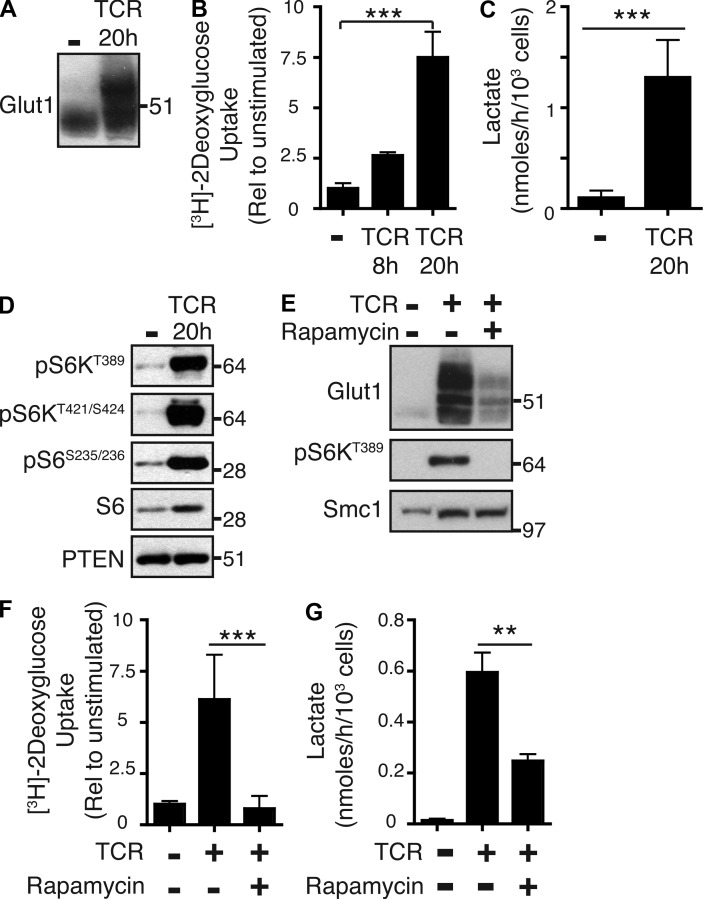
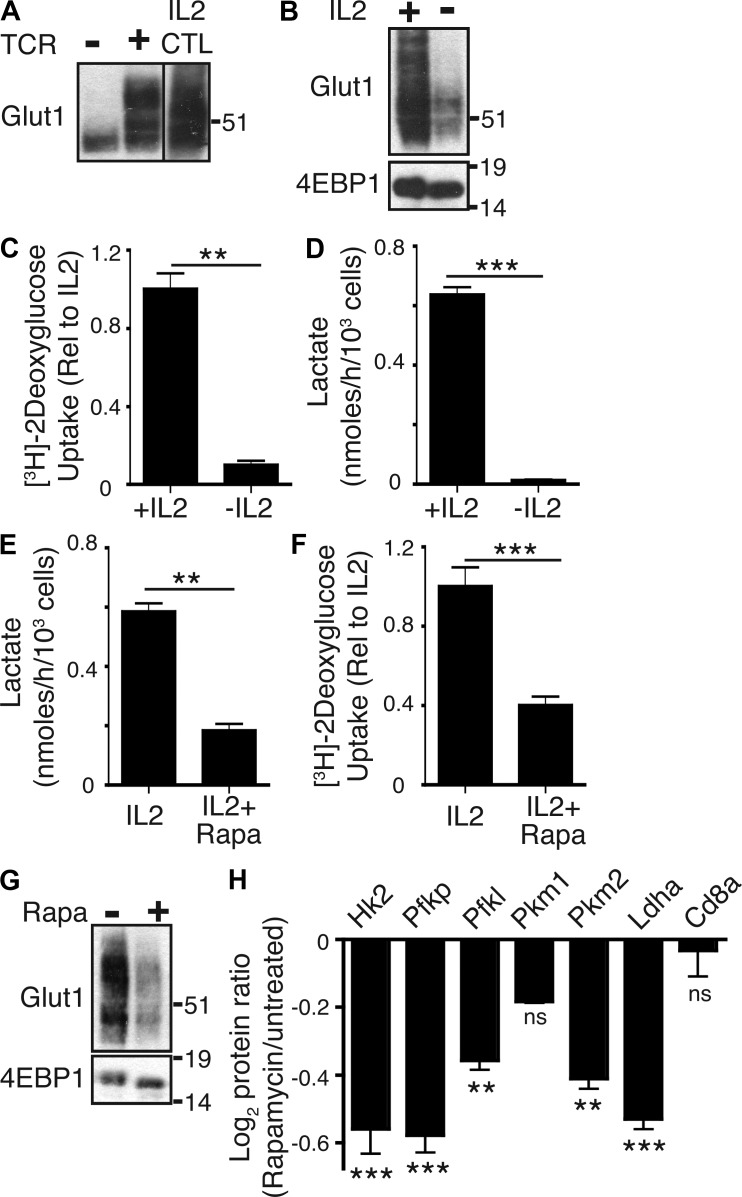
TCR triggering of naive CD8+ T cells with peptide–MHC complexes induced expression of the glucose transporter Glut1 and a concomitant increase in glucose uptake and lactate output (Fig. 1, A–C). TCR triggering also activated mTORC1 activity (Fig. 1 D), as judged by assessing the impact of TCR ligation on phosphorylation of mTORC1 substrate sequences on S6K1 (T389 and S421/424) and the phosphorylation of the S6K1 substrate S6 ribosomal protein (S235/236). The inhibition of mTORC1 activity with rapamycin blocked TCR-induced increases in Glut1 expression and glucose uptake and reduced lactate production (Fig. 1, E–G). TCR-primed CD8+ T cells cultured in IL-2 clonally expanded and differentiated to CTLs. One role for IL-2 is to sustain glucose uptake and glycolysis in TCR-primed T cells. CTLs cultured in IL-2 thus had high levels of Glut1 expression (Fig. 2 A), high levels of glucose uptake, and high levels of lactate output. The expression of Glut1 was lost when CTLs were deprived of IL-2 (Fig. 2 B). Moreover, the removal of IL-2 caused CTLs to decrease glucose uptake and lactate output, i.e., glycolysis (Fig. 2, C and D). Strikingly, when CTLs were treated with rapamycin, they also decreased lactate output, which would be consistent with a model whereby rapamycin treatment inhibits glycolysis (Fig. 2 E). Consistent with such a model, rapamycin-treated T cells showed severely decreased glucose uptake, loss of Glut1 expression, and decreased expression of several essential glycolytic enzymes such as hexokinase 2, phosphofructokinase 1, pyruvate kinases, and lactate dehydrogenase (Fig. 2, F–H). mTORC1 thus integrates both TCR and IL-2 signaling to induce and sustain Glut1 expression and glucose uptake. mTORC1 also controls expression of key glycolytic enzymes in activated CD8+ T cells.
Figure 1.
mTORC1 regulates glucose uptake and glycolysis in TCR-stimulated CD8+ T cells. (A) Immunoblot analysis of Glut1 expression in naive OTI CD8+ T cells ± TCR (SIINFEKL) stimulation for 20 h. (B and C) Naive P14-LCMV CD8+ T cells ± TCR (gp33-41/anti-CD28) stimulation were assayed for glucose uptake (B) and lactate production (C). (D) Immunoblot analysis of naive OTI CD8+ T cells ± TCR (SIINFEKL) stimulation for 20 h. mTORC1 activity was determined by analyzing the phosphorylation of target sequences on S6K1 (T389 and S241/242) and phosphorylation of the S6K1 substrate S6 ribosomal protein. PTEN was used as a loading control. (E–G) Immunoblot analysis (E) and analysis of glucose uptake (F) and lactate production (G) for naive P14-LCMV CD8+ T cells ± TCR (gp33-41/anti-CD28) stimulation with or without rapamycin for 20 h. For all panels, data are mean ± SEM or representative of at least three experiments. All metabolic assays were preformed in triplicate (**, P < 0.01; ***, P < 0.001). Molecular mass is indicated in kilodaltons.
Figure 2.
mTORC1 regulates glucose uptake and glycolysis in IL-2–maintained CTLs. (A) Immunoblot analysis for Glut1 expression in naive OTI CD8+ T cells ± TCR (SIINFEKL) stimulation for 20 h and also mature OTI CTLs. The black line indicates that intervening lanes have been spliced out. (B) Immunoblot analysis for Glut1 expression in CTLs treated with or without IL-2 for 20 h. 4EBP1 was used as a loading control. (C and D) Analysis of glucose uptake (C) and lactate production (D) in P14-LCMV CTLs treated with or without IL-2 for 20 h. (E and F) Analysis of lactate production (E) and glucose uptake (F) in P14-LCMV CTLs treated with or without rapamycin for 20 h. (G) Immunoblot analysis for Glut1 expression in P14-LCMV CTLs treated with or without rapamycin for 20 h. 4EBP1 was used as a loading control. (A–G) Data are mean ± SEM or representative of at least three experiments. All metabolic assays were preformed in triplicate (**, P < 0.01; ***, P < 0.001). Molecular mass is indicated in kilodaltons. (H) SILAC-based proteomic analysis of P14-LCMV CTLs treated with and without rapamycin for 48 h. Shown is the relative expression of rate-limiting glycolytic enzymes (rapamycin/untreated). Data are mean ± SEM for three experiments and were analyzed by ANOVA (**, P < 0.01; ***, P < 0.001). CD8α was used as a control protein with unchanged expression.
mTORC1 controls glucose uptake and glycolysis via HIF1
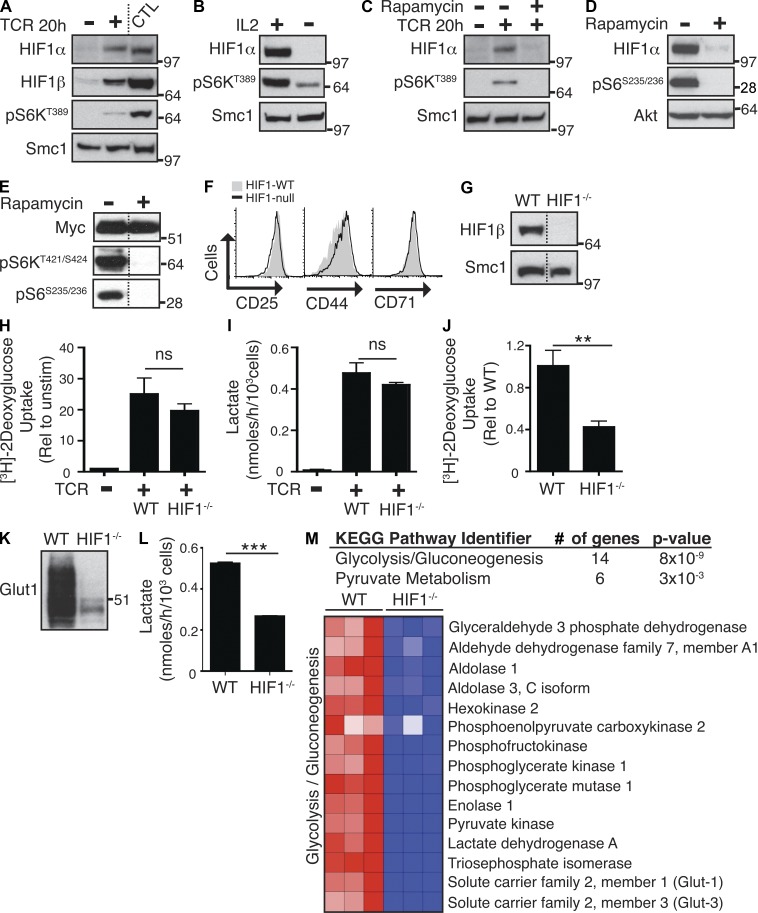
The expression of Glut1 can be controlled by the HIF1α and HIF1β (also known as ARNT or Aryl hydrocarbon receptor [AHR] nuclear translocator) complex (Semenza, 2010). Do CD8+ T cells express HIF1 complexes? Fig. 3 A addresses this issue and shows that TCR-triggered CD8+ T cells expressed both HIF1α and HIF1β. Furthermore, as CD8 T cells differentiated to CTLs, they increased and sustained high levels of HIF1α and HIF1β (Fig. 3 A) in a response that requires sustained IL-2 signaling and mTORC1 activity (Fig. 3 B). TCR-induced HIF1α expression in CD8+ T cells was thus dependent on mTORC1 activity (Fig. 3 C). Similarly, HIF1α protein expression in IL-2–maintained CTLs was dependent on continuous mTORC1 activation (Fig. 3 D). The mTORC1 dependence of HIF1α expression correlates with the requirement for sustained mTORC1 and cytokine signaling to control Glut1 expression and a glycolytic metabolism in CD8+ T cells (Figs. 1 and 2). In this respect, a previous study has implicated c-myc as a major regulator of glucose metabolism in T cells (Wang et al., 2011). However, the expression of c-myc in IL-2–sustained CTLs was not dependent on mTORC1 (Fig. 3 E). The inhibition of mTORC1 with rapamycin thus inhibits Glut1 expression, glucose uptake, and glycolysis in CTLs independently of any effect on c-myc expression.
Figure 3.
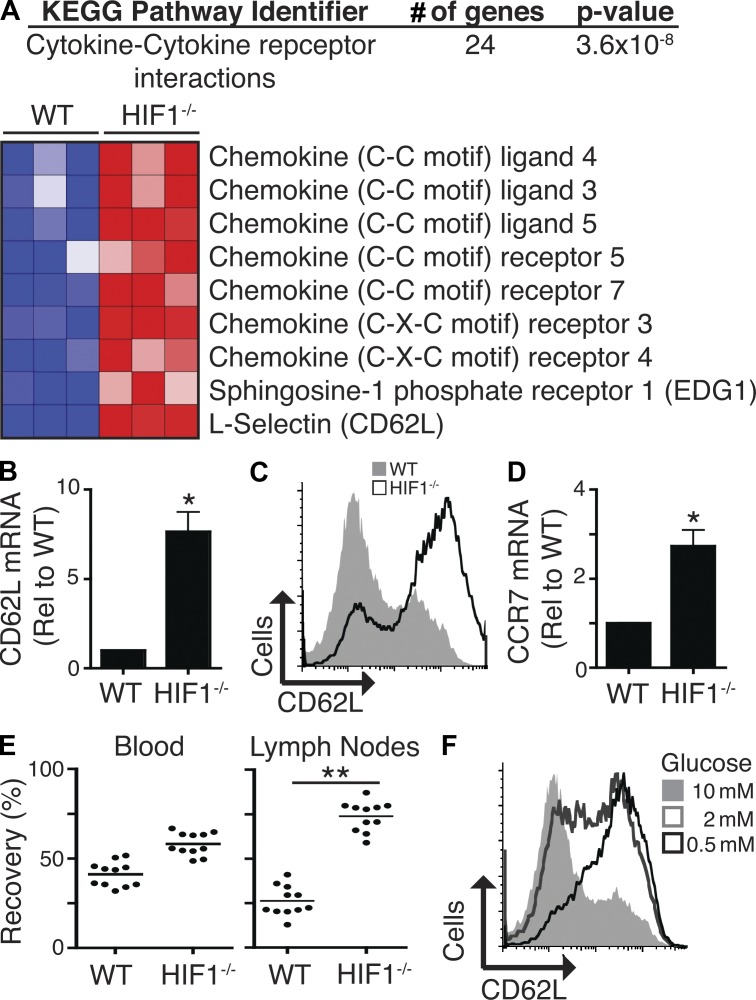
mTORC1 controls glucose uptake and glycolysis via HIF1. (A–D) Immunoblot analysis of HIF1α and HIF1β expression in naive P14-LCMV CD8+ T cells ± TCR (gp33-41/anti-CD28) stimulation for 20 h (A and C) and P14-LCMV CTLs (A, B, and D) treated with and without IL-2 (B) or rapamycin (C and D) for 20 h. Phospho-S6K1 and phospho-S6 were used as a measure of mTORC1 activity. (E) Immunoblot analysis of c-myc expression in CTLs treated with or without rapamycin for 20 h. Phospho-S6K1 and phospho-S6 were used as a measure of mTORC1 activity. (F) Flow cytometric analysis of HIF1βWT/WT CD4Cre (WT) and HIF1βflox/flox CD4Cre (HIF1−/−) CD8+ T cells after TCR (2c11) stimulation for 20 h. (G) Immunoblot analysis of WT and HIF1−/− CTLs. (H and I) Analysis of glucose uptake (H) and lactate production (I) in WT and HIF1−/− naive CD8+ T cells after TCR (2c11/anti-CD28) stimulation for 20 h. Glucose uptake in unstimulated WT naive T cells is also shown (uptake in unstimulated HIF1−/− naive T cells is equivalent to WT; not depicted). (J–L) Analysis of glucose uptake (J), Glut1 expression (K), and lactate production (L) in WT versus HIF1−/− CTLs. (M) A comparison of the transcriptional profile of HIF1 WT versus HIF1−/− CTLs was performed by microarray. Shown here are KEGG pathway analysis of genes down-regulated in HIF1−/− CTLs (top) and a heat map of the relative normalized expression of selected genes that are significantly different in expression in WT versus HIF1−/− CTLs, as determined by microarray. For all panels, data are mean ± SEM or representative of at least three experiments. All metabolic assays were preformed in triplicate (**, P < 0.01; ***, P < 0.001). Molecular mass is indicated in kilodaltons. Dotted lines indicate that intervening lanes have been spliced out.
The HIF1 complex does not initiate but sustains glycolytic metabolism in CD8+ T cells
To explore a causal link between mTORC1, HIF1, and glycolysis, we deleted functional HIF1 transcriptional complexes in CD8+ T cells. In these experiments, mice with floxed HIF1β alleles were backcrossed to transgenic mice expressing Cre recombinase under the control of the CD4 promoter (CD4Cre). These mice produced a normal complement of peripheral α/β T cells in the thymus, lymph nodes, and spleen (not depicted). HIF1β-null CD8+ T cells activated in response to TCR and IL-2 triggering, as judged by their ability to normally up-regulate expression of CD25, CD71, and CD44 and undergo blastogenesis (Fig. 3 F and not depicted). The deletion of HIF1β was confirmed by immunoblot analysis (Fig. 3 G). A previous study has suggested that the TCR-induced glycolytic switch is regulated by c-myc and is not HIF1 dependent (Wang et al., 2011). The present data confirm this and show that TCR-induced glucose uptake and lactate output were normal in HIF1β-null cells (Fig. 3, H and I). However, strikingly, activated HIF1β-null CD8+ T cells could not sustain high levels of glucose uptake as they differentiated to become cytolytic effectors in response to IL-2. HIF1β-null immune-activated CD8+ T cells maintained in the presence of IL-2 thus had greatly decreased Glut1 expression and glucose uptake and produced significantly less lactate compared with control IL-2–maintained CTLs (Fig. 3, J–L). The ability of IL-2 to sustain glucose uptake and glycolysis is thus strictly dependent on HIF1. In this context, HIF1β can complex with AHR. However, CTLs do not express high levels of the AHR, and there is little evidence of functional AHR signaling. Moreover, AHR-null CTLs have normal glucose uptake and lactate output (not depicted).
mTORC1 can regulate glycolysis by controlling expression of Glut1 and the expression of key glycolytic enzymes (Fig. 2, G and H). Is the role of HIF1 in CD8+ T cells restricted to control of Glut1 expression? To explore this issue, we used Affymetrix microarray analysis to transcriptionally profile HIF1β-null CTLs. Approximately 11,517 annotated genes were expressed in CTLs, and the impact of HIF1β loss was a decrease in the expression of <5% of these genes and an increase in the expression of another 6%. The full list of genes changing in expression is detailed in Tables S1 and S2. We then used the functional annotation tools within DAVID Bioinformatics Resources 6.7 (Huang et al., 2009a,b) to perform gene annotation enrichment analysis and KEGG pathway mapping of the significantly changed genes in the HIF1β-null cells. This analysis indicated that one significant impact caused by the loss of HIF1β was down-regulation of multiple genes encoding proteins that control glycolysis and pyruvate metabolism (Fig. 3 M). HIF1β-null cells thus cannot sustain expression of key rate-limiting glycolytic enzymes: hexokinase 2, pyruvate kinase 2, phosphofructokinase, and lactate dehydrogenase. The ability of HIF1 complexes to sustain the T cell glycolytic switch thus extends beyond a simple model of HIF1 regulation of Glut1 and glucose uptake.
The HIF1 complex regulates the CD8+ T cell transcriptional program but is not essential for T cell proliferation
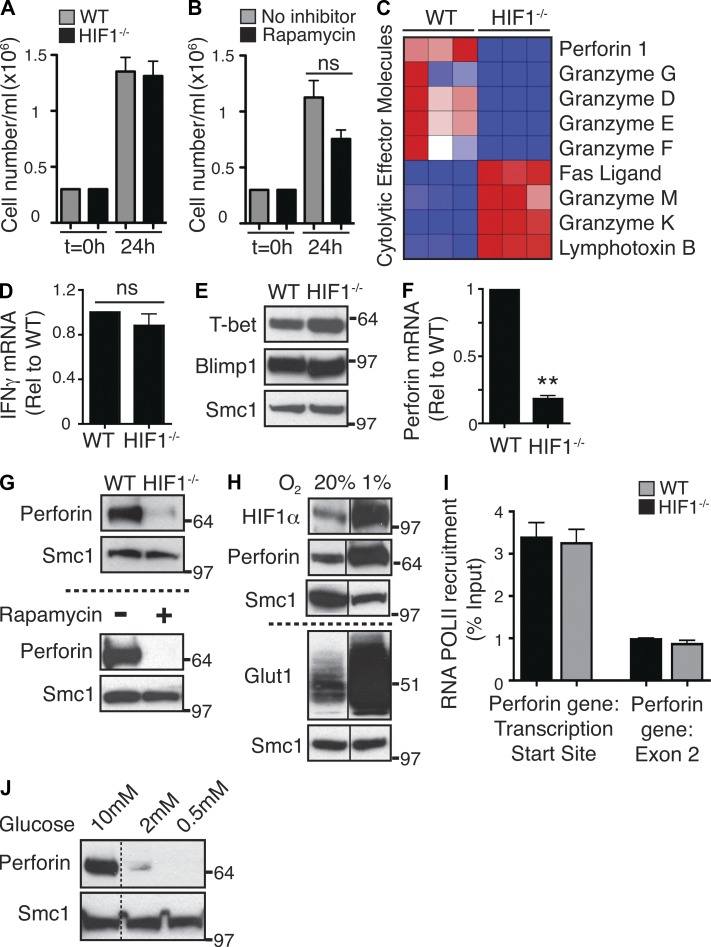
The massive up-regulation of glucose metabolism and the switch to glycolysis under normoxia that accompanies the immune activation of CD8 T cells are thought to be essential to meet the metabolic demands caused by the rapid proliferation of clonally expanding CD8+ T cells. However, CD8+ T cells deleted of HIF1 transcriptional complexes proliferated and clonally expanded normally (Fig. 4 A). Moreover, transcriptional profiling of HIF1β-null CTLs using Affymetrix microarray analysis did not reveal any negative impact of HIF1 deletion on cell cycle progression, cell survival, or mitosis (Tables S1 and S2). The failure to see proliferative defects in HIF1-null T cells is consistent with observations that inhibition of mTORC1 with rapamycin, which down-regulates HIF1 complexes and severely impairs glucose metabolism and glycolysis in CD8+ T cells, does not block the proliferation of CTLs (Fig. 4 B).
Figure 4.
The HIF1 complex regulates the CD8+ T cell transcriptional program but is not essential for T cell proliferation. (A and B) Proliferation analysis of WT and HIF1−/− CTLs (A) and P14-LCMV CTLs treated with and without rapamycin (B). Cells were seeded at 0.3 × 106/ml, and CTL numbers were counted after 24 h. Data are mean ± SEM of five experiments. (C) Heat map showing the relative normalized expression of selected genes that are significantly different in expression in WT versus HIF1−/− CTLs, as determined by microarray. (D) Real-time PCR analysis of IFN-γ expression in WT and HIF1−/− CTLs. Data are mean ± SEM of three experiments in triplicate. (E) Immunoblot analysis of T-bet and Blimp1 expression in WT and HIF1−/− CTLs. Data are representative of two experiments. (F) Real-time PCR analysis of Perforin mRNA expression in WT and HIF1−/− CTLs. Data are mean ± SEM of three experiments in triplicate (**, P < 0.01). (G) Immunoblot analysis of Perforin protein expression in WT and HIF1−/− CTLs (top) and P14-LCMV CTLs treated ± rapamycin for 20 h (bottom). Data are representative of at least three experiments. (H) IL-2–maintained CTLs were placed in either hypoxic (1%) or normoxic (20%) oxygen for 24 h before being subjected to immunoblot analysis for HIF1α, Glut1, and perforin expression. Data are representative of three experiments. (I) ChIP was performed with anti–Pol II, and the changes in Pol II binding to the Perforin transcription start site and the second exon were quantified by real-time PCR. Data were normalized to input DNA amounts and plotted as fold over the values for Pol II binding to the HPRT proximal promoter. Data are mean ± SEM of three experiments performed in duplicate. (J) P14-LCMV T cells were activated for 2 d with gp33-41 and then cultured for a further 4 d with IL-2 in different glucose concentrations. Cells were then subjected to immunoblot analysis for perforin expression. Data are representative of two experiments. Molecular mass is indicated in kilodaltons. Black lines (solid or dotted) indicate that intervening lanes have been spliced out.
A key role for mTORC1 in CD8+ T cells is to promote effector differentiation of CTLs by driving expression of genes encoding cytolytic effector molecules such as perforin, granzymes, and IFN-γ. We therefore interrogated the data to determine whether HIF1 transcriptional complexes mediate mTORC1 control of any of the key molecules that control CTL differentiation. It was thus striking that immune-activated HIF1-null CD8+ T cells had lost expression of perforin and multiple granzymes (Fig. 4 C). This was not a global block in CD8+ T cell differentiation as HIF1 complexes were not required for expression of other CD8 effector molecules such as IFN-γ, Fas ligand, or lymphotoxin (Fig. 4, C and D). Moreover, activated HIF1β-null CD8+ T cells retained expression of the transcription factors T-bet and Blimp-1, which drive T cell differentiation (Fig. 4 E and Tables S1 and S2). The failure of immune-activated HIF1β-null CD8+ T cells to express perforin was confirmed independently by quantitative PCR analysis of perforin mRNA levels and Western blot analysis of perforin protein expression (Fig. 4, F and G). Consistent with a role for mTORC1–HIF1 in controlling perforin expression, protein levels for perforin were also decreased after rapamycin treatment in WT CTLs (Fig. 4 G). We also assessed whether the expression of HIF1 complexes was rate limiting for perforin gene expression by examining the impact of hypoxia on the expression of perforin in WT CTLs. In these experiments, CTLs were switched from normoxic (21%) to hypoxic (1%) oxygen for 24 h. The switch to hypoxia strikingly increased expression of HIF1α and also expression of Glut1, a direct HIF1 transcriptional target (Fig. 4 H). Importantly hypoxia also induced expression of perforin (Fig. 4 H). These data may explain earlier observations that CTLs cultured under hypoxic conditions display increased cytotoxic function (Caldwell et al., 2001).
What is the mechanism for HIF1 control of perforin gene expression? HIF1 regulates the expression of Glut1 by direct binding to the Glut1 promoter. Similarly, HIF1-regulated glycolytic enzymes are direct HIF1 target genes. This raises the question of whether or not HIF1 regulation of perforin gene expression is direct or indirect. There was no evidence for HIF1-binding sites in the perforin promoter. Moreover, there was no change in the recruitment of RNA polymerase II (Pol II) to the transcription start site and distal exons of the perforin gene in HIF1-null CD8+ T cells (Fig. 4 I). Hence the loss of perforin mRNA in HIF1-null CD8+ T cells (Fig. 4 F) was not a consequence of the failure to recruit the appropriate transcription factors to the perforin locus. These data argue that the HIF1 effect on perforin expression is indirect. In this regard, one possibility we considered was that the loss of perforin expression might be an indirect consequence of failed glucose uptake. Here it is relevant that a previous study has shown that glucose deprivation prevents perforin expression in immune-activated CD8+ T cells (Cham et al., 2008). The present data (Fig. 4 J) show that CTLs maintained in 2 mM glucose express substantially lower levels of perforin compared with the control cells maintained in 10 mM glucose. This could explain why an early study noted that glucose deprivation can limit the cytolytic function of effector CTLs (MacDonald and Koch, 1977).
HIF1 regulation of chemokines and chemokine receptors
One unexpected outcome from the microarray experiment came from the bioinformatic pathway analysis of the genes whose expression was increased in HIF1β-null T cells. This analysis indicated that immune-activated HIF1-null CD8+ T cells up-regulated the expression of genes involved in cytokine/cytokine receptor interactions (Fig. 5 A). Closer data interrogation revealed that these HIF1-regulated genes encoded chemokines, chemokine receptors, and adhesion molecules. For example, loss of HIF1 transcriptional complexes increased expression of mRNA encoding the chemokine receptors S1P1, CXCR4, CXCR3, CCR5, and CCR7 in immune-activated CTLs (Fig. 5 A). Moreover, the expression of the gene encoding the cell adhesion molecule CD62L (L-selectin) was increased in HIF1β-null cells (Fig. 5, A and B). Immune-activated CD8+ T cells down-regulate CD62L gene transcription, and thus CTLs normally express low levels of this adhesion molecule (Fig. 5, B and C; Sinclair et al., 2008). However, CD62L mRNA and protein levels were high in activated HIF1β-null CD8+ T cells (Fig. 5, B and C). CCR7 is a chemokine receptor that coordinates the migration of T cells into lymph nodes and is normally down-regulated in CTLs. PCR analysis confirmed the microarray data: immune-activated HIF1β-null T cells retained high levels of CCR7 mRNA compared with normal CTLs (Fig. 5 D).
Figure 5.
HIF1 regulation of chemokines and chemokine receptors. (A) A comparison of the transcriptional profile of WT versus HIF1−/− CTLs was performed by microarray. Shown here are KEGG pathway analysis of genes up-regulated in HIF1−/− CTLs (top) and a heat map showing the relative normalized expression of selected genes that are significantly different in expression in WT versus HIF1−/− CTLs, as determined by microarray. (B) Real-time PCR analysis of CD62L expression in WT and HIF1−/− CTLs. (C) Analysis of CD62L surface expression on WT and HIF1−/− CTLs by flow cytometry. (D) Real-time PCR analysis of CCR7 expression in WT and HIF1−/− CTLs. (E) WT and HIF1−/− CTLs were labeled with CFSE or CellTracker orange (CMTMR) and mixed at a ratio of 1:1 before being injected into C57BL/6 host mice. Values indicate recovery of WT or HIF1−/− cells as a percentage of the total recovered transferred cells from the blood and lymph nodes 4 h after transfer. Each dot indicates a mouse; horizontal bars indicate mean. (F) P14 T cells were activated for 2 d with cognate peptide and then cultured for a further 4 d with IL-2 in different glucose concentrations. Cells were then analyzed for the surface expression of CD62L by flow cytometry. In B and D, mean ± SEM of three experiments performed in triplicate is shown; in C and F, data are representative of at least three experiments (*, P < 0.05; **, P < 0.01).
CD62L and CCR7 are expressed at high levels in naive and memory T cells and are essential for lymphocyte transmigration from the blood into secondary lymphoid tissue. CD62L and CCR7 loss is thus part of the program that redirects effector T cell trafficking away from lymphoid tissue toward sites of inflammation. In this respect, inhibition of mTORC1 with rapamycin can restore expression of CD62L and CCR7 in effector T cells and reprogram their trafficking such that they regain the ability to home to secondary lymphoid organs (Sinclair et al., 2008). The retention of CD62L and CCR7 on immune-activated HIF1β-null CD8+ T cells thus raises the possibility that these cells may retain the migratory properties of naive T cells and preferentially home to secondary lymphoid tissues. However, HIF1β-null CD8+ T cells not only retained expression of the secondary lymphoid organ-homing receptors but also increased expression of inflammatory chemokine receptors such as CXCR3 and CCR5 (Fig. 5 A). These pleiotropic effects make it more difficult to predict the impact of HIF1 loss on T cell trafficking in vivo. Accordingly, the in vivo lymph node–homing ability of activated control or HIF1β-null CD8+ T cells was compared. Strikingly, activated CD8+ T cells that lacked HIF1-mediated transcription retained the capacity to home to secondary lymphoid organs and accumulated in the lymph nodes (Fig. 5 E). There is therefore a dominant requirement for HIF1 transcriptional complexes for the normal programming of effector CD8+ T cell trafficking. Is HIF1 regulation of CD62L gene expression direct, or is the impact of HIF1 on CD62L expression an indirect consequence of the inability of HIF1-null cells to maintain glucose uptake? The experiment in Fig. 5 F addresses this issue and shows that CD8+ T cells stimulated by antigen and IL-2 in culture media with low glucose levels fail to down-regulate CD62L expression.
PI3K- and Akt-independent control of mTORC1 activity and HIF1 expression in CD8+ T cells
The insight that HIF1 transcription factor complexes mediate mTORC1 control of the expression of CD62L, CCR7, and S1P1 raises a question. How does the mTORC1–HIF1 pathway that controls T cell trafficking connect to a well-documented PI3K–Akt–Foxo pathway that also controls the expression of these key trafficking molecules? The retention of high levels of CCR7 and CD62L expression by immune-activated HIF1β-null CD8+ T cells thus phenocopies the impact of inhibiting PI3K–Akt signaling in activated CD8+ T cells (Sinclair et al., 2008; Waugh et al., 2009; Macintyre et al., 2011). PI3K–Akt control of CD62L, CCR7, and S1P1 expression reflects that the expression of these molecules is regulated by the Foxo1 transcription factor (Fabre et al., 2008; Kerdiles et al., 2009). Foxo1 is inactivated by Akt, which phosphorylates Foxo1, resulting in its nuclear exclusion and retention in the cytosol. Akt inhibition results in dephosphorylation of Foxo1 and restores its nuclear location and transcriptional activity and restores expression of CD62L and CCR7 in CTLs (Waugh et al., 2009; Macintyre et al., 2011).
Accordingly, a key question is whether mTORC1 and/or expression of HIF1 complexes regulate Akt activity and Foxo phosphorylation in T cells. It is also pertinent to question whether PI3K and Akt control mTORC1 activity in CD8+ T cells. Lymphocyte signaling models frequently position PI3K–Akt signaling as an upstream obligatory regulator of mTORC1 activity. However, the present results showing that mTORC1 controls glucose metabolism in T cells are inconsistent with these models because they are discrepant with observations that PI3K and Akt do not control glucose uptake in CD8+ T cells (Macintyre et al., 2011). Moreover, the present data herein show that the mTORC1–HIF1 pathway controls expression of the rate-limiting glycolytic enzymes (Figs. 2 H and 3 M). In contrast, microarray analysis of Akt-regulated genes in T cells failed to find any evidence that Akt controlled glucose metabolism or glycolysis (Macintyre et al., 2011).
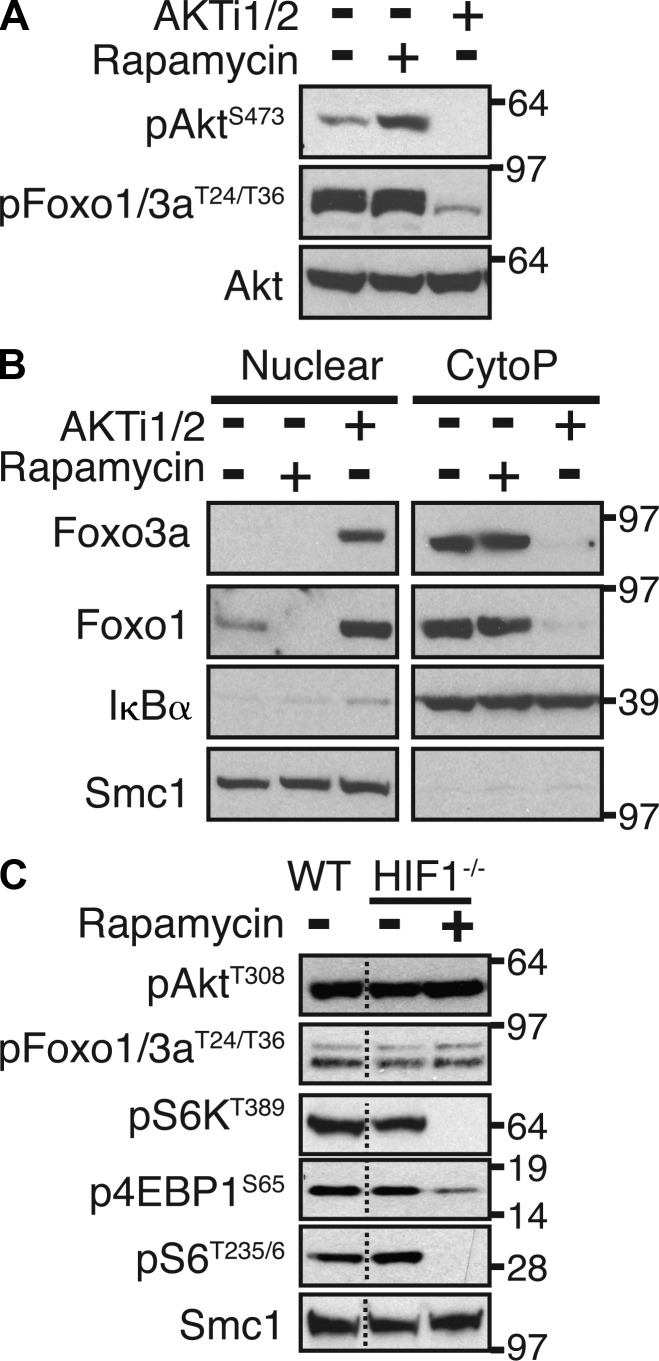
To explore the links between PI3K, Akt, mTORC1, and HIF1, we addressed two questions. Do mTORC1 and HIF1 regulate Akt activity and Foxo phosphorylation? Is PI3K and Akt activity required for mTORC1 activation and HIF1 expression? In the context of the first question, long-term inhibition of mTORC1 can destabilize mTORC2 complexes in some cell systems and suppress Akt function (Sarbassov et al., 2006). Indeed, rapamycin treatment of CD4 T cells activated with CD3 and CD28 antibodies does diminish Akt S473 phosphorylation, although the impact of this on Akt activity has not been fully assessed (Lee et al., 2010; Delgoffe et al., 2011). If this were true in CTLs, then long-term treatment of T cells with rapamycin would result in the loss of Foxo phosphorylation and cause cells to regain Foxo transcriptional activity and thus CD62L and CCR7 expression. We therefore examined the impact of long-term inhibition of mTORC1 with rapamycin on Foxo phosphorylation and localization in CTLs. We also assessed Akt–Foxo and mTORC1 signaling in IL-2–maintained HIF1-null T cells to assess whether loss of HIF1 complexes compromised Foxo phosphorylation/inactivation. Fig. 6 (A and B) shows that inhibition of Akt decreased Foxo phosphorylation (Fig. 6 A) and restored nuclear localization of Foxos in CTLs (Fig. 6 B). In contrast, Akt remained active, i.e., phosphorylated on T308 and S473, and Foxos remained highly phosphorylated and excluded from the nucleus of rapamycin-treated CTLs (Fig. 6, A and B). Additionally, Akt phosphorylation (T308) and the phosphorylation of Foxo transcription factors on Akt substrate sites (T24/32) were unaffected in HIF1β-null CTLs, demonstrating that Akt signaling is not altered by HIF1 deletion (Fig. 6 C). It was also evident that the phosphorylation of mTORC1 substrates S6K1 (p70 S6-kinase 1) and 4EBP1 (eIF4E-binding protein 1), on T389 and S65, respectively, and downstream signaling to S6 ribosomal protein (an S6K1 substrate) were normal in HIF1β-null CD8+ T cells (Fig. 6 C). Therefore, mTORC1 and HIF1 regulate the expression of CD62L and CCR7 and T cell trafficking but do not control Akt–Foxo phosphorylation and localization.
Figure 6.
mTORC1 and HIF1 do not regulate Akt activity or Foxo phosphorylation. (A) Immunoblot analysis of phosphorylated AKT and Foxos in P14-LCMV CTLs treated with and without Akti1/2 or rapamycin for 24 h. (B) P14-LCMV CTLs were treated with and without rapamycin or Akti1/2 for 24 h and subjected to nuclear/cytoplasmic fractionation before immunoblot analysis for Foxo1 and Foxo3a expression. Purity of cytoplasmic and nuclear fractions was confirmed by IκBα and Smc1 expression. (C) Immunoblot analysis of WT or HIF1−/− CTLs for Akt–Foxo and mTORC1 signaling. HIF1−/− CTLs were treated with rapamycin as a negative control for mTORC1 activity. For all panels, data are representative of at least three experiments. Molecular mass is indicated in kilodaltons. Dotted lines indicate that intervening lanes have been spliced out.
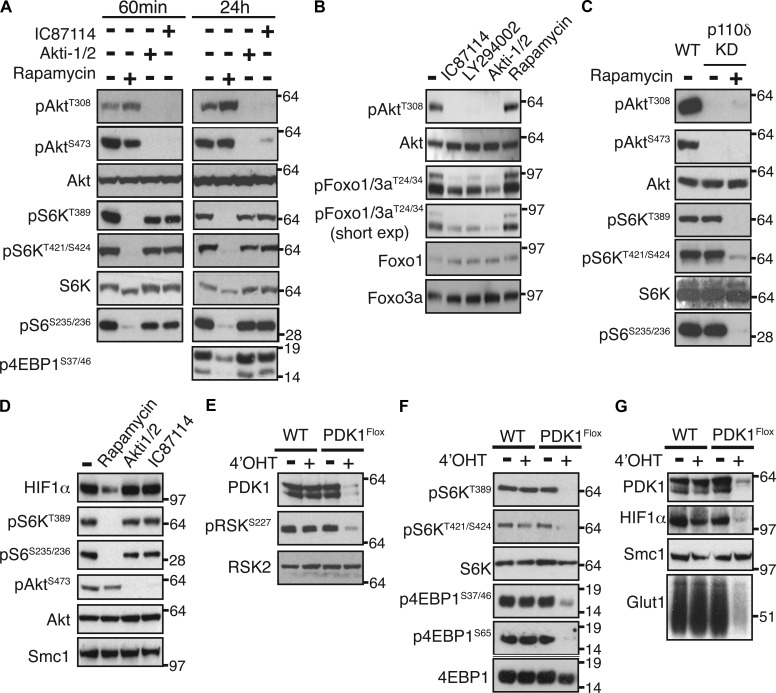
What about the second question? Are PI3K and Akt activity required for mTORC1 activation and HIF1 expression in CD8+ T cells? To address this issue, we used complementary genetic and pharmacological strategies to block PI3K and Akt and then monitored the impact of these perturbations on mTORC1 activity by assessing the phosphorylation of mTORC1 substrate sequences on S6K1 (T389 and S421/424) and 4EBP1 (S35/47) and the phosphorylation of the S6K1 substrate S6 ribosomal protein (S235/236). The data show that IL-2–maintained CTLs contain high levels of active Akt phosphorylated on threonine 308 and high levels of mTORC1 signaling (Fig. 7 A). mTORC1 inhibition with rapamycin abolished the phosphorylation of S6K1 on T389 and S421/424 and blocked S6 phosphorylation. In CD8+ T cells, Akt is activated via a PI3K complex containing the p110δ catalytic subunit (Macintyre et al., 2011). The Akt inhibitor, Akti1/2, or a p110δ inhibitor, IC87114, potently inhibited Akt activity in CTLs as judged by the loss of phosphorylated Akt on T308 and S473 and inhibition of the phosphorylation of the Akt substrates, Foxo transcription factors (Fig. 7, A and B). However, neither Akti1/2 nor IC87114 prevented mTORC1 activity in CD8+ T cells as neither compound blocked the phosphorylation of mTORC1 substrates S6K1 and 4EBP1 or S6 phosphorylation in CTLs (Fig. 7 A). Moreover, CTLs that had WT PI3K p110δ catalytic subunits substituted with a catalytically inactive mutant (p110δD910A) did not activate Akt in response to IL-2 but showed normal rapamycin-sensitive phosphorylation of S6K1 and S6 (Fig. 7 C). Further evidence that mTORC1 activity is independent of PI3K and Akt is that the expression of HIF1α was not regulated by Akt and PI3K inhibitors (Fig. 7 D). These data reveal that PI3K–Akt activity is dispensable for mTORC1 activity and HIF1 expression in CTLs.
Figure 7.
PI3K and Akt do not regulate mTORC1 activity. (A and B) CTLs were cultured in the presence or absence of Akti1/2, IC87114, rapamycin, or LY294002 for 60 min (A and B) or 24 h (A) and subjected to immunoblot analysis with the indicated antibodies. (C) CTLs generated from WT or p110δD910A mice were subjected to immunoblot analysis with or without rapamycin treatment (30 min). Data are representative of two experiments. (D) CTLs were cultured in the presence or absence of Akti1/2, IC87114, or rapamycin for 24 h and subjected to immunoblot analysis with the indicated antibodies. (E–G) CTLs generated from PDK1flox/flox TamoxCre (PDK1Flox) and PDK1WT/WT TamoxCre (WT) mice were treated ± 4′OHT for 3 d to delete PDK1 and subjected to immunoblot analysis. For, A, B, and D–G, data are representative of at least three experiments. Molecular mass is indicated in kilodaltons.
If mTORC1 activity is not mediated by Akt, then what is the alternative pathway? One candidate is the serine/threonine kinase PDK1 because this is known to be an essential regulator of glucose metabolism in CD8+ T cells (Macintyre et al., 2011). We therefore examined whether PDK1 regulates mTORC1 activity and HIF1 expression in activated CD8+ T cells. For these experiments, mice expressing floxed PDK1 alleles were backcrossed with mice that express a tamoxifen-regulated Cre recombinase (Macintyre et al., 2011). PDK1flox/flox TamoxCre CTLs were generated and treated with 4-hydroxytamoxifen (4′OHT) to delete floxed PDK1 alleles. PDK1 deletion was confirmed by analysis of PDK1 protein expression and by the loss of RSK phosphorylation on its PDK1 target site (S227; Fig. 7 E). Importantly, deletion of PDK1 in CTLs prevented the phosphorylation of the mTORC1 substrates S6K1 and 4EBP1 on mTORC1 target residues (Fig. 7 F). PDK1 loss also resulted in loss of expression of HIF1α and the HIF1 target Glut1 (Fig. 7 G). Together these results show that in CD8+ T cells, mTORC1 activity and HIF1α protein expression are controlled by a PI3K–Akt-independent pathway mediated by PDK1.
DISCUSSION
The present study explores the molecular pathways that mediate mTORC1 control of effector CD8+ T cell differentiation. A key finding was that mTORC1 activity is required for immune-activated CD8+ T cells to sustain high rates of glucose uptake. mTORC1 activity is also necessary for CD8+ T cells to initiate and sustain a switch to a glycolytic metabolism. One way in which mTORC1 controls glucose uptake in CD8+ T cells is by controlling expression of the glucose transporter Glut1. CD8+ T cells also show mTORC1 dependence for the expression of hexokinase 2, a key enzyme which phosphorylates glucose to produce glucose-6-phosphate, an essential intermediate in most pathways for glucose metabolism. Importantly, mTORC1 controls expression of key rate-limiting glycolytic enzymes in CD8+ T cells such as phosphofructokinase 1, lactate dehydrogenase, and pyruvate kinase M2.
This ability of mTORC1 to coordinate and sustain expression of glucose transporters and essential glycolytic enzymes during CD8+ T cell differentiation stems from its ability to control expression of the HIF1 transcription factor complex. The initial increase in glucose uptake and switch to glycolysis that immediately follows TCR engagement is not HIF1 dependent. However, HIF1 is essential for antigen-primed T cells to sustain high levels of glucose uptake and glycolytic enzyme expression as they differentiate to cytolytic effector cells. In this respect, CTL differentiation is controlled by the strength and duration of signaling by inflammatory cytokines such as IL-2 (Kalia et al., 2010; Pipkin et al., 2010). The present data now show that the ability of IL-2 to sustain high levels of glucose uptake and maintain a glycolytic metabolism is dependent on mTORC1 induction of HIF1 complexes. However, it was notable that the inhibition of mTORC1 in IL-2–maintained CTLs suppressed glucose uptake and lactate output without having any impact on the expression of c-myc (Fig. 3 E). This is relevant because it has been shown recently that c-myc is required for the initial TCR-induced glycolytic switch in naive T cells (Wang et al., 2011). The present data thus reveal that c-myc expression alone is not sufficient to maintain glucose uptake and glycolysis in CD8+ T cells. Moreover the coordination of c-myc and HIF1 expression must be required for T cells to sustain glucose uptake and glycolysis during CD8+ T cell differentiation.
The ability of HIF1 to link mTORC1 to the control of glucose metabolism in CD8+ T cells made us question whether any other actions of mTORC1 in CD8+ T cell are directed by the HIF1 pathway. In particular, it is well documented that mTORC1 activity is required for expression of CTL effector molecules and for effector CD8+ T cells to switch off expression of the chemokine receptors and adhesion molecules that direct naive and memory T cell entry and egress from secondary lymphoid tissues. For example, mTORC1 signaling causes down-regulation of the adhesion molecule CD62L and the chemokine receptors CCR7 and S1P1 (Sinclair et al., 2008). In this regard, the phenotype of the immune-activated HIF1-null T cells is intriguing. They have many features of effector CD8+ T cells such as normal IFN-γ production and expression of effector molecules such as Fas ligand and lymphotoxin. Also, they are normal in cell size and can rapidly proliferate. However, the HIF1-null cells lack perforin and granzyme expression. Moreover, although they acquire the expression of effector CTL chemokine receptors such as CXCR3 and CCR5, they retain the expression of naive T cell lymph node–homing receptors CD62L, CCR7, and S1P1. They also retain the secondary lymphoid tissue migratory program of naive T cells in vivo. The transcriptional profiling of HIF1β-null immune-activated CD8+ T cells thus affords the insight that mTORC1 regulation of HIF1 controls expression of a subset of CTL effector molecules and controls CD8+ T cell trafficking. These findings reveal a fundamental mechanism linking nutrient sensing and transcriptional control of CD8+ T cell differentiation. Although the HIF1 complex evolved to function as a metabolic sensor of cellular oxygen levels, the present study reveals a novel role for the HIF1 complex in coupling mTORC1 to the control of T cell differentiation.
How does this mTORC1–HIF1 pathway coordinate with other signaling pathways that control CD8+ T cell differentiation? A particular issue is that models of lymphocyte signal transduction invariably link mTORC1 to PI3K–Akt signaling. As discussed, the biochemical experiments that initially proposed this model are flawed because the pharmacological tools used have numerous off-target effects. Nevertheless, the concept of a linear model of PI3K–Akt–mTORC1 signaling is still compelling because the phenotypes of T cells activated in the presence of PI3K–Akt or mTORC1 inhibitors show similarities. For example, the expression of perforin is positively regulated by both Akt (Macintyre et al., 2011) and mTORC1 (Fig. 4 G), and the expression of CD62L, CCR7, and S1P1 is negatively regulated by both Akt and mTORC1 (Sinclair et al., 2008; Macintyre et al., 2011). In a linear model, it would be assumed that PI3K and Akt activity are required for mTORC1 activation. Moreover, there are models proposing mTORC1 control of Akt activity. This latter idea stems from experiments showing that in some cell systems, prolonged inhibition of mTORC1 with rapamycin can disrupt the mTORC2 serine/threonine kinase complex (Sarbassov et al., 2006). mTORC2 phosphorylates Akt on S473, and it has been shown that prolonged rapamycin treatment reduces Akt S473 phosphorylation in CD4 T cells activated with CD3 and CD28 antibodies (Lee et al., 2010; Delgoffe et al., 2011). It is frequently assumed that loss of Akt S473 phosphorylation will block Akt catalytic activity. However, the key rate-limiting phosphorylation on Akt is the PDK1-mediated phosphorylation of T308, and the impact of the reduced Akt S473 phosphorylation on Akt catalytic function in CD4 T cells has not been studied in detail. Moreover, does rapamycin treatment actually disrupt mTORC2 complexes in all T cells? The current study addresses these issues and shows unequivocally that PI3K and Akt activity are not essential for mTORC1 activation or for HIF1 expression in CD8+ T cells. Moreover, rapamycin treatment, even long term, does not prevent Akt phosphorylation or activity, as judged by the normal phosphorylation and nuclear exclusion of the Foxo transcription factors in rapamycin-treated T cells. There is a well-documented Akt–Foxo-mediated pathway to control T cell trafficking in CD8+ T cells, but the mTORC1–HIF1 pathway that controls T cell trafficking described herein is independent of Akt and Foxos. The present study thus indicates that PI3K–Akt and mTORC1 signaling are not linked in a linear pathway. These molecules are activated independently but converge to control the expression of key molecules required for effector CTL function. An essential requirement for these two independent signaling pathways to control T cell differentiation highlights the importance of integrating lymphocyte signal transduction for appropriate immune responses.
MATERIALS AND METHODS
Mice.
All mice used in this study were approved by the University of Dundee ethical review committee and maintained in compliance with UK Home Office Animals (Scientific Procedures) Act 1986 guidelines. PDK1flox/flox TamoxCre mice were generated by breeding C57BL/6GT(ROSA)26tm9(CreEsr1)Arte (TamoxCre), purchased from Taconic, to mice carrying PDK1 floxed alleles (Mora et al., 2003). HIF1βflox/flox mice (Geng et al., 2006) were backcrossed with mice expressing cre recombinase under the control of the CD4 promoter, generating HIF1βflox/flox CD4Cre mice. PI3K p110δD910A/D910A mice carry a point mutation that switches Asp910→Ala (D910A) in the p110δ subunit to inactivate catalytic activity (Okkenhaug et al., 2002). P14-LCMV and OTI transgenic mice have been described previously (Pircher et al., 1989; Kurts et al., 1996).
Cell culture.
CTLs were generated as described previously (Waugh et al., 2009). In brief, lymphocytes isolated from spleens or lymph nodes of P14-LCMV or OTI transgenic mice or nontransgenic mice were activated for 48 h with either 100 ng/ml of soluble LCMV or OTI-specific peptide, gp33-41 and SIINFEKL, respectively, or 0.5 µg/ml anti-CD3 antibody (2c11). Cells were then cultured in 20 ng/ml IL-2 (Proleukin) at 37°C for an additional 6 d. For conditional deletion of PDK1, CTLs were prepared by activating PDK1flox/flox TamoxCre splenocytes with 2c11 for 2 d followed by culturing in IL-2 for 5 d, with 0.6 µM of 4′OHT (Sigma-Aldrich) being added for the final 72 h of culture. Where indicated, cells were treated with various inhibitors: 1 µM Akti1/2 (EMD Millipore), 20 nM rapamycin (EMD Millipore), 10 µM IC87114 (synthesized in-house), and 10 µM LY294002 (Promega).
For short-term (20 h) TCR stimulations, naive CD8+ T cells were purified from lymph nodes of HIF1flox/flox CD4Cre and HIF1WT/WT CD4Cre nontransgenic mice or alternatively from P14-LCMV or OTI transgenic mice by magnetic cell sorting (Miltenyi Biotec). P14-LCMV and OTI naive CD8+ T cells were activated with gp33-41 plus 3 ng/ml anti-CD28 (clone 37.51; eBioscience) and SIINFEKL, respectively. Nontransgenic naive CD8+ T cells were activated with 2c11 plus anti-CD28.
Flow cytometric analysis.
Cells were labeled with APC-efluor780 CD8 (53-6.7), FITC CD25 (7D4), APC CD44 (IM7), PE CD71 (C2), APC CD62L (MEL-14), Alexa Fluor 700 CD45.1 (104), and V450 Horizon CD45.2 (A20) purchased from eBioscience or BD. Live cells were gated according to their forward scatter and side scatter. Data were acquired on either a FACSCalibur or an LSRFortessa (BD) and analyzed using FlowJo software (Tree Star).
Western blot analysis and cytoplasmic nuclear fractionation.
Cells were lysed (2 × 107/ml) in Tris lysis buffer containing 10 mM Tris, pH 7.05, 50 mM NaCl, 30 mM Na pyrophosphate, 50 mM NaF, 5 µM ZnCl2, 10% glycerol, 0.5% Triton, 1 µM DTT, and protease inhibitors (Roche). Lysates were centrifuged (4°C, 16,000 g for 10 min) and separated by SDS-PAGE and transferred to nitrocellulose membrane. Blots were probed with antibodies recognizing pFoxo1/3aT24/34, pS6S235/236, pS6KT389, pS6KT421/S424, pAktT308, pAktS473, p4EBP1S37/46, and p4EBP1S65 (Cell Signaling Technology); pRSKS227 (Santa Cruz Biotechnology, Inc.); and unphosphorylated S6, S6K, Akt, and c-Myc (Cell Signaling Technology); RSK2, IκBα, PTEN, and HIF1β (Santa Cruz Biotechnology, Inc.); HIF1α (R&D Systems); PDK1 (EMD Millipore); Glut1 (a gift from G. Holman, University of Bath, Bath, England, UK; Holman et al., 1990); Perforin (a gift from G. Griffith, Cambridge Institute for Medical Research, Cambridge, England, UK); and Foxo1 and Foxo3a (generated in-house). Immunoblots were probed with an SmcI antibody as a loading control (Bethyl Laboratories, Inc.).
To obtain cytoplasmic and nuclear fractions, cells were lysed in Hepes hypotonic lysis buffer (20 × 106 cells/ml; 10 mM Hepes, pH 7.9, 4 mM MgCl2, 15 mM KCl, 10 mM NaF, 0.1 mM EDTA, 0.15% [vol/vol] Nonidet P-40, 1 mM PMSF, 1 mM Na3VO4, and 1 mM DTT) and centrifuged at 4°C for 1 min at 10,000 g. Cytosolic fractions were removed, and nuclear membrane fractions were suspended in the aforementioned Tris lysis buffer. Immunoblot analysis of IκBα and Smc1 was used to confirm the purity of cytoplasmic and nuclear fractions, respectively.
Glucose uptake.
106 cells were suspended in 400 µl glucose-free media containing 0.5 µCi/ml 2-deoxy-d-[1-3H]glucose ([3H] 2-DG; GE Healthcare) and incubated for 3 (CTLs) or 10 min (TCR-simulated CD8+ T cells). Cells were pelleted, washed, and lysed overnight with 200 µl of 1 M NaOH, and the incorporated 3H radioactivity was quantified via liquid scintillation counting. Measurements were performed in triplicates per condition.
Quantitative real-time PCR.
RNA was extracted using the RNeasy RNA purification mini kit (QIAGEN) according to the manufacturer’s protocol. Purified RNA was reverse transcribed using the qScript cDNA synthesis kit (Quanta). Real-time PCR was performed in triplicates in 96-well plates using iQ SYBR Green–based detection on an iCycler (Bio-Rad Laboratories). For the analysis of mRNA levels, the derived values were averaged and normalized to HPRT mRNA levels.
Primers.
Primers used are as follows: HPRT forward, 5′-TGATCAGTCAACGGGGGACA-3′; HPRT reverse, 5′-TTCGAGAGGTCCTTTTCACCA-3′; CD62L forward, 5′-ACGGGCCCCAGTGTCAGTATGTG-3′; CD62L reverse, 5′-TGAGAAATGCCAGCCCCGAGAA-3′; CCR7 forward, 5′-CAGCCTTCCTGTGTGATTTCTACA-3′; CCR7 reverse, 5′-ACCACCAGCACGTTTTTCCT-3′; Perforin forward, 5′-CGTCTTGGTGGGACTTCAG-3′; Perforin reverse, 5′-GCATTCTGACCGAGGGCAG-3′; IFN-γ forward, 5′-TTACTGCCACGGCACAGTC-3′; and IFN-γ reverse, 5′-AGATAATCTGGCTCTGGCTCTGCGG-3′.
Lactate measurement.
2 × 106/ml CTLs were cultured for 4 h in RPMI 1640 containing 10% dialyzed fetal calf serum, and then the cells were spun and the supernatants were collected. Lactate concentration in the supernatant was quantified using an LDH (lactate dehydrogenase)-based enzyme assay, monitoring the emergence of NADH through increased absorption at 340 nm (the reaction contained 320 mM glycine, 320 mM hydrazine, 2.4 mM NAD+, and 3 U/ml LDH). A standard curve was generated, and the concentration of lactate in the supernatant added to this reaction was calculated.
Adoptive transfer.
CTLs were generated from the spleens of HIF1βflox/flox CD4Cre mice or HIF1βflox/flox mice (as described in Cell culture but with 4 d in IL-2). Cells were then labeled with either CFSE (Invitrogen) or CellTracker Orange (CMTMR; Invitrogen) for 15 min at 37°C, washed, mixed at 1:1 ratio, and suspended in sterile PBS. 5 × 106 mixed cells were injected into the tail vein of C57BL/6 host mice. After 4 h, the mice were sacrificed, and lymph nodes, spleen, and blood were analyzed and the ratio of CFSE- and CMTMR-labeled T cells quantified. Values indicate recovery of HIF1βflox/flox CD4Cre cells versus HIF1βflox/flox cells as a percentage of the total recovered transferred cells.
Chromatin immunoprecipitation (ChIP).
Real-time PCR-based ChIP analysis to measure Pol II binding to the Perforin locus was performed as described previously (Cruz-Guilloty et al., 2009) with minor modifications. In brief, chromatin was immune precipitated with anti–Pol II (Santa Cruz Biotechnology, Inc.) or normal rabbit IgG (Cell Signaling Technology) from 5 × 106 cells in the presence of 0.2 mg/ml BSA. ChIP-grade protein G magnetic beads (Cell Signaling Technology) were used to collect the immune complexes. Chromatin was purified via a NucleoSpin Extract II kit (Macherey-Nagel) and resuspended in TE buffer. Real-time PCR was performed in an iQ5 (Bio-Rad Laboratories) with Perfecta SYBR green FastMix for iQ (Quanta BioSciences). Primers are as follows: Perforin TSS forward, 5′-CAGGGCAGGAAGTAGTAATGATATG-3′; Perforin TSS reverse, 5′-CTTCCTCCTCCTTACCTGAAGTC-3′; Perforin Exon2 forward, 5′-CCAGAGTTTATGACTACTGTG-3′; and Perforin Exon2 reverse, 5′-GTGCTTCTGCTTGCATTCTG-3′.
SILAC.
P14-LCMV CTLs were cultured in SILAC medium as described previously (Navarro et al., 2011) with some minor modifications. In brief, cells were combined and a Thermo Fisher Scientific subcellular protein fractionation kit was used to fractionate CTLs according to the manufacturer’s instructions. Fractions were chloroform methanol precipitated and further separated by molecular weight using denaturing size exclusion chromatography before digestion and LC-MS/MS analysis as described previously (Larance et al., 2012). The peptide mixture was separated by nanoscale C18 reverse phase liquid chromatography (Ultimate 3000 nLC; Dionex) coupled to a LTQ-Orbitrap Velos (Thermo Fisher Scientific). MaxQuant version 1.3.0.5 (Cox and Mann, 2008) was used to process raw MS spectra, using the default settings against the mice Uniprot database (July 2012).
Online supplemental material.
Table S1 lists microarray analysis showing decreased gene expression in HIF1−/− versus HIF1 WT CTLs. Table S2 lists microarray analysis showing increased gene expression in HIF1−/− versus HIF1 WT CTLs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112607/DC1.
Supplementary Material
Acknowledgments
We thank members of the Biological Services Unit, R. Clarke of the Flow Cytometry Facility, members of the “Fingerprints” Proteomics Facility, and members of the D.A. Cantrell laboratory for critical reading of the manuscript. We thank D. Alessi at the University of Dundee for providing mice carrying PDK1 floxed alleles and the Finnish DNA Microarray Centre at the Centre for Biotechnology (Turku, Finland) for the microarray analysis.
This work was supported by a Wellcome Trust Principal Research Fellowship and Program Grant (065975/Z/01/A). E. Rosenzweig and J.L. Hukelmann were supported by Wellcome Trust PhD Studentships.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- 4′OHT
- 4-hydroxytamoxifen
- AHR
- Aryl hydrocarbon receptor
- ChIP
- chromatin immunoprecipitation
- PI3K
- phosphatidylinositol-3 kinase
- Pol II
- RNA polymerase II
References
- Araki K., Turner A.P., Shaffer V.O., Gangappa S., Keller S.A., Bachmann M.F., Larsen C.P., Ahmed R. 2009. mTOR regulates memory CD8 T-cell differentiation. Nature. 460:108–112 10.1038/nature08155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn G.J., Williams J., Sabers C., Wiederrecht G., Lawrence J.C., Jr, Abraham R.T. 1996. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 15:5256–5267 [PMC free article] [PubMed] [Google Scholar]
- Caldwell C.C., Kojima H., Lukashev D., Armstrong J., Farber M., Apasov S.G., Sitkovsky M.V. 2001. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J. Immunol. 167:6140–6149 [DOI] [PubMed] [Google Scholar]
- Cham C.M., Gajewski T.F. 2005. Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 174:4670–4677 [DOI] [PubMed] [Google Scholar]
- Cham C.M., Driessens G., O’Keefe J.P., Gajewski T.F. 2008. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 38:2438–2450 10.1002/eji.200838289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F., Pipkin M.E., Djuretic I.M., Levanon D., Lotem J., Lichtenheld M.G., Groner Y., Rao A. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206:51–59 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Pollizzi K.N., Waickman A.T., Heikamp E., Meyers D.J., Horton M.R., Xiao B., Worley P.F., Powell J.D. 2011. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 12:295–303 10.1038/ni.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre S., Carrette F., Chen J., Lang V., Semichon M., Denoyelle C., Lazar V., Cagnard N., Dubart-Kupperschmitt A., Mangeney M., et al. 2008. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J. Immunol. 181:2980–2989 [DOI] [PubMed] [Google Scholar]
- Fox C.J., Hammerman P.S., Thompson C.B. 2005. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5:844–852 10.1038/nri1710 [DOI] [PubMed] [Google Scholar]
- Geng S., Mezentsev A., Kalachikov S., Raith K., Roop D.R., Panteleyev A.A. 2006. Targeted ablation of Arnt in mouse epidermis results in profound defects in desquamation and epidermal barrier function. J. Cell Sci. 119:4901–4912 10.1242/jcs.03282 [DOI] [PubMed] [Google Scholar]
- Holman G.D., Kozka I.J., Clark A.E., Flower C.J., Saltis J., Habberfield A.D., Simpson I.A., Cushman S.W. 1990. Cell surface labeling of glucose transporter isoform GLUT4 by bis-mannose photolabel. Correlation with stimulation of glucose transport in rat adipose cells by insulin and phorbol ester. J. Biol. Chem. 265:18172–18179 [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kalia V., Sarkar S., Subramaniam S., Haining W.N., Smith K.A., Ahmed R. 2010. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 32:91–103 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kerdiles Y.M., Beisner D.R., Tinoco R., Dejean A.S., Castrillon D.H., DePinho R.A., Hedrick S.M. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10:176–184 10.1038/ni.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurts C., Heath W.R., Carbone F.R., Allison J., Miller J.F., Kosaka H. 1996. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184:923–930 10.1084/jem.184.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance M., Kirkwood K.J., Xirodimas D.P., Lundberg E., Uhlen M., Lamond A.I. 2012. Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Mol. Cell. Proteomics. 11:M111.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Gudapati P., Dragovic S., Spencer C., Joyce S., Killeen N., Magnuson M.A., Boothby M. 2010. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 32:743–753 10.1016/j.immuni.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald H.R., Koch C.J. 1977. Energy metabolism and T-cell-mediated cytolysis. I. Synergism between inhibitors of respiration and glycolysis. J. Exp. Med. 146:698–709 10.1084/jem.146.3.698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre A.N., Finlay D., Preston G., Sinclair L.V., Waugh C.M., Tamas P., Feijoo C., Okkenhaug K., Cantrell D.A. 2011. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 34:224–236 10.1016/j.immuni.2011.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver N.J., Jacobs S.R., Wieman H.L., Wofford J.A., Coloff J.L., Rathmell J.C. 2008. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J. Leukoc. Biol. 84:949–957 10.1189/jlb.0108024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A., Davies A.M., Bertrand L., Sharif I., Budas G.R., Jovanović S., Mouton V., Kahn C.R., Lucocq J.M., Gray G.A., et al. 2003. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. EMBO J. 22:4666–4676 10.1093/emboj/cdg469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M.N., Goebel J., Feijoo-Carnero C., Morrice N., Cantrell D.A. 2011. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat. Immunol. 12:352–361 10.1038/ni.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K., Bilancio A., Farjot G., Priddle H., Sancho S., Peskett E., Pearce W., Meek S.E., Salpekar A., Waterfield M.D., et al. 2002. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 297:1031–1034 [DOI] [PubMed] [Google Scholar]
- Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 32:79–90 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher H., Bürki K., Lang R., Hengartner H., Zinkernagel R.M. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Powell J.D., Delgoffe G.M. 2010. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 33:301–311 10.1016/j.immuni.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.R., Li Q., Odunsi K., Shrikant P.A. 2010. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 32:67–78 10.1016/j.immuni.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22:159–168 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2010. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20:51–56 10.1016/j.gde.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., Chi H. 2011. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208:1367–1376 10.1084/jem.20110278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair L.V., Finlay D., Feijoo C., Cornish G.H., Gray A., Ager A., Okkenhaug K., Hagenbeek T.J., Spits H., Cantrell D.A. 2008. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 9:513–521 10.1038/ni.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., Green D.R. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 35:871–882 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh C., Sinclair L., Finlay D., Bayascas J.R., Cantrell D. 2009. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol. Cell. Biol. 29:5952–5962 10.1128/MCB.00585-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.