Bacterial surface proteins switch the orientation of IgG binding depending on the antibody concentration of their environment.
Abstract
Several of the most significant bacterial pathogens in humans, including Streptococcus pyogenes, express surface proteins that bind IgG antibodies via their fragment crystallizable (Fc) region, and the dogma is that this protects the bacteria against phagocytic killing in blood. However, analysis of samples from a patient with invasive S. pyogenes infection revealed dramatic differences in the presence and orientation of IgG antibodies at the surface of bacteria from different sites. In the throat, IgG was mostly bound to the bacterial surface via Fc, whereas in the blood IgG was mostly bound via fragment antigen-binding (Fab). In infected and necrotic tissue, the Fc-binding proteins were removed from the bacterial surface. Further investigation showed that efficient bacterial IgGFc-binding occurs only in IgG-poor environments, such as saliva. As a consequence, the bacteria are protected against phagocytic killing, whereas in blood plasma where the concentration of IgG is high, the antibodies preferentially bind via Fab, facilitating opsonization and bacterial killing. IgG-poor environments represent the natural habitat for IgGFc-binding bacteria, and IgGFc-binding proteins may have evolved to execute their function in such environments. The lack of protection in plasma also helps to explain why cases of severe invasive infections with IgGFc-binding bacteria are so rare compared with superficial and uncomplicated infections.
It has been estimated that Streptococcus pyogenes, a major bacterial pathogen in humans, causes >700 million cases of throat and skin infections annually. Compared with these very common and mostly uncomplicated infections, the cases where the bacterium invades the blood stream and induces a powerful inflammatory response (Kotb et al., 2002; Herwald et al., 2004; Macheboeuf et al., 2011) are rare (∼0.6 million annually), but connected with high mortality rates (Carapetis et al., 2005; Cole et al., 2011). Apart from the frequent superficial infections mentioned above, there are also hundreds of millions healthy carriers of S. pyogenes (predominantly colonized in the throat). Although a most relevant question, little is known about the mechanisms that convert a nonsymptomatic colonization or a superficial infection into an invasive and life-threatening condition.
Opsonization, the process where antibodies bind to and target microorganisms to facilitate their killing by phagocytes, is vital for the defense against infections. However, many bacterial species, Gram-positive and Gram-negative, aerobic and anaerobic, and pathogenic as well as commensal bacteria (Forsgren and Sjöquist, 1966; Björck and Kronvall, 1984; Lindahl and Kronvall, 1988; Björck, 1988; Mintz and Fives-Taylor, 1994; Sandt and Hill, 2001), interfere with this mechanism by expressing surface proteins that bind antibodies outside the antigen-binding region. Among these proteins some of the most well-known are those that bind to the constant fragment crystallizable (Fc) part of IgG (IgGFc) antibodies with high affinity and specificity, including protein A from Staphylococcus aureus (Forsgren and Sjöquist, 1966), protein G from group G and C streptococci (Björck and Kronvall, 1984; Reis et al., 1984), and the M and M-like proteins from Streptococcus pyogenes (Heath and Cleary, 1989). Apart from these Gram-positive species, IgGFc-binding proteins have been identified also in other pathogenic bacteria such as Pseudomonas maltophilia (Grover et al., 1991), Yersinia pestis (Zav’yalov et al., 1996), and Escherichia coli (Leo and Goldman, 2009).
IgGFc-binding proteins were described in the 1960s (Forsgren and Sjöquist, 1966), but nothing is known about the orientation of IgG antibodies at the surface of IgGFc-binding bacteria under in vivo conditions at different sites, and the consequences this may have for the host–bacterial relationship. It has generally been assumed that, regardless of the setting, bacteria equipped with IgGFc-binding surface proteins preferentially bind IgG via Fc, providing protection against phagocytic killing. However, when analyzing samples of S. pyogenes from a patient with asymptomatic colonization of the throat who developed sepsis and necrotizing fasciitis, the presence and orientation of IgG was markedly different depending on the origin of the samples (throat, blood, or necrotic tissue). This observation and a series of experiments define a biological role for IgGFc-binding bacterial proteins in IgG poor environments, and describe how these proteins in S. pyogenes contribute to toxic invasive disease.
RESULTS
Presence and orientation of IgG antibodies at the surface of S. pyogenes in a patient with severe invasive disease
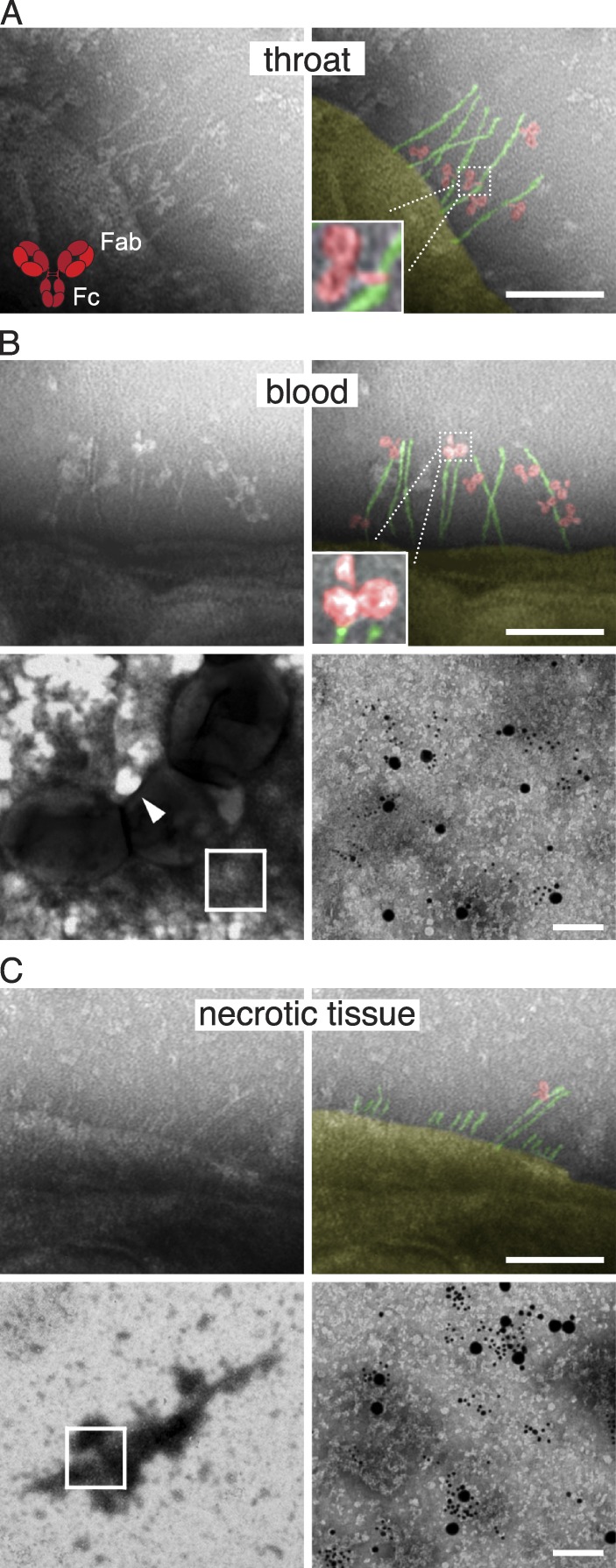
A 31-yr-old woman felt pain in her right thigh after working in her garden. A day later she developed high fever, chills, and profuse vomiting. Her family members had suffered from milder upper respiratory tract infections; she had no such symptoms. 36 h after she fell ill, the patient was admitted to our infectious diseases clinic. At this point she was seriously unwell and in shock, and intravenous fluids and antibiotics were given. There were no clinical signs of pharyngitis, but a throat swab was positive for S. pyogenes, and she had a tender well-demarcated erythema on the right ventral thigh. Throat and blood samples were collected for culture and electron microscopy. The patient was operated on 12 h after admittance (7 h after initial treatment with antibiotics), revealing necrotizing fasciitis. Necrotic tissue was removed from the thigh, and wound fluid and tissue samples were collected. The patient survived and fully recovered (for more details of the case see Doc. S1). There was growth of S. pyogenes (of the M1 serotype) in the throat and necrotic tissue, but not in blood. Negative staining electron microscopy showed the presence of S. pyogenes also in blood, and this technique was used to investigate the presence and orientation of IgG antibodies at the surface of S. pyogenes in the three different samples (Fig. 1). In the throat, IgG was bound predominantly via the Fc region (63%, n = 300), whereas in the blood sample the antibodies mostly interacted with the bacterial surface through fragment antigen-binding (Fab; 61%, n = 300). Notably, most of the bacterial surfaces in blood were covered by aggregates containing IgG and fibrinogen bound to fibrous M1 proteins at the bacterial surface (detected by immunogold labeling). In the necrotic tissue, most of the M1 proteins had been cleaved off the surface, with too few antibodies visible to allow quantification. In this sample the bacteria were surrounded by large protein complexes, which, by immunogold labeling, consisted of M1, fibrinogen and IgG. This indicates that complexes formed at the bacterial surface had been released by proteolytic cleavage of M1 proteins, which is consistent with earlier reports showing that M1 protein can form complexes with fibrinogen and IgG (Kantor, 1965; Åkesson et al., 1994; Kahn et al., 2008). Such released complexes have previously been shown to induce a massive vascular leakage (Herwald et al., 2004), a cardinal symptom of necrotizing fasciitis and streptococcal toxic shock. These human in vivo data indicated that the role of IgGFc-binding proteins, such as the M proteins, is different depending on the phase of infection, and that the IgG concentration in the environment should influence their function.
Figure 1.
Antibodies at the surface of S. pyogenes isolated from a patient with invasive disease. (A–C) Negative staining electron microscopy was used to visualize the localization and orientation of IgG antibodies bound to the surface of bacteria isolated from the throat (A), blood (B), and necrotic tissue fluid in the thigh (C) of a patient suffering from necrotizing fasciitis and streptococcal toxic shock. Bar, 100 nm. The top right images are pseudocolored to better illustrate the single IgG molecules (red) and bacterial IgGFc-binding proteins H/M1 (green). The arrowhead in the bottom left panel of B indicates a typical area from which the top left panel of B was taken. For the tissue sample, most of the M1/H proteins have been cleaved off of the surface, making the analysis of antibody orientation uncertain. The composition of protein complexes found at the bacterial surface in the blood sample (most of the surface was covered by these aggregates) and released from the bacterial surface in the necrotic tissue (B and C, bottom; bars, 50 nm), were analyzed by negative staining electron microscopy (no protein complexes were present in the throat sample). The blown-up regions to the right (indicated with a box to the left) show the presence of gold-labeled IgGFab fragments against H/M1 protein (small bead), IgG (medium bead), and fibrinogen (large bead).
Concentration-dependent bacterial binding of IgG
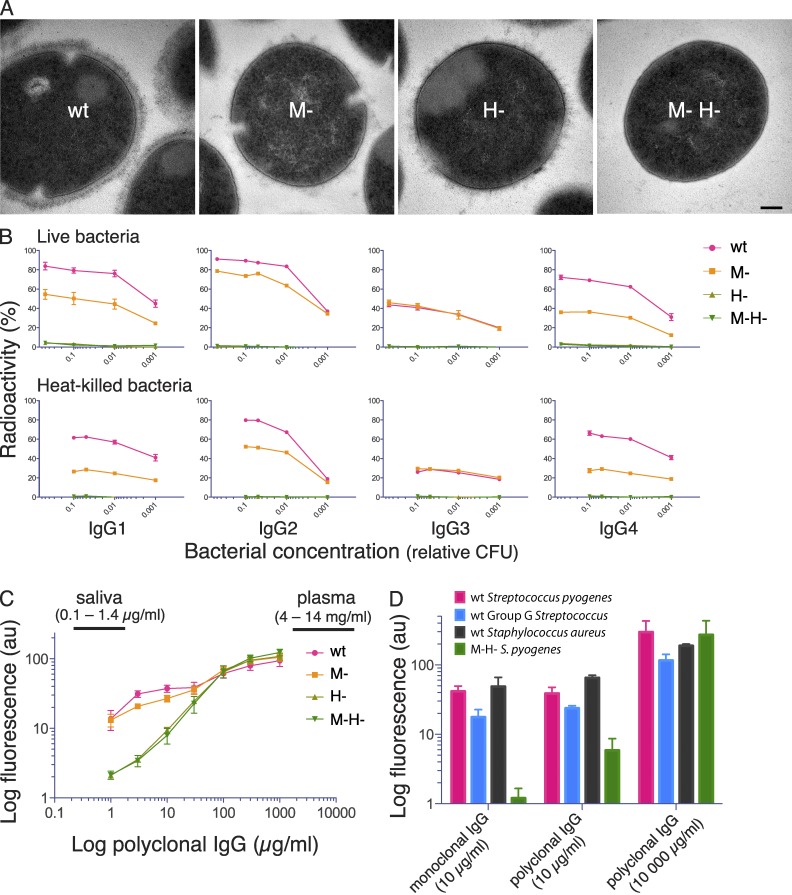
The surface of wild-type S. pyogenes bacteria are dominated by M and M-like proteins (Fischetti, 1989; McNamara et al., 2008). Many of these proteins, including the M1 protein of the strain isolated from the patient described above, bind to the Fc region of human IgG. In addition, some of the M1 strains express an additional IgGFc-binding M-like protein called protein H (Frick et al., 1994; Åkesson et al., 1994), and the strong IgGFc-binding of these strains makes them particularly suitable to study the biological consequences of IgGFc-binding. The bacterial strain infecting the patient described above was of the M1 serotype and expressed protein H (has the emm1 gene and showed high binding of radiolabeled human IgG). We have previously generated mutants in a wild-type S. pyogenes strain (AP1) expressing both M1 and protein H, these mutants were used in the studies described below. On electron micrographs, proteins M1 and H can be seen as long (∼50 nm) hair-like structures at the bacterial surface. Isogenic mutants lacking M1 or protein H have visibly lower amounts of these fibrous structures, whereas they are completely absent in the double mutant (Fig. 2 A).
Figure 2.
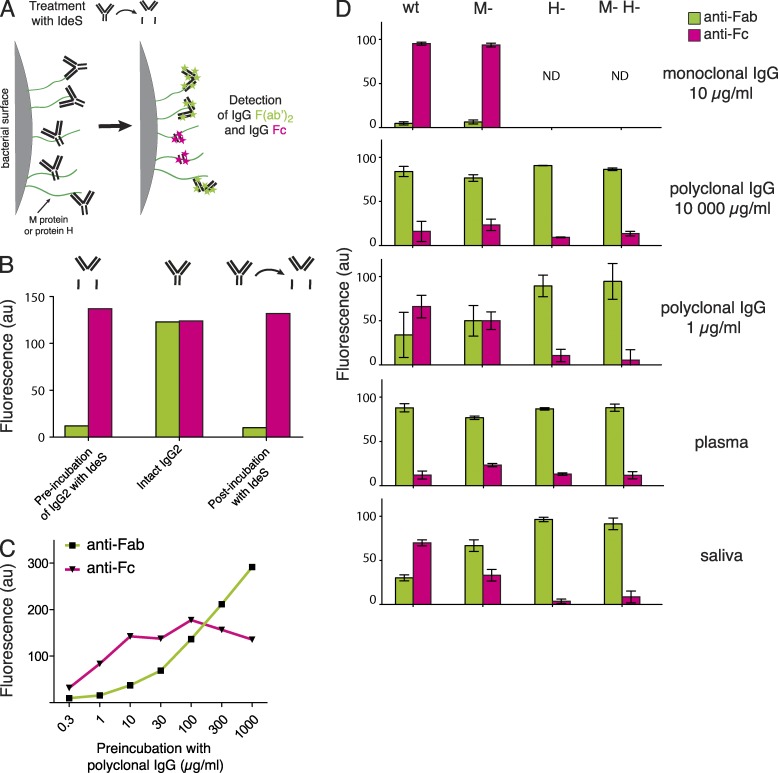
Electron micrographs of S. pyogenes strains and analysis of bacterial IgG binding. (A) Transmission electron microscopy of wild-type S. pyogenes (wt) and isogenic mutants lacking M1 protein (M-), protein H (H-) and both proteins (M- H-). Bar, 100 nm. (B) 125I-labeled monoclonal IgG subclasses were incubated with live or heat-killed S. pyogenes bacteria that had been harvested at exponential growth phase. IgG binding to bacteria is shown as the percentage of added radiolabeled IgG. Bacterial concentration is given as fractions of 4 × 108 CFU. Data are represented as mean ± SEM of three independent experiments. (C) Wild-type and mutant S. pyogenes bacteria were preincubated with increasing concentrations of pooled polyclonal IgG, and surface-associated IgG was measured by flow cytometry. The range of IgG levels in human plasma and saliva are indicated in the figure. Data are represented as mean ± SEM of three independent experiments. (D) Wild-type and the M-H- S. pyogenes mutant, wild-type protein G–expressing group G Streptococcus, and wild-type protein A–expressing S. aureus, were separately preincubated with monoclonal IgG1 (10 µg/ml) or polyclonal IgG (10 or 10, 000 µg/ml), and surface-bound IgG was measured by flow cytometry. Data are represented as mean ± SEM of three independent experiments.
To test the IgGFc-binding properties of proteins M1 and H, human monoclonal IgG of the four subclasses was separately incubated with decreasing numbers of live bacteria, and the radioactivity bound to the bacteria was measured (Fig. 2 B). The results show that the wild-type strain and the M- mutant bound human monoclonal IgG of all four subclasses, whereas the protein H mutant and the double mutant bound poorly, suggesting that protein H is mainly responsible for the IgGFc-binding activity of wild-type bacteria. The results with the H strain indicated that M1 protein itself has a low affinity for IgGFc, but enhances the binding of IgG (except for the IgG3 subclass) to protein H. Very similar results were obtained in binding experiments with heat-killed bacteria (Fig. 2 B, bottom). Next, heat-killed bacteria were incubated with human polyclonal IgG. Throughout this study we used a therapeutic polyclonal IgG preparation (IVIG; Octapharma) pooled from >3,500 healthy individuals with the normal IgG subclass composition, at concentrations ranging from 1 to 1,000 µg per ml. The amount of bound IgG was measured using flow cytometry, and polyclonal IgG bound to all strains in a concentration-dependent manner (Fig. 2 C). Differences in IgG binding between the strains were observed only at low concentrations (<10 µg/ml). With increasing concentration binding also increased for all strains, gradually reducing the differences, and at IgG concentrations >100 µg/ml the strains bound IgG to the same extent. For the wild-type strain, the IgG interaction appeared to be biphasic, with a saturation of binding at lower concentrations (3–30 µg/ml), before aligning with the pattern observed for the mutant strains at high IgG concentrations. As mentioned, many important bacterial pathogens apart from S. pyogenes express IgGFc-binding proteins. Protein A of Staphylococcus aureus and protein G of group C and G streptococci are two well-known examples, and wild-type bacteria expressing these proteins showed the same interaction pattern as S. pyogenes when tested against monoclonal and polyclonal IgG (Fig. 2 D). The continued work was focused on S. pyogenes, but these results suggest that the data obtained could be relevant also for other bacterial species with IgGFc-binding surface proteins.
At low concentrations, IgG is bound to the bacterial surface via Fc
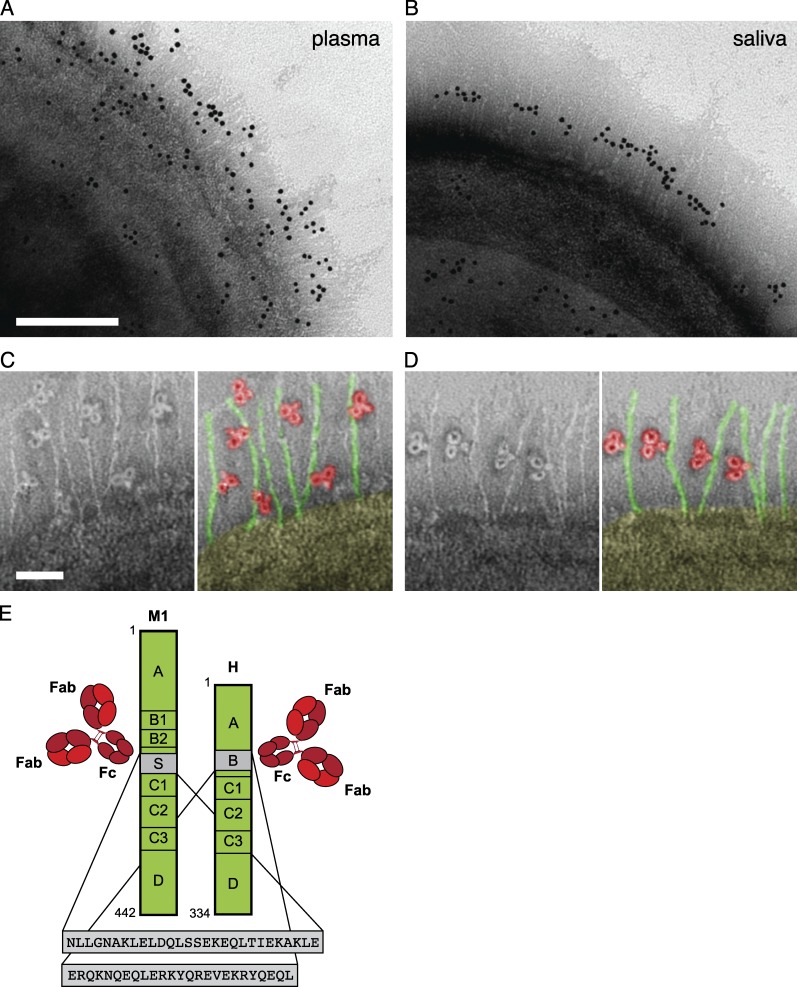
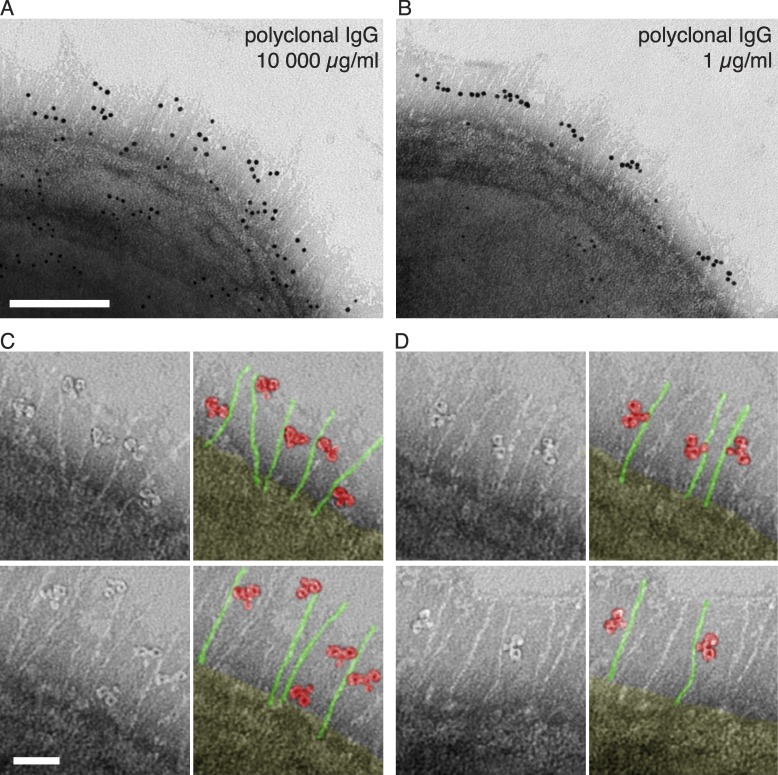
To investigate the localization and orientation of bacteria-bound IgG, we incubated wild-type bacteria with pooled human plasma or saliva samples. The reduced contribution of proteins H and M1 to antibody binding at a high IgG concentration suggests that their function is to be found in settings with low IgG concentration. In the pharynx, S. pyogenes is exposed to saliva where the reported IgG concentration is considerably lower than in blood. To determine the amount of IgG in the samples used here (pooled saliva and plasma from five individuals), we used heavy labeled reference peptides and selected-reaction monitoring mass spectrometry (SRM MS) to quantify the different subclasses of IgG (Fig. 3 and Fig. S1). The results confirmed the very large difference (10,000-fold) in IgG concentration between saliva and plasma, and also showed the distribution of the four IgG subclasses (Table S1). After washing, negative staining electron microscopy with gold-labeled anti-IgG Fab fragments was performed to locate IgG bound to the bacterial surface. After plasma incubation, IgG was found all over the bacterial surface, and to a high degree in the region of the hairlike projections representing proteins M1 and H (Fig. 4 A). In saliva, however, the antibodies strictly interacted with the central region of these surface proteins (Fig. 4 B), a region to which previous studies have mapped the binding site for IgGFc (Frick et al., 1994; Åkesson et al., 1994) (Fig. 4 E). This indicates that specific IgG antibodies in plasma bind to bacterial surface structures via their antigen-binding Fabs, whereas the highly localized binding in saliva suggests binding via Fc. The orientation of individual IgG molecules was visualized by analyzing the negative staining images at higher magnification. The analysis confirmed that the majority of IgG antibodies interacted with the bacterium via Fab in plasma (87%, n = 300; Fig. 4 C) and via Fc in saliva (84%, n = 300; Fig. 4 D). To test whether the orientation at the bacterial surface is directly linked to the IgG concentration, bacteria were incubated with polyclonal IgG at plasma (10,000 µg/ml) and saliva level (1 µg/ml), respectively. The results mimicked those obtained with plasma and saliva, with Fab binding dominating at high IgG concentrations and Fc binding dominating at low concentrations (Fig. 5). Collectively, these experiments suggest that the vast majority of IgG antibodies in plasma that bind to S. pyogenes surface structures, including proteins H and M1, do so through Fab. In contrast, the low total concentration of IgG, and thereby the low number of specific IgG antibodies in saliva, results in predominant binding of IgG via Fc, effectively reversing the normal antibody orientation at the bacterial surface.
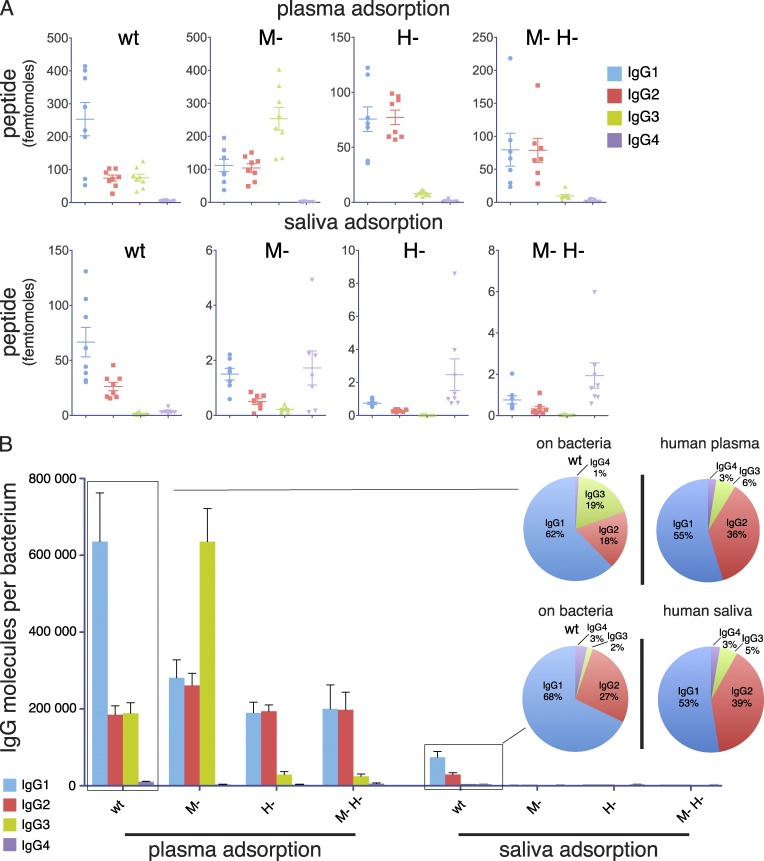
Figure 3.
Profiling of IgG subclass distribution using targeted and quantitative mass spectrometry. (A) Quantification by SRM of IgG bound to the surface of wild-type and mutant S. pyogenes after incubation with either human plasma or saliva. Data are represented as numbers of molecules (femtomole) measured in each paired adsorption experiment (eight independent experiments). (B) Number of IgG molecules per bacterium based on average values from paired samples. The pie charts represent the subclass distribution for IgG bound to wild-type bacteria as compared with the IgG subclass composition of the plasma and saliva samples used for preincubation (eight independent experiments).
Figure 4.
Localization and orientation of IgG at the bacterial surface. (A and B) Negative staining EM was used to visualize the localization and orientation of IgG bound to the bacterial surface. Gold-labeled IgG is found either widely scattered (A, plasma) or in the midsection of the protrusions representing proteins H and M1 (B, saliva) of wild-type S. pyogenes. Bar, 100 nm. Images are representative of two independent experiments. (C and D) High magnification shows single IgG molecules bound either via Fab (c, plasma) or via Fc (d, saliva). Pseudo-color variants are shown to better visualize bound IgG (red) and proteins M and H (green). Bar, 25 nm. 300 bacterial surfaces were analyzed. (E) The wild-type S. pyogenes strain has two surface proteins, M1 protein and protein H, that both bind IgG via the Fc region as indicated in the figure (sequence data adapted from Åkesson et al. [1994] and Frick et al. [1994]).
Figure 5.
Localization and orientation of polyclonal IgG at the bacterial surface. (A and B) Negative staining EM was used to visualize the localization and orientation of IgG bound to the bacterial surface. Gold-labeled IgG is found either widely scattered (A, 10,000 µg/ml IgG) or found along the midsection of the surface proteins (B, 1 µg/ml IgG) of wild-type S. pyogenes. Bar, 100 nm. Images are representative of two independent experiments. (C and D) High magnification shows single IgG molecules bound either via Fab (C, 10,000 µg/ml IgG) or via Fc (D, 1 µg/ml IgG). Two representative images with pseudocolor variants are shown of each experiment. Bar, 25 nm.
IgGFc-binding proteins are required to reverse IgG orientation
The significance of IgGFc-binding proteins for the orientation of surface-bound IgG was further analyzed by a flow cytometry–based method relying on the highly specific activity of the IgG-degrading enzyme IdeS, a cysteine proteinase secreted by S. pyogenes that cleaves human IgG of all four subclasses in the hinge region (von Pawel-Rammingen et al., 2002), generating two Fc halves and one F(ab’)2 fragment (Vincents et al., 2004). By adding IdeS to bacteria with surface-associated IgG, the enzyme will remove the Fc part from IgG that is bound via Fab, and Fab from IgG bound via Fc (Fig. 6 A). In these experiments the bacteria were incubated with different IgG-containing solutions, and after incubation and washing, the samples were split into two groups; one was treated with IdeS and the other served as a paired control. Fc and Fab fragments at the bacterial surface were detected with fluorescently labeled Fab fragments directed toward either human IgGFc or IgGFab. The samples were then subjected to flow cytometry, and the fluorescence was presented as relative to the control sample. To verify the method, we first used monoclonal IgG, which should only be able to bind via the Fc region to the bacterial surface, and confirmed that we could detect bound Fc fragments after treatment with IdeS (Fig. 6 B). When wild-type bacteria were preincubated with increasing concentrations of IgG, the dominating signal shifted from IgGFc to IgGFab (Fig. 6 C). Specific antibodies binding to and blocking the Fc-binding regions of proteins H and M1 could explain the observed decrease of Fc fragments at high IgG concentrations. The concentration-dependent shift from IgGFc to IgGFab was further supported by the experiments summarized in Fig. 6 D, where the IdeS-based method was applied to investigate the interactions of IgG with wild-type and mutant S. pyogenes bacteria preincubated with human monoclonal and polyclonal IgG, plasma, or saliva. As expected, monoclonal IgG bound exclusively via Fc to bacteria expressing protein H, whereas the interaction between monoclonal IgG and the H- and M-H- mutants was below detection limit. At high concentration of polyclonal IgG (including plasma), Fab interactions prevailed for all the strains, whereas at low concentration (1 µg/ml and saliva) especially wild-type and, to a lesser extent, the M- mutant lacking M1 protein but expressing protein H, bound a significant fraction of IgG via Fc. In contrast, the H- and M-H- mutants bound IgG through specific Fab interactions, also at a low concentration of IgG. The results demonstrate that protein H in particular is required to effectively reverse the orientation of IgG antibodies binding to the wild-type S. pyogenes bacteria. It is also clear that this mechanism is functional only in environments with low concentration of specific S. pyogenes IgG antibodies.
Figure 6.
Analysis of IgG orientation at the bacterial surface. (A) Bacteria were preincubated with IgG, washed, and treated with IdeS, a proteinase of S. pyogenes that specifically cleaves IgG in the lower hinge region generating two half-Fc fragments and one F(ab′)2 fragment. Each sample was measured before and after IdeS treatment to determine which fragments of the bound IgG antibodies remained at the bacterial surface after proteolytic cleavage. IgG fragments were identified and measured by flow cytometry with fluorescently labeled Fab fragments raised against human IgGFc or IgGFab fragments. (B) The detection of monoclonal IgG fragments was analyzed after adding precleaved Fc and F(ab′)2 fragments, intact IgG, or the Fc and F(ab′)2 fragments that remain bound after IdeS cleavage. (C) After preincubation with increasing concentrations of polyclonal IgG, wild-type S. pyogenes bacteria were treated with IdeS and the presence of Fc and F(ab′)2 fragments at the bacterial surface was determined. (D) Wild-type and mutant bacteria were preincubated with monoclonal IgG, polyclonal IgG, plasma, or saliva, and the IgG fragments remaining at the bacterial surface after IdeS treatment, compared with paired control samples, were measured. Values are mean ± SEM of three independent experiments.
Binding of IgG subclasses and complement to bacterial surfaces
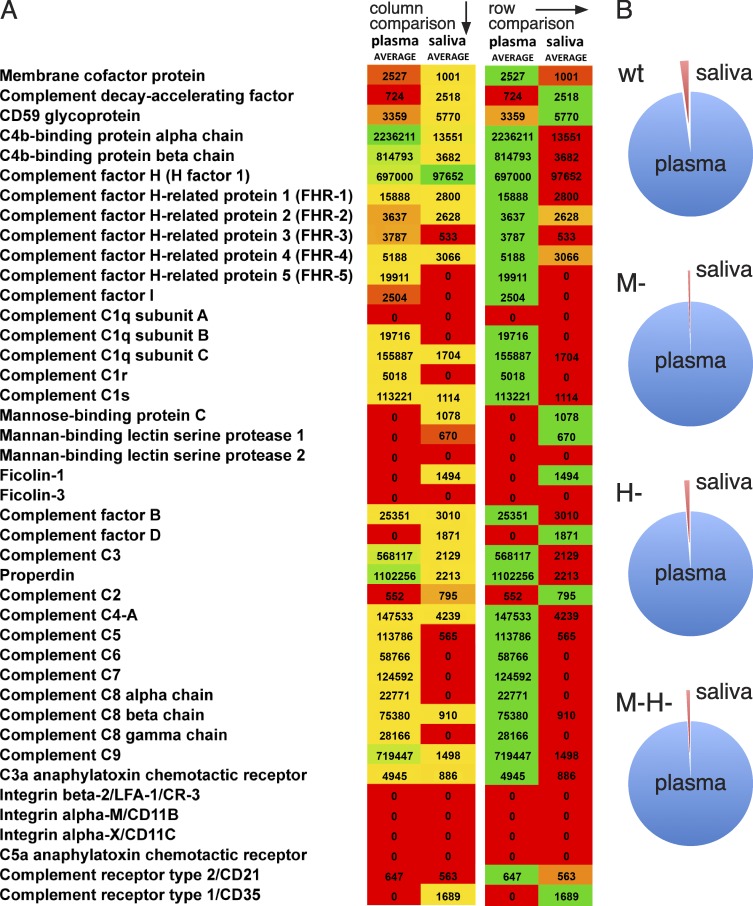
The IgG subclasses have different roles in the human immune response, where IgG1 and IgG3 are the most important for activating the complement system (Brüggemann et al., 1987), as well as triggering Fc-mediated phagocytosis (Nimmerjahn and Ravetch, 2008). The subclass profile of IgG antibodies bound to the bacterial surface may therefore indicate how the bacterium is recognized and handled by the immune system. Thus, S. pyogenes was incubated with pooled and paired plasma and saliva samples from 10 individuals, and the IgG subclasses and complement proteins adsorbed to the bacteria (8 independent experiments) were eluted and quantified by SRM MS. The amounts of IgG (Fig. 3 A) and complement proteins (Fig. 7 and Fig. S1 and S2) bound to the mutant strains after incubation with plasma or saliva were considerably higher for the plasma samples (in most cases >1,000 fold). In contrast, the difference for the number of IgG molecules adsorbed by the wild-type strain in plasma or saliva was only about 10 fold; approximately 106 compared with 105 per bacterium (Fig. 3 B). In plasma, the binding of complement proteins (Fig. 7) showed high levels of C4b-binding protein, which has previously been reported (Carlsson et al., 2003). However, components of the membrane attack complex (C5-C9) and C3 also accumulated, indicating efficient opsonization. Compared with plasma, fewer complement proteins were adsorbed to the bacterial surface in saliva (Fig. 7). In the saliva samples, the dominant protein found at the bacterial surface was complement factor H. This binding has been reported for M protein, and was suggested to be a mechanism for the bacteria to inhibit complement activation (Horstmann et al., 1988), as factor H is a negative regulator of the alternative pathway of complement, and inhibits C3b deposition (Liszewski et al., 1996). The antibody subclass distribution after plasma incubation showed that the levels of bound IgG1 and IgG3 were significantly higher for wild-type bacteria compared with the other strains except for the M- mutant, where the amount of IgG3 was very high (Fig. 3, A and B). The subclass profile of the wild-type strain represents a clear shift from the profile of the pooled human plasma sample (Fig. 3 B). As described above, the vast majority of the IgG bound to the surface of S. pyogenes in plasma are specific antibodies binding via Fab. The considerable increase of IgG3 interacting with the M- mutant indicates that the binding of specific IgG3 antibodies directed against protein H is partially blocked by the simultaneous presence of the M1 protein at the bacterial surface. The subclass distribution was very similar in the plasma and saliva samples, which is in contrast to the IgG subclasses adsorbed to the surface of wild-type bacteria (Fig. 3 B). Especially notable was the difference of bound IgG3, which was threefold higher in plasma versus in saliva. Among the IgG subclasses, the Fc of IgG3 has a lower affinity for bacterial Fc-binding proteins, including protein H (Fig. 2 D). Again, the data in saliva suggest that proteins H and M1 are both required for efficient Fc binding (Fig. 6 D, saliva). These results further underline the observation that the binding of IgG in an environment with a low IgG concentration is mediated mainly through Fc. In plasma, however, IgG is bound via Fab, and these antibodies are predominantly of the most efficiently opsonizing IgG1 and IgG3 subclasses. This orientation and subclass profile indicate that the bacteria normally are phagocytosed and killed when they appear in the blood stream.
Figure 7.
Complement deposition at the bacterial surface in plasma and saliva. (A) Heat map over bacteria-bound complement proteins from SRM adsorption experiments. Data are represented as mean values from paired adsorption experiments (eight biological replicates with four bacterial strains after incubation with plasma or saliva). The heat map was constructed by comparing values between or within either plasma or saliva conditions. All values <500 were difficult to separate from background noise and were set to 0. Sample peptides were prepared as described in the Materials and methods. (B) Relative complement deposition after plasma or saliva adsorption. Data are represented as percentage of total SRM signal for the mean of eight replicate adsorption experiments (plasma/saliva; wt = 7,091,682/159,629; M- = 7,956,055/41,216; H- = 6,719,836/88,078; M-H- = 6,638,641/56,181).
Reversing IgG orientation promotes bacterial survival
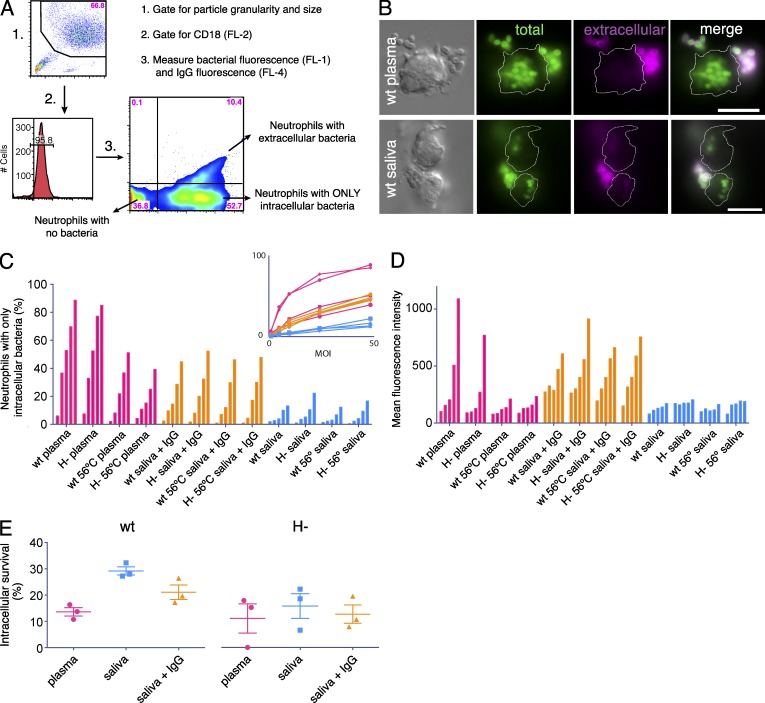
To investigate how IgG orientation and subclass distribution influence uptake and killing by neutrophils, we used a combination of techniques including flow cytometry, fluorescence microscopy, and plating assays, where wild-type and protein H deletion mutant bacteria were analyzed and compared under plasma and saliva conditions. Fig. 8 (C and D) show representative examples of neutrophils that have interacted with wild-type S. pyogenes preincubated with plasma or saliva. Compared with the massive phagocytosis of plasma-treated wild-type bacteria, the uptake after incubation with saliva was much lower (see representative images in Fig. 8 B).
Figure 8.
Phagocytosis and intracellular survival of S. pyogenes. (A) Gating strategy for the flow cytometry-based method to measure phagocytosis. Human neutrophils were gated based on light scatter properties (1) and then gated for CD18 (2) before analysis of Oregon Green/DyLight647-labeled bacteria (3). The top right quadrant represents neutrophils associated with extracellular bacteria; the bottom right quadrant represents neutrophils associated with only intracellular bacteria; and the bottom left represents cells that are not associated with bacteria. (B) Two representative image series from three independent experiments are shown for wild-type bacteria opsonized with either human plasma or saliva (30 min phagocytosis, MOI 25). Differential interference contrast images (top left) show the localization of the neutrophils; this is represented as a thin white line in the other images. All bacteria (Oregon Green–labeled) are shown in green, and extracellular bacteria are shown in magenta (Cy3-labeled anti-IgG). Bars, 10 µm. (C and D) Wild-type and H- mutant bacteria were analyzed after opsonization with either plasma, saliva, heat-inactivated (56°C) plasma and saliva, or saliva samples supplemented with pooled polyclonal IgG (10,000 µg/ml). The bars in groups of five represent MOI 1, 5, 10, 25, and 50 (30 min phagocytosis) and the inset in C shows the same data as a line graph. (C) Percentage of the neutrophil population that has fully internalized at least one bacterium. (D) Mean fluorescence intensity of intracellular bacteria as a relative measure of the number of internalized bacteria per neutrophil. Data are representative of two independent experiments. (E) Intracellular survival of wild-type and H- S. pyogenes was measured after 1 h of phagocytosis by human neutrophils. The bacteria (MOI 10) were opsonized with plasma, saliva, or saliva supplemented with polyclonal IgG (10,000 µg/ml). The data show survival relative to the starting point. Significant differences were recorded when wild-type bacteria preincubated with saliva were compared with either wild-type bacteria treated with plasma (P = 0.023), or to the three samples of H- bacteria preincubated with plasma, saliva, or saliva supplemented with polyclonal IgG (P = 0.034, 0.033, and 0.027, respectively). Data are from three independent experiments.
To characterize the time course of phagocytosis, wild-type bacteria were compared with the H- mutant. The kinetics were stable after 30 min, and the two strains exhibited similar patterns in terms of the percentage of neutrophils with phagocytosed bacteria (not depicted); thus, 30 min was chosen as the time point for the continued phagocytosis experiments. To more accurately account for differences between strains and conditions, the impact of varying the bacterial load (multiplicity of infection [MOI]) was evaluated. The percentages of neutrophils with only intracellular bacteria (Fig. 8 A, bottom right quadrant) were recorded to assess complete phagocytosis (Fig. 8 C). Wild-type and H- bacteria preincubated, respectively, in plasma or saliva (untreated and heat-inactivated for 30 min at 56°C), as well as in saliva samples supplemented with polyclonal IgG (10,000 µg/ml), were tested. The addition of IgG to saliva results in high numbers of correctly (Fab) oriented IgG molecules at the bacterial surface (Figs. 2–4) in the presence of low amounts of complement proteins (Fig. 7 B). With increasing MOIs, three data trends emerged (Fig. 8 C, inset) that were independent of the bacterial strain used: (1) preincubation in plasma resulted in very efficient phagocytosis, (2) preincubation in heat-inactivated plasma or saliva supplemented with IgG lead to less, but still effective, uptake, whereas (3) saliva exhibited a lower degree of phagocytosis. This indicates that phagocytosis under the conditions studied is enhanced by Fab-bound IgG and that the uptake is further increased by complement. To assess the number of internalized bacteria, we analyzed neutrophils with no adherent bacteria and measured their mean fluorescence intensities (Fig. 8 D). The results with plasma-opsonized bacteria (Fig. 8 D, pink bars) stress the importance of complement for the uptake. Moreover, internalization after saliva treatment was low, unless IgG was added. In saliva, which is a less complex environment compared with plasma, high levels of IgG were obviously sufficient to induce efficient phagocytosis.
The large difference in phagocytosis between plasma- and saliva-treated bacteria and the finding that phagocytosis of S. pyogenes is largely independent of IgGFc-binding proteins, left us with an unanswered question: what is the role of IgGFc-binding proteins in an environment with low IgG concentration, such as saliva? Because wild-type S. pyogenes bacteria are known to survive intracellularly in neutrophils (Staali et al., 2003), this was investigated under the aforementioned conditions. Wild-type and H mutant bacteria, preincubated with plasma, saliva, or saliva supplemented with polyclonal IgG (10,000 µg/ml), were phagocytosed by neutrophils and the intracellular survival was determined after 1 h (Fig. 8 E). The results show that wild-type bacteria survived significantly better after opsonization with saliva compared with plasma (P = 0.023), and also compared with H- bacteria preincubated with plasma (P = 0.034), saliva (P = 0.033), or IgG-supplemented saliva (P = 0.027). This suggests that IgGFc-binding proteins in an environment with low IgG concentration promote intracellular survival of S. pyogenes.
DISCUSSION
The patient described in this study was colonized by S. pyogenes in the throat but showed no local symptoms. During an asymptomatic carrier state, it is likely that small numbers of bacteria occasionally invade the microcirculation of the epithelium. Normally such bacteria are rapidly recognized and eliminated, but if they reach a damaged tissue site via the blood stream, this could provide a protected environment rich in nutrients where the pathogen can colonize and grow. After working in the garden, our patient felt pain in her right thigh, probably caused by a muscular bleeding. In this region, the infection was established and rapidly expanded into necrotizing fasciitis and toxic shock. The fact that the patient has diabetes and was under immunosuppressive treatment for rheumatoid arthritis could have contributed to this development. However, in many cases of severe invasive S. pyogenes infections there are no such underlying factors. Whether the bacteria identified in the blood originates from the throat or from necrotic tissue in the thigh remains an open question. However, the access of bacteria-containing samples that could be analyzed directly by electron microscopy from the three sites offered a unique opportunity to follow the presence and orientation of IgG antibodies at the bacterial surface during different phases of infection. The observation that IgG antibodies in the throat sample are associated with the surface mostly via Fc, but in blood through immune Fab-binding, provided a clue to the function of IgGFc-binding proteins. Moreover, and as mentioned above, previous work has indicated that the M protein–fibrinogen–IgG complexes released from the bacterial surface by proteolytic enzymes in the inflamed and necrotic tissue play an important role in the systemic vascular leakage that often has fatal consequences in patients with necrotizing fasciitis and toxic shock. S. pyogenes is an exclusive human pathogen, and for the first time the whole chain of events could now be followed and clarified in a human being. This is important because the IgGFc-binding proteins in S. pyogenes do not bind mouse and rat IgG (Åkesson et al., 1994), which underlines the specificity for the human host and makes it virtually impossible to find a relevant animal model for S. pyogenes infections.
Most individuals develop specific antibodies early in life against the various common bacterial species that are equipped with IgGFc-binding proteins, and the present study demonstrates that at high concentrations of IgG, such as in blood plasma, most of the antibodies bind to S. pyogenes in an opsonizing manner. The reduced contribution of IgGFc-binding could be explained either by saturation of IgGFc-binding sites or by the presence of IgG that bind to their specific antigen with a higher affinity compared with the Fc-binding bacterial protein. Our results favor the latter explanation. The affinity of protein H for IgGFc is high, Kd = 1.6 × 10−9 M (Åkesson et al., 1990), and relatively low for M1 protein, Kd = 3.4 × 10−6 M (Åkesson et al., 1994). Neither is as strong as the affinity reported for many antibodies for their specific antigen (Ka > 1010 M−1). At the bacterial surface there will be a constant equilibrium exchange, and the molecules with the strongest interactions will be enriched over time, likely favoring antibodies binding via Fab. This notion is supported by the binding curves for polyclonal IgG (Fig. 2 C), where the strains lacking protein H exhibit a sigmoidal binding pattern, indicating a single type of specific interaction. In contrast, Fc-binding would generate a linear relationship on a log–log scale. The two protein H–expressing strains exhibit a bi-modal pattern, with an apparent saturation at low-mid concentrations, and then joining the sigmoidal response at higher concentrations. This correlates well with the shift from Fc- to Fab-binding observed in Fig. 6 C. Compared with an IgG-rich environment, the binding pattern at low IgG concentration, such as in saliva, was very different with preferential interaction through Fc.
To obtain a complete and quantitative map of both IgG subclasses and complement proteins binding to the streptococcal surface in plasma and saliva, mass spectrometry techniques were used. Some interesting conclusions can be drawn from the results, especially for wild-type bacteria. In plasma, the bacteria are perfectly opsonized for phagocytosis, as they are coated with Fab-bound IgG1 and IgG3 and large amounts of complement proteins of the classical and alternative pathway (no proteins from the lectin pathway were identified). In contrast, there is only one complement protein present at high levels in saliva; factor H, an inhibitor of the alternative and classical pathways (Liszewski et al., 1996) that was found to interact with M proteins (Horstmann et al., 1988). Otherwise, the wild-type bacteria are covered with Fc-bound IgG1 and IgG2. With this background, the results from the phagocytosis experiments are as expected: efficient neutrophil–bacteria interaction, uptake and killing after opsonization with human plasma, and the opposite after incubation with human saliva from the same donors. It is more difficult to distinguish between the exact contributions of IgG orientation and/or complement inhibition. The protein H mutant behaved similarly in plasma, indicating that IgGFc-binding proteins are superfluous in plasma, probably because of IgG antibodies binding to surface antigens via Fab. In saliva, there is a small increase of phagocytosis of H- bacteria in normal saliva as compared with wild-type, but not in heat-inactivated saliva. This indicates that complement might play a role also in saliva, but the effects are small compared with the plasma samples.
To be able to evaluate phagocytosis and intracellular survival correctly, it is important that the bacterial load is the same when comparing samples. However, we could only make sure that the starting conditions were the same, and in the case of the survival assays, the actual numbers per neutrophil will be different because of differences in the internalization process as shown by the flow cytometry assays. Nevertheless, despite fewer bacteria per neutrophil in the saliva samples, they still survived better than in the plasma samples. This emphasizes that the bacteria are protected in saliva; it should be easier for the neutrophils to kill the few saliva-incubated bacteria that are internalized as compared with the large numbers per neutrophil in plasma samples. Even if the difference is smaller, the same holds true when comparing wild-type with H mutant bacteria. Therefore, the survival of wild-type bacteria is probably underestimated in saliva, showing that bacteria binding factor H and IgG via Fc not only avoid detection and internalization more effectively, they also survive better intracellularly.
Bacterial sepsis is a clinically highly significant condition estimated to kill several million individuals annually (Cohen, 2002; Rittirsch et al., 2008), and a deeper understanding of the molecular and cellular bacterial–host interactions in the blood stream during sepsis will be required to identify novel therapies against one of the leading causes of death world-wide. However, bacteria–host relationships are in general well-balanced and, compared with the healthy carrier state and uncomplicated local infections, sepsis is an extremely rare condition, implicating that the evolution of human bacterial pathogens is taking place in environments other than blood. At the same time, local infections, including bacteria of the normal flora, regularly spill over to the microcirculation in low doses without causing disease. The effective elimination of these should be beneficial to the host, but could also add selective advantages to the bacteria; why kill your host or induce an inflammatory response that may negatively influence your preferred habitat (Sansonetti, 2004)? The present results support this view, and suggest that IgGFc-binding proteins of S. pyogenes have evolved to protect the bacteria in their natural setting. In contrast, they do not provide protection in the blood stream with its high concentration of complement proteins and specific IgG antibodies. Although not studied in such detail as S. pyogenes, the data concerning S. aureus and group G streptococci (Fig. 2 D) suggest that their IgGFc-binding proteins (A and G, respectively) play a similar biological role. These species are also normally found in IgG-poor environments in the skin, the nostrils, and the pharynx (Wertheim et al., 2005). Likewise, E. coli expressing the more recently discovered IgGFc-binding EiB proteins (Leo and Goldman, 2009), are exposed to low concentrations of IgG in the gastrointestinal tract (Kaper et al., 2004). Another fact that may also support a similar function for IgGFc-binding proteins in general, and at the same time shed light on an old observation, is their convergent evolution. Thus, proteins A, G, and H all bind to the Cγ2–Cγ3 interface of IgGFc despite a complete lack of sequence homology in their Fc-binding regions (Frick et al., 1992).
Before this investigation, we and our colleagues in the field had been puzzled by the fact that S. pyogenes is equipped with several highly specific IgG-interacting and -degrading proteins despite the low concentration of IgG in the skin and the pharynx. Apart from the IgGFc-binding proteins of the M protein family, IdeS, which has been such a valuable tool in this study, cleaves IgG with a unique degree of specificity (von Pawel-Rammingen et al., 2002); no other substrate has been identified. In addition, S. pyogenes secretes EndoS, a glucosidase that specifically removes the glycan from IgGFc (Collin and Olsén, 2001). The powerful effect of proteins H and M1 in an IgG-poor environment in contrast to blood plasma helps to clarify this apparent paradox. In more general terms, and perhaps self-evidently, the data indicate that the biological consequences of a molecular microbe–host interaction is to be found in the ecological niche where the microbe has evolved.
MATERIALS AND METHODS
Bacterial strains and culture conditions
S. pyogenes, Group G streptococci (G148; Björck and Kronvall, 1984), and S. aureus (Cowan I; Forsgren and Sjöquist, 1966) were cultured in TH medium (Todd Hewitt Broth; Bacto; BD) at 37°C in an atmosphere supplemented with 5% CO2. The wild-type S. pyogenes AP1 strain (40/58) is of the M1 serotype and was provided by the World Health Organization Streptococcal Reference Laboratory in Prague, Czech Republic. Three isogenic AP1 mutant strains were used; the MC25 strain expresses protein H at the surface but lacks cell wall–anchored M1 protein (Collin and Olsén, 2000), the BM27.6 strain expresses M1 protein but completely lacks protein H (Berge et al., 1997), and the BMJ71 strain is devoid of both surface proteins (Kihlberg et al., 1995). Whenever required, suitable antibiotics were added to the medium at the following final concentrations: 150 µg ml−1 kanamycin (MC25), 1 µg ml−1 erythromycin (BM27.6), and 5 µg ml−1 tetracycline (BMJ71). Bacteria were generally harvested at early log phase and washed three times in PBS. Bacterial concentrations were determined using flow cytometry (FACSCalibur; BD) with CountBright beads (Invitrogen) added as volume standard. The bacteria were then either used immediately or heat-killed at 80°C for 5 min in a heat block (agitation at 800 rpm), followed by rapid cooling of the samples in ice-cold water. The patient strain was isolated from a throat swab and from wound fluid, and its serotype was determined to be of the M1 serotype by PCR and sequencing. The presence of protein H at the surface of the patient strain was determined by measuring binding of radiolabeled human IgG. At a bacterial concentration of 4 × 108 CFU CFU, >50% of the radioactive signal was detected; this represents strong binding in comparison with earlier studies (Åkesson et al., 1990).
Human neutrophil isolation
Whole human blood from healthy donors was layered on Polymorphprep (Axis-Shield) and centrifuged at 400 g for 35 min at 18°C. The neutrophil layer was recovered and suspended in 50 ml Ca2+- and Mg2+-containing PBS. After centrifugation at 350 g for 10 min, erythrocytes were removed by hypotonic lysis for 20 s. The cells were then pelleted at 250 g (5 min), counted using a hemocytometer, and resuspended in Na-medium (5.6 mM glucose, 127 mM NaCl, 10.8 mM KCl, 2.4 mM KH2PO4, 1.6 mM MgSO4, 10 mM Hepes, and 1.8 mM CaCl2; pH adjusted to 7.4 with NaOH) at a concentration of 107 cells ml−1. After purification, the neutrophils were gently rotated end-over-end at room temperature.
Adsorption and opsonization of bacteria
Heparin-treated human plasma and saliva samples from 10 healthy individuals were prepared as described by Rai et al. (2005) and Henson and Wong (2010), respectively. EDTA-free Complete Mini protease inhibitor cocktail (Roche) was added to both sample types before aliquotation and storage at −20°C. Intravenous immunoglobulin (IVIG; Octagam, Octapharma) consists of human IgG from >3,500 pooled plasma samples from healthy individuals. Bacteria were incubated with plasma, saliva, or IVIG for 30 min at 37°C in a shaking heat block (500 rpm), and then washed three times (5,000 g, 5 min, swing-out) with Na-medium. For some experiments, the bacteria were first sonicated (VialTweeter; Hielscher) to disperse large aggregates.
Flow cytometry analysis of bacterium-associated IgG
Bacteria were incubated with DNA-probe Syto9 (Invitrogen) and anti-IgG Fab fragments (DyLight649-conjugated anti–human IgG; Jackson ImmunoResearch Laboratory). Flow cytometric analysis was performed using a FACSCalibur flow cytometer (BD) equipped with 488- and 633-nm lasers. Bacteria were sorted using a side-scatter threshold and gated for bacterial DNA in FL-1. IgG-signal was recorded in FL-4. For each sample at least 10,000 events were recorded, and the results were analyzed using the CellQuest Pro software (BD) and FlowJo 8.8.6 (Tree Star).
Radioligand binding assay
Bacterial overnight cultures were washed twice in PBST (10 mM sodium phosphate buffer, pH 7.2, containing 0.12 M NaCl and 0.05% Tween-20), and resuspended in PBST to a concentration of 2 × 109 CFU/ml. A serial dilution of bacteria (200 µl) was mixed with 25 µl of 125I-labeled protein (10,000 cpm) and incubated at room temperature for 1 h. Cells were spun down, and the radioactivity associated with the pellet was determined in a gamma counter (PerkinElmer).
Antibody orientation assay
Bacteria were incubated with IgG-containing solutions as described above and washed. The samples were split and one fraction was incubated with IdeS (0.5 µg/ml, 2 h, 37°C), with the other serving as internal control. Both fractions were washed and split once more so that each bacteria condition generated four samples. These samples were analyzed with flow cytometry as described above, except with detection of IgGFc and IgGFab (DyLight649-conjugated anti–human IgGFc or IgGFab; Jackson ImmunoResearch Laboratory) instead of whole IgG. The data are presented as percentage signal of the internal control.
Phagocytosis
Bacteria were incubated with IgG-containing solutions as described above and stained with 20 µM Oregon Green 488-X succinimidyl ester (Invitrogen) for 30 min in the dark at room temperature. Excess fluorochrome was then removed by washing the bacteria three times in Na-medium. Neutrophils were incubated with f-MLF (1 µM, 10 min, 37°C) before bacteria were added at different MOIs (1, 5, 10, 25, or 50). The samples were put on over-end rotation in a 37°C incubator, and aliquots were withdrawn at indicated time points and placed on ice. Samples were kept on ice until flow cytometry analysis or fixed with 2% PFA for microscopy analysis. Samples were incubated with PE-conjugated anti-CD18 (integrin β2 subunit) and DyLight649-conjugated anti-IgG Fab fragments. Gating was performed using forward scatter versus side scatter, and gating for cells positive for phagocytosis was performed by selecting for Oregon Green–positive and DyLight649-negative cells in the gate corresponding to neutrophils (CD18-positive). For each sample, 10,000 events were analyzed using FlowJo (Tree Star).
Analysis of intracellular bacterial survival
Intracellular survival of S. pyogenes was analyzed similarly to what has been described previously (Nordenfelt et al., 2009). In brief, before presentation to cells, bacteria were opsonized (see above) and then gently centrifuged (200 g, 2 min, swing-out) to remove aggregates (checked by microscopy). After equilibration at 37°C, neutrophils were presented to the bacteria by centrifugation (12,000 g, 30 s, fixed angle). The phagocytic process was halted by placing the samples on ice, and extracellular bacteria were removed by thorough washing (200 g, 2 min, swing-out). After washing, the temperature was raised to 37°C to allow continued phagosomal maturation. Samples were withdrawn after 30 min, and the neutrophils were lysed by incubating the samples for 5 min at pH 11.0 with water as previously described (Decleva et al., 2006). The samples were serially diluted in TH, and then plated on TH-agar plates. After an overnight incubation at 37°C, the number of CFUs was determined.
Mass spectrometry
SRM transition lists were generated using a previously published method (Lu et al., 2007; Picotti et al., 2010). Protein samples were denatured using 8 M urea, reduced with 5 mM TCEP, and alkylated with 10 mM iodoacetamide before overnight trypsinization. Trypsin was inactivated by lowering the pH to 2 and the peptides were immobilized onto C18 columns (UltraMicro Spin Silica; Vydac). After multiple washes, the peptides were eluted (acetonitrile/formic acid) and solvents were evaporated in a SpeedVac centrifuge. After resupension, the samples were briefly sonicated before storage at −80°C. The samples were loaded onto an RP-HPLC column for peptide separation and subsequently introduced into the mass spectrometer through electrospray ionization. The peptide ions are filtered through the three quadropoles guided by the previously generated SRM assays to collect measurements for each selected peptide. Identified IgG peptides (Fig. S1) were synthesized (Aqua QuantPro; Thermo Fisher Scientific), and the known concentrations were used to absolutely quantify the peptides used for IgG measurements (DTLMISR, NQVSLTCLVK, GPSVFPLAPSSK, GLPAPIEK, SCDTPPPCPR, and GLPSSIEK).
The SRM measurements were performed on a TSQ Vantage triple-stage quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). Chromatographic separations of peptides were performed on an Eksigent 2D NanoLC system (Eksigent) with a PicoTip Emitter (10-µm tip, 75 µm × 12 cm; New Objective) packed with Reprosil-Pur C18-AQ resin (3 µm; Dr. Maisch GmbH). The mobile phase consisted of solvent A, 0.1% aqueous formic acid, and solvent B, acetonitrile with 0.1% formic acid. Peptides were eluted with a flow rate of 300 nl/min with a gradient of 97% solvent A at 0–5 min, 85% solvent A at 8 min, 65% solvent A at 42 min, and 10% solvent A at 45–50 min. The mass spectrometer was operated in SRM mode with Q1 and Q3 set at unit resolution (FWHM 0.7 D). A spray voltage of +1,700 V was used with a heated ion transfer setting of 270°C for desolvation. Data were acquired using the Xcalibur software (version 2.1.0). The dwell time was set to 10 ms, and the scan width to 0.01 m/z. All collision energies were calculated using the formula: CE = (Parent m/z) × 0.034 + 3.314.
Electron microscopy
Patient sample preparation.
A scrape from the throat was obtained with a plastic wire loop. The material at tip of the loop was resuspended and fixed in 100 μl EM fix (2.5% glutaraldehyde, 0.15 M Na-Cacodylate, pH 7.2). 10 ml of blood was centrifuged at 200 g for 10 min. The supernatant was collected and centrifuged at 3,000 g for another 10 min, and the resulting pellet was fixed and resuspended in 50 μl EM fix. Wound fluid (200 μl) was resuspended in EM fix (800 μl).
Negative staining.
The location of IgG antibodies binding to bacterial surface proteins was analyzed by negative staining and transmission electron microscopy, as previously described (Engel and Furthmayr, 1987). IgG samples were either used directly or conjugated with 10 nm colloidal gold. Suspensions of bacteria (1%) were mixed with of IgG, or IgG-Au conjugates, and then incubated for 1 h at room temperature. 5-µl aliquots were adsorbed onto carbon-coated grids for 1 min, washed with two drops of water, and stained on two drops of 0.75% uranyl formate. The grids were rendered hydrophilic by glow discharge at low pressure in air.
Ultrathin sectioning.
Bacterial suspensions were fixed (1.5% paraformaldehyde and 0.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4) for 1 h at room temperature, followed by washing with 0.1 M phosphate buffer, pH 7.4. The fixed and washed samples were subsequently dehydrated in ethanol and further processed for Lowicryl embedding (Carlemalm et al., 1985). Sections were cut with an LKB Ultratome and mounted on gold grids. Sections were washed with distilled water and post-stained with 2% uranyl acetate and lead citrate. All samples were examined with a JEOL JEM 1230 electron microscope operated at 80-kV accelerating voltage. Images were recorded with a Gatan Multiscan 791 charge-coupled device camera.
Fluorescence microscopy
Acquisition of images was performed using a fluorescence microscope (Nikon Eclipse TE300 equipped with a Hamamatsu C4742-95 cooled charge-coupled device camera, using a Plan Apochromat 100× objective with NA 1.4) and a N.A 1.4 oil condenser. The acquisition software used was Nikon NIS-Elements 3. Images were processed using Adobe Photoshop CS5 and ImageJ. In all figures, acquisition of images was made using the same exposure time for each fluorophore. During post-processing (only linear changes), the images were treated identically to maintain the relative intensity.
Statistics
In Fig. 8, GraphPad Prism 5 was used for repeated measure one-way ANOVA with Bonferroni multiple comparison correction.
Online supplemental material
Fig. S1 shows SRM analysis of IgG subclass distribution. Fig. S2 shows a heat map over bacteria-bound complement system molecules from SRM adsorption experiments. Table S1 shows IgG levels in saliva and plasma. The supplemental text details the patient’s case history. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120325/DC1.
Acknowledgments
We thank Ingbritt Gustafsson, Monica Heidenholm, Maria Baumgarten, and Kristoffer Sjöholm for technical assistance.
The work was funded by the Swedish Research Council (project 7480 and 2008:3356), the Swedish Foundation for Strategic Research (grant no. FFL4), Crafoordska Stiftelsen (grant no. 20090802 and 20100892), the Swedish Government Funds for Clinical Research (ALF), Hansa Medical AB, the Medical Faculty at Lund University, the Foundations of Knut and Alice Wallenberg, Torsten and Ragnar Söderberg, Alfred Österlund, Greta and Johan Kock, Lars Hiertas Minne, and the Royal Physiographic Society.
The sample collection was approved by the ethics committee of Lund University Hospital (project number LU 790/2005 [2006-01-17]) and informed consent was obtained from the patient.
We declare no competing financial interests.
Author contributions: P. Nordenfelt, J. Malmström, and L. Björck designed the study, analyzed the data, and wrote the manuscript with input from the other co-authors. P. Nordenfelt performed flow cytometry, binding assays, adsorption experiments, antibody orientation assays, heavy peptide selection, fluorescence microscopy, phagocytosis, and intracellular survival studies. S. Waldemarson performed and analyzed SRM quantification experiments. A. Linder identified patients with potential invasive S. pyogenes infection and collected clinical samples. C. Karlsson participated in heavy peptide selection and analysis. M. Mörgelin performed the electron microscopy.
Footnotes
Abbreviations used:
- Fab
- fragment antigen-binding
- Fc
- fragment crystallizable
- IgGFab
- Fab region of IgG
- IgGFc
- Fc region of IgG
- MOI
- multiplicity of infection
- SRM
- selected-reaction monitoring
References
- Åkesson P., Cooney J., Kishimoto F., Björck L. 1990. Protein H—a novel IgG binding bacterial protein. Mol. Immunol. 27:523–531 10.1016/0161-5890(90)90071-7 [DOI] [PubMed] [Google Scholar]
- Åkesson P., Schmidt K.H., Cooney J., Björck L. 1994. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem. J. 300:877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge A., Kihlberg B.M., Sjöholm A.G., Björck L. 1997. Streptococcal protein H forms soluble complement-activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J. Biol. Chem. 272:20774–20781 10.1074/jbc.272.33.20774 [DOI] [PubMed] [Google Scholar]
- Björck L. 1988. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J. Immunol. 140:1194–1197 [PubMed] [Google Scholar]
- Björck L., Kronvall G. 1984. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol. 133:969–974 [PubMed] [Google Scholar]
- Brüggemann M., Williams G.T., Bindon C.I., Clark M.R., Walker M.R., Jefferis R., Waldmann H., Neuberger M.S. 1987. Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J. Exp. Med. 166:1351–1361 10.1084/jem.166.5.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis J.R., Steer A.C., Mulholland E.K., Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685–694 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- Carlemalm E., Villiger W., Hobot J.A., Acetarin J.D., Kellenberger E. 1985. Low temperature embedding with Lowicryl resins: two new formulations and some applications. J. Microsc. 140:55–63 10.1111/j.1365-2818.1985.tb02660.x [DOI] [PubMed] [Google Scholar]
- Carlsson F., Berggård K., Stålhammar-Carlemalm M., Lindahl G. 2003. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J. Exp. Med. 198:1057–1068 10.1084/jem.20030543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. 2002. The immunopathogenesis of sepsis. Nature. 420:885–891 10.1038/nature01326 [DOI] [PubMed] [Google Scholar]
- Cole J.N., Barnett T.C., Nizet V., Walker M.J. 2011. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9:724–736 10.1038/nrmicro2648 [DOI] [PubMed] [Google Scholar]
- Collin M., Olsén A. 2000. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 36:1306–1318 10.1046/j.1365-2958.2000.01942.x [DOI] [PubMed] [Google Scholar]
- Collin M., Olsén A. 2001. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20:3046–3055 10.1093/emboj/20.12.3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decleva E., Menegazzi R., Busetto S., Patriarca P., Dri P. 2006. Common methodology is inadequate for studies on the microbicidal activity of neutrophils. J. Leukoc. Biol. 79:87–94 10.1189/jlb.0605338 [DOI] [PubMed] [Google Scholar]
- Engel J., Furthmayr H. 1987. Electron microscopy and other physical methods for the characterization of extracellular matrix components: laminin, fibronectin, collagen IV, collagen VI, and proteoglycans. Methods Enzymol. 145:3–78 10.1016/0076-6879(87)45003-9 [DOI] [PubMed] [Google Scholar]
- Fischetti V.A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren A., Sjöquist J. 1966. “Protein A” from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J. Immunol. 97:822–827 [PubMed] [Google Scholar]
- Frick I.M., Wikström M., Forsén S., Drakenberg T., Gomi H., Sjöbring U., Björck L. 1992. Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc. Natl. Acad. Sci. USA. 89:8532–8536 10.1073/pnas.89.18.8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick I.M., Åkesson P., Cooney J., Sjöbring U., Schmidt K.H., Gomi H., Hattori S., Tagawa C., Kishimoto F., Björck L. 1994. Protein H—a surface protein of Streptococcus pyogenes with separate binding sites for IgG and albumin. Mol. Microbiol. 12:143–151 10.1111/j.1365-2958.1994.tb01003.x [DOI] [PubMed] [Google Scholar]
- Grover S., McGee Z.A., Odell W.D. 1991. Isolation of a 30 kDa immunoglobulin binding protein from Pseudomonas maltophilia. J. Immunol. Methods. 141:187–197 10.1016/0022-1759(91)90145-6 [DOI] [PubMed] [Google Scholar]
- Heath D.G., Cleary P.P. 1989. Fc-receptor and M-protein genes of group A streptococci are products of gene duplication. Proc. Natl. Acad. Sci. USA. 86:4741–4745 10.1073/pnas.86.12.4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson B.S., Wong D.T. 2010. Collection, storage, and processing of saliva samples for downstream molecular applications. Methods Mol. Biol. 666:21–30 10.1007/978-1-60761-820-1_2 [DOI] [PubMed] [Google Scholar]
- Herwald H., Cramer H., Mörgelin M., Russell W., Sollenberg U., Norrby-Teglund A., Flodgaard H., Lindbom L., Björck L. 2004. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 116:367–379 10.1016/S0092-8674(04)00057-1 [DOI] [PubMed] [Google Scholar]
- Horstmann R.D., Sievertsen H.J., Knobloch J., Fischetti V.A. 1988. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. USA. 85:1657–1661 10.1073/pnas.85.5.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn F., Mörgelin M., Shannon O., Norrby-Teglund A., Herwald H., Olin A.I., Björck L. 2008. Antibodies against a surface protein of Streptococcus pyogenes promote a pathological inflammatory response. PLoS Pathog. 4:e1000149 10.1371/journal.ppat.1000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor F.S. 1965. Fibrinogen precipitation by streptococcal M protein. I. Identity of the reactants, and stoichometry of the reaction. J. Exp. Med. 121:849–859 10.1084/jem.121.5.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J.B., Nataro J.P., Mobley H.L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Kihlberg B.M., Cooney J., Caparon M.G., Olsén A., Björck L. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19:299–315 10.1016/S0882-4010(96)80003-9 [DOI] [PubMed] [Google Scholar]
- Kotb M., Norrby-Teglund A., McGeer A., El-Sherbini H., Dorak M.T., Khurshid A., Green K., Peeples J., Wade J., Thomson G., et al. 2002. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat. Med. 8:1398–1404 10.1038/nm1202-800 [DOI] [PubMed] [Google Scholar]
- Leo J.C., Goldman A. 2009. The immunoglobulin-binding Eib proteins from Escherichia coli are receptors for IgG Fc. Mol. Immunol. 46:1860–1866 10.1016/j.molimm.2009.02.024 [DOI] [PubMed] [Google Scholar]
- Lindahl G., Kronvall G. 1988. Nonimmune binding of Ig to Clostridium perfringens. Preferential binding of IgM and aggregated IgG. J. Immunol. 140:1223–1227 [PubMed] [Google Scholar]
- Liszewski M.K., Farries T.C., Lublin D.M., Rooney I.A., Atkinson J.P. 1996. Control of the complement system. Adv. Immunol. 61:201–283 10.1016/S0065-2776(08)60868-8 [DOI] [PubMed] [Google Scholar]
- Lu P., Vogel C., Wang R., Yao X., Marcotte E.M. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25:117–124 10.1038/nbt1270 [DOI] [PubMed] [Google Scholar]
- Macheboeuf P., Buffalo C., Fu C.-Y., Zinkernagel A.S., Cole J.N., Johnson J.E., Nizet V., Ghosh P. 2011. Streptococcal M1 protein constructs a pathological host fibrinogen network. Nature. 472:64–68 10.1038/nature09967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara C., Zinkernagel A.S., Macheboeuf P., Cunningham M.W., Nizet V., Ghosh P. 2008. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 319:1405–1408 10.1126/science.1154470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz K.P., Fives-Taylor P.M. 1994. Identification of an immunoglobulin Fc receptor of Actinobacillus actinomycetemcomitans. Infect. Immun. 62:4500–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Ravetch J.V. 2008. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 8:34–47 10.1038/nri2206 [DOI] [PubMed] [Google Scholar]
- Nordenfelt P., Bauer S., Lönnbro P., Tapper H. 2009. Phagocytosis of Streptococcus pyogenes by all-trans retinoic acid-differentiated HL-60 cells: roles of azurophilic granules and NADPH oxidase. PLoS ONE. 4:e7363 10.1371/journal.pone.0007363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. 2010. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods. 7:43–46 10.1038/nmeth.1408 [DOI] [PubMed] [Google Scholar]
- Rai A.J., Gelfand C.A., Haywood B.C., Warunek D.J., Yi J., Schuchard M.D., Mehigh R.J., Cockrill S.L., Scott G.B.I., Tammen H., et al. 2005. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 5:3262–3277 10.1002/pmic.200401245 [DOI] [PubMed] [Google Scholar]
- Reis K.J., Ayoub E.M., Boyle M.D. 1984. Streptococcal Fc receptors. I. Isolation and partial characterization of the receptor from a group C streptococcus. J. Immunol. 132:3091–3097 [PubMed] [Google Scholar]
- Rittirsch D., Flierl M.A., Ward P.A. 2008. Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 8:776–787 10.1038/nri2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandt C.H., Hill C.W. 2001. Nonimmune binding of human immunoglobulin A (IgA) and IgG Fc by distinct sequence segments of the EibF cell surface protein of Escherichia coli. Infect. Immun. 69:7293–7303 10.1128/IAI.69.12.7293-7203.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P.J. 2004. War and peace at mucosal surfaces. Nat. Rev. Immunol. 4:953–964 10.1038/nri1499 [DOI] [PubMed] [Google Scholar]
- Staali L., Mörgelin M., Björck L., Tapper H. 2003. Streptococcus pyogenes expressing M and M-like surface proteins are phagocytosed but survive inside human neutrophils. Cell. Microbiol. 5:253–265 10.1046/j.1462-5822.2003.00272.x [DOI] [PubMed] [Google Scholar]
- Vincents B., von Pawel-Rammingen U., Björck L., Abrahamson M. 2004. Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry. 43:15540–15549 10.1021/bi048284d [DOI] [PubMed] [Google Scholar]
- von Pawel-Rammingen U., Johansson B.P., Björck L. 2002. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 21:1607–1615 10.1093/emboj/21.7.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim H.F.L., Melles D.C., Vos M.C., van Leeuwen W., van Belkum A., Verbrugh H.A., Nouwen J.L. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- Zav’yalov V.P., Abramov V.M., Cherepanov P.G., Spirina G.V., Chernovskaya T.V., Vasiliev A.M., Zav’yalova G.A. 1996. pH6 antigen (PsaA protein) of Yersinia pestis, a novel bacterial Fc-receptor. FEMS Immunol. Med. Microbiol. 14:53–57 10.1111/j.1574-695X.1996.tb00267.x [DOI] [PubMed] [Google Scholar]