Abstract
Lampreys and hagfish are primitive jawless vertebrates capable of mounting specific immune responses. Lampreys possess different types of lymphocytes, akin to T and B cells of jawed vertebrates, that clonally express somatically diversified antigen receptors termed variable lymphocyte receptors (VLRs), which are composed of tandem arrays of leucine-rich repeats. The VLRs appear to be diversified by a gene conversion mechanism involving lineage-specific cytosine deaminases. VLRA is expressed on the surface of T-like lymphocytes; B-like lymphocytes express and secrete VLRB as a multivalent protein. VLRC is expressed by a distinct lymphocyte lineage. VLRA-expressing cells appear to develop in a thymus-like tissue at the tip of gill filaments, and VLRB-expressing cells develop in hematopoietic tissues. Reciprocal expression patterns of evolutionarily conserved interleukins and chemokines possibly underlie cell-cell interactions during an immune response. The discovery of VLRs in agnathans illuminates the origins of adaptive immunity in early vertebrates.
Keywords: jawless vertebrate, antigen receptor, leucine-rich repeat, somatic diversification, lymphocyte lineage
INTRODUCTION
When the phylogenetic basis for adaptive immunity was probed using classical methods for assessing cellular and humoral responses, adaptive immunity was found to be a vertebrate innovation. All the jawed vertebrates are capable of cell-mediated and humoral immune responses. Even the cartilaginous fish representatives, i.e., sharks, skates, and rays, have T and B lymphocyte lineages that use V(D)J recombination to generate diverse T cell receptor (TCR) and B cell receptor (BCR) repertoires in the thymus and hematopoietic tissues, respectively (1, 2).
In early studies of the jawless vertebrates, lampreys were found to accept skin autografts but to reject first-set allografts slowly and second-set allografts with accelerated kinetics (3, 4). In a mixed leukocyte reaction system demonstrating alloreactivity for hagfish, the adherent myeloid fraction of blood cells was shown to be largely responsible for driving the alloresponse, whereas responder cells were restricted to the nonadherent lymphocyte fraction (5). Furthermore, immunization of lampreys and hagfish with a variety of bacteria, viruses, and xenogeneic cells induced the production of circulating antigen-specific agglutinins (3, 6, 7). The relatively high molecular weight of these agglutinins was consistent with the then-emerging view that basal vertebrates rely predominantly on oligomeric IgM antibodies for humoral immunity (8). However, attempts to confirm that IgM was the high-molecular-weight agglutinin of lampreys and hagfish failed (9). With the elucidation of central roles for the Ig, TCR, and MHC (major histocompatibility complex) genes in the adaptive immune system of jawed vertebrates, the conspicuous elusiveness of these genes in jawless vertebrates began to raise doubt about the validity of the earlier phenomenological observations indicating that the jawless vertebrates possess an adaptive immune system (10). The advent of modern molecular genetic tools thus led to a renewed search for the roots of adaptive immunity in jawless vertebrates (11–13).
The discovery of variable lymphocyte receptors (VLRs), which are the subject of this review, provided an answer to this puzzle; it is now clear that jawless vertebrates developed a unique form of adaptive immunity that does not rely on BCR, TCR, or MHC molecules.
DISCOVERY OF VLR
While searching for lamprey genes whose expression is upregulated in antigen- and mitogen-stimulated lymphocytes, Pancer and colleagues (14) identified a large number of cDNA clones encoding multiple leucine-rich repeat (LRR) modules. The sequence of individual LRR modules was highly diverse, and the number of LRR modules was also variable; by contrast, the sequences flanking LRR modules were invariant. Genomic analysis revealed that all the cDNA sequences were derived from a single gene lacking sequences coding for most of the LRR modules, although it had many LRR-encoding modules in its vicinity (Figure 1a). Surprisingly, in lymphocytes, adjacent gene segments coding for LRR modules were incorporated into this single gene, enabling it to encode functional proteins. The extensive diversity generated by this lymphocyte-specific recombination suggested strongly that this gene encodes the long-sought antigen receptors, which were termed VLRs. The expression of only one assembled VLR gene by individual lymphocytes was indicative of a clonally diverse VLR repertoire (14). Subsequent studies provided ample evidence that the VLRs are agnathan antigen receptors (15–17).
Figure 1.
Organization and assembly of VLR genes. (a) Assembly of VLR genes. Multiple LRRNT-, LRR1-, LRRV-, LRRVe-, CP-, and LRRCT-encoding modules are located adjacent to the germ-line VLR gene. During the development of lymphoid cells, these modules are incorporated into the VLR gene by a gene conversion–like mechanism. The assembled VLR gene encodes a GPI-anchored or transmembrane protein. LRR1 and LRRVe denote LRRV modules located at the N and C termini, respectively. The figure is intended to emphasize the essential features of VLR assembly and does not reproduce the organization of a specific VLR locus. (b) Comparison of the germ-line structures of known VLR genes. The organization of the VLR gene shows minor variations depending on loci and species. (Abbreviations: CP, connecting peptide; CT, C terminal; GPI, glycosyl-phosphatidylinositol; LRR, leucine-rich repeat; NT, N terminal; VLR, variable lymphocyte receptor.)
The discovery of a VLR gene in the lamprey allowed the identification of two VLR genes in hagfish: One corresponded to the previously identified lamprey VLR gene, and the other was a novel VLR gene (18). These genes were termed VLRB and VLRA, respectively. A VLRA gene was subsequently identified in lampreys (19), demonstrating that both lampreys and hagfish have VLRA and VLRB genes. More recently, a third VLR gene, termed VLRC, was identified in the lamprey (20). Currently, it is not known whether hagfish possess a VLRC gene.
VLR PROTEIN STRUCTURES
The VLRs are expressed as membrane-bound proteins, composed of an N-terminal cap (LRRNT), LRR1, multiple LRRV modules, a connecting peptide (CP) LRR, a C-terminal cap (LRRCT), and an invariant stalk region rich in threonine and proline residues (14). Each of the LRRV modules is composed of 24 amino acids with the consensus sequence XLXXLXXLXLXXNXLXXLPXXXFX (where X stands for any amino acid). The most N-terminal LRR module, designated LRR1, is shorter than other LRR modules and is composed of 18 residues; the most C-terminal LRR module, LRRVe, is also unique, with a distinct sequence signature (15). Sequence diversity is primarily found in 3′-LRRNT (3′-part of LRRNT), LRR1, LRRV, LRRVe, CP, and 5′-LRRCT (5′-part of LRRCT) portions of the VLRs. In the case of VLRB, the stalk region has a glycosyl-phosphatidylinositol (GPI) cleavage site and is anchored to the cell membrane by GPI linkage (14). VLRA and VLRC are likely transmembrane proteins and are not secreted. VLRB is also secreted as a multimeric antibody by mature plasmacytes, as described below (17).
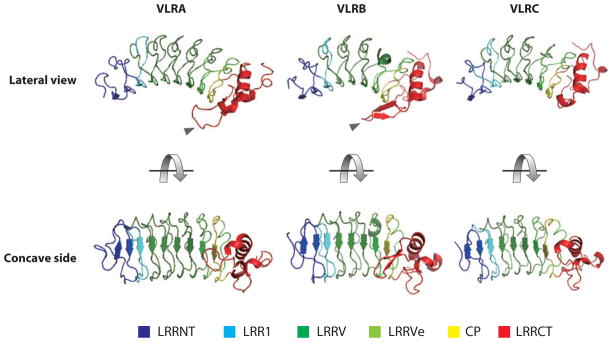
Use of the lamprey VLRB sequences to model the LRR portion of the protein predicted a curved solenoid structure (14), which has since been elucidated by solving the crystal structure for recombinant VLRB and VLRA proteins of both hagfish and lamprey origin (21–24). As for other typical LRR proteins, such as the Toll-like receptors, the LRRNT and LRRCT cap the ends of the solenoid to protect the hydrophobic core. The concave surface is formed by parallel beta sheets that contain most of the variable residues (Figure 2).
Figure 2.
Three-dimensional structures of variable lymphocyte receptor (VLR) molecules. Crystal structures of lamprey VLRA (PDB ID 3M18) and VLRB (PDB ID 3E6J) molecules and a predicted three-dimensional structure of lamprey VLRC (accession number BAJ14926). Prediction was made using the crystal structure of lamprey VLRB as a template. Protrusions located in the C-terminal leucine-rich repeat (LRRCT) of VLRA and VLRB are indicated by gray arrowheads.
VLR GENE STRUCTURES
Organization of germ-line VLR genes differs slightly depending on genes and animal species, but cardinal features are conserved (Figure 1b). They all have structures incapable of encoding any functional proteins, invariably lacking sequences coding for 3′-LRR1, LRRV, LRRVe, and 5′-CP (14, 18–20). Sequence modules coding for LRRNT, LRRV, LRRVe, CP, and LRRCT are located in multiple copies (some of them in several hundred copies) adjacent to the germ-line gene. Among the three VLR genes, hagfish VLRB and lamprey VLRC have the most and least incomplete structures, respectively. The structure of VLRA, but not VLRB, genes is conserved between hagfish and lampreys.
VLR GENE ASSEMBLY
During lymphocyte development, the intervening sequence of the germ-line VLR gene is replaced by a gene conversion–like mechanism in a stepwise manner, beginning either from its 5′- or 3′-ends, by adding flanking LRR sequences, eventually forming a completely assembled VLR gene (15, 19, 25) (Figure 1a). Short stretches of nucleotide homology (10–30 base pairs) are found between donor and acceptor sequences; thus, the sequences located at the ends of the most newly copied LRR sequences determine which flanking LRR sequences are copied into the germ-line gene in the next step. Unlike the recombination-activating gene (RAG)-mediated recombination of BCR/TCR genes, this process does not involve gene rearrangement, but rather it is thought to be gene conversion mediated by cytosine deaminases (CDA) of the AID-APOBEC family (19) (discussed below).
VLR assembly occurs in only one locus in each lymphocyte, with one notable exception as discussed below (20, 26, 27). Thus, a single lymphocyte expresses either VLRA, VLRB, or VLRC proteins. Furthermore, VLR assembly generally occurs monoallelically, thus enabling expression of a single type of functional VLR on each lymphocyte, analogous to the allelic exclusion observed for BCR/TCR genes (14, 20, 26). One study of hagfish lymphocytes showed that diallelic assembly occurs in approximately 5–10% of the cells (27); however, in most cases, only one assembled allele was functional, and the other allele underwent defective assembly, suggesting the presence of a feedback mechanism that prevents diallelic assembly of functional VLR genes.
Fluorescence in situ hybridization studies showed that hagfish VLRA and VLRB genes are physically distant from each other, albeit located on the same chromosome, suggesting that they function as independent recombination units (28). Consistent with this, hagfish VLRA and VLRB genes do not use LRR modules with identical nucleotide sequences. No information is available on the chromosomal localization of lamprey VLR genes. However, lamprey VLRA and VLRB genes also seem to function as independent units, given that they do not share LRR modules with identical nucleotide sequences. It remains to be seen whether the same holds true for lamprey VLRC; most of the currently available VLRC and VLRA/B cDNA sequences are derived from different species of lampreys, precluding large-scale comparison of VLRC versus VLRA/B LRRV sequences in a single species.
VLR GENE EXPRESSION
In lampreys, VLRB transcripts are ~60-fold more abundant than are VLRA or VLRC transcripts, both of which are expressed at comparable levels (20). This probably accounts for the fact that only VLRB transcripts were initially identified in a library enriched for cDNAs expressed by antigen- and mitogen-stimulated lymphoblasts (14). By contrast, expressed sequence tag analysis suggested that VLRA transcripts were more abundant than VLRB transcripts in unstimulated hagfish lymphocytes (18). It remains to be confirmed whether the relative expression levels of VLRA and VLRB genes indeed differ between lampreys and hagfish.
LAMPREY CDA GENES
It has been proposed that the incomplete VLR germ-line genes in agnathan lymphocyte progenitors undergo assembly by a gene conversion–like process (15, 19, 25, 29). Gene conversion, which also underlies the generation of antibody diversity in birds (30, 31) as well as rabbits and cattle (32, 33), requires the activity of activation-induced cytosine deaminase (AID) (34, 35). AID belongs to the AID-APOBEC family of CDAs; APOBEC3 is its closest relative in jawed vertebrates (36). Interestingly, two genes encoding presumptive AID-APOBEC CDAs, designated CDA1 and CDA2, were detected in the lamprey genome (19). Both genes are expressed in lymphocytes (19); CDA1 expression is predominantly observed in lamprey lymphocytes expressing VLRA (26), whereas CDA2 expression is associated with the VLRB-expressing lymphocyte lineage (26, 37). The mechanism controlling lineage-specific expression of CDA genes is unknown. Interestingly, however, the mutually exclusive expression of VLRA and VLRB genes in lymphocytes of the hagfish Eptatretus burgeri (27) and of VLRA and VLRB genes and proteins in lymphocytes of the lamprey Petromyzon marinus (26) suggests that the expression of these two antigen receptors is also regulated in a lineage-specific fashion. These findings raise the interesting possibility that both VLRA/CDA1 and VLRB/CDA2 gene pairs are subject to lineage-specific transcriptional regulation. This situation is unlike that observed in the lymphocytes of jawed vertebrates in which expression of RAG1 and RAG2 is lineage independent, as it occurs in both T and B lymphocyte lineages (38). Notably, however, AID is preferentially expressed by B lineage cells of jawed vertebrates, where it contributes to somatic hypermutation of the variable regions of the BCR (35).
Owing to the lack of suitable immunological reagents, analysis of CDA1 and CDA2 expression in lamprey tissues is currently possible only by means of RNA in situ hybridization. CDA1 expression has been found exclusively by cells in the thymoid, the presumptive thymus equivalent in the lamprey larvae (see below), which is located at the tip of the gill filaments; by contrast, CDA2 expression is not present in the thymoid tissue but was found in cells of the typhlosole, a major hematopoietic site of lamprey larvae, and in blood (37). To date, if CDA1 or CDA2 expression occurs in VLRC-expressing lymphocytes remains unknown. Because the predicted protein sequences of VLRA and VLRC are more closely related to each other than either is to VLRB, and given the presence of VLRA and VLRC assemblies in one of many cells (the former is nonfunctional, and the latter is predicted to be functional) (20), CDA1, rather than CDA2, may be expressed in the VLRC lineage of lymphocytes.
The patterns and levels of CDA1 and CDA2 expression do not change after immunization or mitogenic stimulation, as determined by RNA in situ hybridization analysis (37). Somatic hypermutation has been proposed to occur in VLRA-expressing lymphocytes (39); however, time-resolved analysis of protein expression during an immune response is necessary before definitive conclusions can be reached.
Phylogenetic analysis and secondary structure predictions indicate that AID and CDA1 are related more closely to each other than are AID and CDA2 (19). Furthermore, expression of CDA1 in Escherichia coli and yeast conferred a mutagenic phenotype and its expression in yeast diploids induced an increase in the rate of intragenic recombination (19). A detailed comparison of mutagenized sequences in bacteria and yeast with those observed in assembled VLRs suggested that somatic mutations play a minor role (if any) in the diversification of potential antigen-contact residues in lamprey VLRs (19); similarly, gene conversion is the major mechanism of somatic diversification of chicken Ig genes (30, 31).
Interaction of AID with replication protein A (RPA) is one mechanism by which AID is proposed to gain access to transcribed double-stranded DNA (40). Interaction of mouse AID and RPA depends on phosphorylation at serine 38 (S38); in zebrafish, which features a glycine residue at the corresponding position 42, a phosphomimetic aspartic acid residue at position 44 (D44) serves an equivalent function (41). Interestingly, in lamprey CDA1, the position equivalent to mouse S38 is occupied by glutamic acid (E44), which may serve as a phosphomimetic residue in a manner similar to D44 in zebrafish. Hence, CDA1/RPA interaction may be an evolutionarily conserved feature of antigen receptor diversification in vertebrates, and lamprey CDA1 may be capable of functionally replacing mouse AID. Notably, no information exists as yet to support the prediction that CDA2 is a functional CDA.
MAGNITUDE OF SOMATIC DIVERSIFICATION
Although the mechanism of antigen receptor diversification in agnathans differs markedly from that of gnathostomes, a similar level of diversification is achieved through the use of a large number of variable LRR donor cassettes (14, 19, 25). LRRV and LRRVe represent the most abundant module types in the germ line of the sea lamprey in which 513 unique modules flank the incomplete VLRA and 820 flank the VLRB gene (19). If the LRRs were incorporated as discrete modules into a functional VLR, a conservative estimate of the combinatorial diversity would be >1014. However, an additional stochastic element is introduced into functional VLRs by fusional diversity, because the VLR genes are not assembled by discrete insertion of individual modules. Instead, multiple donor LRRs can contribute to the creation of a fused hybrid-assembled LRR module (14, 19, 25). This patchwork design increases the possible number of permutations for VLR assembly to suggest a lower boundary of 1014 to 1017 different receptors of VLRA and VLRB types (19), numbers comparable to the potential diversity of the TCR and BCR repertoires in mammals. Notably, there is no evidence for N-nucleotide addition at the junction of inserted LRR modules during VLR assembly (25). Hence, junctional diversity appears to be a unique feature of BCR and TCR antigen receptors of jawed vertebrates.
Because AID plays an essential role in the somatic hypermutation of the Ig-variable region of jawed vertebrates (42), the expression of CDA1 and CDA2 by lymphocytes in jawless vertebrates theoretically could contribute to tertiary VLR diversification via somatic hypermutation. However, no direct evidence for somatic hypermutation or affinity maturation following antigen stimulation currently exists. In a study in which high-affinity VLRs were created by in vitro affinity maturation, 13 highly similar VLRA sequences were isolated from lymphocytes of a single immunized lamprey (39). However, whether these sequences were derived in lineal fashion by somatic hypermutation in a clonal lymphocyte precursor or were derived from multiple lymphocytes in the primary repertoire cannot be resolved for these data. Indeed, CDA1 is selectively expressed in the thymoid region where assembly occurs, and neither antigenic nor mitogenic stimulation modifies this pattern of CDA1 expression (37). If, similar to CDA1, CDA2 proves to have CDA activity, it will be interesting to investigate the possibility of hypermutation in the context of VLRB-expressing lymphocytes, where the production of higher affinity VLRB antibodies may be expected.
ANTIGEN BINDING BY VLRS
The prediction of antigen binding by the concave surface residues has been confirmed by site-directed mutagenesis (17) and elucidated precisely by solution of the structures of recombinant VLRB antibodies in complex with antigens of both a carbohydrate and a protein nature (22, 24). A protruding loop at the base of the concave surface, which is coded by variable inserts in the C-terminal cap, also contributes directly to antigen binding. The antigen-binding pocket of a VLRB antibody thus can be envisioned as the cupped palm of a hand with a thumb-like protrusion at its base. The thumb-like loop is more flexible and shares the unique feature of being able to bind to residues in antigen clefts [such as the catalytic region of hen egg lysozyme (HEL)] with unusual heavy-chain antibodies made by camels and sharks (43, 44). Lamprey VLRB antibodies are secreted as multivalent proteins with four or five pairs of identical chains; the antigen-binding LRR regions are located at the ends of the highly flexible stalk regions that are linked by disulfide bonds in their hydrophobic C terminus (17). In addition to the remarkable avidity attributable to their 8–10 antigen-binding sites, the VLRB antibodies are relatively resistant to denaturation by heat as well as very acidic and alkaline conditions (17).
The VLRA receptors are probably expressed on the lymphocyte surface as transmembrane proteins, and in contrast to VLRB, the VLRA proteins are not released or secreted after antigenic stimulation (26). So far, the structure of only one recombinant VLRA molecule in complex with antigen has been solved (21). In an effort to isolate antigen-specific VLRA molecules, VLRA cDNAs were derived after immunization with HEL from lymphocyte mRNA; these cDNAs directed the expression of a structurally diverse collection of VLRA molecules on the cell surface of yeast cells (21). One monomeric recombinant form of VLRA binds HEL protein with a dissociation constant of 180 pM, a value close to that of Ig/antigen complexes but substantially higher than that generally observed for TCR/pMHC complexes (45). The ligand-binding site spans the entire concave surface, from the LRRNT to the LRRCT modules. It appears, however, that LRRNT-mediated contacts are not required for binding; by contrast, the contact of antigen with LRRCT is essential for complex formation. The structure of VLRA molecules suggests that VLRA binding is based on conformational selection from a dynamic equilibrium of preexisting isomers, effectively increasing the antigen-binding capacity of VLRA molecules (21). Whereas the unique insert found in the LRRCT module packs against one side of the antigen in the VLRA-HEL complex, the analogous part of the VLRB protein projects into a pocket of the antigen in the VLRB-HEL complex (24). At least for the two HEL-specific VLR molecules whose structures have been solved in complex with native antigens, it appears that the epitopes recognized by VLRB and VLRA do not overlap. Nonetheless, it remains to be seen whether direct antigen binding is a general feature of VLRA molecules.
A crystal structure is currently not available for VLRC. However, three-dimensional modeling predicts that the overall structure of VLRC is similar to those of VLRA and VLRB molecules (Figure 2).
Several structural features distinguishing the three VLR isotypes suggest that they may employ unique antigen-binding modes. The average number of LRRV modules is larger in VLRA and VLRC than in VLRB molecules (15, 18, 20). Hence, the surfaces provided by LRRV modules in VLRAs and VLRCs are expected to contribute proportionally more to antigen binding than their LRRNT and LRRCT modules; furthermore, the functionality of VLRA and VLRC molecules may depend on a minimum distance of LRRNT and LRRCT modules. Sequence diversity of LRRNT is much less pronounced in VLRA and VLRC than in VLRB, compatible with the possibility that this module forms part of the antigen-binding surface only in VLRB. All VLRB molecules exhibit sizable protrusions in the LRRCT module that significantly contribute to antigen binding, at least for the few examples in which the structures of VLRB-antigen complexes are known (22, 24). VLRA molecules, however, are distinguished by considerable variation in the lengths of the LRRCT insert, suggesting greater variability in antigen-interaction modes. Interestingly, VLRA.R2.1, which binds its antigen directly as discussed above (21), exhibits a long protrusion in the LRRCT region akin to typical VLRB sequences, perhaps facilitating its direct binding capacity. The lengths of LRRCT inserts in VLRCs are much shorter than those in VLRBs and VLRAs, with the LRRCT of VLRC essentially incapable of forming protrusions; moreover, sequence variation in the LRRCT region of VLRC is much less pronounced than in that of the other two isotypes (20).
These structural features suggest that LRRNT regions of VLRAs and VLRCs (and perhaps additionally LRRCT modules of VLRCs) may engage in ternary complex formation on the cell surfaces of T-like cells and/or on those of other cells. In this manner, the comparatively invariant parts of VLRA and VLRC could participate in lymphocyte activation (by transmitting conformational changes after antigen binding to accessory molecules) and/or antigen recognition (by binding to antigen-presenting molecules).
LAMPREY LYMPHOCYTE LINEAGES
T-like and B-like lymphocyte lineages have been defined in lampreys on the basis of the cell-surface expression of VLRA or VLRB, expression of CDA1 or CDA2 during VLR assembly, ability to bind antigens that induce proliferative responses, ability to differentiate into VLR-secreting cells, responsiveness to the classical T cell mitogen PHA, and their gene expression profiles. CDA1 expression coincides with the productive assembly and expression of VLRA by lymphocytes in the thymoid region of the gill filaments (37). The VLRA+ cells are similar to T cells in that they respond to antigenic or PHA stimulation with proliferation, and they fail to undergo differentiation into VLRA-secreting cells; moreover, it has not been possible to show that they bind complex antigens to which they respond with proliferation (26). The VLRB+ cells are similar to B cells in that they bind cognate antigens, respond with proliferation, and undergo differentiation into plasmacytes that secrete multimeric VLRB antibodies (26).
GENE EXPRESSION PROFILES IN DIFFERENT LYMPHOCYTE LINEAGES
Informative gene expression profiles have been obtained for the VLRA+ and VLRB+ lymphocyte populations by analysis of a limited number of orthologous genes typically expressed by mammalian T and B lineage cells (26). Lamprey VLRA+ cells preferentially express orthologs of genes that are typically, although not exclusively, expressed by T lineage cells, whereas the VLRB+ cells preferentially express genes that are orthologous to ones typically expressed by B cells of jawed vertebrates. The VLRA+ cells express orthologs for several transcription factor genes that are associated with T cell differentiation, including GATA2/3, crel, aryl hydrocarbon receptor, and BCL11b. They also preferentially express transcripts for the chemokine receptor CCR9 that lymphocyte progenitors use for homing to the thymus, the T cell fate-determining molecule Notch 1, and the CD45 tyrosine phosphatase receptor protein that is essential for T cell development in mice and humans. Conversely, the VLRB+ cells preferentially express orthologous genes for the hematopoietic progenitor homing receptor CXCR4; Toll-like receptor family members TLR2, TLR7, and TLR10, the ligation of which can enhance B cell activation; the BCR-triggered signaling elements Syk; and B cell adaptor protein. These gene expression patterns reinforce the remarkably similar behavioral characteristics that lamprey VLRA+ and VLRB+ cells respectively share with mammalian T and B cells, and they suggest a common evolutionary origin.
SITES OF DEVELOPMENT OF LYMPHOCYTE LINEAGES
The hematopoietic tissues of lamprey undergo significant morphological changes when the ammocoete larva metamorphs into a young adult. It then develops further to the parasitizing stage (for approximately half of the >40 species of lamprey) and finally to a sexually mature adult (46). By morphological criteria, the typhlosole and the kidneys are the major sites of hematopoiesis in ammocoetes (47); in adult lampreys, the majority of hematopoietic activity is ascribed to the supraneural body (46). Owing to the lack of suitable functional assays (for instance, transplantation procedures), it is not clear whether these tissues harbor hematopoietic precursors. Among the more differentiated progeny of hematopoietic precursors, myeloid and erythroid lineages can easily be identified using morphological criteria. Lymphocyte development is, however, much more difficult to assess by cytology alone. New genetic tools have enabled a more detailed analysis of lymphocyte development in lamprey larvae. In jawed vertebrates, B cell development takes place in several hematopoietic sites, including the spleen and the spiral valve in elasmobranchs, the kidney in teleosts, and the bone marrow in amphibians and other tetrapods. Using CDA2 expression as a marker of ongoing B cell differentiation, Bajoghli et al. (37) demonstrated that the VLRB lineage of lamprey lymphocytes probably develops in the typhlosole and/or the kidney. By contrast, in all jawed vertebrates, T cell development is confined to the thymus (48); this lympho-epithelial tissue is distinguished by a unique pattern of gene expression, namely the thymopoietic transcription factor Foxn1 in stromal cells and the TCR and Rag genes in lymphocytes. By analogy, the thymus equivalent in lamprey was predicted to exhibit coexpression of the lamprey FOXN1 homolog, the VLRA antigen receptor gene and the CDA1 gene. Conversely, no expression of VLRB and CDA2 was predicted at this site. Using these criteria, candidate thymopoietic tissues, termed thymoid, were identified in lamprey larvae at the tip of the gill filaments (37) (Figure 3). Histologically, the hematopoietic tissue in the thymoid appears to be divided into various zones: CDA1 expression is confined to the outer edge of thymoid structures facing the pharyngeal cavity, whereas the expression of VLRA is more widespread and also occurs in the inner area of the thymoid (37). This suggests the presence of spatially segregated and possibly functionally distinct lymphopoietic microenvironments in the thymoid; it is unclear, however, whether this pattern is functionally equivalent to the cortical and medullary regions of the thymus in jawed vertebrates.
Figure 3.
The thymoid structure at the tip of gill filaments of lamprey larvae. (a) Lympho-epithelial structures at the tip of gill filaments as revealed by light microscopy after fixation in Bouin’s solution and hematoxylin/eosin (H&E) staining. The blood vessel (BV) underneath the thymoid is indicated; secondary lamellae emanate from the filament stem. The so-called cavernous bodies (CB) are located at the base of the gill filaments. (b) RNA in situ hybridization after fixation in paraformaldehyde with a CDA1-specific probe reveals the presence of developing T-like cells in the lymphoepithelial tissues at the tip of the gill filaments.
The spatially segregated development of lymphocyte lineages that is implied by the nonoverlapping expression patterns of VLRA/CDA1 and VLRB/CDA2 genes suggests the presence of guidance cues that direct developing hematopoietic precursor cells to their required destinations. It is unknown at present whether lamprey lymphocytes also require trophic signals, such as those mediated by a presumptive c-kit homolog (11) for survival and/or proliferation. Indeed, a functional requirement of the stem cell factor (KitL)/c-kit system has not yet been demonstrated in jawed fishes; by contrast, genetic evidence suggests a role for IL-7 in the lymphocyte development of teleost fishes (49). Interestingly, genes encoding proteins resembling stem cell factor and IL-7 have not yet been identified in jawless fishes; this reflects either the difficulties associated with recognizing highly diverged yet functionally equivalent cytokines or the presence of alternative cytokine networks.
Apart from cytokines, chemokines and their receptors are likely to play an important role in lymphocyte differentiation. Owing to its exceptional degree of sequence conservation, CXCR4 is the only chemokine receptor that has been unequivocally defined in lampreys (50); however, evidence for a number of additional chemokine receptors has been obtained, some with lineage-specific expression patterns (26, 51). More work is required to resolve the uncertainties concerning the functional assignment of these chemokine receptors. Unlike their receptors, chemokines are more difficult to recognize by means of sequence homology alone. Hence, only limited information exists on the complexity of the chemokine family in jawless fishes and on the functional correspondence, if any, to chemokines of jawed vertebrates. This is particularly true for constitutively expressed chemokines, although a number of putative homologs of inducible chemokines have been detected (11, 26, 51, 52).
While evidence for spatially segregated primary lymphoid tissues in lamprey larvae is accumulating, it is unclear whether secondary lymphoid organs are also present. Although lymphoid aggregations in the wall of the pharynx and in the hypo- and epipharyngeal folds of lamprey larvae have been variably considered as possible thymus equivalents (for detailed discussion, see Reference 37), they more likely represent secondary lymphoid tissues. Their reassessment in naive and immunized animals by means of molecular and antibody probes promises to shed light on this issue. The same applies to the supraneural body in adult lamprey, whose cellular content drastically changes during the course of an immune response (3).
IMMUNE RESPONSE IN LAMPREYS
In the past 50 years, several attempts have been made to analyze immune responses in jawless fishes. In early experiments using adult hagfish, no evidence for specific immune responses was forthcoming: Neutralizing antibodies could not be detected, delayed sensitivity was not elicited, and rejection of autologous and allogeneic skin grafts could not be established (53). Hence, attention shifted to the analysis of lamprey, in which evidence for specific immune responses was readily obtained. Early experiments established that, although antigen-specific responses could not be detected after immunization with soluble antigens, robust production of specific agglutinins could be observed after immunization with Brucella and the titers increased upon repeated immunization (3). Such immune responses can now be explained by the generation of antigen-specific VLRB antibodies (16). A delayed-type immune response could be observed after injection of bovine gamma globulin in complete Freund’s adjuvant, although the cell type associated with induration and swelling of the skin was not identified. Concomitantly, the tissue of the supraneural body becomes infiltrated by lymphoid cells (3), although their nature has not yet been studied using molecular tools or VLR-specific antibodies. In contrast to autologous skin grafts, allografts were rejected; early signs of rejection were seen at 11 days after transplantation and full rejection occurred after three to six weeks (3). Although morphological assessment suggested that polymorphic mononuclear cells rather than lymphocytes predominate the infiltrate during graft rejection (54), this finding needs to be investigated further with molecular probes for the three VLR isotypes and for other cell types.
The mechanisms for antigen presentation to the various types of lymphocytes in the jawless vertebrates are unresolved. As already mentioned, when immunized with bacterial or foreign blood cells, lampreys produce VLRB antibodies against iterative protein and carbohydrate determinants on the surface of these complex antigens. It is difficult to elicit VLRB antibody responses by immunizing lampreys with soluble or aggregated proteins, however, even when administered with adjuvants that effectively enhance antibody responses in jawed vertebrates (16). It is even more difficult to elicit VLRB antibody responses in hagfish, although antigen-specific agglutinin responses to bacteria were reported when animals were immunized under warmer than normal conditions (7). Since it has not been possible to demonstrate antigen binding by the VLRA+ lymphocytes, even for antigens that induce them to proliferate, it is possible that these cells see only processed antigen like their T cell counterparts in jawed vertebrates (26). Whether or not these lamprey T-like cells cooperate with the B-like cells to achieve effective antibody responses is also unresolved at present.
The discovery of lamprey T- and B-like lymphocyte lineages implied the existence of lymphokines regulating their interaction during an immune response. Indeed, a remarkable complementarity of lymphokine and lymphokine receptor expression has been demonstrated for VLRB- and VLRA-expressing lymphocytes (26). For instance, a lamprey homolog of the macrophage migration inhibitory factor, which is inducibly expressed by T cells and myelomonocytic cells in jawed vertebrates, has been identified (52) and shown to be expressed predominantly by VLRA-expressing lymphocytes (26). The same is true for an IL-17 homolog, which is a proinflammatory cytokine that plays a key role in regulating the immune response of jawed vertebrates; interestingly, the presumptive IL-17R is expressed by VLRB-expressing lymphocytes (26). A homolog of IL-8, which is expressed by VLRB-type lymphocytes (26), may be involved in attracting VLRA-expressing cells (expressing an IL-8R homolog) to sites where VLRB cells are activated during an immune response. Collectively, these findings suggest that B and T cells in lamprey may be able to communicate during the execution of an immune response.
EVOLUTION OF ANTIGEN RECOGNITION IN VERTEBRATES
It is remarkable that jawed and jawless vertebrates use structurally unrelated receptors for antigen recognition. However, deployment of structurally unrelated immune receptors for the same purpose is not unprecedented. For recognition of MHC class I molecules by natural killer (NK) cells, humans use killer Ig-like receptors, which are members of the Ig superfamily (55). In contrast, mouse NK cells use the Ly49 C-type lectin-like receptors for recognition of the same ligands (56, 57). In this case, convincing evidence indicates that a common ancestor of primates and rodents had both types of NK receptor genes (58–60) and that, in the course of speciation, primates and rodents adopted distinct families of molecules as their NK receptors for classical MHC class I molecules.
Comparative genome analyses indicate that a common ancestor of jawed and jawless vertebrates had both VLR-like and TCR/BCR-like molecules (Figure 4). On the basis of the similarity of gene structures and a characteristic insertion in LRRCT, it has been proposed that an ancestor of VLR emerged from a glycoprotein Ibα-like protein, a component of the platelet glycoprotein receptor complex conserved in all vertebrates (19). Similarly, searches for Ig superfamily proteins in jawless vertebrates identified molecules thought to be related to the evolutionary precursors of BCR/TCR. These include the lamprey “TCR-like gene,” which encodes an immunoreceptor tyrosine-based inhibition motif-bearing membrane protein that has a V-C2-type domain organization (61), and hagfish agnathan-paired receptors resembling antigen receptors, which have a single extra-cellular V-type Ig-like domain with a canonical J segment (62). The occurrence of these receptors in jawless vertebrates suggests that a common ancestor of all vertebrates had V-type Ig-like domains that could evolve into those of BCR/TCR (63). Thus, analogous to the NK receptors discussed above, an Ig superfamily protein was adopted as an antigen receptor in the jawed vertebrate lineage in which RAG insertion occurred, leading to the emergence of BCR and TCR. By contrast, an appropriate RAG insertion did not take place in the jawless vertebrate lineage, and a VLR-like LRR family protein instead was chosen as an antigen receptor. A question that remains to be answered (and may never be answered) is whether the common ancestor of jawed and jawless vertebrates had only precursors of VLR and BCR/TCR or a primitive, but functional, VLR or Ig superfamily–based antigen recognition system. In the latter case, switching of receptors from VLR to Ig superfamily proteins (or vice versa) must have occurred after the jawed and jawless vertebrates diverged.
Figure 4.
Evolution of adaptive immunity in vertebrates. A common ancestor of jawed and jawless vertebrates is hypothesized to have possessed primordial versions of BCR/TCR and VLR. Bifurcation of lymphocytes also seems to have taken place in the vertebrate ancestor. Acquisition of RAG recombinase activities likely occurred after the emergence of cytosine deaminase activities; this event together with a second round of WGD appear to have played a crucial role in the emergence of the jawed vertebrate-type adaptive immune system. After the split of jawless and jawed vertebrates, further diversification at the genetic and cellular levels occurred independently in these two groups of vertebrates. Cells of the T- and B-lymphocyte lineages are indicated in light green and orange, respectively. Abbreviations: BCR, B cell receptor; RAG, recombination-activating gene; TCR, T cell receptor; VLR, variable lymphocyte receptor; WGD, whole-genome duplication.
Jawless vertebrates appear to lack many paralogous genes with critical roles in jawed vertebrate–type adaptive immunity; these paralogs, known as ohnologs, are a consequence of two rounds of whole-genome duplication thought to have taken place in a common ancestor of vertebrates and then in a common ancestor of jawed vertebrates (1, 64, 65). Besides BCR, TCR, and MHC molecules, jawless vertebrates also lack the components of the classical as well as lytic pathways of complement activation (66), with the notable exceptions of C1q, C3, and factor B (66, 67). Thus, the distinction between the two forms of adaptive immunity in vertebrates clearly goes beyond the differential use of antigen receptors. Nevertheless, the overall designs of adaptive immune systems in jawed and jawless vertebrates are strikingly similar (as discussed in the preceding section), reflecting their common evolutionary origin (1, 68, 69). Future work should allow us to assess more accurately the extent of similarity and dissimilarity of the two forms of adaptive immunity. Undoubtedly, such studies will further illuminate both flexible and conserved aspects of immune system evolution.
CONCLUSIONS
The discovery of VLR-type antigen receptors in lamprey has provided unprecedented insight into the origins of adaptive immunity at the early stages of vertebrate evolution. Although the molecular details of antigen receptor structure and their modes of somatic diversification differ between jawless and jawed vertebrates, the dichotomy of functionally distinct yet interactive lymphocyte lineages is conserved and hence appears to be an ancestral feature of the vertebrates. Given these striking similarities in design, the immune system of jawed vertebrates will guide future studies of agnathan adaptive immunity. Conversely, studies of the alternative adaptive immune system in jawless vertebrates may yield insight into aspects of adaptive immunity in jawed vertebrates that remain elusive. The parallelism of vertebrate lymphocyte lineages also provides fresh impetus to examine the origins of the different hematopoietic lineages in nonvertebrate species.
SUMMARY POINTS.
The organization of lamprey and hagfish lymphocytes into T- and B-like lineages raises basic questions about the evolution of adaptive immune systems.
Antigen receptors of agnathans comprise somatically diversified LRR proteins that are clonally expressed.
Three types of VLRs are known: VLRA and VLRC are more similar to each other than either is to VLRB.
VLRA is expressed on the surface of T-like lymphocytes that appear to develop in a thymus-like structure at the tip of gill filaments.
VLRB is expressed on the surface of B-like lymphocytes and secreted as a multivalent antibody molecule.
The unique physicochemical properties of VLRB antibodies suggest their usefulness for biomedical purposes.
Reciprocal expression of interleukins, chemokines, and their receptors on T- and B-like cells suggests an evolutionarily conserved mode of cellular interactions during an immune response.
FUTURE ISSUES.
How do agnathan lymphocytes functionally interact with each other and with other cell types?
In which tissues do agnathan lymphocytes develop from their progenitors?
Is it possible to define and isolate agnathan hematopoietic stem cells?
Does selection of VLR repertoires occur, and if so, where and how?
How is self/nonself-discrimination achieved?
What is the mechanism, if any, of antigen presentation?
Are VLRC+ cells similar to T cells, and, if so, how do VLRA+ cells and VLRC+ cells divide their labor?
Among the methodological advances required are derivation of lamprey cell lines, generation of inbred lamprey lines, and genetic-modification strategies.
Acknowledgments
We thank the members of our laboratories for their contributions to the original studies reviewed here. Research in the Boehm laboratory is supported by the Max-Planck Society and the Deutsche Forschungsgemeinschaft. Research in the Kasahara laboratory is supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Uehara Memorial Foundation. Research in the Cooper laboratory is supported by the National Institutes of Health (RO1 AI072435) and the Georgia Research Alliance.
Glossary
- Jawed vertebrates (gnathostomes)
all vertebrates, except the jawless vertebrates, including cartilaginous fish, bony fish, amphibians, reptiles, birds, and mammals
- V(D)J recombination
a gene rearrangement process involving RAG-mediated recombination in the variable regions of TCR and BCR genes in jawed vertebrates
- TCR
T cell receptor
- BCR
B cell receptor
- Jawless vertebrates (agnathans)
the most basal group of vertebrates, comprising approximately 100 species of lampreys and hagfish
- Ig
immunoglobulin
- MHC
major histocompatibility complex
- VLR
variable lymphocyte receptor
- LRR
leucine-rich repeat
- Gene conversion
a form of homologous recombination that is initiated by DNA double-strand breaks and results in nonreciprocal transfer of genetic information
- RAG
recombination-activating gene
- CDA
cytosine deaminase
- AID
activation-induced cytosine deaminase
- Somatic diversification
a process resulting in changes to the germ-line sequence of genes in individual cells and that are retained by cell progeny
- HEL
hen egg lysozyme
- NK
natural killer
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Thomas Boehm, Email: boehm@immunbio.mpg.de.
Nathanael McCurley, Email: nmccurl@emory.edu.
Yoichi Sutoh, Email: ysuto@med.hokudai.ac.jp.
Michael Schorpp, Email: schorpp@immunbio.mpg.de.
Masanori Kasahara, Email: mkasaha@med.hokudai.ac.jp.
Max D. Cooper, Email: mdcoope@emory.edu.
LITERATURE CITED
- 1.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol. 2010;10:543–53. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finstad J, Good RA. The evolution of the immune response. III Immunologic responses in the lamprey. J Exp Med. 1964;120:1151–68. doi: 10.1084/jem.120.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perey DY, Finstad J, Pollara B, Good RA. Evolution of the immune response. VI First and second set skin homograft rejections in primitive fishes. Lab Invest. 1968;19:591–97. [PubMed] [Google Scholar]
- 5.Raison RL, Gilbertson P, Wotherspoon J. Cellular requirements for mixed leucocyte reactivity in the cyclostome, Eptatretus stoutii. Immunol Cell Biol. 1987;65(Pt. 2):183–88. doi: 10.1038/icb.1987.20. [DOI] [PubMed] [Google Scholar]
- 6.Fujii T, Nakagawa H, Murakawa S. Immunity in lamprey. I Production of haemolytic and haemag-glutinating antibody to sheep red blood cells in Japanese lampreys. Dev Comp Immunol. 1979;3:441–51. doi: 10.1016/s0145-305x(79)80040-3. [DOI] [PubMed] [Google Scholar]
- 7.Linthicum DS, Hildemann WH. Immunologic responses of Pacific hagfish. III Serum antibodies to cellular antigens. J Immunol. 1970;105:912–18. [PubMed] [Google Scholar]
- 8.Pollara B, Litman GW, Finstad J, Howell J, Good RA. The evolution of the immune response. VII Antibody to human “O” cells and properties of the immunoglobulin in lamprey. J Immunol. 1970;105:738–45. [PubMed] [Google Scholar]
- 9.Litman GW, Finstad FJ, Howell J, Pollara BW, Good RA. The evolution of the immune response. VIII Structural studies of the lamprey immunoglobulin. J Immunol. 1970;105:1278–85. [PubMed] [Google Scholar]
- 10.Klein J, Sato A, Mayer WE. Jaws and AIS. In: Kasahara M, editor. Major Histocompatibility Complex: Evolution, Structure, and Function. Heidelberg: Springer-Verlag; 2000. pp. 3–26. [Google Scholar]
- 11.Mayer WE, Uinuk-ool T, Tichy H, Gartland LA, Klein J, Cooper MD. Isolation and characterization of lymphocyte-like cells from a lamprey. Proc Natl Acad Sci USA. 2002;99:14350–55. doi: 10.1073/pnas.212527499. Provides an early characterization of gene expression patterns by lymphocyte-like cells in lamprey. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki T, Shin-I T, Kohara Y, Kasahara M. Transcriptome analysis of hagfish leukocytes: a framework for understanding the immune system of jawless fishes. Dev Comp Immunol. 2004;28:993–1003. doi: 10.1016/j.dci.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Uinuk-ool T, Mayer WE, Sato A, Dongak R, Cooper MD, Klein J. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci USA. 2002;99:14356–61. doi: 10.1073/pnas.212527699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–80. doi: 10.1038/nature02740. Originally identified in lamprey lymphocytes a novel type of antigen receptor (i.e., VLRB) exhibiting somatic diversification and clonal expression. [DOI] [PubMed] [Google Scholar]
- 15.Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–73. doi: 10.1126/science.1119420. Provides direct evidence that a mechanism similar to gene conversion is used for VLR assembly. [DOI] [PubMed] [Google Scholar]
- 16.Alder MN, Herrin BR, Sadlonova A, Stockard CR, Grizzle WE, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol. 2008;9:319–27. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 17.Herrin BR, Alder MN, Roux KH, Sina C, Ehrhardt GR, et al. Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA. 2008;105:2040–45. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102:9224–29. doi: 10.1073/pnas.0503792102. Identifies a second VLR gene, VLRA, now known to distinguish a T-like lymphocyte lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–56. doi: 10.1038/ni1463. Identifies two homologs of AID in lamprey with characteristics suggesting their involvement in the assembly of VLR genes by gene conversion. [DOI] [PubMed] [Google Scholar]
- 20.Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M. Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci USA. 2010;107:14304–8. doi: 10.1073/pnas.1001910107. Introduces VLRC, which is more similar to VLRA than VLRB and may be expressed in a separate T-like lymphocyte lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L, Velikovsky CA, Xu G, Iyer LM, Tasumi S, et al. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci USA. 2010;107:13408–13. doi: 10.1073/pnas.1005475107. Provides the structure of an in vitro–selected VLRA molecule in complex with a native protein antigen, HEL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–37. doi: 10.1126/science.1162484. Demonstrates the first structure of a VLRB antibody in complex with a carbohydrate antigen, illustrating how LRR modules contribute to the antigen-binding surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HM, Oh SC, Lim KJ, Kasamatsu J, Heo JY, et al. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282:6726–32. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 24.Velikovsky CA, Deng L, Tasumi S, Iyer LM, Kerzic MC, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat Struct Mol Biol. 2009;16:725–30. doi: 10.1038/nsmb.1619. Provides the first structure of VLB complexed with a protein antigen, HEL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagawa F, Kishishita N, Shimizu K, Hirose S, Miyoshi M, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8:206–13. doi: 10.1038/ni1419. Provides a detailed mechanistic investigation of VLR assembly, concluding that it bears similarity to gene conversion. [DOI] [PubMed] [Google Scholar]
- 26.Guo P, Hirano M, Herrin BR, Li J, Yu C, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. Identifies two lymphocyte lineages in lamprey, characterized by T-like and B-like cells with distinct gene expression patterns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishishita N, Matsuno T, Takahashi Y, Takaba H, Nishizumi H, Nagawa F. Regulation of antigen-receptor gene assembly in hagfish. EMBO Rep. 2010;11:126–32. doi: 10.1038/embor.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasamatsu J, Suzuki T, Ishijima J, Matsuda Y, Kasahara M. Two variable lymphocyte receptor genes of the inshore hagfish are located far apart on the same chromosome. Immunogenetics. 2007;59:329–31. doi: 10.1007/s00251-007-0200-3. [DOI] [PubMed] [Google Scholar]
- 29.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–22. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Thompson CB, Neiman PE. Somatic diversification of the chicken immunoglobulin light chain gene is limited to the rearranged variable gene segment. Cell. 1987;48:369–78. doi: 10.1016/0092-8674(87)90188-7. [DOI] [PubMed] [Google Scholar]
- 31.Reynaud C-A, Anquez V, Grimal H, Weill J-C. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–88. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 32.Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–97. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 33.Parng C-L, Hansal S, Goldsby RA, Osborne BA. Gene conversion contributes to Ig light chain diversity in cattle. J Immunol. 1996;157:5478–86. [PubMed] [Google Scholar]
- 34.Arakawa H, Buerstedde J-M. Immunoglobulin gene conversion: insights from bursal B cells and the DT40 cell line. Dev Dyn. 2004;229:458–64. doi: 10.1002/dvdy.10495. [DOI] [PubMed] [Google Scholar]
- 35.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 36.Conticello SG, Langlois M-A, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 37.Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, et al. A thymus candidate in lamprey. Nature. 2011;470:90–94. doi: 10.1038/nature09655. Identifies a presumptive thymus equivalent at the tip of gill filaments. [DOI] [PubMed] [Google Scholar]
- 38.Schatz DG, Oettinger MA, Schlissel MS. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–83. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 39.Tasumi S, Velikovsky CA, Xu G, Gai SA, Wittrup KD, et al. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA. 2009;106:12891–96. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–11. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 41.Basu U, Wang Y, Alt FW. Evolution of phosphorylation-dependent regulation of activation-induced cytidine deaminase. Mol Cell. 2008;32:285–91. doi: 10.1016/j.molcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 43.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA. 2006;103:4586–91. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science. 2004;305:1770–73. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 45.van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–84. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 46.Ardavin CF, Gomariz RP, Barrutia MG, Fonfria J, Zapata A. The lympho-hemopoietic organs of the anadromous sea lamprey, Petromyzon marinusA comparative study throughout its life span. Acta Zool. 1984;65:1–15. [Google Scholar]
- 47.Ardavin CF, Zapata A. Ultrastructure and changes during metamorphosis of the lympho-hemopoietic tissue of the larval anadromous sea lamprey Petromyzon marinus. Dev Comp Immunol. 1987;11:79–93. doi: 10.1016/0145-305x(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 48.Boehm T, Bleul CC. The evolutionary history of lymphoid organs. Nat Immunol. 2007;8:131–35. doi: 10.1038/ni1435. [DOI] [PubMed] [Google Scholar]
- 49.Iwanami N, Mateos F, Hess I, Riffel N, Soza-Ried C, et al. Genetic evidence for an evolutionarily conserved role of IL-7 signaling in T cell development of zebrafish. J Immunol. 2011;186:7060–66. doi: 10.4049/jimmunol.1003907. [DOI] [PubMed] [Google Scholar]
- 50.Kuroda N, Uinuk-ool TS, Sato A, Samonte IE, Figueroa F, et al. Identification of chemokines and a chemokine receptor in cichlid fish, shark, and lamprey. Immunogenetics. 2003;54:884–95. doi: 10.1007/s00251-002-0531-z. [DOI] [PubMed] [Google Scholar]
- 51.Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–97. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Sato A, Uinuk-ool TS, Kuroda N, Mayer WE, Takezaki N, et al. Macrophage migration inhibitory factor (MIF) of jawed and jawless fishes: implications for its evolutionary origin. Dev Comp Immunol. 2003;27:401–12. doi: 10.1016/s0145-305x(02)00136-2. [DOI] [PubMed] [Google Scholar]
- 53.Papermaster BW, Condie RM, Good RA. Immune response in the California hagfish. Nature. 1962;196:355–57. doi: 10.1038/196355a0. [DOI] [PubMed] [Google Scholar]
- 54.Fujii T, Hayakawa I. A histological and electron-microscopic study of the cell types involved in rejection of skin allografts in ammocoetes. Cell Tissue Res. 1983;231:301–12. doi: 10.1007/BF00222182. [DOI] [PubMed] [Google Scholar]
- 55.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, et al. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–48. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 56.Gumperz JE, Parham P. The enigma of the natural killer cell. Nature. 1995;378:245–48. doi: 10.1038/378245a0. [DOI] [PubMed] [Google Scholar]
- 57.Yokoyama WM. Natural killer cell immune responses. Immunol Res. 2005;32:317–25. doi: 10.1385/IR:32:1-3:317. [DOI] [PubMed] [Google Scholar]
- 58.Barten R, Trowsdale J. The human Ly-49L gene. Immunogenetics. 1999;49:731–34. doi: 10.1007/s002510050675. [DOI] [PubMed] [Google Scholar]
- 59.Hoelsbrekken SE, Nylenna Ø, Saether PC, Slettedal IÖ, Ryan JC, et al. Cutting edge: molecular cloning of a killer cell Ig-like receptor in the mouse and rat. J Immunol. 2003;170:2259–63. doi: 10.4049/jimmunol.170.5.2259. [DOI] [PubMed] [Google Scholar]
- 60.Welch AY, Kasahara M, Spain LM. Identification of the mouse killer immunoglobulin-like receptor-like (Kirl) gene family mapping to chromosome X. Immunogenetics. 2003;54:782–90. doi: 10.1007/s00251-002-0529-6. [DOI] [PubMed] [Google Scholar]
- 61.Pancer Z, Mayer WE, Klein J, Cooper MD. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc Natl Acad Sci USA. 2004;101:13273–78. doi: 10.1073/pnas.0405529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki T, Shin IT, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol. 2005;174:2885–91. doi: 10.4049/jimmunol.174.5.2885. [DOI] [PubMed] [Google Scholar]
- 63.Kasahara M, Kasamatsu J, Sutoh Y. Two types of antigen receptor systems in vertebrates. Zool Sci. 2008;25:969–75. doi: 10.2108/zsj.25.969. [DOI] [PubMed] [Google Scholar]
- 64.Kasahara M. Genome duplication and T cell immunity. Prog Mol Biol Transl Sci. 2010;92:7–36. doi: 10.1016/S1877-1173(10)92002-4. [DOI] [PubMed] [Google Scholar]
- 65.Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–71. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 66.Kimura A, Ikeo K, Nonaka M. Evolutionary origin of the vertebrate blood complement and coagulation systems inferred from liver EST analysis of lamprey. Dev Comp Immunol. 2009;33:77–87. doi: 10.1016/j.dci.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Matsushita M, Matsushita A, Endo Y, Nakata M, Kojima N, et al. Origin of the classical complement pathway: lamprey orthologue of mammalian C1q acts as a lectin. Proc Natl Acad Sci USA. 2004;101:10127–31. doi: 10.1073/pnas.0402180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boehm T. Design principles of adaptive immune systems. Nat Rev Immunol. 2011;11:307–17. doi: 10.1038/nri2944. [DOI] [PubMed] [Google Scholar]
- 69.Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–57. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]